Abstract
Candida albicans is the major invasive fungal pathogen of humans, causing diseases ranging from superficial mucosal infections to disseminated, systemic infections that are often life-threatening. Resistance of C. albicans to antifungal agents and limited antifungal agents has potentially serious implications for management of infections. As a famous multiherb prescription in China, Huanglian Jiedu Decoction (HLJJD, Orengedokuto in Japan) is efficient against Trichophyton mentagrophytes and C. albicans. But the antifungal mechanism of HLJDD remains unclear. In this study, by using RNA-seq technique, we performed a transcriptomics analysis of gene expression changes for C. albicans under the treatment of HLJDD. A total of 6057 predicted protein-encoding genes were identified. By gene expression analysis, we obtained a total of 735 differentially expressed genes (DEGs), including 700 upregulated genes and 35 downregulated genes. Genes encoding multidrug transporters such as ABC transporter and MFS transporter were identified to be significantly upregulated. Meanwhile, by pathway enrichment analysis, we identified 26 significant pathways, in which pathways of DNA replication and transporter activity were mainly involved. These results might provide insights for the inhibition mechanism of HLJDD against C. albicans.
1. Introduction
Candida albicans is the most prevalent opportunistic fungal pathogen implicated in superficial mucosal infections as well as invasive disseminated infections, especially in immunocompromised patients [1, 2]. C. albicans infections are usually treated with antifungal agents, such as azoles, echinocandins, and polyene drugs. Limited by the number of available antifungal targets, the antifungal agents still remain restricted. The azoles are the most widely used drugs for treating pathogenic fungal infections. Sterol 14α-demethylase (ERG11) is an ancestral activity of the cytochrome P450 superfamily, which is required for ergosterol biosynthesis in fungi and cholesterol biosynthesis in mammals [3]. As a key enzyme of sterol biosynthesis, Erg11 is the main target for therapeutic azole antifungal drugs [4, 5].
Widespread overuse of azole drugs for decades has led to the occurrence of drug-resistant isolates [6–8]. The prolonged and repeated treatment of OPC (oropharyngeal candidiasis) in AIDS patients has resulted in an increasing frequency of therapy failures caused by the emergence of fluconazole-resistant C. albicans strains. In one study, the levels of fluconazole resistance of a series of 17 clinical isolates taken from a single HIV-infected patient who was treated with azoles over 2 years increased over 200-fold [9]. In recent years, the incidence of azole-resistant strains of C. albicans has increased, especially the rapid emergence of fluconazole-resistant strains. In the vast majority of countries, far less than 10% of C. albicans strains isolated from 1997 to 2001 are resistant to fluconazole [10]. But recent study in China showed that the rate of fluconazole resistance in C. albicans was almost 14.1% [11]. In USA, compared with 2008, the proportion of cases identified from 2008 to 2013 from Georgia and Maryland with fluconazole resistance decreased (GA: 8.0% to 7.1%, −10%; MD: 6.6% to 4.9%, −25%), but the proportion of cases with an isolate resistant to an echinocandin increased (GA: 1.2% to 2.9%, +147%; MD: 2.0% to 3.5%, +77%) [12]. So far, several resistance mechanisms of C. albicans have been well characterized: alterations in the sterol biosynthesis pathway, mutations in the ERG11 gene encoding the drug target enzyme, overexpression of the ERG11 gene, and overexpression of genes encoding efflux pumps [13]. Resistance of C. albicans to antifungal agents and limited antifungal agents has potentially serious implications for management of infections.
As a famous multiherb prescription in China, Huanglian Jiedu Decoction (HLJJD, Orengedokuto in Japan) is an aqueous extract of 4 herbal materials, Coptidis Rhizoma, Scutellariae Radix, Phellodendri Cortex, and Gardeniae Fructus with the ratio of 3 : 2 : 2 : 3. HLJJD was first mentioned in the book Wai-Tai-Mi-Yao compiled by Wang Tao in the Tang dynasty (about 752 AD), and it has been widely used in the clinical practice in China and officially listed in the Chinese Pharmacopoeia [14]. HLJJD has been widely used in the treatment of gastrointestinal disorders, inflammation and cardiovascular diseases, and Alzheimer's disease in China [15–17]. Modern pharmacological research also demonstrated multiple biological activities of HLJDD: decreasing levels of plasma glucose and blood lipid in type 2 diabetes mellitus [18, 19]; increasing the cerebral blood flow, inhibiting the platelet aggregation; reducing hypertension and altering the gene expression profiles of spontaneous hypertensive rats [20–23]; reducing hepatic triglyceride accumulation, restraining the preadipocyte differentiation and lipid accumulation, inhibiting the lipid peroxidation, and preventing atherosclerosis [15, 24–26]. Anti-inflammatory effects of HLJDD were also investigated in some papers [18, 27–29]. Moreover, in Mugil cephalus, 1% modified HLJDD feeding for 28 days may be an optimal dose to prevent Lactococcus garvieae infection and could be used in aquaculture industries. In in vitro study, the modified HLJDD also activated the plasma bactericidal activities [30].
Each herb of HLJDD contains many chemical components. Some papers had reported the content determinations of components contained in HLJDD [31, 32]. HLJDD contains multiple bioactive secondary metabolites, mainly including alkaloids from Coptidis Rhizoma and Phellodendri Cortex, flavonoids from Scutellariae Radix, and terpenes from Gardeniae Fructus. There are 4 typical compounds from HLJDD: geniposide, baicalin, berberine, and baicalein [27]. Further study showed that the combination of fluconazole and baicalein or berberine produced potently synergistic action in vitro, while baicalein and berberine showed weak antifungal activity when they were tested alone [33, 34]. Our preliminary work showed that HLJDD is efficient against Trichophyton mentagrophytes and C. albicans [35]. HLJDD showed its impressive antifungal effect by multitarget and multichannel actions, due to the multiple components. For that reason, the use of HLJDD may be more beneficial to human health in fungal infection treatment as diverse mechanisms showed complementary effects between herbs. But the antifungal mechanism of HLJDD remains unclear.
RNA-seq (deep-sequencing of cDNA) has been used successfully to identify and quantify gene expression at a genome scale level under different conditions or in different cell types. Moreover, it is significantly more sensitive than microarray hybridization approaches [36]. This approach has already been used in C. albicans to generate a high-resolution map of the C. albicans transcriptome under several different environmental conditions [37]. The effect of berberine chloride on Microsporum canis infection was analyzed by the construction of a transcriptome of the M. canis cellular responses upon berberine treatment [38]. Therefore, in this study, by using RNA-seq technique, we performed a large-scale analysis of gene expression changes when C. albicans was exposed to HLJDD, to better understand how HLJDD inhibits the growth of C. albicans.
2. Materials and Methods
2.1. Strain and Culture Conditions
The C. albicans strain used in this study is SC5314 [39]. C. albicans strains were routinely grown on YPD (1% yeast extract, 2% peptone, and 2% glucose) medium.
2.2. Preparation of the Extract of HLJDD
The herbal medicines of modified HLJDD were dried at 40°C for 24 h and then pulverized to powder using a mechanical blender. 0.5%, 1%, and 2% w/w of the powder was prepared and boiled for 30 min with 200 mL of deionized water, and the aqueous extracts were filtered through Whatman number 1 filter paper. The HLJDD residues were also boiled with another 200 mL of deionized water.
2.3. Determination of Sensitivity of the SC5314 Strain to HLJDD
Antifungal susceptibility testing was performed by using the CLSI M27-A3 microbroth dilution method [40]. MICs were determined after growth at 30°C for 24 h for HLJDD. MICs were read as the lowest drug concentration producing a prominent decrease in turbidity translating to 100% growth reduction compared with the drug-free control.
2.4. Total RNA Extraction
To identify genes in the early response of C. albicans to HLJDD, we treated the isolate with HLJDD at 20 mg/mL, the lowest drug concentration producing a prominent decrease in turbidity translating to 100% growth reduction compared with the drug-free control determined above. To extract total RNA, the cells of SC5314 were inoculated into YPD medium and cultured at 30°C overnight. Before SC5314 were harvested for RNA extraction, the culture was treated with HLJDD at 20 mg/mL for 3 h. The untreated culture was used as the control. Total RNA was isolated according to the protocol described by Alison et al. [41].
2.5. RNA Sequencing and Assembling
Three independent experiments were performed for the study of either control C. albicans or C. albicans with HLJDD treatment. Shear cDNA into 300–500 bp fragments using ultrasonic apparatus (Fisher) and purify it with Ampure beads (Agencourt, America). Library of all the samples was constructed according to the procedure of NEBNext® UltraTM RNA Library Prep Kit for Illumina (NEB, America). Sequencing library was checked with Onedrop quantitation, 2% agarose gel electrophoresis detection, and high sensitivity of DNA chip detection. Paired-end sequencing of cDNA was carried out with Illumina Hiseq TM2000. Raw data was filtered by removing reads with adaptor sequences, as well as low quality reads. Then, clean reads were obtained and mapped to reference sequences using SOAP (2.21) [42].
2.6. Gene Prediction and Annotation
Trinity software was used to assemble the clean reads into contigs and BLAST (2.2.23) was used to do gene prediction. Predicted sequences (e-value < 1.0e − 05) were annotated with information from GenBank NR, GO, and KEGG using BLAST2GO (2.2.5). GO classification was conducted using WEGO [43].
2.7. Analysis of Differential Expressed Genes
The expression level for each gene is determined by the numbers of reads uniquely mapped to the specific gene and the total number of uniquely mapped reads in the sample. The gene expression level is calculated by using RPKM (Reads Per kb per Million reads) method [36]. Then, NOI seq method was applied to screen differentially expressed genes between two groups, with the threshold of significance as fold change of RPKM ≥ 3 and Probability ≥ 0.8 [44].
2.8. Enrichment Analysis of GO and KEGG Pathways
Enrichment analysis was performed by hypergeometric test to find significantly enriched GO terms and KEGG pathways in DEGs. False discovery rate (FDR) of pathways was calculated. The threshold of significance of pathways was set as FDR < 0.05.
2.9. Real-Time Quantitative Reverse Transcription- (qRT-) PCR
To evaluate the validation of RNA-seq results, we conducted quantitative real-time (RT) PCR assays for determination of expression of 8 genes. Gene expression levels were calculated using the 2−ΔΔCt method [45]. For each sample, PCR amplifications with primer pair actin-F and actin-R for the quantification of expression of actin gene were performed as a reference. The experiment was repeated 3 times.
3. Results
3.1. RNA Sequencing and Gene Prediction
Approximately 12,000,000 raw reads were obtained from each sample. After filtering by quality, about 96% clean reads were mapped. Summary of mapping result was shown in Table 1. Using the longest sequence of a subgroup as the unigene as the reference sequence, we got 6057 predicted protein-encoding genes totally. The data have been submitted to NCBI under BioProject accession number PRJNA314910.
Table 1.
Summary of reads in C. albicans with or without HLJDD treatment.
| Sample | Total Reads | Total mapped reads | Mapping percentage |
|---|---|---|---|
| Ca_CK_1 | 12,377,083 | 11,840,278 | 95.66% |
| Ca_CK_2 | 11,758,367 | 11,313,372 | 96.22% |
| Ca_CK_3 | 12,283,182 | 11,830,964 | 96.32% |
| Ca_HT_1 | 12,406,841 | 11,924,995 | 96.12% |
| Ca_HT_2 | 11,803,831 | 11,338,558 | 96.06% |
| Ca_HT_3 | 12,212,721 | 11,727,140 | 96.02% |
3.2. Identification and Verification of Differentially Expressed Genes
By using the threshold of significance as fold change of RPKM ≥ 3 and Probability ≥ 0.8, we obtained a total of 735 differentially expressed genes (DEGs), including 700 upregulated genes and 35 downregulated genes (Supporting Information Table S1 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/3198249). The 20 most upregulated genes in response to HLJDD are listed in Table 2.
Table 2.
The 20 most upregulated genes in response to HLJDD treatment.
| Standard or systematic name in CGD | ID in GenBank | Annotation | Size | log2 ratio | Probability |
|---|---|---|---|---|---|
| C7_01060W_A | XP_720301.1 | Hypothetical protein | 142 aa | 11.25 | 0.81 |
| C5_04240C_A | XP_721977.1 | Hypothetical protein | 103 aa | 11.21 | 0.80 |
| C1_03880C_A | XP_711956.1 | Hypothetical protein | 120 aa | 9.85 | 0.95 |
| C7_01130C_A | XP_712469.1 | Hypothetical protein | 146 aa | 9.42 | 0.92 |
| PGA39 | EEQ43586.1 | Predicted protein | 288 aa | 9.32 | 0.85 |
| C1_12040W_A | XP_716393.1 | Hypothetical protein | 143 aa | 9.31 | 0.92 |
| CR_01870C_A | XP_718251.1 | Hypothetical protein | 196 aa | 8.90 | 0.85 |
| CR_04980C_A | XP_711981.1 | Hypothetical protein | 193 aa | 8.87 | 0.92 |
| LIP10 | XP_723508 | Secretory lipase 10 | 465 aa | 8.85 | 0.81 |
| C3_01010W_A | XP_718606.1 | Hypothetical protein | 102 aa | 8.67 | 0.90 |
| C4_06150C_A | EEQ44911.1 | Tat binding protein 1-interacting | 175 aa | 8.27 | 0.99 |
| C7_03900W_A | XP_715240.1 | Hypothetical protein | 109 aa | 7.96 | 0.94 |
| CR_07970C_A | XP_714226.1 | Hypothetical protein | 119 aa | 7.91 | 0.81 |
| CR_06990W_A | XP_712676.1 | Transcription activator | 865 aa | 7.84 | 0.88 |
| C5_02090W_A | EEQ43139.1 | Predicted protein | 100 aa | 7.84 | 0.84 |
| SPO22 | XP_718811.1 | Meiosis specific protein | 566 aa | 7.73 | 0.96 |
| CR_07550C_A | XP_710398.1 | Hypothetical protein | 101 aa | 7.72 | 0.82 |
| C3_02250C_A | XP_721699.1 | Hypothetical protein | 162 aa | 7.71 | 0.85 |
| C7_01060W_A | XP_718305.1 | Hypothetical protein | 111 aa | 7.70 | 0.89 |
A total of 8 genes including 7 upregulated and 1 downregulated gene from DGE libraries were selected for real-time PCR analysis to validate the DGE data. The results showed that 8 genes were demonstrated to have a consistent change for both DGE and real-time PCR while actin genes had no significant difference in real-time PCR (Supporting Information Table S2).
3.3. Effects of HLJDD Treatment on the Genes Involved in Sterol Biosynthesis
As the most widely used antifungal drugs, azoles can block fungal sterol biosynthesis pathway. Thus, effects of HLJDD on the genes involved in sterol biosynthesis were analyzed in detail. Expression of 23 genes involved in sterol biosynthesis was detected in the RNA-seq analysis, and expression of 8 genes showed a more than 2-fold increase after C. albicans was treated with HLJDD; only the genes encoding sterol 24-C-methyltransferase (ERG6) and C-8 sterol isomerase (ERG2) were upregulated by more than 3 times (Table 3). None of these 23 genes was downregulated significantly (Probability > 0.8) by HLJJD.
Table 3.
Response to HLJDD of the genes involved in ergosterol biosynthesis.
| Standard or systematic name in CGD | ID in GenBank | Annotation | log2 ratio | Probability |
|---|---|---|---|---|
| ERG1 | XP_711894.1 | Squalene monooxygenase | 2.13 | 0.95 |
| ERG2 | XP_718886.1 | C-8 sterol isomerase | 3.23 | 0.94 |
| ERG3 | XP_713577.1 | C-5 sterol desaturase | 1.84 | 0.94 |
| ERG4 | XP_717662.1 | Sterol C-24 (28) reductase | −0.34 | 0.65 |
| ERG5 | XP_716933.1 | Cytochrome P450 61 | 2.02 | 0.97 |
| ERG6 | XP_721588.1 | Sterol 24-C-methyltransferase | 3.33 | 0.97 |
| ERG7 | XP_722471.1 | 2,3-Oxidosqualene-lanosterol cyclase | −0.23 | 0.38 |
| ERG8 | XP_722678.1 | Phosphomevalonate kinase | −0.01 | 0.03 |
| ERG9 | XP_714460.1 | Squalene synthetase | −0.60 | 0.77 |
| ERG10 | XP_710124.1 | Acetyl-CoA acetyltransferase IA | 0.96 | 0.91 |
| ERG11 | XP_716761.1 | Cytochrome P450 51 | 2.02 | 0.97 |
| ERG12 | XP_723305.1 | Mevalonate kinase | 0.97 | 0.85 |
| ERG13 | XP_716446.1 | Hydroxymethylglutaryl-CoA synthase | 2.30 | 0.97 |
| MVD1/ERG19 | XP_718960.1 | Diphosphomevalonate decarboxylase | 0.04 | 0.12 |
| ERG24 | XP_710205.1 | Delta(14)-sterol reductase | 2.53 | 0.93 |
| ERG25 | XP_713420.1 | C-4 methylsterol oxidase | 1.31 | 0.91 |
| XP_722703.1 | C-4 methylsterol oxidase | 1.02 | 0.92 | |
| ERG26 | XP_715564.1 | C-3 sterol dehydrogenase/C-4 decarboxylase | 0.23 | 0.39 |
| ERG27 | XP_717865.1 | 3-Keto sterol reductase | 1.67 | 0.90 |
| ERG28 | XP_717865.1 | Hypothetical protein | 0.38 | 0.68 |
| HMG1 | XP_713636.1 | Hydroxymethylglutaryl-CoA reductase | 2.36 | 0.96 |
| IDI1 | XP_720295.1 | Isopentenyl-diphosphate delta-isomerase | −0.09 | 0.24 |
| CYB5 | XP_720295.1 | Cytochrome b5 | −0.27 | 0.62 |
3.4. Effects of HLJDD Treatment on the Genes Encoding Multidrug Transporters
In C. albicans, upregulation of multidrug transporter genes is one of the well-documented mechanisms of resistance to azole antifungal agents [9, 46–48]. Two families of multidrug transporters, the ABC (ATP-binding cassette) transporter family (Cdr1p and Cdr2p) and the major facilitator superfamily (MFS, CaMdr1p), have been shown to be involved in resistance to azole antifungal agents [47, 48]. Thus, we also paid attention to the multidrug transporter genes. In genome sequences of C. albicans, a total of 36 genes are annotated as multidrug transporters. In this study, expression of 32 genes was detected by the RNA-seq, and 7 genes were identified to be significantly upregulated more than 3 times by HLJDD treatment (Table 4), including CDR2 (Candida Drug Resistance) from the family of ABC transporters. Cdr2 has been shown as the principal mediators of resistance to azoles due to transport phenomena [47, 48].
Table 4.
Response to HLJDD of the genes involved in multidrug resistance of C. albicans.
| Standard or systematic name in CGD | ID in GenBank | Annotation | log2 ratio | Probability |
|---|---|---|---|---|
| CDR1 | XP_723062.1 | Multidrug resistance protein CDR1 | 2.42 | 0.99 |
| CDR2 | XP_723022.1 | Multidrug resistance ABC transporter | 5.32 | 0.99 |
| CDR3 | XP_441615.1 | N terminal 2/3 of opaque-specific ABC transporter | 0.75 | 0.67 |
| CDR4 | XP_717543.1 | Potential ABC transporter | −2.49 | 0.99 |
| ATM1 | XP_712090.1 | Potential mitochondrial ABC transporter similar to S. cerevisiae ATM1 | 0.79 | 0.76 |
| HST6 | XP_716101.1 | Potential ABC transporter similar to S. cerevisiae STE6 | 5.44 | 0.88 |
| MDL1 | XP_718280.1 | Potential ABC transporter similar to S. cerevisiae mitochondrial inner membrane MDL1 | 0.81 | 0.79 |
| MLT1 | XP_717637.1 | Vacuolar multidrug resistance ABC transporter | 1.75 | 0.93 |
| MDR1 | XP_719165.1 | Major Facilitator Transporter | 0.63 | 0.77 |
| CR_04620C_A | XP_717510.1 | MFS transporter, DHA1 family, multidrug resistance protein | 4.46 | 0.91 |
| SGE11 | XP_715705.1 | Potential MFS-MDR transporter | 1.31 | 0.84 |
| C1_10710C_A | XP_714012.1 | MFS transporter, DHA2 family, multidrug resistance protein | 5.1 | 0.90 |
| C3_03070W_A | XP_720131.1 | MFS transporter, DHA2 family, multidrug resistance protein | 0.47 | 0.66 |
| NAG4 | XP_712435.1 | MFS transporter, DHA1 family, multidrug resistance protein | 5.83 | 0.77 |
| TPO41 | XP_717426.1 | MFS transporter, DHA1 family, multidrug resistance protein | 2.34 | 0.95 |
| C6_01870C_A | XP_716705.1 | MFS transporter, DHA1 family, multidrug resistance protein | 2.59 | 0.94 |
| NAG3 | XP_712434.1 | MFS transporter, DHA1 family, multidrug resistance protein | 2.8 | 0.85 |
| C1_10200C_A | XP_723572.1 | MFS transporter, DHA1 family, multidrug resistance protein | 1.23 | 0.91 |
| C2_02570W_A | EEQ45693.1 | MFS transporter, DHA1 family, multidrug resistance protein | 1.34 | 0.78 |
| TPO3 | XP_723233.1 | MFS transporter, DHA1 family, multidrug resistance protein | −0.95 | 0.85 |
| HOL1 | XP_721489.1 | MFS transporter, DHA1 family, multidrug resistance protein | 2.0 | 0.85 |
| CR_01340W_A | XP_718285.1 | MFS transporter, DHA1 family, multidrug resistance protein | 3.93 | 0.93 |
| HOL4 | XP_712971.1 | MFS transporter, DHA1 family, multidrug resistance protein | 0.88 | 0.81 |
| C3_03440C_A | XP_720169.1 | Potential drug or polyamine transporter | 3.44 | 0.95 |
| TPO2 | XP_715197.1 | Potential drug or polyamine transporter | 2.31 | 0.82 |
| QDR3 | XP_714342.1 | Potential multidrug resistance transporter | 2.33 | 0.87 |
| C2_00540W_A | XP_719644.1 | Potential MATE family drug/sodium antiporter | −0.28 | 0.56 |
| C7_03590C_A | EEQ47129.1 | Multidrug resistance protein, MATE family | 0.16 | 0.26 |
| C1_00830W_A | XP_718985.1 | Potential MATE family drug/sodium antiporter | 0.44 | 0.34 |
| CR_10640W_A | XP_719407.1 | Multidrug resistance protein, MATE family | 3.15 | 0.96 |
| QDR2 | XP_714698.1 | Potential quinidine/multidrug transporter | 1.63 | 0.94 |
| FLU1 | XP_721413.1 | Multidrug efflux transporter | 1.76 | 0.91 |
3.5. Enrichment Analysis of GO and KEGG Pathways
GO and KEGG assignments were used to classify the genes in the response of C. albicans to HLJDD. By GO classification analysis, the percentage and distribution of top-level GO terms were portrayed in the 3 categories: (A) cellular component; (B) molecular function, and (C) biological process (Figure 1). A high percentage of genes were assigned to “cell,” “cell part,” “binding,” “catalytic,” “cellular process,” and “metabolic process” (Figure 1).
Figure 1.
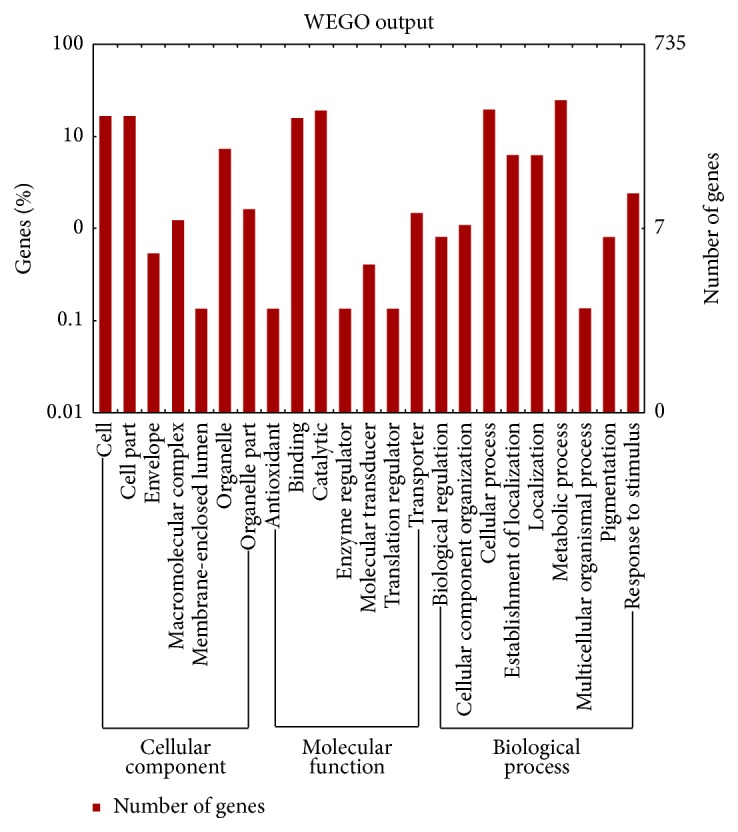
Functional categories of genes in C. albicans in response to HLJDD.
By enrichment analysis, with FDR < 0.05, 23 significant GO terms and 3 significant KEGG pathways were identified (Supporting Information Table S3). These significant pathways were mainly associated with DNA replication and transporter activity. The maps with highest unigene representation were meiosis (cal04113; 23 unigenes), followed by cell cycle (cal04111; 23 unigenes), and DNA replication (cal03030; 11 unigenes).
4. Discussion
C. albicans is the most prevalent opportunistic fungal pathogen causing superficial to systemic infections in immunocompromised individuals [1, 2]. The concomitant use of drugs and the lack of available drugs frequently result in the occurrence of drug-resistant isolates and strains display multidrug resistance (MDR). In search of novel fungicides, efficiency of medicinal plants against fungi has been reported, but studies on their underlying mechanisms are very few [49]. In this study, we explored a famous multiherb prescription in China, Huanglian Jiedu Decoction (HLJJD, Orengedokuto in Japan), for its antifungal potential. Our preliminary work showed that HLJDD is efficient against Trichophyton mentagrophytes and C. albicans [35]. HLJDD showed its impressive antifungal effect by multitarget and multichannel actions, but studies on the underlying mechanisms are very few. To determine the antifungal mechanism of HLJDD against C. albicans, we performed a large-scale analysis of gene expression changes when C. albicans was exposed to HLJDD, to better understand how HLJDD inhibits the growth of C. albicans. Due to the multiple components of HLJDD, it is most likely that the antifungal effect is multitarget and multichannel actions. KEGG analysis suggested that 3 cellular functions were affected in C. albicans upon HLJDD treatment, including meiosis, cell cycle, and DNA replication. Most genes (56 genes) involved in the 3 cellular functions were upregulated excepted for 1 gene, potential hexose transporter (XP_719596.1). Among these genes, Spo22 (also called Zip4) (XP_718811.1) was upregulated obviously upon HLJDD treatment. Zip4/Spo22 was shown to be a central protein of the SICs (synapsis initiation complexes), from which the polymerization of the transverse filament proceeds. In S. cerevisiae, Zip4/Spo22 was identified as a member of the ZMM group of proteins that also includes Zip1, Zip2, Zip3, Msh4, Msh5, and Mer3 which together control the formation of class I COs [50–52]. In Arabidopsis thaliana, Zip4/Spo22 function in class I CO formation is conserved with budding yeast. However, mutation in AtZIP4 does not prevent synapsis, showing that both aspects of the Zip4 function (i.e., class I CO maturation and synapsis) can be uncoupled [51].
Azoles are the most widely used antifungal drugs, which target on cytochrome P450 lanosterol 14α-demethylase encoded by the ERG11 gene. In Fusarium graminearum, using a deep serial analysis of gene expression (DeepSAGE) sequencing approach, the transcriptional response of F. graminearum to tebuconazole (a widely used azole fungicide) was profiled. Expression of 23 genes involved in sterol biosynthesis was detected in the DeepSAGE analysis, and expression of 9 genes showed a more than 5-fold increase after the fungus was treated with tebuconazole. None of these 23 genes was downregulated by more than 5 times by tebuconazole [53]. Thus, effects of HLJDD on the genes involved in sterol biosynthesis were analyzed in detail. Expression of 23 genes involved in sterol biosynthesis was detected in the RNA-seq analysis, and expression of 8 genes showed a more than 2-fold increase after the fungus was treated with HLJDD, only the genes encoding sterol 24-C-methyltransferase (ERG6) and C-8 sterol isomerase (ERG2) were upregulated by more than 3 times (Table 3). None of these 23 genes was downregulated significantly (Probability > 0.8) by HLJJD. These results indicate that HLJDD might also affect sterol biosynthesis of C. albicans.
Overexpression of multidrug resistance efflux transporter genes in several fungi was found to be correlated with azole resistance [54]. In C. albicans, a number of efflux transporter genes have been cloned and characterized. Two families of multidrug transporters, the ABC (ATP-binding cassette) transporter family (Cdr1p and Cdr2p) and the major facilitator superfamily (MFS, CaMdr1p), have been shown to be involved in resistance to azole antifungal agents [47, 48]. Expression of 32 genes out of 36 genes annotated as multidrug transporters in genome sequences of C. albicans was detected by the RNA-seq sequencing. Expression of 13 genes was upregulated by more than 2 times by HLJDD; meanwhile, only 2 genes were significantly downregulated including CDR4. In addition, expression of only 4 genes was upregulated by more than 3 times by HLJDD, including CDR2, which plays an important role in azole resistance (Table 4). The upregulated expression of these genes may be related to efflux of HLJJD, which provides supporting evidence to previous studies on expression level.
Previous study examined changes in the gene expression profile of C. albicans following exposure to representatives of the 4 currently available classes of antifungal agents, the azoles (ketoconazole), polyenes (amphotericin B), echinocandins (caspofungin), and nucleotide analogs (5-flucytosine). And the data showed that none of the differentially regulated genes found exhibited similar changes in expression for all 4 classes of drugs. Thus, the response of C. albicans to different drugs seems to be highly specific [55]. Ketoconazole exposure increased the expression of genes involved in lipid, fatty acid, sterol metabolism, and several genes associated with azole resistance, including CDR1 and CDR2 [56]. It is surprising that HLJDD increased the expression of genes involved in sterol metabolism and azole resistance (CDR1 and CDR2). Considering the similarity of expression changing pattern, it is possible that HLJDD affects sterol metabolism. And further experiments are required to confirm this hypothesis.
5. Conclusions
In conclusion, we performed a transcriptomics analysis of gene expression changes for C. albicans under treatment of HLJDD using RNA-seq technique. Overall, a total of 6057 predicted protein-encoding genes were identified. Further gene expression analysis revealed a total of 735 differentially expressed genes (DEGs), including 700 upregulated genes and 35 downregulated genes. Intensive bioinformatics analysis identified 26 significant pathways, and DNA replication and transporter activity were mainly involved. In addition, genes encoding multidrug transporters such as ABC transporter and MFS transporter were identified to be significantly upregulated. Overall, the results from this study might provide insights in understanding of the mechanisms for the response of C. albicans to HLJJD. Furthermore, this work demonstrates the potential utility of the RNA-seq technique in antifungal studies.
Supplementary Material
Table S1: 735 differentially expressed genes (DEGs).
Table S2: A list of primers used in real-time PCR analysis and comparison in the changes of gene expression.
Table S3: Significant pathways of DEGs.
Acknowledgments
The research was supported by the National Science Foundation (81173271 and 31540034), Zhejiang Provincial Natural Science Foundation (LQ14C010005), and Open Project Program of Key Laboratory of Mental Health, Institute of Psychology, Chinese Academy of Sciences (KLMH2014G03).
Competing Interests
The authors declare that they have no competing interests.
Authors' Contributions
Qianqian Yang and Lei Gao equally contributed to this work.
References
- 1.Linares C. E. B., Giacomelli S. R., Alves S. H., Schetinger M. R. C., Altenhofen D., Morsch V. M. Fluconazole and amphotericin-B resistance are associated with increased catalase and superoxide dismutase activity in Candida albicans and Candida dubliniensis . Revista da Sociedade Brasileira de Medicina Tropical. 2013;46(6):752–758. doi: 10.1590/0037-8682-0190-2013. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L., Yan K., Zhang Y., et al. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(11):4606–4611. doi: 10.1073/pnas.0609370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly S. L., Lamb D. C., Jackson C. J., Warrilow A. G. S., Kelly D. E. The biodiversity of microbial cytochromes P450. Advances in Microbial Physiology. 2003;47:131–186. doi: 10.1016/S0065-2911(03)47003-3. [DOI] [PubMed] [Google Scholar]
- 4.Parker J. E., Merkamm M., Manning N. J., Pompon D., Kelly S. L., Kelly D. E. Differential azole antifungal efficacies contrasted using a Saccharomyces cerevisiae strain humanized for sterol 14α-demethylase at the homologous locus. Antimicrobial Agents and Chemotherapy. 2008;52(10):3597–3603. doi: 10.1128/aac.00517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warrilow A. G. S., Martel C. M., Parker J. E., et al. Azole binding properties of Candida albicans sterol 14-α demethylase (CaCYP51) Antimicrobial Agents and Chemotherapy. 2010;54(10):4235–4245. doi: 10.1128/aac.00587-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher P. J., Bennett D. E., Henman M. C., et al. Reduced azole susceptibility of oral isolates of Candida albicans from HIV-positive patients and a derivative exhibiting colony morphology variation. Journal of General Microbiology. 1992;138(9):1901–1911. doi: 10.1099/00221287-138-9-1901. [DOI] [PubMed] [Google Scholar]
- 7.Redding S., Smith J., Farinacci G., et al. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clinical Infectious Diseases. 1994;18(2):240–242. doi: 10.1093/clinids/18.2.240. [DOI] [PubMed] [Google Scholar]
- 8.White T. C., Marr K. A., Bowden R. A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clinical Microbiology Reviews. 1998;11(2):382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White T. C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate, with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrobial Agents and Chemotherapy. 1997;41(7):1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazen K. C., Baron E. J., Colombo A. L., et al. Comparison of the susceptibilities of Candida spp. to fluconazole and voriconazole in a 4-year global evaluation using disk diffusion. Journal of Clinical Microbiology. 2003;41(12):5623–5632. doi: 10.1128/jcm.41.12.5623-5632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W., Tan J., Sun J., et al. Invasive candidiasis in intensive care units in China: in vitro antifungal susceptibility in the China-SCAN study. The Journal of Antimicrobial Chemotherapy. 2014;69(1):162–167. doi: 10.1093/jac/dkt330. [DOI] [PubMed] [Google Scholar]
- 12.Cleveland A. A., Harrison L. H., Farley M. M., et al. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008–2013: results from population-based surveillance. PLoS ONE. 2015;10(3) doi: 10.1371/journal.pone.0120452.e0120452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morschhäuser J. The genetic basis of fluconazole resistance development in Candida albicans . Biochimica et Biophysica Acta. 2002;1587(2-3):240–248. doi: 10.1016/s0925-4439(02)00087-x. [DOI] [PubMed] [Google Scholar]
- 14.Coifman R. R., Weiss G. Analyse Harmonique Non-Commutative sur Certains Espaces Homogenes. Vol. 242. Berlin, Germany: Springer; 2010. (Lecture Notes in Mathematics). [Google Scholar]
- 15.Ohta Y., Kobayashi T., Nishida K., Sasaki E., Ishiguro I. Preventive effect of Oren-gedoku-to (Huanglian-Jie-Du-Tang) extract on the development of stress-induced acute gastric mucosal lesions in rats. Journal of Ethnopharmacology. 1999;67(3):377–384. doi: 10.1016/s0378-8741(99)00093-8. [DOI] [PubMed] [Google Scholar]
- 16.Wang L. M., Mineshita S. Preventive effects of Unsei-in and Oren-gedoku-to, Chinese traditional medicines, against rat paw oedema and abdominal constriction in mice. Journal of Pharmacy and Pharmacology. 1996;48(3):327–331. doi: 10.1111/j.2042-7158.1996.tb05927.x. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y. H., Jiang P., Wang S. P., et al. Plasma pharmacochemistry based approach to screening potential bioactive components in Huang-Lian-Jie-Du-Tang using high performance liquid chromatography coupled with mass spectrometric detection. Journal of Ethnopharmacology. 2012;141(2):728–735. doi: 10.1016/j.jep.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X.-J., Deng Y.-X., Shi Q.-Z., He M.-Y., Chen B., Qiu X.-M. Hypolipidemic effect of the Chinese polyherbal Huanglian Jiedu decoction in type 2 diabetic rats and its possible mechanism. Phytomedicine. 2014;21(5):615–623. doi: 10.1016/j.phymed.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Yu Y.-L., Lu S.-S., Yu S., et al. Huang-Lian-Jie-Du-Decoction modulates glucagon-like peptide-1 secretion in diabetic rats. Journal of Ethnopharmacology. 2009;124(3):444–449. doi: 10.1016/j.jep.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 20.Yue G.-H., Zhuo S.-Y., Xia M., Zhang Z., Gao Y.-W., Luo Y. Effect of Huanglian Jiedu Decoction on thoracic aorta gene expression in spontaneous hypertensive rats. Evidence-Based Complementary and Alternative Medicine. 2014;2014:9. doi: 10.1155/2014/565784.565784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu X., Xu Z., He Y., Chi H. Protective function of Huanglian Jiedu Tang on rats after focal cerebral ischemic reperfusion and its influence on caspase-3 expression. Modern Journal of Integrated Traditional Chinese and Western Medicine. 2009;18(14):1598–1599. [Google Scholar]
- 22.Hwang Y. S., Shin C. Y., Huh Y., Ryu J. H. Hwangryun-Hae-Dok-tang (Huanglian-Jie-Du-Tang) extract and its constituents reduce ischemia-reperfusion brain injury and neutrophil infiltration in rats. Life Sciences. 2002;71(18):2105–2117. doi: 10.1016/s0024-3205(02)01920-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q., Ye Y.-L., Yan Y.-X., et al. Protective effects of Huanglian-Jiedu-Tang on chronic brain injury after focal cerebral ischemia in mice. Journal of Zhejiang University Medical sciences. 2009;38(1):75–80. doi: 10.3785/j.issn.1008-9292.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi T., Ohta Y., Inagaki S., Harada N. Inhibitory action of Oren-gedoku-to extract on enzymatic lipid peroxidation in rat liver microsomes. Biological and Pharmaceutical Bulletin. 2001;24(10):1165–1170. doi: 10.1248/bpb.24.1165. [DOI] [PubMed] [Google Scholar]
- 25.Ikarashi N., Tajima M., Suzuki K., et al. Inhibition of preadipocyte differentiation and lipid accumulation by orengedokuto treatment of 3T3-L1 cultures. Phytotherapy Research. 2012;26(1):91–100. doi: 10.1002/ptr.3493. [DOI] [PubMed] [Google Scholar]
- 26.Sekiya N., Kainuma M., Hikiami H., et al. Oren-gedoku-to and Keishi-bukuryo-gan-ryo inhibit the progression of atherosclerosis in diet-induced hypercholesterolemic rabbits. Biological and Pharmaceutical Bulletin. 2005;28(2):294–298. doi: 10.1248/bpb.28.294. [DOI] [PubMed] [Google Scholar]
- 27.Lu J., Wang J.-S., Kong L.-Y. Anti-inflammatory effects of Huang-Lian-Jie-Du decoction, its two fractions and four typical compounds. Journal of Ethnopharmacology. 2011;134(3):911–918. doi: 10.1016/j.jep.2011.01.049. [DOI] [PubMed] [Google Scholar]
- 28.Fukutake M., Miura N., Yamamoto M., et al. Suppressive effect of the herbal medicine Oren-gedoku-to on cyclooxygenase-2 activity and azoxymethane-induced aberrant crypt foci development in rats. Cancer Letters. 2000;157(1):9–14. doi: 10.1016/S0304-3835(00)00432-8. [DOI] [PubMed] [Google Scholar]
- 29.Zeng H., Dou S., Zhao J., et al. The inhibitory activities of the components of Huang-Lian-Jie-Du-Tang (HLJDT) on eicosanoid generation via lipoxygenase pathway. Journal of Ethnopharmacology. 2011;135(2):561–568. doi: 10.1016/j.jep.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 30.Choi W. M., Lam C. L., Mo W. Y., et al. Effects of the modified Huanglian Jiedu decoction on the disease resistance in grey mullet (Mugil cephalus) to Lactococcus garvieae . Marine Pollution Bulletin. 2014;85(2):816–823. doi: 10.1016/j.marpolbul.2014.04.043. [DOI] [PubMed] [Google Scholar]
- 31.Liu L., Yu S., Xie L. Simultaneous determination of geniposide and 4 flavonoids in the traditional Chinese medicinal preparation Huanglian Jiedu decoction by HPLC with programmed wavelength UV detection. Chinese Journal of Pharmaceutical Analysis. 2008;28(2):182–186. [Google Scholar]
- 32.Li Q., Wang L., Dai R., Bi K. Determination of berberine hydrochloride in coptis decoction by HPLC. Northwest Pharmaceutical Journal. 2004;19:51–52. [Google Scholar]
- 33.Huang S., Cao Y. Y., Dai B. D., et al. In vitro synergism of fluconazole and Baicalein against clinical isolates of Candida albicans resistant to fluconazole. Biological and Pharmaceutical Bulletin. 2008;31(12):2234–2236. doi: 10.1248/bpb.31.2234. [DOI] [PubMed] [Google Scholar]
- 34.Quan H., Cao Y.-Y., Xu Z., et al. Potent in vitro synergism of fluconazole and berberine chloride against clinical isolates of Candida albicans resistant to fluconazole. Antimicrobial Agents and Chemotherapy. 2006;50(3):1096–1099. doi: 10.1128/aac.50.3.1096-1099.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao M., Xia X., Cao Y. In vitro antifungal effect of coptidis decoction for detoxification or combined with western medicine. Chinese Archives of Traditional Chinese Medicine. 2009;27(3):585–587. [Google Scholar]
- 36.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruno V. M., Wang Z., Marjani S. L., et al. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Research. 2010;20(10):1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao C.-W., Ji Q.-A., Wei Q., Liu Y., Pan L.-J., Bao G.-L. Digital gene expression analysis of Microsporum canis exposed to berberine chloride. PLoS ONE. 2015;10(4) doi: 10.1371/journal.pone.0124265.e0124265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillum A. M., Tsay E. Y. H., Kirsch D. R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Molecular and General Genetics. 1984;198(1):179–182. doi: 10.1007/bf00328721. [DOI] [PubMed] [Google Scholar]
- 40.Standards NCfCL. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard. National Committee for Clinical Laboratory Standards; 2002. [Google Scholar]
- 41.Alison A., Gottschling D. E., Kaiser C. A. Methods in Yeast Genetics. New York, NY, USA: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 42.Li R., Yu C., Li Y., et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 43.Ye J., Fang L., Zheng H., et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Research. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Research. 2011;21(12):2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Perea S., López-Ribot J. L., Wickes B. L., et al. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrobial Agents and Chemotherapy. 2002;46(6):1695–1703. doi: 10.1128/aac.46.6.1695-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanglard D., Ischer F., Monod M., Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143(2):405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 48.Sanglard D., Kuchler K., Ischer F., Pagani J.-L., Monod M., Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrobial Agents and Chemotherapy. 1995;39(11):2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao C. W., Ji Q. A., Rajput Z. I., Wei Q., Liu Y., Bao G. L. Antifungal efficacy of Phellodendron amurense ethanol extract against Trichophyton mentagrophytes in rabbits. Pakistan Veterinary Journal. 2014;34(2):219–223. [Google Scholar]
- 50.Whitby M. C. Making crossovers during meiosis. Biochemical Society Transactions. 2005;33(6):1451–1455. doi: 10.1042/BST20051451. [DOI] [PubMed] [Google Scholar]
- 51.Chelysheva L., Gendrot G., Vezon D., Doutriaux M.-P., Mercier R., Grelon M. Zip4/Spo22 is required for class ICO formation but not for synapsis completion in Arabidopsis thaliana . PLoS Genetics. 2007;3(5):802–813. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perry J., Kleckner N., Börner G. V. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(49):17594–17599. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X., Jiang J., Shao J., Yin Y., Ma Z. Gene transcription profiling of Fusarium graminearum treated with an azole fungicide tebuconazole. Applied Microbiology and Biotechnology. 2010;85(4):1105–1114. doi: 10.1007/s00253-009-2273-4. [DOI] [PubMed] [Google Scholar]
- 54.Lupetti A., Danesi R., Campa M., Tacca M. D., Kelly S. Molecular basis of resistance to azole antifungals. Trends in Molecular Medicine. 2002;8(2):76–81. doi: 10.1016/s1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- 55.Liu T. T., Lee R. E. B., Barker K. S., et al. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans . Antimicrobial Agents and Chemotherapy. 2005;49(6):2226–2236. doi: 10.1128/aac.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Backer M. D., Ilyina T., Ma X.-J., Vandoninck S., Luyten W. H. M. L., Bossche H. V. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrobial Agents and Chemotherapy. 2001;45(6):1660–1670. doi: 10.1128/aac.45.6.1660-1670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: 735 differentially expressed genes (DEGs).
Table S2: A list of primers used in real-time PCR analysis and comparison in the changes of gene expression.
Table S3: Significant pathways of DEGs.


