Abstract
Animal models have contributed to our understanding of breast cancer, with publication of results in high-impact journals almost invariably requiring extensive in vivo experimentation. As such, many laboratories hold large collections of surplus animal material, with only a fraction being used in publications relating to the original projects. Despite being developed at considerable cost, this material is an invisible and hence an underutilised resource, which often ends up being discarded. Within the breast cancer research community there is both a need and desire to make this valuable material available for researchers. Lack of a coordinated system for visualisation and localisation of this has prevented progress. To fulfil this unmet need, we have developed a novel initiative called Sharing Experimental Animal Resources: Coordinating Holdings—Breast (SEARCHBreast) which facilitates sharing of archival tissue between researchers on a collaborative basis and, de facto will reduce overall usage of animal models in breast cancer research. A secure searchable database has been developed where researchers can find, share, or upload materials related to animal models of breast cancer, including genetic and transplant models. SEARCHBreast is a virtual compendium where the physical material remains with the original laboratory. A bioanalysis pipeline is being developed for the analysis of transcriptomics data associated with mouse models, allowing comparative study with human and cell line data. Additionally, SEARCHBreast is committed to promoting the use of humanised breast tissue models as replacement alternatives to animals. Access to this unique resource is freely available to all academic researchers following registration at https://searchbreast.org.
Keywords: Breast cancer, Animal models, Animal material, 3Rs
Introduction
Pre-clinical studies in animals have translated to human benefit in breast cancer. A key example is the pharmaceutical tamoxifen, which has saved the lives of millions of people diagnosed with oestrogen receptor positive (ER+) breast cancer. Its approval as a clinical therapy was aided by elegant work with rodents conducted in the 1970s [1, 2]. This work also demonstrated the benefits of tamoxifen in breast cancer chemoprevention [3, 4]. Other examples resulting from initial pre-clinical studies in rodents include work leading to the development of trastuzumab [5, 6], and aromatase inhibitors [7, 8] as targeted therapies in clinical breast cancer. Results generated using in vitro models do not translate directly into clinical trials hence a stage of animal experimentation is involved in the development of novel breast cancer therapies. In addition, publication of studies in high-impact journals most often requires that data are verified in at least one (and sometimes in several) in vivo model. As an example, [9] used the MMTV-v-Ha-ras transgenic mouse model, developed originally by the Leder [10] and Jolicoeur groups [11] to examine the potential role of farnesyltransferase inhibitors on tumour regression. Similarly, the efficacy of dipyridamole in preventing breast cancer initiation, progression and metastasis was tested in MMTV-PyMT transgenic mice [12], which were developed originally by the Muller lab [13]. Of these two examples (there are many more, as reviewed by [14]) both required extensive breeding programmes and subsequent hypothesis testing. Hence, a single project may involve the use of a large number of animals, each generating surplus tissues and other material that potentially could be utilised in future studies by other research groups.
Animal research includes syngeneic, xenograft and genetically modified (GEM) models. These are now an integral part of thousands of original publications each year, yielding significant advances across many research fields. However, this results in significant animal sacrifice; in 2013 alone, an estimated 20 million animals were used in scientific studies in the European Union and the United States. Scientists are encouraged to consider alternatives before embarking on new animal experiments, including due diligence on the validity of the model being considered in recapitulating human disease and/or phenotype, as well as employing the principles of the 3Rs—Replacement, Reduction and Refinement. The 3Rs were developed over 50 years ago as a framework for humane animal research [15] and is now part of national and international legislation regulating the use of animals in experiments. Not generally considered in this context is the potential wastage of surplus material from animal studies, or whether material is already available that can be used to answer the research question. Typically, research groups tend to collect materials from their projects, with the surplus sometimes being stored indefinitely, but also frequently being discarded at the end of the project. While the collection, storage, archiving and access policies for human clinical samples and transcriptomic data are routinely included in grant applications and often manuscript submissions, there is no obligation for investigators using animal models to do the same. This has economic, scientific and ethical shortcomings, resulting in considerable waste including duplication of experiments. This potentially lost material represents a valuable resource that could be used productively if there was a way for other researchers to identify it. Furthermore, if pre-existing models or materials were accessible elsewhere, obtaining these instead of recreating them would lead to fewer animals being used, as well as saving both time and money [16], illustrated schematically in Fig. 1. Currently, researchers lack the resources (money, databases, space, time) to generate and maintain systems that would catalogue their archival material and make it visible to other interested parties. SEARCHBreast therefore provides the missing link between those who have material available and those who would like to access existing samples, rather than repeating studies that have already been done. In particular, investigators without access to animal facilities may benefit from joining SEARCHBreast, providing them with an opportunity to carry out the type of studies that would increase the impact of their research.
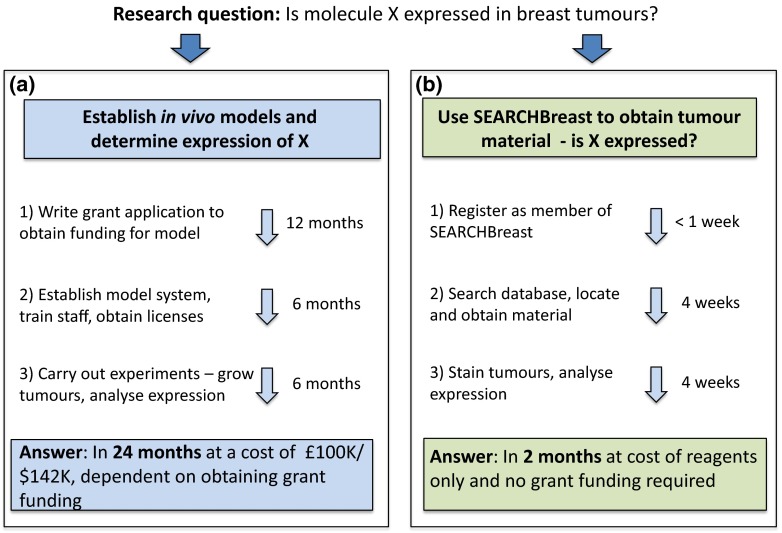
Fig. 1.
An example of the benefits of using SEARCHBreast to source material, to study if molecule X is expressed in breast tumour models. The cost/time required for generation of material from scratch is shown in a while in b the same process using SEARCHBreast is much faster and at minimal cost to the researcher. This has the added advantage of identifying the most relevant model for future studies from a more extensive range of models than may be available to the researcher locally
Establishing SEARCHBreast for more efficient use of materials derived from breast cancer studies in animals
Recognising that the majority of available animal resources physically store and provide live animals (e.g. The Jackson Laboratory, [17] or The Knockout Mouse Project (KOMP) Repository [18] which uses embryonic stem cells to create new genetic models), we considered that a virtual resource which made pre-existing archived materials already generated from animal models, more visible and accessible to the breast cancer research community, would be attractive to researchers. This would provide scientists with the opportunity of fostering greater collaborations, while at the same time reducing the numbers of animals used and encouraging better use of animal models. To confirm the need for such a resource, a widespread consultation was conducted with the UK breast cancer research community, resulting in overwhelming support for the initiative. As a result, Sharing Experimental Animal Resources: Coordinating Holdings—Breast (SEARCHBreast) was created, with funding secured through an Infrastructure for Innovation award from the National Centre for the Replacement, Refinement & Reduction of Animals in Research (NC3Rs). The resource provides scientists with the opportunity for surplus animal tissue to be utilised by other breast cancer investigators to further their studies, after the initial holder of the materials has finished their work. Thus, scientists requiring animal models or derived materials are provided with a mechanism for finding and accessing those generated by expert groups, avoiding the need to create in vivo models from scratch and/or validating a pathway of interest in accurate models prior to embarking on time consuming and expensive breeding regimes. Through this, SEARCHBreast aims to address each of the Replacement, Reduction and Refinement aspects of the 3Rs, while also accelerating breast cancer research.
SEARCHBreast: a comprehensive database of breast cancer mouse models
SEARCHBreast does not physically store material; the database is a virtual online resource available through the website, https://searchbreast.org. The database consists of descriptions of animal models (syngeneic, xenograft (including PDX) and GEM) and associated tissue and materials (mainly FFPE tissues and histological slides), which are available for sharing between academics within the breast cancer research community on a collaborative basis.
The SEARCHBreast visual identity and strapline; connect, share, discover (Fig. 2) embraces our ethos of connecting breast cancer researchers to encourage sharing of resources, knowledge and ideas thereby increasing the quality of research outputs to discover more about breast cancer. We developed the resource by inviting key UK breast cancer researchers with a track record in the use of complex model systems to attend two initial workshops. Their input helped shape the structure and content of the SEARCHBreast database. This was an iterative process to develop a system that contained sufficient information to facilitate the localisation of the material that was quick and easy to use for scientists. The database contains key information about the material available (e.g. tumour type, metastasis, fixation method), and additional details (e.g. therapeutic doses, time points, group sizes, analyses already performed) are exchanged once a collaboration is agreed. This approach aims to make the material entry process as simple as possible, minimising the time it takes to register material. The types of information captured and the materials available are shown in Table 1.
Fig. 2.
SEARCHBreast logo and strapline
Table 1.
Information about the type of material available on the SEARCHBreast portal
| Mouse information | Tissue availablea | Phenotypes | Analysis performed |
|---|---|---|---|
| Model name, cell line, Jax stock number, GEM allele, transplantation site, strain, sex | Tumour, lung, mammary fat pad, lymph node, heart, spleen, liver, long bones, skull, serum, vertebrae, circulating DNA embryos, sperm, live animals, | ER/PR/HER2 status, metastasis sites metastatic penetrance CK5/CK8/p63/SMA status | Tumour volume, general histology, vascularisation, immune infiltrate, apoptosis, proliferation, transcriptome analysis, metastatic assay, microenvironment analysis |
aMany of the tissues available are stored as formalin-fixed paraffin-embedded, frozen, or as histological slides, enabling user-friendly samples to be sent ready for immediate use by scientists
The website and underlying database are designed for security. They have access restrictions and password encryption, and additional security features have been incorporated. All users of SEARCHBreast wishing to view the materials available are first required to register. Once registered, they can upload models they may have available to share, or search the database for material that may augment their research. Having identified material they wish to use, researchers can request it from the holder, via an email enquiry to the SEARCHBreast coordinator. The name of the holder and their institution are not identified when the database is viewed. If the owner chooses to share, they contact the requestor and the collaboration proceeds according to the regulations of the institutions involved (Fig. 3). Importantly, there is no obligation to share the material registered in SEARCHBreast. If the holder of the material considers that a proposed collaboration is not of interest/benefit/sufficient quality etc., they are entitled to decline the invitation to share. The response is communicated by the SEARCHBreast coordinator. No costs are associated with using SEARCHBreast; registration, depositing and searching are free of charge. Subsequent costs for material exchanged is by arrangement between the collaborating researchers. SEARCHBreast only ask that they are acknowledged in publications arising from productive exchanges.
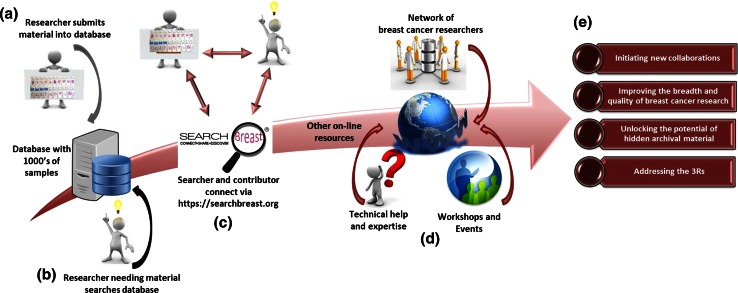
Fig. 3.
Schematic of information flow through the SEARCHBreast website and database. Researchers enter models into the database via an interface at the website (a). Complemented by bioinformatics software, it can be searched by scientists looking for models to further their research (b). Once material is identified this can be requested via SEARCHBreast (c). The SEARCHBreast website provides access to other useful online information plus upcoming workshops and events (d). The expected outputs from SEARCHBreast are shown (e)
Resource tools to improve the use of animals in research
As well as widening access to archival animal materials, the SEARCHBreast website can be used to assist researchers apply the 3Rs to their work. Links to online technical information written by experts provide standard operating procedures (SOPs) that give guidelines on experimental procedures for animal models.
Although scientists should consider the 3Rs in their research, an unintended consequence is the increasing prevalence of underpowered results that invalidate studies through few samples being used. This leads to wasted material through ill-considered experimental design [19]. The SEARCHBreast website provides links to additional resources to assist researchers in, for example, determining the correct sample size during the design of their experiments using Experimental Design Assistant [20]
A bioinformatics pipeline is being integrated into the SEARCHBreast website for the analysis of transcriptomics data associated with the mouse models within the database. This will allow users to identify models that are known to reflect changes in certain genes, such as specific oncogenes. Users will consequently be able to find models that work best for the molecular investigations they wish to pursue. To support this, a comparative large-scale analysis of gene expression profiles from breast cancer mouse models, human breast cancer samples and cell lines is underway, with the results being incorporated into the search facility of the SEARCHBreast database.
Other features on the website include explanations of the principles of the 3Rs and links to associated resources, including the NC3Rs website [21] and the NC3Rs Arrive guidelines [22] which have been developed to improve the reporting of animal research, plus information about upcoming workshops and events hosted by, or attended by, the SEARCHBreast team.
Humanised 3D models as alternatives to animal models
A further aim of SEARCHBreast is to promote the use of alternative humanised models of breast cancer by aligning with another resource, the Breast Cancer Now Tissue Bank (BCNTB) [23, 24]. The BCNTB provides human breast cancer and normal breast tissue for research, and specialises in providing primary cell cultures derived from these cells. SEARCHBreast encourages scientists to consider developing 3D cell culture models as an alternative to animal models. Researchers who apply for human breast material from the BCNTB will, via SEARCHbreast, have a mechanism for matching their in vitro work to the most relevant in vivo model.
In respect of this aim, SEARCHBreast organised a workshop dedicated to the replacement of animals in breast cancer research using 3D models. This was a networking opportunity which focused on practical considerations on handling humanised cell and tissue models; providing solutions to the perceived barriers of access to samples; and tips on establishing primary cultures with a view to using these in 3D breast cancer models, and introduced new technology allowing 3D tissue modelling of breast cancer using virtual pathology [25]. Many of the lectures from this event are available to view on the SEARCHBreast website (https://searchbreast.org/) with the workshop proceedings recently published [26].
Discussion
For breast cancer researchers to publish their work in higher tier scientific journals it is becoming increasingly necessary to include an in vivo component, placing pressure on scientists to include animal experimentation. However, many researchers do not have the expertise or the financial resources to carry out complex in vivo studies. SEARCHBreast has begun to address this by making materials that might otherwise go to waste more visible and accessible to researchers, potentially increasing the quality and impact of their research through translation/validation in an in vivo model. This may benefit early career researchers in particular, who may require pilot data from in vivo studies in order to obtain independent funding, as well as scientists without access to animal facilities or the necessary legislative approval which in itself can take months to put in place in some countries. In contrast to the emphasis put on safe storage, cataloguing, sample tracking and shared utilisation through tissue access committees for material generated from human studies, there has been little interest in developing similar best practise for animal studies, despite the considerable investment they represent both in terms of time and money. It is unlikely to remain acceptable that this potentially valuable resource is discarded due to lack of storage facilities, and funding bodies may in the future request a plan for storage and sharing of any excess material. Joining SEARCHBreast represents an easy and accessible way by which researchers can address this, as well as increasing the impact of their work.
We anticipate that SEARCHBreast will encourage funding bodies to include consideration of what to do with any surplus material at the end of a funded study, and to stimulate researchers to become members of SEARCHBreast and register their available material. Prior to the establishment of SEARCHBreast, no significant organised system had been made to enhance visibility and facilitate sharing of material from animal models of breast cancer. This pioneering approach aims to produce more efficient and effective use of these models, opening up new opportunities in breast cancer research. Importantly, both parties will benefit from collaborating through sharing material; the researcher seeking material potentially avoiding having to do in vivo studies, and the holder of the material gaining new and unplanned collaborative studies, through making the otherwise unused resource available. SEARCHBreast is the initiator and driver of this novel way of viewing unused material as an untapped enabler of new research, at the same time firmly addressing the 3Rs. Furthermore, in the future, this platform may be extended beyond breast cancer to apply these principles in other diseases.
Acknowledgments
We thank all workshop participants and SEARCHBreast members for helping shape the SEARCHBreast resource and for using this to deposit their models. SEARCHBreast was developed by an Infrastructure for Impact Award from NC3Rs (Grant Ref: NC/L001004/1).
Compliance with ethical standards
Conflict of interest
None.
Footnotes
Karen Blyth and Phil Carter have contributed equally to this work.
References
- 1.Jordan VC, Jaspan T. Tamoxifen as an anti-tumour agent: oestrogen binding as a predictive test for tumour response. J Endocrinol. 1976;68:453–460. doi: 10.1677/joe.0.0680453. [DOI] [PubMed] [Google Scholar]
- 2.Jordan VC, Koerner S. Tamoxifen (ICI 46,474) and the human carcinoma 8S oestrogen receptor. Eur J Cancer. 1975;11:205–206. doi: 10.1016/0014-2964(75)90119-X. [DOI] [PubMed] [Google Scholar]
- 3.Jordan VC. Effect of tamoxifen (ICI 46,474) on initiation and growth of DMBA-induced rat mammary carcinomata. Eur J Cancer. 1976;12:419–424. doi: 10.1016/0014-2964(76)90030-X. [DOI] [PubMed] [Google Scholar]
- 4.Jordan VC. Tamoxifen (ICI46,474) as a targeted therapy to treat and prevent breast cancer. Br J Pharmacol. 2006;147(Suppl 1):S269–S276. doi: 10.1038/sj.bjp.0706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepard HM, Lewis GD, Sarup JC, Fendly BM, Maneval D, Mordenti J, Figari I, Kotts CE, Palladino MA, Jr, Ullrich A, et al. Monoclonal antibody therapy of human cancer: taking the HER2 protooncogene to the clinic. J Clin Immunol. 1991;11:117–127. doi: 10.1007/BF00918679. [DOI] [PubMed] [Google Scholar]
- 6.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chumsri S, Howes T, Bao T, Sabnis G, Brodie A. Aromatase, aromatase inhibitors, and breast cancer. J Steroid Biochem Mol Biol. 2011;125:13–22. doi: 10.1016/j.jsbmb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chumsri S, Brodie A. Aromatase inhibitors and breast cancer. Horm Mol Biol Clin Investig. 2012;9:119–126. doi: 10.1515/hmbci-2012-0001. [DOI] [PubMed] [Google Scholar]
- 9.Barrington RE, Subler MA, Rands E, Omer CA, Miller PJ, Hundley JE, Koester SK, Troyer DA, Bearss DJ, Conner MW, Gibbs JB, Hamilton K, Koblan KS, Mosser SD, O’Neill TJ, Schaber MD, Senderak ET, Windle JJ, Oliff A, Kohl NE. A farnesyltransferase inhibitor induces tumor regression in transgenic mice harboring multiple oncogenic mutations by mediating alterations in both cell cycle control and apoptosis. Mol Cell Biol. 1998;18:85–92. doi: 10.1128/MCB.18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 11.Tremblay PJ, Pothier F, Hoang T, Tremblay G, Brownstein S, Liszauer A, Jolicoeur P. Transgenic mice carrying the mouse mammary tumor virus ras fusion gene: distinct effects in various tissues. Mol Cell Biol. 1989;9:854–859. doi: 10.1128/MCB.9.2.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Schwab LP, Fan M, Seagroves TN, Buolamwini JK. Chemoprevention activity of dipyridamole in the MMTV-PyMT transgenic mouse model of breast cancer. Cancer Prev Res (Phila) 2013;6:437–447. doi: 10.1158/1940-6207.CAPR-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/MCB.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menezes ME, Das SK, Emdad L, Windle JJ, Wang XY, Sarkar D, Fisher PB. Genetically engineered mice as experimental tools to dissect the critical events in breast cancer. Adv Cancer Res. 2014;121:331–382. doi: 10.1016/B978-0-12-800249-0.00008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell WMS, Burch RL. The principles of humane experimental technique. London: Methuen; 1959. [Google Scholar]
- 16.Speirs V. Animal research: share surplus animal tissue. Nature. 2015;522:156. doi: 10.1038/522156c. [DOI] [PubMed] [Google Scholar]
- 17.Jackson-Laboratory (1999) The Jackson Laboratory: The Mutant Mouse Resource and Research Center (MMRRC). https://www.mmrrc.org/. Accessed 08 Feb 2016
- 18.Lloyd KC. A knockout mouse resource for the biomedical research community. Ann NYAcad Sci. 2011;1245:24–26. doi: 10.1111/j.1749-6632.2011.06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cressey D. UK funders demand strong statistics for animal studies. Nature. 2015;520:271–272. doi: 10.1038/520271a. [DOI] [PubMed] [Google Scholar]
- 20.NC3RS (2015) The NC3Rs Experimental Design Assistant. https://eda.nc3rs.org.uk/. Accessed 08 Feb 2016
- 21.NC3Rs (2005) The National Centre for the Replacement Refinement of Animals in Research. http://www.nc3rs.org.uk/. Accessed 08 Feb 2016
- 22.NC3Rs (2010) The NC3Rs Arrive Guidelines. https://www.nc3rs.org.uk/arrive-guidelines. Accessed 08 Feb 2016
- 23.Speirs V, Morgan A. Breast cancer: Investment biobanking--increased returns from tissue samples. Nat Rev Clin Oncol. 2013;10:128–129. doi: 10.1038/nrclinonc.2013.19. [DOI] [PubMed] [Google Scholar]
- 24.BCNTB (2010) The Breast Cancer Now Tissue Bank. https://www.breastcancertissuebank.org/. Accessed 31 March 2016
- 25.Booth ME, Treanor D, Roberts N, Magee DR, Speirs V, Hanby AM. Three-dimensional reconstruction of ductal carcinoma in situ with virtual slides. Histopathology. 2015;66:966–973. doi: 10.1111/his.12561. [DOI] [PubMed] [Google Scholar]
- 26.Morrissey B, Blyth K, Carter P, Chelala C, Holen I, Jones L, Speirs V. SEARCHBreast Workshop Proceedings: 3D Modelling of Breast Cancer. Altern Lab Anim. 2015;43:367–3375. doi: 10.1177/026119291504300604. [DOI] [PubMed] [Google Scholar]