Main conclusion
SCARECROW controls Arabidopsis root meristem size from the root endodermis tissue by regulating the DELLA protein RGA that in turn mediates the regulation ofARR1levels at the transition zone.
Coherent organ growth requires a fine balance between cell division and cell differentiation. Intriguingly, plants continuously develop organs post-embryonically thanks to the activity of meristems that allow growth and environmental plasticity. In Arabidopsis thaliana, continued root growth is assured when division of the distal stem cell and their daughters is balanced with cell differentiation at the meristematic transition zone (TZ). We have previously shown that at the TZ, the cytokinin-dependent transcription factor ARR1 controls the rate of differentiation commitment of meristematic cells and that its activities are coordinated with those of the distal stem cells by the gene SCARECROW (SCR). In the stem cell organizer (the quiescent center, QC), SCR directly suppresses ARR1 both sustaining stem cell activities and titrating non-autonomously the ARR1 transcript levels at the TZ via auxin. Here, we show that SCR also exerts a fine control on ARR1 levels at the TZ from the endodermis by sustaining gibberellin signals. From the endodermis, SCR controls the RGA REPRESSOR OF ga1-3 (RGA) DELLA protein stability throughout the root meristem, thus controlling ARR1 transcriptional activation at the TZ. This guarantees robustness and fineness to the control of ARR1 levels necessary to balance cell division to cell differentiation in sustaining coherent root growth. Therefore, this work advances the state of the art in the field of root meristem development by integrating the activity of three hormones, auxin, gibberellin, and cytokinin, under the control of different tissue-specific activities of a single root key regulator, SCR.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-016-2471-0) contains supplementary material, which is available to authorized users.
Keywords: ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1), Differentiation, Gibberellin, Hormones, Root, SCARECROW (SCR)
Introduction
The ability of plants to generate organs continuously depends on the activity of meristems, localized regions where plant stem cells reside (reviewed in Heidstra and Sabatini 2014). The root meristem is responsible for generating the root system, which anchors the plant to the soil while providing all the necessary nutrients. Longitudinally, the root meristem can be divided into three functionally distinct zones: a distal stem cell niche (SCN) generating transit-amplifying cells, which divide in the proximal meristem (PM) and differentiate in the elongation/differentiation zone (EDZ) (reviewed in Heidstra and Sabatini 2014). The boundary where cells leave the meristem and enter the EDZ is called transition zone (TZ). To maintain root meristem activity and thus root growth, the activities of these zones must be well coordinated, and different plant hormones have been shown to play a crucial role in ensuring this coordination. For example, the balance between cell division in the PM and cell differentiation in the EDZ depends on the antagonistic interaction between cytokinin, promoting cell differentiation, and auxin, prompting cell division (Dello Ioio et al. 2007, 2008). Cytokinin promotes cell differentiation via the cytokinin-dependent response transcription factor ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) (Dello Ioio et al. 2007; Taniguchi et al. 2007), which in turn activates the auxin signaling inhibitor gene SHY2 (Tian et al. 2002; Dello Ioio et al. 2008). Another plant hormone, gibberellin (GA) (Olszewski et al. 2002; Hedden and Sponsel 2015), has been reported to control cell division in the PM acting from the root endodermis (Achard et al. 2009; Ubeda-Tomás et al. 2009). In particular, it was shown that GA sustains cell division by suppressing the ARR1 transcript level at the TZ via the DELLA protein REPRESSOR OF GA1-3 (RGA) (Silverstone et al. 1998; Moubayidin et al. 2010).
A single gene, SCARECROW (SCR) (Di Laurenzio et al. 1996), is responsible for the spatial coordination between stem cell activity in the SCN and differentiation of their daughters at the TZ (Moubayidin et al. 2013). The SCR protein, a member of the GRAS family of transcription factors, is specifically expressed in the stem cell niche organizing cells, the quiescent center (QC), and in the endodermis (Di Laurenzio et al. 1996; Pysh et al. 1999; Wysocka-Diller at al. 2000; Lee et al. 2008). Besides its role in sustaining the stem cells’ self-renewal activity (Sabatini et al. 2003; Moubayidin et al. 2013) SCR is known to be involved in many other root developmental events such as regulation of formative stem cell division (Heidstra et al. 2004; Cui et al. 2007; Sozzani et al. 2010; Cruz-Ramírez et al. 2012) and protoxylem formation (Carlsbecker et al. 2010).
SCR sustains stem cell activities by directly suppressing ARR1 expression in the QC cells, thus titrating auxin production (Moubayidin et al. 2013). Auxin produced in the QC not only controls stem cell division activity but at the same time acts, via polar transport, as a long-distance signal to fine-tune the level of ARR1 transcription in the TZ (Moubayidin et al. 2013). In this way, ARR1 is positioned at the TZ where cell differentiation is initiated (Moubayidin et al. 2013).
Here, we report that in addition to controlling ARR1 position and activity via auxin (Moubayidin et al. 2013) SCR controls ARR1 levels also through GA from the endodermis, thus contributing to the fine-tuning of the ARR1-mediated cell differentiation commitment which is necessary to control root meristem size.
Materials and methods
Plant materials, growth conditions, and treatments
The Arabidopsis thaliana ecotypes Columbia (Col-0), Wassilewskija (Ws) and Landsberg erecta (Ler) were used. arr1-3 is in Col-0 background (Dello Ioio et al. 2007), scr-1 and sne-1 are in Ws background (Cui and Benfey 2009; Moubayidin et al. 2013), rga-24 is in Ler background (Moubayidin et al. 2010), pRGA:rga-Delta17 and pRGA:GFP:rga-Delta17 are in Ler background (Dill et al. 2001), ARR1:GUS, scr-1 ARR1:GUS and RGA:GFP were previously described (Dill et al. 2001; Mason et al. 2004; Moubayidin et al. 2010, 2013). Seeds were sterilized and grown as described previously (Perilli and Sabatini 2010). Gibberellin (GA3-Sigma) treatments were performed using 10 μM GA dissolved in ethanol, for 24 h on 5-day-old seedlings as previously described (Moubayidin et al. 2010). Experiments with scr-3 N9094 UAS::SCR:GR and relative control plants were performed by germinating seeds on plates containing 2 μM Dexamethasone (Dex-Sigma) for 5 days and then transferring the seedlings on plates containing 10 μM GA and 2 μM Dex for 24 h. pRGA:rga-Delta17 and pRGA:GFP:rga-Delta17 were obtained from Prof Tai-Ping Sun, Department of Biology, Duke University, Durham, NC, USA. sne-1 mutant (FLAG_461E03) was obtained from the Versailles Arabidopsis Stock Center, INRA, France.
Root length and meristem size analysis
For root length measurements, plates were photographed and the resulting images were analyzed using the ImageJ software available online (http://rsbweb.nih.gov/ij/) as described in Perilli and Sabatini (2010). Root meristem size is expressed as the number of cortex cells in a file extending from the quiescent center to the first elongated cortex cell, as described previously (Dello Ioio et al. 2007; Perilli and Sabatini 2010).
At the optical microscope, the distinction between dividing and differentiating root meristematic cells belonging to the cortex file is performed by the operator taking into account multiple parameters, i.e., the smaller size, the presence of a large central vacuole, and the denser cytoplasm of dividing meristematic cells versus differentiating ones. Moreover, both the position of the TZ in the outward tissue, the epidermis, and the extension of the lateral root cap are also evaluated as helpful references to univocally identify the cortex TZ and, hence, to measure meristem size. For each experiment, a minimum of 90 plants was analyzed using an Axio Imager.A2 (Zeiss) light microscopy and average and standard deviation were calculated. To measure root development over time, root length and meristem size were analyzed at different days post-germination (dpg): 3 dpg, 5 dpg, 7 dpg, 9 dpg, 12 dpg, and 15 dpg.
GUS histochemical assay
To visualize ARR1:GUS line in wild-type and mutant backgrounds, GUS histochemical assay was performed using the β-glucuronidase substrate X-gluc (5-bromo-4-chloro-3-indolyl glucuronide, Duchefa) as previously described (Perilli and Sabatini 2010; Moubayidin et al. 2013). Five-day-old seedlings were incubated for 16 h at 37 °C in the dark and imaged using the Axio Imager.A2 (Zeiss) microscopy.
qRT-PCR experiments
Total RNA was extracted from 5 days-old roots using the TRIsure reagent (Bioline), and the first strand cDNA was synthesized using the Superscript® III First Strand Synthesis System (Invitrogen). PCR amplification was carried out in the presence of the double-strand DNA-specific dye SYBR Green (Sigma). Amplification was monitored in real time with the 7300 Real-Time PCR System (Applied Biosystems). Experiments were performed in triplicates from RNA of root tissue. Amplification of ACTIN 2 served as control. Data are expressed in Log2 (2−ΔΔCt) ratio. qRT-PCR was performed three times on three independent RNA batches, and results were comparable in all experiments. Student t test was performed to know the actual significance of these data, the site used is: http://graphpad.com/quickcalcs/ttest2.cfm. Quantitative RT-PCR analysis was conducted using the following gene-specific primers:
ARR1 FW: 5′-TTGAAGAAACCGCGTGTCGTCT-3′ and ARR1 RV: 5′-CCTTCTCAACGCCGAGCTGATTAA-3′ for ARR1 (Moubayidin et al. 2013)
SNE FW: 5′-GTTCACCATGTCGTCGGAGA-3′ and
SNE RV: 5′-GAGCTCTGTTTCCGACAAGTG-3′ for SNEEZY (Sozzani et al. 2010)
RGA FW: 5′-CATGTTCCTCCACCGTCTTC-3′ and
RGA RV: 5′-AAAAAGGCAAAACCCTAGATC-3′ for RGA
ACT FW: 5′-CCTTCTCAACGCCGAGCTGATTAA-3′ and
ACT RV: 5′-GTGGATTCCAGCAGCTTCCAT-3′ for ACTIN2
Molecular cloning and genotyping
The SCR coding sequence lacking the stop codon was combined with the glucocorticoid receptor (GR) and cloned in the upstream activating sequence (UAS) cassette in between the 6xUAS and 35STerminator. Using the flanking NotI sites the UAS::SCR cassette was subsequently cloned into the pGreenII (Hellens et al. 2000)-based pGII124 binary vector (methotrexate resistance in planta) and transformed into scr-3 N9094 plants by floral dip.
The primers for genotyping plants for rga-24 deletion line were as follows:
RGA n212: 5′-GGTGATTTTCACGGTGGTTG-3′
rga-24 n205: 5′-TCGCTTAGTAGTTAGTACTC-3′
rga-24 n253: 5′-CATAGACCATAGTATTCGTGA-3′
RGA n212 and rga-24 n205 pair was used for amplifying the wild-type DNA copy of RGA, while rga-24 n205 and rga-24 n253 were used to amplify DNA from the mutant as described in de Lucas et al. (2008).
Confocal image processing
Confocal images of median longitudinal sections of 5-day-old roots were taken using a Zeiss LSM 780 microscope. A 10 mg/ml propidium iodide (Sigma) solution was used to visualize the cell wall.
Results
SCR mediates GA-dependent control of root meristem size from the endodermis
Tissue-specific complementation experiments where the SCR protein was reintroduced in the scr mutant only in specific districts of its expression domain suggest that activity of SCR in the QC is necessary and sufficient to control QC and stem cell function: in scr mutant plants where SCR was expressed only in the QC, stem cell activities and root growth were reestablished (Sabatini et al. 2003; Moubayidin et al. 2013). However, roots were shorter and root meristems smaller than wild type (Sabatini et al. 2003, Fig. 1a). Ground tissue (cortex and endodermis)-specific complementation could not sustain root meristem indeterminate growth but roots grew longer (Sabatini et al. 2003) and meristems were bigger compared to the scr mutant roots (Fig. 1b), indicating that SCR expression in the endodermis, besides regulating root radial patterning (Di Laurenzio et al. 1996; Heidstra et al. 2004) contributes also to root meristem size determination and overall root growth.
Fig. 1.
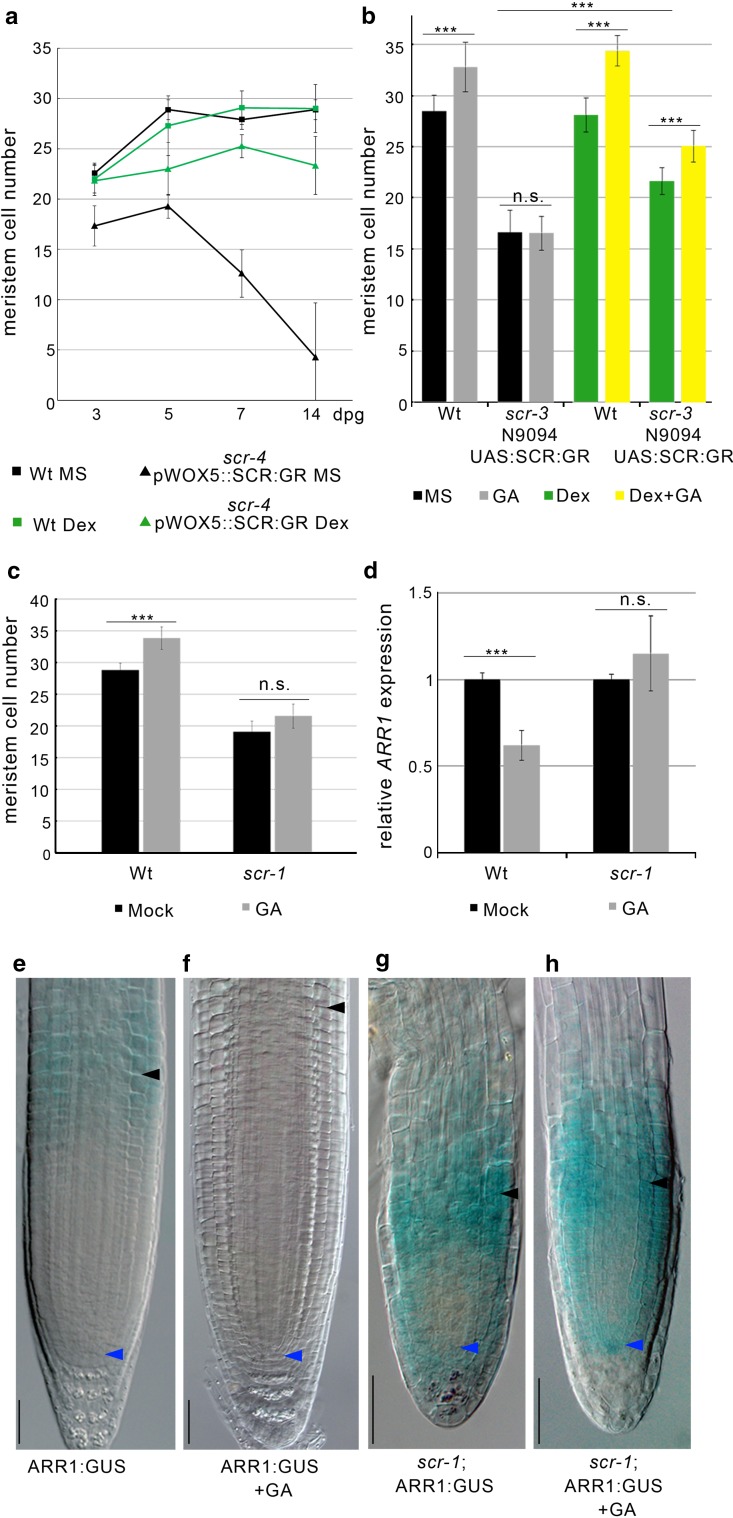
SCR controls ARR1 expression via gibberellin from the endodermis. a Root meristem cell number of wild type (Wt) and scr-4 pWOX5:UGU-GFP UAS:SCR:GR (scr-4 pWOX5::SCR:GR) treated (green lines) or untreated (black lines) with Dexamethasone (Dex) measured over time. dpg days post-germination. Error bars indicate standard deviation. b Root meristem cell number of Wt and scr-3 N9094 UAS::SCR::GR grew on MS (black columns) for 5 days treated for 24 h with 10 µM Gibberellin (GA, gray columns), 2 µM Dex (green columns), and both (Dex + GA, yellow columns). Error bars indicate standard deviation. Student’s t test; ***P < 0.001; n.s. not significant. c, d Root meristem cell number (c) and qRT-PCR of ARR1 expression (d) in 5 dpg wild-type and scr-1 roots grown 24 h on 10 µM gibberellin (GA) (gray columns) or mock (black columns). Error bars indicate standard deviation. Student’s t test; ***P < 0.001; n.s., not significant (n = 3 for qRT-PCR experiments). e–h Expression of the pARR1:ARR1:GUS construct in root meristems at 5 dpg of wild type (e, f) and of scr-1 (g, h) grown on mock (e, g) and 24 h on 10 µM GA (f, h). Blue and black arrowheads indicate the quiescent center and the cortex transition zone, respectively. Scale bars represent 50 µm
GA is predicted to regulate root meristem size from the endodermis (Achard et al. 2009; Ubeda-Tomás et al. 2009) and in this tissue, SCR is known to directly activate the SNEEZY (SNE) gene (Cui et al. 2007), one of the F-box proteins necessary for gibberellin-mediated degradation of the RGA protein (Ariizumi and Steber 2011; Ariizumi et al. 2011). We thus wondered whether SCR controls root meristem size from the endodermis sustaining GA activity. To this aim we first exposed wild-type and scr-1 mutant seedlings to exogenous GA application, monitoring root meristem size after 24 h.
Root meristem size was measured as the number of meristematic cortex cells using manual labeling at the optical microscope (see “Materials and methods”) rather than the program Cell-o-Tape (French et al. 2012). In fact, while automating and speeding up operations, this program does not take into account changes neither in the density of the cytoplasm nor in the vacuole size of differentiating cells, important characteristics that allow, in our experience, a more precise positioning of the transition zone (Perilli and Sabatini 2010).
Upon GA treatments wild-type root meristem were enlarged, while no increase in meristem size was observed in the scr-1 mutant (Fig. 1c, e–h) suggesting that SCR activity is necessary to mediate GA-dependent control on root meristem size. To understand whether SCR functions in the endodermis to control root meristem size via GA we expressed the SCR gene in this tissue of the scr-3 mutant roots using a GAL4/UAS trans-activation system. In wild-type roots the N9094 (GAL4,UAS∷GFP) enhancer trap line from the J. Haseloff (http://www.plantsci.cam.ac.uk/Haseloff) collection, GFP, is expressed specifically in the cortex and endodermis tissues (Suppl. Fig. S1a) while in the scr mutant it is expressed in the monolayer typical of this mutant (Suppl. Fig. S1b) (Sabatini et al. 2003). The N9094 driver was introduced in scr-3 mutant plants carrying a UAS::SCR::GR construct where the SCR protein activities are inducible by Dexamethasone (Dex). In the resulting scr-3 N9094 UAS::SCR::GR(N9094≫SCR:GR) plants, GFP expression monitored GAL4 activity and hence ectopic SCR::GR expression in the mutant background. scr-3N9094≫SCR:GR plants were germinated on Dex and were transferred on GA for 24 h. Analysis of the root meristem size of these plants revealed that the endodermis-specific induction of SCR was sufficient to enlarge the scr-3 meristem (Fig. 1b). Moreover, GA sensitivity was restored, as the meristem enlarged compared to GA-treated scr-3 roots (Fig. 1b). These data substantiate the notion that SCR mediates GA-dependent control of root meristem size functioning from the endodermis.
SCR mediates GA-dependent ARR1 transcriptional suppression
We have previously shown that GA controls root meristem size by lowering the expression of ARR1 at the TZ (Moubayidin et al. 2010) (Fig. 1c–f). We thus wondered whether SCR promotes the GA-mediated downregulation of ARR1 expression at the TZ, in addition to the regulation it has been shown to exert from the QC via auxin (Moubayidin et al. 2013).
Indeed, while GA negatively controls ARR1 transcription in the wild type (Fig. 1d–f) (Moubayidin et al. 2010), no downregulation of ARR1 expression was observed in the scr-1 mutant upon GA treatment as visualized both by quantitative real-time PCR (qRT-PCR) (Fig. 1d) and the analysis of scr-1 plants harboring a pARR1:ARR1:GUS (Fig. 1g, h). These data suggest that SCR is necessary to mediate the GA-dependent transcriptional downregulation of ARR1 in the control of root meristem size.
SCR mediates ARR1 transcriptional suppression via the RGA protein
Since GA acts via RGA to suppress ARR1 (Moubayidin et al. 2010) and SCR has been shown to directly control transcription of the specific RGA F-box protein SNE (Cui et al. 2007; Ariizumi and Steber 2011; Ariizumi et al. 2011), we hypothesized that the inability of GA to down-regulate ARR1 in the scr-1 mutant background could be due to increased levels of the RGA protein in the mutant.
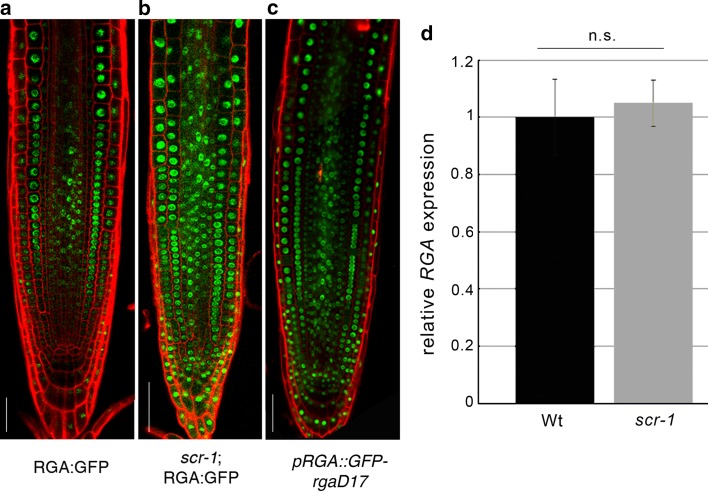
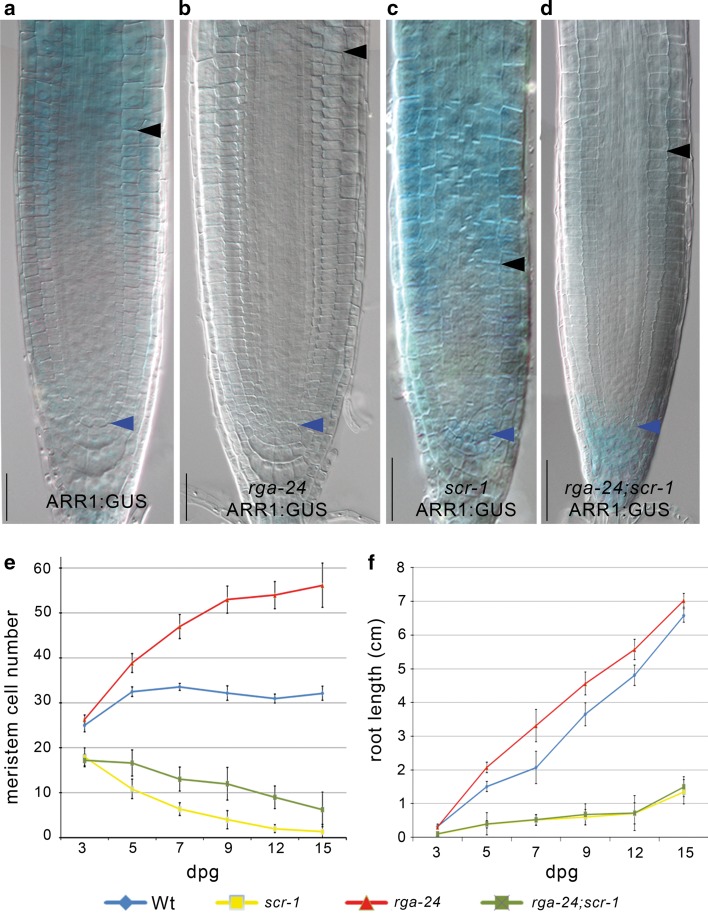
To test this hypothesis, we monitored RGA expression in the scr-1 mutant using a pRGA:RGA:GFP translational fusion (Silverstone et al. 2001). As predicted, the level of the RGA protein was higher than in wild-type roots, and the domain of RGA expression was extended to the whole meristem in scr-1, including the stem cell niche (Fig. 2a, b). In contrast, the level of the RGA transcript was not increased in the scr-1 mutant as revealed by qRT-PCR (Fig. 2d), suggesting that SCR controls RGA at the protein level. Accordingly, the translational fusion of the GA-insensitive semi-dominant mutant version of RGA protein under its own promoter and fused to the GFP, named rga-Delta17(pRGA:GFP:rga-Delta17) (Dill et al. 2001), was found to be expressed throughout the meristem (Fig. 2c), thus phenocopying the ectopic expression of RGA shown in scr-1 mutant (Fig. 2b). We next set to demonstrate how, in the scr-1 mutant, the high level of RGA (Fig. 2b) contributes to the high level of ARR1 expression (Figs. 1g and 3c) (Moubayidin et al. 2013). To this end, we generated rga-24;scr-1 double-mutant plants—where rga-24 is the loss of function mutant of RGA (Dill and Sun 2001)—carrying a pARR1:ARR1:GUS construct. As previously shown ARR1 levels in the rga-24 mutant background were low compared to the wild-type roots (Moubayidin et al. 2010) (Fig. 3a, b), while ARR1 was ectopically expressed in the QC of the scr-1 mutant (Fig. 3c) (Moubayidin et al. 2013). Compared to scr-1, ARR1 expression in rga-24;scr-1 plants was lower in the TZ but unaltered in the QC (Fig. 3c, d). This pattern of expression of ARR1 is consistent with the notion that, in the QC, SCR directly controls ARR1 (Moubayidin et al. 2013), while at the TZ, SCR modulates ARR1 expression cell non-autonomously via RGA. Interestingly, rga-24;scr-1 plants displayed a slightly longer root meristem than scr-1 plants but could not sustain indeterminate root growth (Fig. 2e, f), resembling scr mutant plants where SCR function was reintroduced only in the endodermis (Sabatini et al. 2003). Indeed, the stem cell niche was not rescued as visualized by the presence of differentiation markers in place of columella stem cells (Suppl. Figs S1c-S1f).
Fig. 2.
SCR controls RGA protein stability in the root meristem. a, b Expression of the pRGA:RGA:GFP (RGA:GFP) construct in wild-type roots (Wt) (a) and in the scr-1 mutant (b). c Root meristem expression of pRGA:GFP:rga-Delta17 (pRGA::GFP-rgaD17). Scale bars represent 50 µm. d qRT-PCR of RGA expression in 5 dpg wild-type and scr-1 roots. Error bars indicate standard deviation. Student’s t test; n.s., not significant, n = 3
Fig. 3.
SCR controls ARR1 expression at the transition zone via the DELLA protein RGA. a–d Expression of the pARR1:ARR1:GUS construct in Wt (a), rga-24 (b), scr-1 (c), and rga-24;scr-1 (d) mutants. Roots were analyzed 5 days post-germination (dpg). Scale bars represent 50 µm. e, f Root meristem cell number (e) and root length (f) measured over time of Wt, scr-1, rga-24, and rga-24;scr-1
Post-translational regulation of RGA controls root meristem size
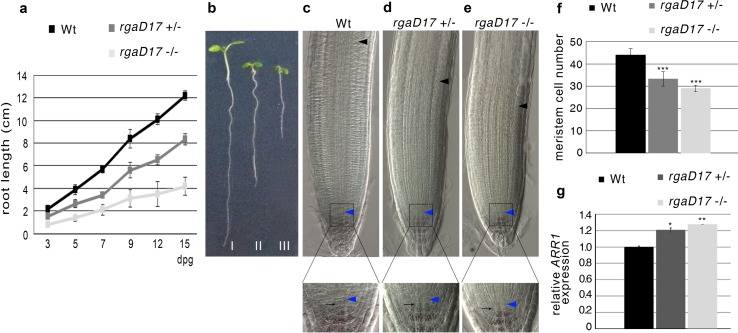
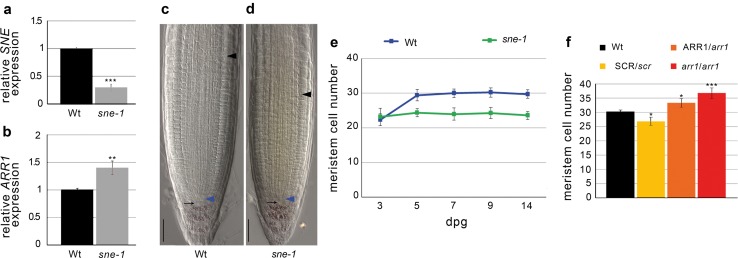
It has been shown that SCR directly activates SNE (Cui et al. 2007), an F-box responsible for DELLA protein degradation (Ariizumi and Steber 2011; Ariizumi et al. 2011). Moreover, we found that the degradation-resistant protein version of the RGA protein leads to an ectopic expression of RGA as shown in scr-1 (Fig.s 2a-2c). Therefore, we wondered whether SCR could regulate the RGA-dependent control of root meristem size by acting on RGA protein stability. First, we analyzed the rate of root growth and the meristem size of pRGA::rga-Delta17 (rga-Delta17) (Dill et al. 2001). rga-Delta17 heterozygote and homozygote seedlings, showed a dose-dependent phenotype compared to the control plants, as the roots grew shorter with increasing doses of rga-Delta17 copies (Fig. 4a, b). Accordingly, the root meristem of 5-day-old heterozygote and homozygote rga-Delta17 mutants showed a reduction in their size in a dose-dependent manner respect to the control, without affecting SCN activity (Fig. 4c–f). To link this gradual reduction in root length and meristem size to an enhancement in cell differentiation rate, we analyzed mRNA ARR1 levels in heterozygote and homozygote rga-Delta17 roots. We found that the content of ARR1 was inversely proportional to the root lengths, as ARR1 increased with increasing copies of the mutated version of RGA (Fig. 4g). All together these data showed that the GA-mediated degradation control of RGA is sufficient to fine-regulate root meristem size and root growth. Consequently, we predicted a similar effect on root meristem size and root growth control in plant mutated in the SNE gene. To this end, we used the sne-1 loss of function mutant (Cui and Benfey 2009) where the transcriptional level of SNE is severely reduced (Fig. 5a), and analyzed the ARR1 mRNA level by qRT-PCR. As visualized in Fig. 5b, the ARR1 transcript level was higher in the sne-1 mutant background than in the wild type, supporting the idea that the SNE protein is necessary for an indirect control of ARR1 expression. We then wondered whether the high level of ARR1 correlates with an increased differentiation rate at the TZ resulting in a shorter root meristem of the sne-1 mutant. Indeed the root meristem of the sne-1 mutant was smaller compared to wild type (Fig. 5c d). Moreover, sne-1 root meristem size is precociously imposed (Fig. 5e) according to the role of GA in controlling ARR1 transcript levels (Moubayidin et al. 2010), suggesting that the SNE protein is involved in root meristem size determination, presumably by controlling ARR1 activity.
Fig. 4.
Post-translational regulation of RGA controls root meristem size. a Root length measured over time of Wild type (Wt), pRGA::rga-Delta17 (rgaD17) heterozygote (±) and homozygote (−/−) mutants. b Seedlings of Wt (I), rgaD17 ± (II), and rgaD17 −/− (III) mutants. c–e Root meristem of 5-day-old Wt, rgaD17 heterozygote (±) and homozygote (−/−) mutants. Blue and black arrowheads indicate the QC and the cortex TZ, respectively. Blow up of the stem cell niches and lugol staining is shown in the bottom panel. Black arrows indicate the columella stem cells. f, g Root meristem cell number (f) and qRT-PCR of ARR1 expression (g) in 5 dpg Wt, rgaD17 heterozygote (±) and homozygote (−/−) mutants roots. Error bars indicate standard deviation. Student’s t test; ***P < 0.001; **P < 0.01; *P < 0.05 (n = 3 for qRT-PCR experiments)
Fig. 5.
Modulation of ARR1 level affects root meristem size. a, b qRT-PCR of SNE (a) and ARR1 (b) expression in 5dpg roots in wild-type and sne-1 background. Error bars indicate standard deviation. Student’s t test; **P < 0.01; ***P < 0.001, n = 3. c, d Root meristems of wild type (c) and sne-1 mutant (d) at 5 dpg. Blue and black arrowheads indicate the QC and the cortex TZ, respectively. e Root meristem cell number of wild type and sne-1 measured over time. Error bars indicate standard deviation. f Root meristem cell number measured at 5dpg of Wt roots, heterozygote SRC/scr-1 and ARR1/arr1-3 roots and homozygote arr1-3 roots. Error bars indicate standard deviation. Student’s t test; *P < 0.05; ***P < 0.001
ARR1 controls root meristem size in a dose-dependent manner
The data presented above suggest that the levels of ARR1 must be very finely regulated to determine root meristem size. We, therefore, set to establish whether a dose-dependent relation exists between the level of ARR1 and root meristem size by measuring the meristems of plant heterozygous for the scr-1 and arr1 mutations obtained by backcrossing the scr and arr1 mutants with the wild type. Five-day-old scr-1 heterozygous plants displayed root meristems shorter than Wt (Fig. 5f) whose roots kept growing over time. On the other hand, arr1 heterozygous plants displayed root meristems whose size was intermediate between wild-type plants and arr1 homozygous mutants (Fig. 5f), providing evidence for a dose-dependent control of meristem size by ARR1.
Discussion
The number of transit-amplifying cells, which undergo a finite number of cell divisions in the proximal meristem (the division zone), can influence the rate of the overall root growth. Therefore, a tight control of the cell number within the meristem tunes the driving force of the root growth.
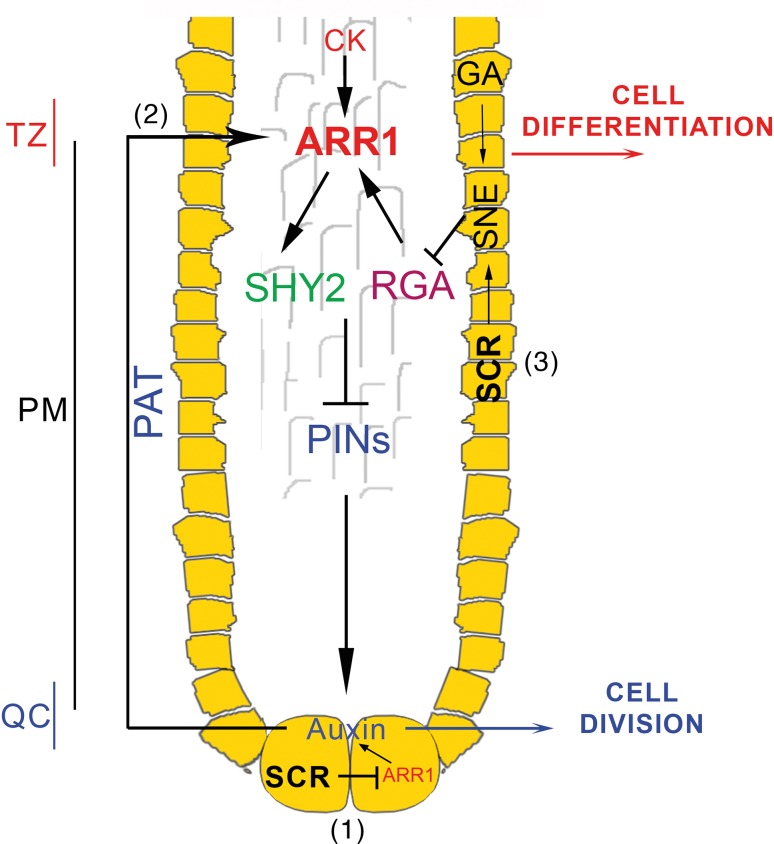
Our data unveil a novel, SCR-based, regulatory circuit that controls meristem size from the root endodermis: in this tissue SCR directly activates the gene encoding the F-box protein SNE (Cui et al. 2007) that in the presence of GA mediates the degradation of the DELLA protein RGA (Ariizumi and Steber 2011; Ariizumi et al. 2011) that in turn mediates the regulation of ARR1 levels at the TZ (Moubayidin et al. 2010) (Fig. 6). This integrates the current view of the control of the Arabidopsis root meristem size by SCR, and allows to propose a general model where the gene SCR is a key regulator of root meristem size acting from two different meristematic tissues on the level of ARR1: in the QC, SCR controls QC and stem cell activities (Sabatini et al. 2003) by directly suppressing the ARR1 gene (Moubayidin et al. 2013); in addition from the QC, SCR controls stem cell daughters’ differentiation at the TZ by controlling ARR1 expression at the TZ via auxin (Moubayidin et al. 2013) and from the endodermis via gibberellins (this work, Fig. 6). How, mechanistically, SCR controls ARR1 expression in different ways from different root tissues will require much further work. An intriguing possibility is that SCR needs different cell/tissue-specific co-factor to sustain its activity as already observed during asymmetric cell division processes necessary to generate the root ground tissues (Cruz-Ramìrez et al. 2012).
Fig. 6.
Model for the SCR-based regulatory circuits that control root meristem size. The model shows different tissue-specific activities of SCR on the control of ARR1 expression. Yellow cells represent SCR expression domain (endodermis and quiescent center, QC). From the QC, SCR represses ARR1 in two different ways: (1) SCR directly represses ARR1 in the QC cells, which in turn controls auxin production, thus enabling stem cell division. (2) With the same molecular mechanism SCR, via polar auxin transport (PAT), also exerts a long-distance control on ARR1 at the transition zone (TZ) enabling cytokinin to sustain cell differentiation via the SHY2/PINs (polar auxin transporters) pathway (Dello Ioio et al. 2008; Moubayidin et al. 2013). (3) From the root endodermis SCR positively transduces gibberellin (GA) signaling, presumably by controlling the F-box SNEEZY (SNE) (Cui et al. 2007), thus in turn controlling the DELLA protein RGA degradation (Ariizumi and Steber 2011; Ariizumi et al. 2011). In this way SCR tightly controls the level of ARR1-mediated differentiation, via RGA (Moubayidin et al. 2010 and this work)
Author contribution statement
L.M. conceived the research, planned and performed experiments, analyzed the data and wrote the manuscript; E.S. planned and performed experiments; L.G. performed experiments; I.T. constructed and provided scr-3 N9094 UAS::SCR::GR (N9094≫SCR:GR) seeds; R.H. supervised experiments and provided critical review of the manuscript; P.C. discussed experiments and provided critical review of the manuscript; S.S. conceived the research, analyzed the data and wrote the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Prof Tai-Ping Sun for providing pRGA:rga-Delta17 and pRGA:GFP:rga-Delta17 mutants. We thank Prof Lars Østergaard, Dr Raffaele Dello Ioio and Dr Marta Del Bianco for reading the manuscript. This work was supported by The European Research Council (Grant number 260368 to S.S., L.M., E.S., L.G.), and by MIUR, Progetti di Ricerca di Interesse Nazionale (Grant number 2010HEBBB8 to P.C.).
Abbreviations
- ARR1
ARABIDOPSIS RESPONSE REGULATOR 1
- DEX
Dexamethasone
- DPG
Days post-germination
- GA
Gibberellin
- QC
Quiescent center
- RGA
REPRESSOR OF ga1-3
- SCN
Stem cell niche
- SCR
SCARECROW
- SNE
SNEEZY
- TZ
Transition zone
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interests.
Contributor Information
Laila Moubayidin, Phone: +441603452135, Email: laila.moubayidin@jic.ac.uk.
Sabrina Sabatini, Phone: +390649917916, Email: sabrina.sabatini@uniroma1.it.
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19:1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Steber CM. Mutations in the F-box gene SNEEZY result in decreased Arabidopsis GA signaling. Plant Signal Behav. 2011;6:831–833. doi: 10.4161/psb.6.6.15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Lawrence PK, Steber CM. The role of two f-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 2011;155:765–775. doi: 10.1104/pp.110.166272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, Campilho A, Sebastian J, Bowman JL, Helariutta Y, Benfey PN. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, Caballero-Pérez J, Horvath B, Long Y, Mähönen AP, Zhang H, Xu J, Murray JA, Benfey PN, Bako L, Marée AF, Scheres B. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell. 2012;150:1002–1015. doi: 10.1016/j.cell.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Benfey PN. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 2009;58:1016–1027. doi: 10.1111/j.1365-313X.2009.03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;24:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/S0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Wilson MH, Kenobi K, Dietrich D, Voß U, Ubeda-Tomás S, Pridmore TP, Wells DM. Identifying biological landmarks using a novel cell measuring image analysis tool: Cell-o-Tape. Plant Methods. 2012;8:7. doi: 10.1186/1746-4811-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Sponsel V. A century of gibberellin research. J Plant Growth Regul. 2015;2015:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S. Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol. 2014;15:301–312. doi: 10.1038/nrm3790. [DOI] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/A:1006496308160. [DOI] [PubMed] [Google Scholar]
- Lee MH, Kim B, Song SK, Heo JO, Yu NI, Lee SA, Kim M, Kim DG, Sohn SO, Lim CE, Chang KS, Lee MM, Lim J. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 2004;135:927–937. doi: 10.1104/pp.103.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol. 2010;20:1138–1143. doi: 10.1016/j.cub.2010.05.035. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sozzani R, Pacifici E, Salvi E, Terpstra I, Bao D, van Dijken A, Dello Ioio R, Perilli S, Ljung K, Benfey PN, Heidstra R, Costantino P, Sabatini S. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell. 2013;26:405–415. doi: 10.1016/j.devcel.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14:S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Sabatini S. Analysis of root meristem size development. Methods Mol Biol. 2010;655:177–187. doi: 10.1007/978-1-60761-765-5_12. [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313X.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/tpc.13.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007;48:263–277. doi: 10.1093/pcp/pcl063. [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Federici F, Casimiro I, Beemster GT, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 2009;19:1194–1199. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.