Abstract
Objective
The role and mechanisms of insulin receptor internalization remain incompletely understood. Previous trafficking studies of insulin receptors involved fluorescent protein tagging at their termini, manipulations that may be expected to result in dysfunctional receptors. Our objective was to determine the trafficking route and molecular mechanisms of functional tagged insulin receptors and endogenous insulin receptors in pancreatic beta-cells.
Methods
We generated functional insulin receptors tagged with pH-resistant fluorescent proteins between domains. Confocal, TIRF and STED imaging revealed a trafficking pattern of inter-domain tagged insulin receptors and endogenous insulin receptors detected with antibodies.
Results
Surprisingly, interdomain-tagged and endogenous insulin receptors in beta-cells bypassed classical Rab5a- or Rab7-mediated endocytic routes. Instead, we found that removal of insulin receptors from the plasma membrane involved tyrosine-phosphorylated caveolin-1, prior to trafficking within flotillin-1-positive structures to lysosomes. Multiple methods of inhibiting caveolin-1 significantly reduced Erk activation in vitro or in vivo, while leaving Akt signaling mostly intact.
Conclusions
We conclude that phosphorylated caveolin-1 plays a role in insulin receptor internalization towards lysosomes through flotillin-1-positive structures and that caveolin-1 helps bias physiological beta-cell insulin signaling towards Erk activation.
Keywords: Insulin receptor internalization, Insulin resistance, Pancreatic islet beta-cells, Autocrine insulin signaling
Highlights
-
•
Insulin receptors are tagged between domains to maintain the functionality and endogenous intracellular trafficking patterns.
-
•
Insulin receptors have virtually no co-localization with classical endocytosis markers, but co-localize with Cav11.
-
•
Insulin receptor internalization and anti-apoptotic Erk signaling are modulated by phosphorylated Cav1.
1. Introduction
The pathogenesis of type 2 diabetes is associated with defects in both insulin signaling and pancreatic beta-cell function. Evidence has emerged that insulin itself has powerful anti-apoptotic and pro-proliferative effects in pancreatic beta-cells via the Raf1/Mek/Erk pathway [1], [2], [3], [4], [5], [6], [7], but the mechanisms that bias towards this arm of the insulin signaling cascade have not been identified. Insulin receptors, like other tyrosine kinases, are internalized upon ligand binding [8]. The most studied mode of endocytosis is facilitated by a coat protein called clathrin, but alternate routes involving caveolin and cholesterol dependent lipid rafts have also been proposed [9], [10]. Depending on the cell type, studies have implicated either of these pathways in insulin receptor internalization [11], [12], [13]. It has also been demonstrated that dynamin, a key GTPase in endocytic vesicle synthesis, is required for insulin-stimulated Erk activation [14]. However, since dynamin is required for the formation of both clathrin- or caveolin-coated endocytic vesicles, this finding does not distinguish between specific pathways [10]. Caveolin-1 knockout mice have been reported to exhibit impaired insulin signaling in multiple tissues [15], [16], [17], pointing to a possible role for Cav1 in insulin receptor dynamics and signaling. Little is known about the mechanisms involved in insulin receptor internalization in pancreatic beta-cells, and what has been reported [18] differs from findings in other cell types [12], [13].
Two insulin receptor (InsR) splice variants are present in most tissues, and previous studies in beta-cells pointed to their localization in distinct membrane domains and roles in specific signaling processes [2], [18], [19]. Notwithstanding, one would expect a significant amount of colocalization based on the ability of InsRA and InsRB to form heterodimers [20], [21]. One challenge in the study of insulin receptor trafficking is the discrepancy between the behavior of fluorescently tagged insulin receptors and the localization of endogenous insulin receptors. Previous studies have fused fluorescent proteins to the N-termini or C-termini of the insulin receptor [19], [22], which are critical domains for insulin binding and tyrosine kinase signaling [21]. These tagging manipulations could result in non-functional receptors.
The goal of our study was to investigate the internalization and trafficking of functional insulin receptors by labeling InsR proteins between domains with pH-resistant fluorescent proteins and by using antibodies to assess endogenous proteins. Using super-resolution, total internal reflection fluorescence (TIRF), and confocal imaging, we identified a Cav1-Flot1 trafficking pathway for insulin receptors in pancreatic beta-cells. Cav1-dependent InsR internalization has the potential to be a bifurcation point between Erk and Akt arms of the insulin signaling pathway.
2. Materials and methods
2.1. Cell culture
Primary mouse or human islets were dispersed and cultured as described [23] in 5 mM glucose CMRL medium (Life Technologies) supplemented with penicillin/streptomycin (100 μg/ml) and 10% (v/v) FBS, unless otherwise stated. MIN6 cells and HEK 293T cells were cultured in DMEM media containing 25 mM glucose (Sigma–Aldrich, St Louis, MO) supplemented with 10% (v/v) FBS (Life Technologies, Burlington, ON) and penicillin/streptomycin (100 μg/ml; Life Technologies). 25 mM glucose DMEM culture media for NIH-3T3 cells was supplemented with 10% (v/v) new born calf serum (Life Technologies) and penicillin/streptomycin (100 μg/ml). All cells were cultured at 37 °C and 5% CO2.
For FITC-insulin (Life Technologies) uptake, MIN6 cells were cultured in glass bottom 24 well plates (MatTek Corporation, Ashland, MA) in 5 mM glucose DMEM containing 10% (v/v) FBS for 24 h. Prior to treatment, cells were cultured for 2 h in 5 mM glucose DMEM media without serum. Subsequently, this media was replaced by 5 mM glucose DMEM containing 200 nM FITC-insulin and cells were incubated for 1 h at 37 °C and 5% CO2. Prior to imaging, cells were washed twice with PBS. Live cells were imaged in Ringer's buffer (119 mM NaCl, 4.7 mM KCl, 25 mM NaHCO3, 2.5 mM CaCl2, 1.2 mM MgSO4, 1.2 mM H2PO4) containing 5 mM glucose. For Alexa 488 labeled EGF uptake, InsRA-TagRFP transfected MIN6 cells grown on coverslips were pulsed for 1 min with fluorescently labeled A488-EGF (final concentration 600 ng/ml, Life Technologies) in 25 mM glucose DMEM media supplemented with 10% (v/v) FBS. After the indicated chase times, cells were fixed in 4% (w/v) paraformaldehyde (Sigma–Aldrich) for 20 min. Coverslips were mounted using Prolong Gold Antifade reagent (Life Technologies).
For insulin treatment prior to immunoblot analysis, MIN6 cells were cultured in 5 mM glucose DMEM containing 0.5% (v/v) FBS for 24 h. Subsequently, the media was replaced with Ringer's buffer supplemented with 0.2% (w/v) bovine serum albumin (Sigma–Aldrich, St Louis, MO) and cells were incubated for 2 h at 37 °C and 5% CO2. Human recombinant insulin (Sigma–Aldrich) was diluted in Ringer's buffer supplemented with 0.2% (w/v) bovine serum albumin and cells were treated for the time periods as indicated at 37 °C and 5% CO2.
MIN6 cell lines were produced after native cells were transfected with Cav1-wt-RFP, Cav1-Y14F-RFP or Cav1-Y14D using the NEON system and then cultured in media with 400 μg/ml G418 (Life technologies) for 10 passages. For subsequent passages, the G-418 concentration was reduced to 50 μg/ml.
2.2. Plasmids and molecular cloning
CAV1-mRFP, CAV1-Y14F-mRFP and CAV1-Y14D-mRFP plasmids have been described [24]. CAV1-eGFP was purchased from Addgene (#14433). LAMP2-eGFP, Flotillin-1-eGFP, InsR-A/B-C-eGFP plasmids were generous gifts from J. Lippincott-Schwartz (Bethesda, MD, USA), Ai Yamamoto (New York, NY, USA), and Ingo Leibiger (Stockholm, Sweden), respectively. Igf1r-TagGFP2 was produced by Life Technologies gene synthesis service. PCR amplifications of DNA fragments were performed using AccuPrime Pfx polymerase (Life Technologies) according to the manufacturer's instructions. Prior to ligation with target vectors, all PCR products were sub-cloned into pCR2.1 TOPO vector using the TOPO TA cloning kit (Life Technologies). Subsequently, the PCR products were excised from the pCR2.1 TOPO vector, gel purified and ligated with the digested target vector using T4 DNA ligase according to the manufacturer's instruction. The primer sequences and PCR-generated restriction sites are available on request.
2.3. Transfection
Plasmid DNAs (2 μg) were transfected into 1 million MIN6 cells using the NEON system (Life Technologies) using two 1200 V pulses and a pulse width of 20 ms. Cav1 RNA interference was achieved by transfecting MIN6 cells with Dharmacon SMARTpool siRNA against Cav1 (Cat. L-058415-00-0005) using Lipofectamine RNAiMAX (Life technologies) according to the manufacturer's instructions achieving a final siRNA pool concentration of 100 nM. Dharmacon SMARTpool scrambled siRNA was used at 100 nM as a control. Cells were analyzed 48 h after transfection.
2.4. Immunofluorescence
For immunofluorescence analysis, cells were fixed in 4% (w/v) paraformaldehyde (Sigma–Aldrich) for 20 min and permeabilized subsequently in 0.1% (v/v) Triton X-100 (Sigma–Aldrich) for 15 min. Primary antibodies targeting InsR (Cat. #3020S), Clathrin (Cat. # 4796), Cav1 (Cat. #3267), Rab7 (Cat. #9367), and Igf1r (Cat. #3027) from Cell Signalling (Danvers, MA), Flotillin-1 (Cat. #3868-1) from Epitomics (Burlingame, CA), Rab4a (Cat. #sc-26562), Rab5a (Cat. # sc-166600) from Santa Cruz Biotechnology (Dallas, TX), LAMP2 (Cat. #PRS3627) from Sigma–Aldrich (St. Louis, MO), and Na/K pump (Cat. # ab76020) from Abcam (Eugene, OR) were applied for 12 h at 4 °C. Coverslips were washed in PBS, and secondary antibodies (Alexa 488, Alexa 594 Life Technologies; Atto 425, OG 488 ATTO-TEC GmbH, Siegen, Germany) were applied for 2 h at room temperature. After washing in PBS, coverslips were mounted with Prolong Gold Antifade (Life Technologies).
For pancreatic tissue staining, tissue sections were de-paraffinized with three xylene washes and rehydrated with ethanol solutions (100%, 95%, 70%), followed by a PBS wash. Epitope retrieval was performed by incubating sections in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween-20, pH 6.0) for 15 min at 95 °C. The sections were briefly washed in PBS and blocked with DAKO Protein Block solution for 30 min. The sections were incubated with primary antibodies overnight at 4 °C. Following PBS washes, the sections were incubated with secondary antibodies for 1 h at room temperature. The sections were washed and mounted using Prolong Gold Antifade.
2.5. Imaging
Prior to fixation, cells were cultured in the conditions described above unless otherwise noted. For live cell imaging, cells were in Ringer's buffer supplemented with 0.2% (w/v) bovine serum albumin and 20 mM glucose unless otherwise indicated. TIRF live-cell imaging was performed on a Zeiss Axiovert 200M with a 100x Alpha-Plan-Fluar NA 1.45 oil objective (Zeiss, Germany) and a TIRF laser angle modifier (Zeiss). Cells were kept at 37 °C in an incubation chamber (Harvard Apparatus, Holliston, MA). Images were acquired with a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ). 445 nm and 532 nm diode pumped crystal lasers were used to excite mTFP and TagRFP respectively. The TIRF system was upgraded during the study to a 405 nm, 488 nm, 561 nm solid state diode laser system which was also used to excite TagRFP and eGFP tagged proteins. Confocal imaging of fixed or live samples was performed with an Olympus Fluoview FV1000 laser scanning confocal microscope. 488 nm Argon, 543, 633 nm HeNe and 405 nm diode lasers were used to excite fluorescent proteins or dyes. UPlan 60X NA 1.35 Oil and U-Plan S Apo 100X NA 1.40 Oil objectives (Olympus, Tokyo, Japan) were used to acquire images. G-STED and CW-STED super-resolution imaging was conducted on Leica Microsystems setups [25]. Images were acquired through an HCX PlanAPO 100x OIL, NA 1.40 objective (Leica Microsystems). A 496 nm or 488 nm laser was used to excite Alexa 488 and a 458 nm laser was applied to excite mTurquoise. A 592 nm laser was used for stimulated emission depletion to reach STED resolution.
2.6. Image analysis
The ImageJ distribution Fiji was used to process confocal micrographs [26]. Chromatic aberration between individual channels was corrected using the Fiji plugin StackReg [27]. Subsequently, the local background was subtracted using the rolling ball method [28]. Object based colocalization analysis of endosomes was performed based on the distance of the object centers between the different channels using the Fiji plugin JaCoP [29]. Images were manually thresholded to ensure an appropriate detection of puncta. The same threshold was applied throughout a set of images for a given channel. Subsequently, all objects larger than 10 pixels and smaller than 200 pixels were used for the colocalization analysis. The distances between the centers of the previously identified objects between channels were used to determine colocalization based on the resolution of the microscope setup. A distance of 1 pixel or less between object centers was defined as a colocalization event. The Venn diagrams represent the sizes of the analyzed object populations relative to each other as well as their colocalization. CellProfiler software and its standard modules were used to track organelle dynamics including life time and intensity in live cell recordings [30].
2.7. Immunoblot
MIN6 cells were washed with Ringers buffer prior to lysis with RIPA buffer (50 mM β-glycerol phosphate, 10 mM HEPES, 1% Triton X-100, 70 mM NaCl, 2 mM EGTA, 1 mM Na3VO4, and 1 mM NaF) supplemented with complete EDTA-free protease inhibitor cocktail (Roche, Laval, QC). Protein quantification was determined by the Bradford method. Membranes were probed with antibodies against pERK1/2 (Cat. #4370), ERK1/2 (Cat. #4695), pAkt (Cat. #9271), Akt (Cat. #9272), INSR (Cat. #3020S), CAV1 (Cat. #3267), all from Cell Signalling, and β-actin (Cat. #NB600-501, Novus Biologicals, Littleton, CO) following standard procedures. For sub-cellular fractionation, cells were processed using the Plasma Membrane Extraction kit (Abcam) following the manufacturers instructions.
2.8. Immunoprecipitation
Immunoprecipitation was performed by incubating 1 mg of protein lysate with InsR antibodies (Cat. #3020S, Cell Signalling) at 4 °C for 12 h. Subsequently, the solution was incubated with PureProteome™ Protein G Magnetic Beads (Millipore, Billerica, Massachusetts, USA) and subsequently washed according to the manufacturer's instructions. InsR protein complexes were separated from the beads by 10 min incubation at 95 °C. The resulting protein solution was analyzed by standard immunoblot procedures.
2.9. In vivo experiments
The caveolin-1 knockout (Cav1−/−) mice used in this study were originally produced by T.V. Kurzchalia (MPI-CBG, Dresden, Germany) [31]. They were housed in the facility at the Université catholique de Louvain in Belgium. Cav1+/+ controls generated through heterozygous mating were used to control for genetic background.
2.10. Statistics
All studies were repeated at least 3 times. Data are expressed as mean with S.E.M. Means were considered statistically significantly different if p ≤ 0.05. Student's t-tests were used for comparison between two means.
3. Results
3.1. Subcellular localization of endogenous insulin receptors in pancreatic beta-cells
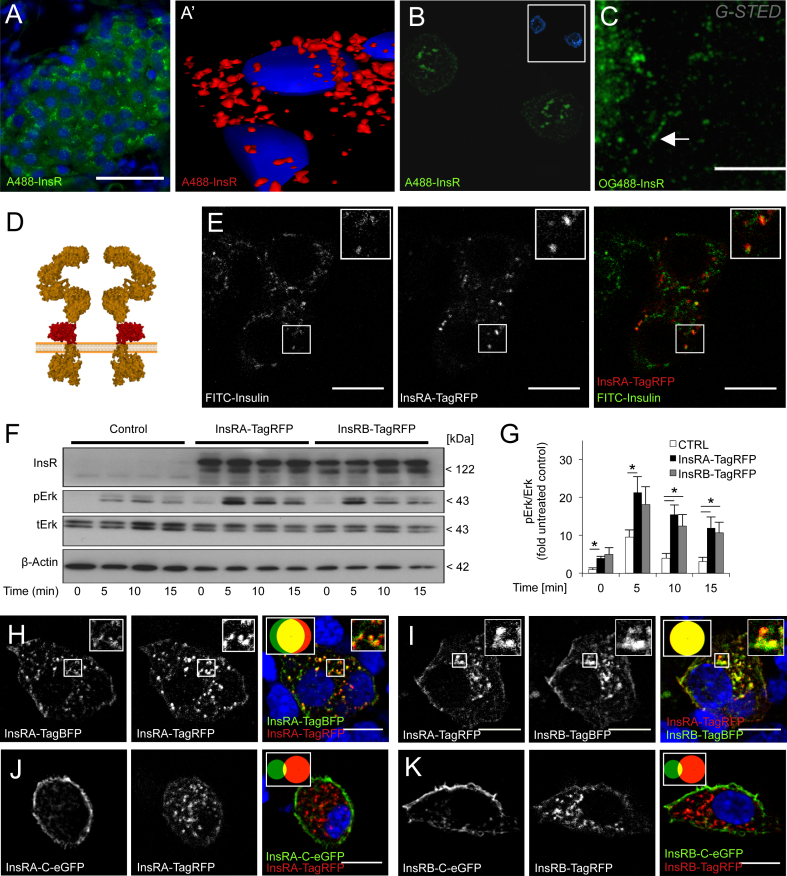
In most cell types, insulin receptors have been found in multiple cellular compartments, including the plasma membrane, endosomes, lysosomes, and perhaps secretory granules and the nucleus [8], [32], [33], [34]. In vivo, we observed that the majority of insulin receptors were within the beta-cell cytoplasm in mouse pancreatic sections (Figure 1A, A′). In vitro, confocal imaging of dispersed cells demonstrated that insulin receptors were located on the membranes of cytoplasmic vesicles in isolated human and mouse beta-cells (Figure 1B, Figure S1A), pointing to a possible role for receptor internalization in this cell type [35]. The size of the insulin receptor-containing structures was near the limit of conventional light microscopy. Using stimulated emission depletion (STED) super-resolution microscopy [36], we found that insulin receptors in the membrane were clustered in domains that were ∼60–80 nm and confirmed the intracellular location of a large fraction of the endogenous insulin receptors (Figure 1C). These imaging data were confirmed by sub-cellular fractionation of primary human islets, which showed that insulin receptors were in fractions enriched for markers of intracellular membranes (Figure S1B). Thus, in vivo and in vitro data indicate that endogenous insulin receptors exist primarily in an internalized state in beta-cells.
Figure 1.
Design and validation of functional fluorescent protein-tagged insulin receptors that mimic endogenous insulin receptor localization. (A) Islets in pancreatic tissue sections from 24 week-old mice are labeled with mouse monoclonal antibody to the insulin receptor and the DRAQ5 DNA stain (blue). Scale bar = 50 μm. (A′) 3D reconstruction of InsR staining in a pancreatic mouse tissue section. (B) Confocal imaging of endogenous insulin receptor localization in dispersed, fixed primary human beta-cells. Insets show AMCA-insulin immunofluorescence. Scale bar = 10 μm. (C) STED super resolution microscopy identifies small ∼80 nm clusters of plasma membrane insulin receptors in fixed MIN6 cells (arrow). Scale bar = 5 μm (D) Schematic of inter-domain tagging strategy used in the present study. Orange structure = InsR; Red structure = TagRFP. (E) Representative confocal image of colocalization between internalized FITC-insulin (200 nM, 1 h) and interdomain-tagged InsRA-TagRFP in live MIN6 cells cultured at 0 mM glucose. Scale bar = 10 μm. (F, G) Expression of InsRA-TagRFP and InsRB-TagRFP sustains Erk phosphorylation in HEK 293T cells stimulated with 50 nM insulin (n = 4) at 0 mM Glucose. (H) Colocalization of identical insulin receptor isoforms tagged with different fluorescent proteins in fixed MIN6 cells (n = 10). Scale bar = 10 μm. Inset Venn diagrams, here and throughout, can be used visualize the colocalization of the color-coded proteins and represent the relative size of the puncta pools and their overlap. (I) Colocalization of fluorescently labeled insulin receptor A and B isoforms in fixed MIN6 cells (n = 10). Identical results were obtained with InsRA-TagBFP and InsRB-TagRFP (not shown). Scale bar = 10 μm. (J, K) C-terminal-tagged insulin receptors have a primarily plasma membrane localization that does not substantially colocalize with interdomain-tagged insulin receptors in fixed MIN6 cells (n = 10). Scale bar = 10 μm
3.2. Development of functional fluorescent insulin receptor fusion proteins to monitor trafficking
To track the route of insulin receptors in living cells, we employed a novel tagging strategy whereby pH-resistant fluorescent proteins were placed at the extracellular domain of the insulin receptor in between the furin-like region and the transmembrane domain, avoiding the interruption of known functional domains [21] (Figure 1D). TagRFP and TagBFP were chosen because they are bright, monomeric, and, most importantly, pH-resistant, making them ideal for a tagging location that resides within the lumens of potentially acidic organelles [37], [38].
Control experiments were conducted to ensure the functionality of these interdomain-tagged insulin receptors and determine whether their cellular distribution matched endogenous insulin receptor proteins. We confirmed first that interdomain-tagged InsRA-TagRFP (Figure 1E) and InsRB-TagRFP (not shown) bound and internalized FITC-labeled insulin in cultured beta-cells. To further validate that InsRA-TagRFP or InsRB-TagRFP were functional, we over-expressed them in HEK 293T cells, which, unlike beta-cells, do not constantly secrete insulin, thereby permitting the assessment of insulin signal transduction while controlling dose and temporal stimulation. Indeed, interdomain-tagged insulin receptors mediated Erk activation in a temporal pattern that was similar to endogenous receptors (Figure 1F, G). These data indicate that interdomain-tagged InsRs are suitable tools to monitor the dynamics of functional insulin receptors.
Next, we assessed the subcellular localization of tagged insulin receptors by confocal microscopy and stringent object-based colocalization. As a positive control, cells were transfected with InsRA-TagRFP and InsRA-TagBFP (i.e. the same InsR isoform with different fluorescent tags) to determine maximum biological colocalization using our analyses routines (Figure 1H). As illustrated by the Venn diagram (inset of Figure 1H), co-localization of InsRA-TagRFP objects with InsRA-TagBFP objects was not 100%. This is due to the existence of puncta with sub-threshold expression of one InsR sub-unit, as well as different signal-to-noise ratios between TagRFP and TagBFP. Similarly, we found significant intracellular colocalization between InsRA-TagRFP and InsRB-TagRFP (Figure 1I), suggesting similar trafficking routes for each isoform, and between insulin receptor homodimers and heterodimers [20]. Conservative, object-based colocalization analysis clearly showed that the majority of these interdomain insulin receptor fusion proteins (InsRA-TagRFP, InsRB-TagRFP) localized to vesicle-like cytoplasmic structures, with a minority in the plasma membrane of MIN6 cells (Figure 1H, I and throughout). This mirrors the pattern observed for endogenous beta-cell insulin receptors (Figure 1A–C and throughout) and is in contrast to the near exclusive plasma membrane localization of insulin receptors tagged at their C-termini (InsRA-C-eGFP) published previously [19] (Figure 1J, K). We also found that clusters of C-terminal tagged insulin receptors exhibit dramatically reduced mobility in the MIN6 cell plasma membrane compared to interdomain-tagged InsRA-TagRFP (10.5 s ± 2.6 s vs. 3.4 s ± 0.05 s domain life time, p < 0.05, Supplemental Movie 1). Our data using inter-domain-tagged insulin receptors indicate that they are functional and traffic in a pattern that is indistinguishable from endogenous insulin receptors.
Supplementary video related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.01.009.
The following is the supplementary data related to this article:
Movie S1: Time lapse TIRF imaging of HEK-293T cells expressing either InsRA-TagRFP (inter-domain tagged) or InsRA-C-eGFP (C-terminal label) constructs recorded at 25 mM glucose.
We also briefly investigated the distribution of Igf1 receptors in beta-cells and found them to also be mostly intracellular (Figure S2). Indeed, interdomain-tagged Igf1 receptors exhibited a strong degree of co-localization with interdomain-tagged insulin receptors (Supplemental Movie 2), consistent with their ability to heterodimerize with insulin receptors. Future studies are warranted to fully investigate the trafficking of Igf1 receptors using this new approach.
Supplementary video related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.01.009.
The following is the supplementary data related to this article:
Movie S2: Live cell imaging of a MIN6 cell co-transfected with IGF1R-TagGFP2 (left) and InsRA-TagRFP (middle) (both interdomain tagged). TagGFP2 and TagRFP was inserted between two domains at bp 2809/aa 937, and bp 2812/aa 938, respectively.
3.3. Insulin receptors do not travel in vesicles marked with clathrin, Rab5a, Rab7, or Rab11a
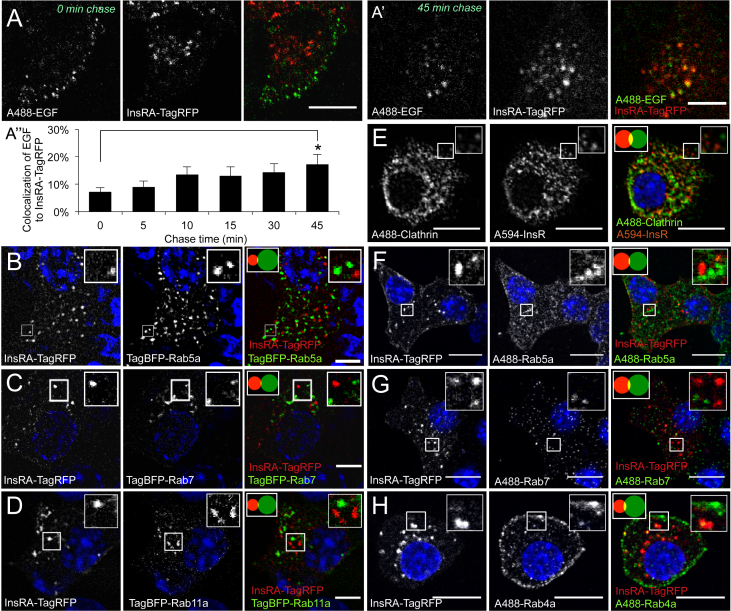
We examined the trafficking route of these functional insulin receptors by comparing their localization to endosomal pathway markers. First, we examined whether insulin receptors colocalized with epidermal growth factor (EGF), an established endosomal cargo of the clathrin pathway, in a pulse chase experiment. Fluorescent Alexa-488 labeled EGF showed very limited colocalization (<5%) to InsRA-TagRFP at short-chase times, indicating that the proximal routes of endocytosis were likely distinct (Figure 2A). The significantly increased colocalization at longer chase times indicated that the vesicles of the two different internalization routes eventually merged into more mature compartments such as lysosomes (Figure 2A′,A″). Next, we assessed the colocalization of InsRA-TagRFP with Rab5a, a marker associated with clathrin-dependent endocytosis [39], as well as Rab7-labeled late endosomes and Rab11a-labeled recycling endosomes and found virtually no colocalization (Figure 2B–D). Similarly, endogenous insulin receptors showed minimal colocalization with endogenous clathrin in primary mouse beta-cells (Figure 2E), and tagged insulin receptors did not appreciably colocalize with endogenous Rab5a, Rab7, or Rab4A (Figure 2F–H). Clathrin loss-of-function experiments resulted in gross changes in beta-cell plasma membrane morphology (not shown), so we were unable to rule out some small role for clathrin in insulin receptor trafficking. Notwithstanding, these data demonstrate that insulin receptors spend little, if any, time in endocytic vesicles labeled with these Rab-GTPases, or clathrin.
Figure 2.
Insulin receptors do not associate with markers of clathrin-dependent endocytosis. (A, A′) Pulse-chase colocalization analysis of Alexafluor488-labeled EGF relative to InsRA-TagRFP indicates separate entry pathways but eventual fusion into a shared mature pool of vesicles in MIN6 cells cultured in 0 mM glucose. A significant increase in the degree of colocalization was observed at a 45 min chase time compared to 0 min chase time (n = 4 images per time point). Scale bar = 5 μm. (B–D) Colocalization analysis of InsRA-TagRFP with TagBFP-tagged Rab5a (early endosomes), TagBFP-Rab7 (late endosomes), and TagBFP-Rab11a (recycling endosomes) in fixed MIN6 cells (n = 10). Similar results observed with InsRB-TagRFP. Scale bar = 5 μm. (E) Colocalization analysis of immunolabeled endogenous insulin receptors with immunolabeled endogenous clathrin in MIN6 cells (n = 10). Scale bar = 10 μm. (F–H) Colocalization analysis of tagged insulin receptors with immunolabeled endogenous Rab5a, Rab7, and Rab4a (alternate marker for recycling endosomes) in MIN6 cells (n = 10). Scale bar = 10 μm. (B–H) Prior fixation, MIN6 cells were cultured in 25 mM glucose.
3.4. Cav1 participates in the dislocation of insulin receptors from the plasma membrane
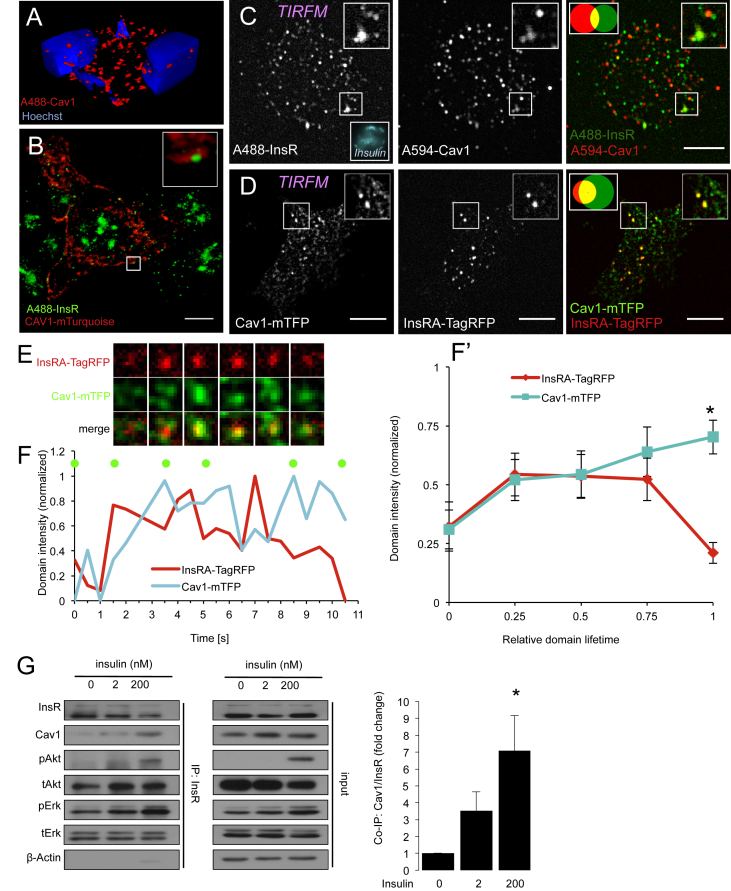
Given the observation that insulin receptors did not significantly colocalize with proteins or cargo that mark ‘classical endosomes’ in beta-cells, we examined alternative internalization and trafficking routes. We investigated caveolin-1 (Cav1) because insulin receptors colocalize with Cav1 in adipocytes, and Cav1 knockout mice exhibit impaired insulin signaling in other tissues [15], [16], [17]. Cav1 is prominently expressed in primary beta-cells, in vivo and in vitro (Figure S3A, B), and, to a lesser extent, in beta-cell lines (Figure S3C) [40], [41]. Confocal imaging illustrated a primarily cytoplasmic distribution of Cav1 in vivo within mouse pancreas sections, and in vitro in isolated mouse beta-cells (Figures. 3A, S3A, B), consistent with previous results [40], [41]. Endogenous Cav1 exhibited robust colocalization with endogenous insulin receptors in isolated mouse beta-cells (Figure S3B). STED super-resolution imaging revealed that Cav1 could be found as structures appearing to surround insulin receptor containing complexes at the plasma membrane (Figure 3B). TIRF imaging revealed colocalization of endogenous insulin receptors and Cav1 in the <200 nm near plasma membrane space of isolated human beta-cells (Figure 3C).
Figure 3.
Caveolin-1 is associated with insulin receptor internalization. (A) 3D reconstruction of subcellular Cav1 staining in murine islets cells of a pancreatic section. (B) STED super-resolution imaging of mTurquoise-tagged Cav1 and endogenous insulin in MIN6 cells. Inset shows Cav1 surrounding a cluster of insulin receptors. Scale bar = 5 μm. (C) Immunolabeling and TIRF microscopy of dispersed human islet cells cultured in 5 mM glucose demonstrates colocalization of endogenous Cav1 to endogenous insulin receptors at the plasma membrane (n = 20 cells). Scale bar = 5 μm. (D) TIRF imaging reveals a high degree of colocalization of Cav1-mTFP to InsRA-TagRFP at the plasma membrane of live MIN6 cells cultured in 20 mM glucose (n = 10). Scale bar = 5 μm. (E, F) Live-cell TIRF imaging in MIN6 cells cultured in 20 mM glucose reveals the reciprocal recruitment of InsRA-TagRFP and Cav1-mTFP to membrane domains prior to the internalization of insulin receptors. Pixel size = 0.129 μm (F, F′) Intensity analysis of a single (F) InsRA-TagRFP positive membrane domain during the process of Cav1 mediated vesicle budding. Green dots correspond to the time points of the images in E. (F′) shows the averaged and normalized intensities of 15 analyzed InsRA-TagRFP positive membrane domains during Cav1 mediated vesicle budding from the plasma membrane of MIN6 cells cultured in 20 mM glucose. *p < 0.05. (G) Co-immunoprecipitation of insulin receptors from NIH-3T3 cells reveals an insulin dependent binding of Cav1 to the insulin receptor (n = 3). NIH-3T3 cells were cultured in 0 mM glucose during insulin treatments for 5 min *p < 0.05
To examine the dynamic relationship between Cav1 and insulin receptors in living cells, we overexpressed fluorescence protein-tagged Cav1 in MIN6 cells to levels that were comparable to those found in primary beta-cells (Figure S3C). Static TIRF microscopy demonstrated high colocalization between CAV1-mTFP and InsRA-TagRFP at the plasma membrane (Figure 3D), with 60% ± 13% of insulin receptors colocalizing with CAV1. Time-lapse TIRF imaging revealed that CAV1-mTFP and InsRA-TagRFP co-accumulated immediately prior to the internalization of the insulin receptors into the intracellular space (Figure 3E, F, F′ Supplemental Movie 3). Taken together, these data suggest that Cav1 is a key coat protein involved in the dislocation of functional insulin receptors from the beta-cell plasma membrane.
Supplementary video related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.01.009.
The following is the supplementary data related to this article:
Movie S3: Time lapse TIRF imaging of a Cav1 MIN6 cell expressing InsRA-TagRFP and Cav1-mTFP recorded at 25 mM glucose.
Given the functional relationship between Cav1 phosphorylation and insulin receptor signaling [12], [13], [42], we examined whether both proteins interact as part of a physical complex upon insulin stimulation using co-immunoprecipitation. Indeed, insulin dose-dependently increased Cav1 binding to insulin receptors in NIH-3T3 cells (Figure 3G). Together with our live-cell imaging (Figure 3E, F), these experiments suggest that insulin can stimulate the recruitment of Cav1 to insulin receptor positive membrane domains.
3.5. Insulin receptors are present in flotillin-I-positive vesicles and LAMP2 labeled lysosomes
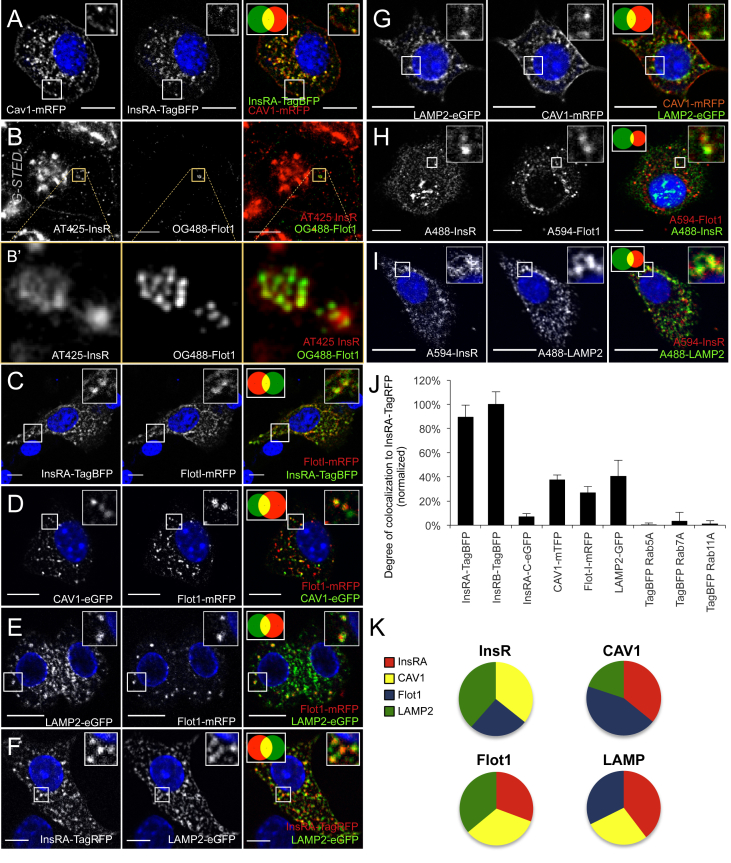
Studies in other cell types indicate that insulin receptors travel to lysosomes [8], and we have published data illustrating the colocalization of endogenous InsR with Lysotracker in human beta-cells [23], but the intracellular route of beta-cell insulin receptors has not been reported. Confocal imaging demonstrated that tagged CAV1 colocalized with tagged InsRA on intracellular vesicular structures, but to a lesser extent than the colocalization between these proteins at the plasma membrane (Figure 3, Figure 4A). Flotillin-I (Flot1) has been described in clathrin-independent endocytosis [43], associates with caveolin-containing structures [43], [44], and has been implicated in insulin signaling in other cell types [45], [46]. Endogenous Flot1 was expressed in primary mouse beta-cells (Figure 4H). Using super-resolution imaging, we found that endogenous Flot1 was localized on intracellular vesicles containing insulin receptors (Figure 4B, B′). Similarly, InsRA-TagBFP showed robust colocalization with Flot1-mRFP positive vesicles by confocal imaging (Figure 4C). Flot1-mRFP also colocalized with both Cav1-eGFP and LAMP2-eGFP (Figure 4D, E), suggesting a distribution of internalized insulin receptors to early and late endolysosomal compartments.
Figure 4.
InsR-TagRFP is transported to lysosomes in Cav-1 and Flot-1 positive vesicles. (A) Confocal imaging of Cav1-mRFP and InsRA-TagBFP in MIN6 cells (n = 10). Scale bars = 10 μm. (B) G-STED microscopy in MIN6 cells illustrates the presence of Flot1 on vesicles containing endogenous insulin receptors. Scale bars = 5 μm. (B′) Insets show flotillin-1 positive vesicles harboring insulin receptors. (C–G) Colocalization between InsRA-TagBFP and Flot1-mRFP, Flot1-mRFP and CAV1-eGFP, Flot1-mRFP and LAMP2-eGFP, InsRA-TagRFP and LAMP2-eGFP, CAV1-mRFP and LAMP2-eGFP in MIN6 cells demonstrates a Cav1-Flot1-LAMP2 endocytic trafficking route to lysosomes (n = 10). Scale bars = 10 μm; F: Scale bar = 5 μm. (H, I) Immunolabeling of endogenous insulin receptors, endogenous Flot1, and endogenous LAMP2 (lysosomes) in primary 12 week-old mouse islet cells (n = 10). Scale bars = 10 μm. (J) Summary of object-based colocalization of tagged proteins compared with InsRA-TagRFP in MIN6 cells. Note that InsRA controls represent the maximum colocalization in this system (n = 10 cells per condition). (K) Summary of colocalization within vesicular pools in the Cav1/Flot1/Lamp2 pathway of MIN6 cells (n = 10 cells per condition). (A–I) MIN6 cells were cultured in 25 mM glucose and primary cells were cultured in 11 mM glucose prior fixation.
A substantial proportion of endogenous or tagged insulin receptors were found in LAMP2-marked lysosomes (Fig. 4F, I). Some colocalization between CAV1-mRFP and LAMP2-eGFP was also observed (Figure 4G), consistent with previous reports [47], [48]. The concept that insulin receptors are transported in Flot1-positive vesicles to lysosomes was supported by immunofluorescence staining of endogenous proteins in isolated primary mouse beta-cells (Figure 4H–K).
3.6. Cav1 phosphorylation modulates InsR internalization and Erk signaling in vitro
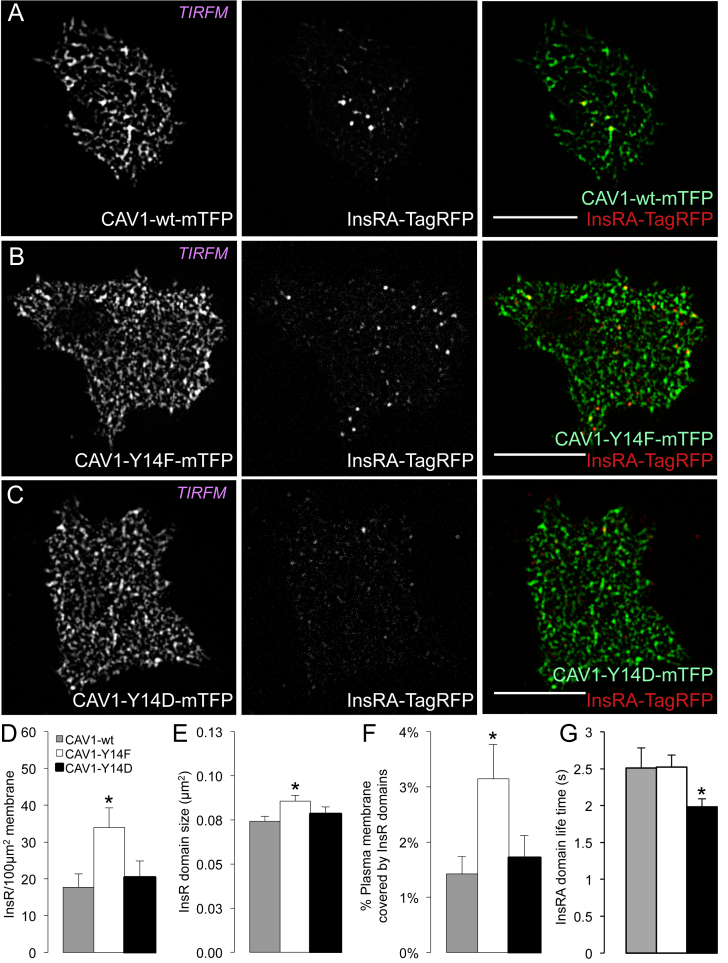
Colocalization studies showed a clear association between Cav1 and insulin receptors, but loss-of-function studies were required to determine whether Cav1 drives the process of insulin receptor internalization. Thus, we co-expressed mTFP-labeled Cav1 mutants, a constitutively active form harboring phosphomimetic at tyrosine 14 or a dominant negative form harboring non-phosphorylatable mutations at tyrosine 14 [24], [49], along with TagRFP-labeled insulin receptors. TIRF microscopy demonstrated that expression of the dominant negative Cav1-Y14F mutant increased InsRA-TagRFP density at the plasma membrane when compared to cells expressing wildtype Cav1 (Figure 5A–F). Subsequently, we tested the effects of modified Cav1 phosphorylation on InsR internalization with time-lapse TIRF microscopy. For this experiment, we cloned an eGFP interdomain-tagged InsR construct (InsR-lum-eGFP) to take advantage of the superior photostability of eGFP (eGFP pH-sensitivity was not a confounder in the extracellular pH neutral environment). Stable MIN6 cell lines expressing Cav1 mutants showed that the expression of the phospho-mimetic Cav1-Y14D mutant leads to a significantly shorter lifetime of InsRA domains at the plasma membrane compared to controls (Figure 5G, Supplemental Movie 4). These data are consistent with the concept that Cav1 phosphorylation promotes insulin receptor internalization in beta-cells.
Figure 5.
Caveolin-1 phosphorylation modulates insulin receptor domain size and internalization. (A–C) TIRF microscopy of live MIN6 cells expressing InsRA-TagRFP and Cav1 mutant proteins. Scale bar = 10 μm. (D–F) Overexpression of the Cav1-Y14F mutant leads to increased InsRA-TagRFP domain density, increased InsRA-TagRFP domain size, and a higher content of InsRA-TagRFP at the plasma membrane of fixed MIN6 cells (n = 10 cells per condition). (G) Quantification of TIRF time lapse imaging of InsRA-lum-eGFP domain lifetime at the plasma membrane of stable MIN6 cell lines expressing various Cav1 mutants. *p < 0.05, n > 4 cells per condition. (A–G) MIN6 cells were cultured in 20 mM glucose during image acquisition.
Supplementary video related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.01.009.
The following is the supplementary data related to this article:
Movie S4: Time lapse TIRF imaging of Cav1 MIN6 cell lines expressing various Cav1 mutants and inter-domain tagged InsRA-lum-eGFP constructs recorded at 25 mM glucose.
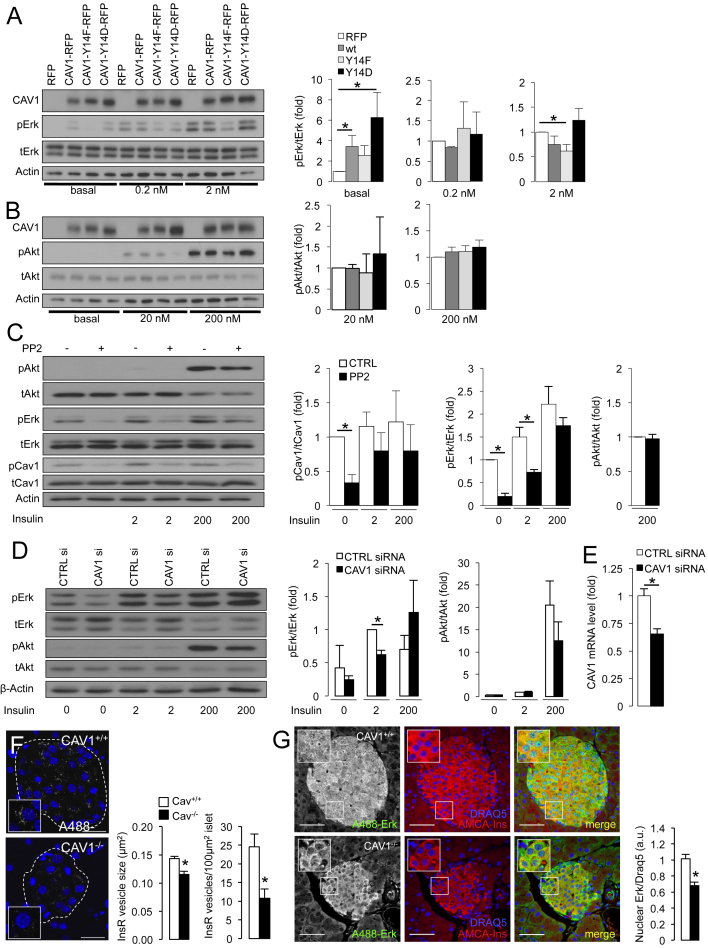
Next, we sought to establish the functional consequences of altered Cav1 phosphorylation in insulin signaling in beta-cells, which even under basal conditions have insulin present in their cultures [50]. Overexpressing wildtype Cav1 or the constitutively active Cav1-Y14D mutant was sufficient to significantly increase Erk phosphorylation in beta-cells in the absence of added insulin (Figure 6A). In the presence of 2 nM added insulin, the dominant negative Cav1-Y14F mutant significantly suppressed Erk activation (Figure 6A). On the other hand, neither basal nor exogenous insulin-stimulated Akt phosphorylation was affected by the expression of wildtype or mutant Cav1 (Figure 6B). Similar results were obtained in experiments where endogenous Cav1 phosphorylation was inhibited in NIH-3T3 and MIN6 cells by the Src kinase inhibitor PP2 (Figs 6C, S4). While Src has multiple cellular targets, collectively these data suggest a role for Cav1 phosphorylation in insulin receptor internalization and Erk signaling.
Figure 6.
Caveolin-1 enhances insulin-stimulated Erk, but not Akt, signaling in vitro. (A, B) Overexpression of wildtype Cav1-mRFP or the phosphomimetic mutant Cav1-Y14D significantly increases basal insulin signaling to Erk. Erk signaling stimulated by 2 nM added insulin is significantly reduced in cells overexpressing the dominant negative mutant Cav1-Y14F. Akt signaling is unaffected by the overexpression of Cav-1 mutants. Experiments were performed in MIN6 cells treated for 5 min with the indicated doses of insulin (n = 10). (C) Insulin stimulated (5 min) Erk, but not Akt signaling is significantly reduced in NIH-3T3 cells previously treated for 2 h with 10 μM of Src kinase inhibitor PP2 (n = 4). *p < 0.05. (D) Erk, but not Akt signaling is significantly reduced in MIN6 cells treated with Cav1-siRNA and a physiological dose of insulin for 5 min (n = 4). *p < 0.05. (A–D) MIN6 cells and NIH-3T3 cells were incubated in 0 mM glucose during 5 min of insulin treatments. (E) Knockdown efficiency in MIN6 cells cultured at 25 mM glucose and treated with a siRNA pool against murine Cav1 (n = 6 independent samples per condition). *p < 0.05. (F) Reduced insulin receptor immunolabeling of intracellular vesicles in islets of Cav1−/− mice. *p < 0.05. (n = 9 islets). Cav1+/+ = white, Cav1−/− = black, throughout this figure. Scale bar = 25 μm. (G) Reduced Erk signaling visualized by nuclear translocation in islet cells from 12 week old Cav1−/− mice (n = 3 mice, 22 analyzed islets). Scale bar = 50 μm.
3.7. Cav1 loss reduces Erk signaling in vitro and in vivo
We sought to further define the role of Cav1 using in vitro and in vivo gene targeting. Significantly reducing Cav1 expression by ∼30% in MIN6 cells with siRNA led to a proportional reduction in Erk phosphorylation stimulated by 2 nM added insulin (Figure 6D, E). Interestingly, Erk phosphorylation was not affected by Cav1 knockdown in the context of a super-physiological insulin dose, which likely activates Igf1 receptors [21]. Akt signaling was not affected at any dose of added insulin in these experiments (Figure 6D).
To complement these in vitro results, we studied pancreatic tissue sections from Cav1−/− mice. Cav1−/− beta-cells exhibited significantly reduced insulin receptor puncta (Figure 6F) and a significant reduction of nuclear Erk (Figure 6G), consistent with a down-regulation of Erk signaling in vivo. Akt phosphorylation did not appear to be altered in Cav1−/− beta-cells (data not shown). Collectively, these data demonstrate that caveolin-1 modulates insulin signaling in beta-cells, in vitro and in vivo.
4. Discussion
In the present study, we used a novel tagging strategy and multiple imaging modalities to implicate caveolin-1 and flotillin-1 in insulin receptor trafficking in pancreatic beta-cells, a critical insulin target cell type [1], [2], [3], [4], [5], [7], [23], [51]. We provide evidence that phospho-cav1-mediated endocytosis is a bifurcation point biasing signaling towards the Erk pathway. We find that insulin receptors largely bypass clathrin-, Rab5a- or Rab7-positive endosomes, distinguishing this pathway from the reported endocytic route of other tyrosine kinase receptors in other tissues. Our data provide new molecular insights into the mechanisms of insulin receptor trafficking, in general, and lay the groundwork for therapeutic efforts to harness insulin signaling in beta-cells.
The pathogenesis of type 2 diabetes is characterized by alterations in insulin signaling in many tissues. The role of insulin signaling defects in pancreatic beta-cells remains underappreciated, although conceptually it links the two most widely recognized hallmarks of diabetes progression, ‘insulin resistance’ and beta-cell dysfunction (including inappropriate insulin hyper-secretion) [4]. Indeed, reduced insulin receptor expression and loss of insulin signaling have been observed in islets from patients with type 2 diabetes [52], [53]. Evidence from animal models also suggests that the loss of beta-cell insulin signaling is sufficient for diabetes progression [54]. Although the Akt arm of the insulin signaling pathway has received the most attention, we have previously shown that physiological levels of insulin [55] have little effect on Akt in beta-cells [5], but instead appear to selectively signal through the Raf1/Mek/Erk pathway [5], [6], [23]. Our data permit us to speculate that Erk and Akt signaling may occur from insulin receptors localized in different nanodomains, with Akt signaling occurring mostly at the plasma membrane [56] and Erk signaling occurring preferentially from internalized (or internalizing) receptors in this cell type, and perhaps others [57]. As Erk shuttles to the nucleus upon activation [58], it may be advantageous for Erk signaling to occur from endosomes or lysosomes located closer to the nucleus. Our study also sheds light on the fate of many internalized insulin receptors in beta-cells, by revealing that many insulin receptors are found in LAMP2-positive lysosomes and by excluding a major role for Rab11a or Rab4 positive-recycling endosomes. However, this finding does not exclude recycling of insulin receptors via a pathway independent of Rab11a. Notwithstanding, if the majority of insulin receptors are degraded before returning to the plasma membrane, it could represent a way of limiting autocrine insulin signaling in the face of elevated local insulin [50].
Caveolin-1 has previously been implicated in beta-cell physiology, specifically in insulin secretion and cytokine-induced death [40], [41], [59], but its roles in endocytosis or insulin signaling had not been demonstrated. Allelic variation in the human caveolin-1 gene (CAV1 – rs926198, rs3807989) has been linked to fasting hyperinsulinemia and insulin resistance as measured by HOMA-IR and hyperinsulinemic–euglycemic clamps [60]. Similarly, Cav1 knockout mice have been reported to display fasting hyperinsulinemia, insulin resistance and glucose intolerance [60], [61]. A previous report indicated that Cav1 loss impaired insulin signaling in adipocytes but not liver or skeletal muscle [62]. Phosphorylated Cav1 is associated with focal adhesion signaling [24], and our data support a novel role for phospho-Cav1 in the regulation of insulin signaling in beta-cells. Electron microscopy illustrated that Cav1-positive adipocyte membrane pits harbor insulin-receptors [63]. The concept of caveolae as nanodomains for insulin receptors at the plasma membrane has been suggested for several tissues [64], [65], [66]. Cav1 and their associated cavins are generally negative regulators of raft-dependent endocytosis [67] while Cav1 tyrosine phosphorylation has been reported to influence pinocytosis and promote integrin internalization [68]. The demonstration here that phospho-Cav1 promotes InsR internalization defines a novel role for Cav1 tyrosine phosphorylation. Cav1 delivery to endolysosomal compartments is enhanced by modulation of lysosomal pH and cholesterol content and by disruption of the assembly of Cav1-contained structures that have been named caveolae by some investigators [47]. Similarly, flotillin-I is associated with endosomes and lysosomes [43]. Our data support the concept that Cav1 and flotillin-1 define a receptor trafficking endosomal route to lysosomes. Indeed, the observation that insulin receptors do not colocalize to Rab5a-positive early endosomes or Rab7-positive late endosomes suggested that they traffic via a pathway that is relatively independent of the well studied pathway downstream of clathrin. However, it is not possible to fully rule out a mechanistic role for clathrin itself without molecular loss-of-function studies under conditions with otherwise preserved normal beta-cell physiology.
The present study also illustrates technical considerations for the analysis of tyrosine kinase receptor trafficking. Attaching fluorescent proteins to key domains has been shown to disrupt the function of some proteins [69], [70]. To the best of our knowledge, N-terminal- or C-terminal-tagged insulin receptors have not been shown to localize with endogenous insulin receptors or to signal with similar efficiency to downstream targets, associating a caveat to previous studies and perhaps leading to controversial results [12], [13], [18], [71]. We overcame this technical challenge by tagging insulin receptors between known functional domains, a method applied to a few other proteins [72], [73]. We demonstrated that interdomain-tagged insulin receptors have similar functionality when compared to untagged, endogenous receptors. We also employed monomeric fluorescent proteins that, unlike weak dimers such as GFP, do not accelerate the formation of dimers between tagged proteins. These fluorescent proteins were highly pH-resistant, reducing signal loss in the acidic endo/lysosomal lumens. Our tagging strategy is likely to be broadly applicable to other tyrosine kinase receptors and to insulin receptors in other tissues. This information will be critical as we seek to understand the molecular mechanisms of insulin resistance in order to develop approaches to modulate insulin sensitivity in obesity and early stage type 2 diabetes.
Funding
This work was funded by a JDRF grant (17-2013-4) and a CIHR grant (MOP-133692) to JDJ, and a CIHR grant (MOP-126029) to IRN.
TB was supported by the Karl-Heinz Frenzen-Stiftung.
GEL was supported by post-doctoral fellowships from JDRF, CIHR, MSFHR, and CDA.
Author contributions
JDJ designed the study, analyzed the data and co-wrote the manuscript. JDJ is the guarantor of this work.
TB designed the study, performed experiments, analyzed the data and co-wrote the manuscript.
GEL, HC, SS, performed experiments and analyzed the data.
IRN analyzed the data and provided critical reagents.
PG coordinated experiments with Cav1−/− mice.
MP and SL performed experiments.
Acknowledgments
We thank R.E. Campbell, A. Ullrich, I.B. Leibiger, Ai Yamamoto and J. Lippincott-Schwartz for plasmids. We thank C. Dessy (Université Catholique de Louvain, Brussels, Belgium) for providing the pancreatic sections from Cav1−/− mice, and N. Antoine (Université Catholique de Louvain, Brussels, Belgium) for performing insulin secretion experiments. We thank R. Sidhu and U. Schwarz (Leica Microsystems, Germany) for access to CW-STED and G-STED systems, for advice on sample preparation, and for some image acquisition. R. Slaaby and M. Andersen (Novo-Nordisk A/S, Denmark) for advice and parallel studies on insulin receptor processing.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.01.009.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Figure S1: (A) Confocal imaging of endogenous insulin receptor localization in dispersed primary mouse beta-cells. Insets show AMCA-insulin immunofluorescence. Scale bar = 10 μm. (B) Subcellular localization of primary human beta-cells (60% purity) on plasma membranes and intracellular membranes demonstrated by cellular fractionation.
Figure S2: Immunofluorescence staining illustrating the localization of endogenous insulin like growth factor 1 receptor (green) in a fixed MIN6 cell achieved by confocal imaging. Scale bar = 10 μm.
Figure S3: (A) Endogenous caveolin-1 has a primarily intracellular distribution in murine pancreatic beta-cells in vivo. Tissue section obtained from a 12 week-old mouse. Scale bar = 10 μm. (B) Immunolabeling of dispersed islet cells obtained from a 12 week old mouse demonstrates colocalization of endogenous Cav1 to endogenous insulin receptors. Scale bar = 10 μm. (C) Quantification of endogenous and over-expressed levels of Cav1 relative to endogenous levels in primary mouse islets.
Figure S4: Insulin stimulated Erk, but not Akt signaling is significantly reduced in MIN6 cells treated with the Src kinase inhibitor PP2 (n = 4). *p < 0.05. Treatments performed in 0 mM glucose.
References
- 1.Okada T., Liew C.W., Hu J., Hinault C., Michael M.D., Krtzfeldt J. Insulin receptors in beta-cells are critical for islet compensatory growth response to insulin resistance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8977–8982. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leibiger I.B., Leibiger B., Berggren P.O. Insulin signaling in the pancreatic beta-cell. Annual Review of Nutrition. 2008;28:233–251. doi: 10.1146/annurev.nutr.28.061807.155530. [DOI] [PubMed] [Google Scholar]
- 3.Johnson J.D., Alejandro E.U. Control of pancreatic beta-cell fate by insulin signaling: the sweet spot hypothesis. Cell Cycle. 2008;7:1343–1347. doi: 10.4161/cc.7.10.5865. [DOI] [PubMed] [Google Scholar]
- 4.Mehran A.E., Templeman N.M., Brigidi G.S., Lim G.E., Chu K.Y., Hu X. Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metabolism. 2012;16:723–737. doi: 10.1016/j.cmet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Johnson J.D., Bernal-Mizrachi E., Alejandro E.U., Han Z., Kalynyak T.B., Li H. Insulin protects islets from apoptosis via Pdx1 and specific changes in the human islet proteome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19575–19580. doi: 10.1073/pnas.0604208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alejandro E.U., Lim G.E., Mehran A.E., Hu X., Taghizadeh F., Pelipeychenko D. Pancreatic beta-cell Raf-1 is required for glucose tolerance, insulin secretion, and insulin 2 transcription. FASEB Journal. 2011;25:3884–3895. doi: 10.1096/fj.10-180349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beith J.L., Alejandro E.U., Johnson J.D. Insulin stimulates primary beta-cell proliferation via Raf-1 kinase. Endocrinology. 2008;149:2251–2260. doi: 10.1210/en.2007-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Posner B.I., Kahn M.N., Bergeron J.J. Internalization of insulin: structures involved and significance. Advances in Experimental Medicine and Biology. 1985;189:159–173. doi: 10.1007/978-1-4757-1850-8_10. [DOI] [PubMed] [Google Scholar]
- 9.Lajoie P., Nabi I.R. Lipid rafts, caveolae, and their endocytosis. International Review of Cell and Molecular Biology. 2010;282:135–163. doi: 10.1016/S1937-6448(10)82003-9. [DOI] [PubMed] [Google Scholar]
- 10.Parton R.G., del Pozo M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nature Reviews Molecular Cell Biology. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 11.Smith R.M., Harada S., Smith J.A., Zhang S., Jarett L. Insulin-induced protein tyrosine phosphorylation cascade and signalling molecules are localized in a caveolin-enriched cell membrane domain. Cell Signal. 1998;10:355–362. doi: 10.1016/s0898-6568(97)00170-8. [DOI] [PubMed] [Google Scholar]
- 12.Kabayama K., Sato T., Saito K., Loberto N., Prinetti A., Sonnino S. Dissociation of the insulin receptor and caveolin-1 complex by ganglioside GM3 in the state of insulin resistance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13678–13683. doi: 10.1073/pnas.0703650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagerholm S., Ortegren U., Karlsson M., Ruishalme I., Strålfors P. Rapid insulin-dependent endocytosis of the insulin receptor by caveolae in primary adipocytes. PLoS ONE. 2009;4:e5985. doi: 10.1371/journal.pone.0005985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceresa B.P., Kao A.W., Santeler S.R., Pessin J.E. Inhibition of clathrin-mediated endocytosis selectively attenuates specific insulin receptor signal transduction pathways. Molecular and Cellular Biology. 1998;18:3862–3870. doi: 10.1128/mcb.18.7.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen A.W., Razani B., Wang X.B., Combs T.P., Williams T.M., Scherer P.E. Caveolin-1-deficient mice show insulin resistance and defective insulin receptor protein expression in adipose tissue. American Journal of Physiology. Cell Physiology. 2003;285:C222–C235. doi: 10.1152/ajpcell.00006.2003. [DOI] [PubMed] [Google Scholar]
- 16.Razani B., Combs T.P., Wang X.B., Frank P.G., Park D.S., Russell R.G. Caveolin-1-deficient mice are lean, resistant to diet-induced obesity, and show hypertriglyceridemia with adipocyte abnormalities. Journal of Biological Chemistry. 2002;277:8635–8647. doi: 10.1074/jbc.M110970200. [DOI] [PubMed] [Google Scholar]
- 17.Trajkovski M., Hausser J., Soutschek, Bhat B., Akin A., Zavolan M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 18.Uhles S., Moede T., Leibiger B., Berggren P.-O., Leibiger I.B. Selective gene activation by spatial segregation of insulin receptor B signaling. FASEB Journal. 2007;21:1609–1621. doi: 10.1096/fj.06-7589com. [DOI] [PubMed] [Google Scholar]
- 19.Uhles S., Moede T., Leibiger B., Berggren P.-O., Leibiger I.B. Isoform-specific insulin receptor signaling involves different plasma membrane domains. The Journal of Cell Biology. 2003;163:1327–1337. doi: 10.1083/jcb.200306093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belfiore A., Frasca F., Pandini G., Sciacca L., Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocrine Reviews. 2009;30:586–623. doi: 10.1210/er.2008-0047. [DOI] [PubMed] [Google Scholar]
- 21.De Meyts P., Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nature Reviews Drug Discovery. 2002;1:769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 22.Gerdes J.M., Christou-Savina S., Xiong Y., Moede T., Moruzzi N., Karlsson-Edlund P. Ciliary dysfunction impairs beta-cell insulin secretion and promotes development of type 2 diabetes in rodents. Nature Communications. 2014;5:5308. doi: 10.1038/ncomms6308. [DOI] [PubMed] [Google Scholar]
- 23.Alejandro E.U., Kalynyak T.B., Taghizadeh F., Gwiazda K.S., Rawstron E.K., Jacob K.J. Acute insulin signaling in pancreatic beta-cells is mediated by multiple Raf-1 dependent pathways. Endocrinology. 2010;151:502–512. doi: 10.1210/en.2009-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goetz J.G., Lajoie P., Wiseman S.M., Nabi I.R. Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Reviews. 2008;27:715–735. doi: 10.1007/s10555-008-9160-9. [DOI] [PubMed] [Google Scholar]
- 25.Vicidomini G., Moneron G., Han K.Y., Westphal V., Ta H., Reuss M. Sharper low-power STED nanoscopy by time gating. Nature Methods. 2011;8:571–573. doi: 10.1038/nmeth.1624. [DOI] [PubMed] [Google Scholar]
- 26.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thevenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Transactions on Image Processing: A Publication of the IEEE Signal Processing Society. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 28.Sternberg S.R. Biomedical Image-Processing. Computer. 1983;16:22–34. [Google Scholar]
- 29.Bolte S., Cordelieres F.P. A guided tour into subcellular colocalization analysis in light microscopy. Journal of Microscopy. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 30.Lamprecht M.R., Sabatini D.M., Carpenter A.E. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 31.Drab M., Verkade P., Elger M., Kasper M., Lohn M., Lauterbach B. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 32.Nelson J.D., LeBoeuf R.C., Bomsztyk K. Direct recruitment of insulin receptor and ERK signaling cascade to insulin-inducible gene loci. Diabetes. 2011;60:127–137. doi: 10.2337/db09-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saltiel A.R., Pessin J.E. Insulin signaling in microdomains of the plasma membrane. Traffic. 2003;4:711–716. doi: 10.1034/j.1600-0854.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- 34.McClain D.A. Mechanism and role of insulin receptor endocytosis. American Journal of the Medical Sciences. 1992;304:192–201. doi: 10.1097/00000441-199209000-00009. [DOI] [PubMed] [Google Scholar]
- 35.Leibiger I.B., Brismar K., Berggren P.-O. Novel aspects on pancreatic beta-cell signal-transduction. Biochemical and Biophysical Research. 2010;396:111–115. doi: 10.1016/j.bbrc.2010.02.174. [DOI] [PubMed] [Google Scholar]
- 36.Huang B., Babcock H., Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merzlyak E.M., Goedhart J., Shcherbo D., Bulina M.E., Shcheglov A.S., Fradkov A.F. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nature Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 38.Subach O.M., Gundorov I.S., Yoshimura M., Subach F.V., Zhang J., Grüenwald D. Conversion of red fluorescent protein into a bright blue probe. Chemistry & Biology. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeigerer A., Gilleron J., Bogorad R.L., Marsico G., Nonaka H., Seifert S. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485:465–470. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
- 40.Veluthakal R., Chvyrkova I., Tannous M., McDonald P., Amin R., Hadden T. Essential role for membrane lipid rafts in interleukin-1beta-induced nitric oxide release from insulin-secreting cells: potential regulation by caveolin-1+ Diabetes. 2005;54:2576–2585. doi: 10.2337/diabetes.54.9.2576. [DOI] [PubMed] [Google Scholar]
- 41.Nevins A.K., Thurmond D.C. Caveolin-1 functions as a novel Cdc42 guanine nucleotide dissociation inhibitor in pancreatic beta-cells. The Journal of Biological Chemistry. 2006;281:18961–18972. doi: 10.1074/jbc.M603604200. [DOI] [PubMed] [Google Scholar]
- 42.Kimura A., Mora S., Shigematsu S., Pessin J.E., Saltiel A.R. The insulin receptor catalyzes the tyrosine phosphorylation of caveolin-1. Journal of Biological Chemistry. 2002;277:30153–30158. doi: 10.1074/jbc.M203375200. [DOI] [PubMed] [Google Scholar]
- 43.Glebov O.O., Bright N.A., Nichols B.J. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nature Cell Biology. 2006;8:46–54. doi: 10.1038/ncb1342. [DOI] [PubMed] [Google Scholar]
- 44.Bickel P.E., Scherer P.E., Schnitzer J.E., Oh P., Lisanti M.P., Lodish H.F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. Journal of Biological Chemistry. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 45.Baumann C.A., Ribon V., Kanzaki M., Thurmond D.C., Mora S., Shigematsu S. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 46.Liu J., Deyoung S.M., Zhang M., Dold L.H., Saltiel A.R. The stomatin/prohibitin/flotillin/HflK/C domain of flotillin-1 contains distinct sequences that direct plasma membrane localization and protein interactions in 3T3-L1 adipocytes. Journal of Biological Chemistry. 2005;280:16125–16134. doi: 10.1074/jbc.M500940200. [DOI] [PubMed] [Google Scholar]
- 47.Mundy D.I., Li W.P., Luby-Phelps K., Anderson R.G. Caveolin targeting to late endosome/lysosomal membranes is induced by perturbations of lysosomal pH and cholesterol content. Molecular Biology of the Cell. 2012;23:864–880. doi: 10.1091/mbc.E11-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayer A., Stoeber M., Ritz D., Engel S., Meyer H.H., Helenius A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. Journal of Cell Biology. 2010;191:615–629. doi: 10.1083/jcb.201003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S., Seitz R., Lisanti M.P. Phosphorylation of caveolin by src tyrosine kinases. The alpha-isoform of caveolin is selectively phosphorylated by v-Src in vivo. Journal of Biological Chemistry. 1996;271:3863–3868. [PubMed] [Google Scholar]
- 50.Wang M., Li J., Lim G.E., Johnson J.D. Is dynamic autocrine insulin signaling possible? A mathematical model predicts picomolar concentrations of extracellular monomeric insulin within human pancreatic islets. PLoS ONE. 2013;8:e64860. doi: 10.1371/journal.pone.0064860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohsugi M., Cras-Meneur C., Zhou Y., Bernal-Mizrachi E., Johnson J.D., Luciani D.S. Reduced expression of the insulin receptor in mouse insulinoma (MIN6) cells reveals multiple roles of insulin signaling in gene expression, proliferation, insulin content, and secretion. Journal of Biological Chemistry. 2005;280:4992–5003. doi: 10.1074/jbc.M411727200. [DOI] [PubMed] [Google Scholar]
- 52.Gunton J.E., Kulkarni R.N., Yim S., Okada T., Hawthorne W.J., Tseng Y.H. Loss of ARNT/HIF1beta Mediates Altered Gene Expression and Pancreatic-Islet Dysfunction in Human Type 2 Diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 53.Folli F., Okada T., Perego C., Gunton J., Liew C.W., Akiyama M. Altered insulin receptor signalling and beta-cell cycle dynamics in type 2 diabetes mellitus. PLoS ONE. 2011;6:e28050. doi: 10.1371/journal.pone.0028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., Zhang N., He M., Yang Z., Tong W., Wang Q. Increased beta-cell apoptosis and impaired insulin signaling pathway contributes to the onset of diabetes in OLETF rats. Cellular Physiology and Biochemistry. 2008;21:445–454. doi: 10.1159/000129637. [DOI] [PubMed] [Google Scholar]
- 55.Wang M., Li J., Lim G.E., Johnson J.D. Is dynamic autocrine insulin signaling possible? A mathematical model predicts picomolar concentrations of extracellular monomeric insulin within human pancreatic islets. PLoS ONE. 2013 doi: 10.1371/journal.pone.0064860. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao X., Lowry P.R., Zhou X., Depry C., Wei Z., Wong G.W. PI3K/Akt signaling requires spatial compartmentalization in plasma membrane microdomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14509–14514. doi: 10.1073/pnas.1019386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang P.Y., Weng J., Anderson R.G. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 58.Lidke D.S., Huang F., Post J.N., Rieger B., Wilsbacher J., Thomas J.L. ERK nuclear translocation is dimerization-independent but controlled by the rate of phosphorylation. Journal of Biological Chemistry. 2010;285:3092–3102. doi: 10.1074/jbc.M109.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kepner E.M., Yoder S.M., Oh E., Kalwat M.A., Wang Z., Quilliam L.A. Cool-1/betaPIX functions as a guanine nucleotide exchange factor in the cycling of Cdc42 to regulate insulin secretion. American journal of physiology. Endocrinology and Metabolism. 2011;301:E1072–E1080. doi: 10.1152/ajpendo.00312.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pojoga L.H., Underwood P.C., Goodarzi M.O., Williams J.S., Adler G.K., Jeunemaitre X. Variants of the caveolin-1 gene: a translational investigation linking insulin resistance and hypertension. The Journal of Clinical Endocrinology and Metabolism. 2011;96:E1288–E1292. doi: 10.1210/jc.2010-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asterholm I.W., Mundy D.I., Weng J., Anderson R.G., Scherer P.E. Altered mitochondrial function and metabolic inflexibility associated with loss of caveolin-1. Cell Metabolism. 2012;15:171–185. doi: 10.1016/j.cmet.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohen A.W., Razani B., Schubert W., Williams T.M., Wang X.B., Iyengar P. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 2004;53:1261–1270. doi: 10.2337/diabetes.53.5.1261. [DOI] [PubMed] [Google Scholar]
- 63.Gustavsson J., Parpal S., Karlsson M., Ramsing C., Thorn H., Borg M. Localization of the insulin receptor in caveolae of adipocyte plasma membrane. FASEB Journal. 1999;13:1961–1971. [PubMed] [Google Scholar]
- 64.Gomez-Ruiz A., de Miguel C., Campion J., Martinez J.A., Milagro F.I. Time-dependent regulation of muscle caveolin activation and insulin signalling in response to high-fat diet. FEBS Letters. 2009;583:3259–3264. doi: 10.1016/j.febslet.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 65.Oh Y.S., Khil L.Y., Cho K.A., Ryu S.J., Ha M.K., Cheon G.J. A potential role for skeletal muscle caveolin-1 as an insulin sensitivity modulator in ageing-dependent non-obese type 2 diabetes: studies in a new mouse model. Diabetologia. 2008;51:1025–1034. doi: 10.1007/s00125-008-0993-0. [DOI] [PubMed] [Google Scholar]
- 66.Wang H., Wang A.X., Barrett E.J. Caveolin-1 is required for vascular endothelial insulin uptake. American Journal of Physiology Endocrinology and Metabolism. 2011;300:E134–E144. doi: 10.1152/ajpendo.00498.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le P.U., Guay G., Altschuler Y., Nabi I.R. Caveolin-1 is a negative regulator of caveolae-mediated endocytosis to the endoplasmic reticulum. Journal of Biological Chemistry. 2002;277:3371–3379. doi: 10.1074/jbc.M111240200. [DOI] [PubMed] [Google Scholar]
- 68.Cheng Z.-J., Singh R.D., Holicky E.L., Wheatley C.L., Marks D.L., Pagano R.E. Co-regulation of caveolar and Cdc42-dependent fluid phase endocytosis by Phosphocaveolin-1. Journal of Biological Chemistry. 2010;285:15119–15125. doi: 10.1074/jbc.M109.069427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baens M., Noels H., Broeckx V., Hagens S., Fevery S., Billiau A.D. The dark side of EGFP: defective polyubiquitination. PLoS ONE. 2006;1:e54. doi: 10.1371/journal.pone.0000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ketelaar T., Anthony R.G., Hussey P.J. Green fluorescent protein-mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin depolymerizing factor's actin depolymerizing activity in vitro. Plant Physiology. 2004;136:3990–3998. doi: 10.1104/pp.104.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matthews L.C., Taggart M.J., Westwood M. Modulation of caveolin-1 expression can affect signalling through the phosphatidylinositol 3-kinase/Akt pathway and cellular proliferation in response to insulin-like growth factor I. Endocrinology. 2008;149:5199–5208. doi: 10.1210/en.2007-1211. [DOI] [PubMed] [Google Scholar]
- 72.Baehler P.J., Biondi R.M., van Bemmelen M., Veron M., Reymond C.D. Random insertion of green fluorescent protein into the regulatory subunit of cyclic adenosine monophosphate-dependent protein kinase. Methods in Molecular Biology. 2002;183:57–68. doi: 10.1385/1-59259-280-5:057. [DOI] [PubMed] [Google Scholar]
- 73.Rocheleau J.V., Edidin M., Piston D.W. Intrasequence GFP in class I MHC molecules, a rigid probe for fluorescence anisotropy measurements of the membrane environment. Biophysical Journal. 2003;84:4078–4086. doi: 10.1016/S0006-3495(03)75133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Time lapse TIRF imaging of HEK-293T cells expressing either InsRA-TagRFP (inter-domain tagged) or InsRA-C-eGFP (C-terminal label) constructs recorded at 25 mM glucose.
Movie S2: Live cell imaging of a MIN6 cell co-transfected with IGF1R-TagGFP2 (left) and InsRA-TagRFP (middle) (both interdomain tagged). TagGFP2 and TagRFP was inserted between two domains at bp 2809/aa 937, and bp 2812/aa 938, respectively.
Movie S3: Time lapse TIRF imaging of a Cav1 MIN6 cell expressing InsRA-TagRFP and Cav1-mTFP recorded at 25 mM glucose.
Movie S4: Time lapse TIRF imaging of Cav1 MIN6 cell lines expressing various Cav1 mutants and inter-domain tagged InsRA-lum-eGFP constructs recorded at 25 mM glucose.