Abstract
Flowering Locus C (FLC), a floral repressor, plays an important role in flowering. The mechanisms regulating FLC gene expression and protein function have been studied extensively; however, post-translational regulation of FLC remains unclear. Here, we identified Arabidopsis HIGH PLOIDY2 (HPY2) as an E3 SUMO ligase for FLC. In vitro and vivo pull-down assays showed that FLC physically interacts with HPY2. In vitro assays showed that the stimulation of FLC sumoylation by HPY2 was dependent on SUMO-activating enzyme E1 and -conjugating enzyme E2, indicating that HPY2 was an E3 SUMO ligase for FLC. In transgenic plants, inducible HPY2 overexpression increased the concentration of FLC, indicating that HPY2 stabilized FLC through direct sumoylation. Flowering time in hpy2-2 mutants was shorter than in wild-type plants under long- and short-day conditions, with a greater effect under short-day conditions, and FLC was downregulated in hpy2-2 mutants. These data indicate that HPY2 regulates FLC function and stability at both the transcriptional and post-translational levels through its E3 SUMO ligase activity.
Keywords: E3 SUMO ligase, FLC, HPY2, post-translational modification, SUMO, sumoylation
Introduction
Post-translational modification is an important mechanism for the regulation of protein function and stability. To date, more than 200 types of protein modification have been reported (Castro et al., 2012). One of these protein modifications, sumoylation, targets lysine for addition of a small ubiquitin-related modifier (SUMO; Wilkinson and Henley, 2010). SUMO is a small peptide with a molecular mass of approximately 11 kDa. In yeast and animal systems, SUMO regulates various cellular processes such as stress and defense responses, nitrogen metabolism, hormone signaling, epigenetic gene expression, growth, and flowering (Hotson et al., 2003; Kurepa et al., 2003; Lois et al., 2003; Murtas et al., 2003; Miura et al., 2005, 2007a; Yoo et al., 2006; Catala et al., 2007; Lee et al., 2007; Conti et al., 2008; Park et al., 2011; Son et al., 2014; Kim D.Y. et al., 2015; Kim S.I. et al., 2015).
Although, conjugation of SUMO to target proteins can occur independently of ligases, modification of target proteins with SUMO is usually catalyzed by E3 SUMO ligases (Wilkinson and Henley, 2010). The four types of E3 SUMO ligases identified to date are RanGAP1-binding protein 2 (RanBP2), polycomb group 2 (Pc2), non-SMC element/methyl methanesulfonate sensitive 1 (NES2/MMS21), and SAP and MIZ/protein inhibitor of activated STAT (SIZ/PIAS; Johnson and Gupta, 2001; Kahyo et al., 2001; Rose and Meier, 2001; Pichler et al., 2002; Kagey et al., 2003; Verger et al., 2003). PIAS/SIZ-type E3 SUMO ligase has five domains: SAP, PINIT, SP-RING finger, SXS, and PHD (Sharrocks, 2006; Miura et al., 2007a; Garcia-Dominguez et al., 2008; Cheong et al., 2009). To date, two PIAS-type SUMO E3 ligases, SIZ1 (Miura et al., 2005, 2007a) and High Ploidy 2 (HPY2; Huang et al., 2009; Ishida et al., 2009, 2012), have been reported in Arabidopsis.
AtSIZ1 is involved in nutrient assimilation, hormone signaling, growth, flowering, and stress responses (Miura et al., 2005, 2007b, 2010; Yoo et al., 2006; Catala et al., 2007; Lee et al., 2007; Garcia-Dominguez et al., 2008; Jin et al., 2008; Miura and Ohta, 2010; Park et al., 2011; Son et al., 2014; Kim D.Y. et al., 2015; Kim S.I. et al., 2015). AtSIZ1 has several target proteins, including the nitrate reductases NIA1 and NIA2, INDUCER OF CBF EXPRESSION 1 (ICE1), the R2R3-type transcription factor MYB30, FLOWERING LOCUS C (FLC), SLEEPY1 (SLY1), and chromomethylase 3 (CMT3; Elrouby and Coupland, 2010; Miller et al., 2010; Miura and Hasegawa, 2010; Park et al., 2011; Zheng et al., 2012; Conti et al., 2014; Son et al., 2014; Kim D.Y. et al., 2015; Kim S.I. et al., 2015).
AtHPY2 is much smaller than AtSIZ1, comprising 249 amino acids and only a single domain (SP-RING zinc finger domain). Little is known about the role of HPY2, but studies suggest it is involved in regulating cell cycle progression, meristem development, and auxin signaling (Ishida et al., 2009; Okushima et al., 2014). AtSIZ1 and HPY2 exhibit different expression patterns, and their mutants display distinct dwarf phenotypes (Ishida et al., 2012). In addition, their reciprocal expression does not complement the single-mutant phenotypes (Ishida et al., 2012). To date, there have been no reports of sumoylation of specific target proteins by HPY2.
Flowering Locus C, a MADS-box transcription factor, participates in the control of flowering time (Samach et al., 2000; Simpson and Dean, 2002). Expression of FLC is negatively regulated by components of the autonomous pathway and by vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999; He and Amasino, 2005; Krichevsky et al., 2006; Greb et al., 2007), and FLC transcription is modulated by epigenetic methylation (Swiezewski et al., 2009; Heo and Sung, 2011). Ubiquitination via E3 ubiquitin ligase activity of SINAT5 is thought to regulate FLC stability (Park et al., 2007). FLC function and stability are also controlled by sumoylation, although the associated E3 SUMO ligase remains unidentified (Son et al., 2014). These findings suggest that post-translational mechanisms are involved in the regulation of floral transition by FLC. However, although transcriptional control of FLC is relatively well-characterized, the mechanisms underlying post-translational modification of FLC remain unclear.
Here, we provide the first evidence that HPY2 functions as an E3 SUMO ligase for FLC, directly interacting with FLC to catalyze its sumoylation. Sumoylation stabilizes FLC, and loss of HPY2 causes early flowering. These findings indicate that HPY2 positively controls FLC-mediated flowering repression through FLC sumoylation.
Materials and Methods
Plant Materials and Growth Conditions
Columbia-background ecotype Arabidopsis thaliana [wild-type (WT)] and the T-DNA insertion hpy2-2 mutant were used in this study. To sterilize the seeds, WT and hpy2-2 mutant seeds were treated with 5% sodium hypochlorite and 0.1% Triton X-100 for 10 min. For germination and growth in medium, the seeds were thoroughly washed with distilled water and then stored at 4°C in the dark. After 4 days, the seeds were plated on Murashige and Skoog (MS) agar medium including 2% sucrose. To prepare soil-grown plants, WT and hpy2-2 mutant seeds were directly sown into vermiculite soil. Plants, including seedlings, were grown at 22°C under a 16 h light/8 h dark cycle (long day) or under an 8 h light/16 h dark cycle (short day) in a growth chamber.
Construction of Recombinant Plasmids
To produce His6-FLC and GST-HPY2, full-length FLC and HPY2 cDNAs were amplified by PCR and cloned into the pET28a (Novagen) and pGEX4T-1 (Amersham Biosciences) vectors, respectively. To prepare GST-FLC-Myc, full-length FLC cDNA was amplified using primers to add a Myc tag and cloned into pGEX4T-1. The FLC mutant proteins GST-FLCm1(K5R)-Myc, GST-FLCm2(K135R)-Myc, and GST-FLCm3(K154R)-Myc were prepared as previously described (Son et al., 2014). Numbers indicate the positions of the lysine (K) residues, and R indicates replacement of lysine with arginine residues. For production of His6-AtSUMO1, Arabidopsis full-length SUMO1 cDNA was amplified by PCR and cloned into pET28a. Primer sequences are described in Supplementary Table S1.
Purification of Recombinant Proteins
All recombinant proteins were expressed in Escherichia coli strain BL21 and purified as previously described (Colby et al., 2006; Son et al., 2014). Briefly, His6-AtSUMO1, His6-FLC, SUMO-activating enzyme E1 (His6-AtSAE1b and His6-AtSAE2), and SUMO-conjugating enzyme E2 (His6-AtSCE1) were purified with Ni2+-nitrilotriacetate (Ni2+-NTA) resins (Qiagen). GST, GST-FLC-Myc, GST-FLCm1(K5R)-Myc, GST-FLCm2(K135R)-Myc, GST-FLCm3(K154R)-Myc, and GST-HPY2 were purified with glutathione resins (Pharmacia).
In Vitro and In Vivo Interaction Assays
In vitro pull-down assays to assess interactions between GST-HPY2 and His6-FLC were carried out using 2 μg each of GST-HPY2 bait and His6-FLC prey as previously described (Son et al., 2014). His6-FLC was detected by Western blot analysis using anti-His antibody (Santa Cruz Biotechnology).
Plant expression plasmids were constructed to investigate the direct interaction between HPY2 and FLC in vivo. To express FLC, the corresponding full-length cDNA was amplified using a forward primer to add a hexameric Myc (Myc6) and a reverse primer and cloned into the pBA002 vector. To express HPY2, the corresponding full-length cDNA was amplified by PCR using a forward primer and a reverse primer to add a FLAG3 tag and cloned into the pBA002 vector. Hexameric Myc-tagged or trimeric FLAG-tagged primers were synthesized by DNA synthesizer (Bioneer). Wild-type Arabidopsis plants were infiltrated with different combinations of Agrobacterium transformed with 35S-Myc6-FLC or 35S-HPY2-FLAG3 constructs. Total protein was extracted from each sample 2 days after infiltration, and Myc6-FLC and HPY2-FLAG3 were detected by Western blot analysis with anti-Myc and anti-FLAG antibodies, respectively. Finally, total protein was extracted from each sample and immunoprecipitated with anti-FLAG antibody (1 μg/mL, Santa Cruz Biotechnology) in a buffer containing 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, 1 mM PMSF, and a protease inhibitor cocktail (Promega). FLC was detected in immunoprecipitated samples by Western blot analysis with anti-Myc antibody (0.5 mg/mL, Sigma-Aldrich).
Sumoylation Assays
In vitro sumoylation was performed in 30 μL of reaction buffer [200 mM Hepes (pH 7.5), 5 mM MgCl2, 2 mM ATP] with 50 ng of His6-AtSAE1b, 50 ng of His6-AtSAE2, 50 ng of His6-AtSCE1, 8 μg of His6-AtSUMO1, and 100 ng of GST-FLC-Myc, with or without 200 ng of GST-HPY2. After incubation for 3 h at 30°C, the reaction mixtures were separated on 11% SDS-PAGE gels. Sumoylated GST-FLC-Myc was detected by Western blot analysis using anti-Myc antibody (Santa Cruz Biotechnology). Sumoylation reactions were also performed as described above to identify the lysine residue modified by SUMO using WT FLC (GST-FLC-Myc) and mutant FLC [GST-FLCm1(K5R)-Myc, GST-FLCm2(K135)-Myc, or GST-FLCm3(K154R)-Myc].
Preparation of Single and Double Transgenic Plants
Flowering Locus C- or mFLC(K154R)-overexpressing plants were prepared by floral dipping as previously described (Clough and Bent, 1998; Son et al., 2014). Double transgenic Arabidopsis plants overexpressing HPY2 and FLC or HPY2 and mFLC were produced by introduction of XVE-HA3-HPY2 and 35S-FLC-FLAG3, or XVE-HA3-HPY2, and 35S-mFLC-FLAG3. Recombinant plasmid XVE-HA3-HPY2 was generated by amplification of HPY2 cDNA with a forward primer tagged with trimeric HA3 (HA3) and a reverse primer, and insertion of the resultant fragment into the pET8 vector. Trimeric HA-tagged primers were synthesized by DNA synthesizer (Bioneer).
Examination of HPY2 Effect on FLC Stability In Vivo
Two independent transgenic 35S-FLC-FLAG3 and XVE HA3-HPY2 lines and two independent transgenic 35S-mFLC-FLAG3 and XVE HA3-HPY2 lines were used for the experiment. Two-weeks-old double transgenic plants carrying XVE HA3-HPY2 and 35S-FLC-FLAG3, or XVE HA3-HPY2, and 35S-mFLC-FLAG3, were treated with 10 μM β-estradiol. After 15 h, samples were ground in liquid nitrogen, and lysates were separated by 11% SDS-PAGE. FLC-FLAG3 and mFLC-FLAG3 levels were examined by Western blot analysis with anti-FLAG antibody. HA3-HPY2 induction was analyzed by Western blot analysis with anti-HA antibody.
Examination of Flowering Time in hpy2-2 Mutants
Flowering time was investigated under long- and short-day conditions. WT and hpy2-2 mutant plants were grown, and rosette leaves were counted when inflorescences appeared. Twenty plants of each type (WT or hpy2-2 mutant) were used in the experiment.
Quantitative Real-time RT-PCR Analysis
Wild type and hpy2-2 mutants were grown in long- and short-day conditions as described above. Total RNA was extracted from the leaves of WT and hpy2-2 mutants, and quantitative real-time RT-PCR was used to assess transcript levels of FLC, SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), FLOWERING LOCUS T (FT), and TWIN SISTER OF FT (TSF) as described previously (Park et al., 2011). Tubulin amplification was used as an internal control. All reactions were repeated three times. Primer sequences are listed in Supplementary Table S1.
Results
FLC Interacts with HPY2 In Vitro and In Vivo
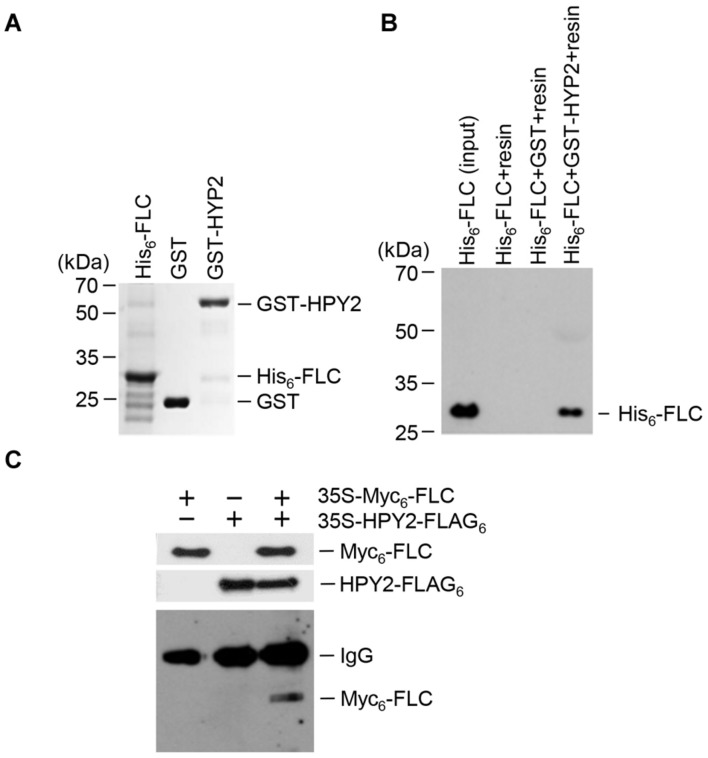
Previous results showed that E3 SUMO ligase AtSIZ1 interacted with FLC to inhibit sumoylation (Son et al., 2014). Here, we aimed to identify the E3 SUMO ligase that stimulated FLC sumoylation. HPY2, an Arabidopsis E3 SUMO ligase possessing an SP-RING (SIZ/PIAS-RING) domain, was chosen as a candidate. An in vitro pull-down assay was used to examine the interaction of HPY2 with FLC. Recombinant proteins GST-HPY2 and His6-FLC were overexpressed in E. coli and purified with glutathione or Ni2+-NTA resins (Figure 1A). The results showed that GST-HPY2 was able to pull down His6-FLC (Figure 1B). The interaction between HPY2 and FLC was also examined by immunoprecipitation (IP). Two constructs, 35S-Myc6-FLC and 35S-HPY2-FLAG3, were coinfiltrated into the leaves of WT plants. After IP with anti-FLAG antibody, Myc6-FLC was examined by Western blot analysis using anti-Myc antibody. Myc6-FLC was clearly detected (Figure 1C), indicating a strong interaction between HPY2 and FLC, consistent with the results of the in vitro pull-down experiment.
FIGURE 1.
Interaction of HPY2 with FLC. (A) His6-FLC and GST-HPY2 were overexpressed in Escherichia coli and purified with Ni2+-NTA or glutathione affinity columns. (B) The His6-FLC protein was pulled down with the GST-HPY2 protein, separated on 11% SDS-polyacrylamide gels, and analyzed by Western blotting with anti-His antibody. (C) In vivo interaction of HPY2 and FLC. Wild-type (WT) plants were infiltrated with different combinations of 35S-Myc6-FLC or 35S-HPY2-FLAG3 constructs. Total protein was extracted from each sample and immunoprecipitated with anti-FLAG antibody. After immunoprecipitation, Myc6-FLC was detected by Western blotting with anti-Myc antibody. Myc6-FLC and HPY2-FLAG3 expression was also examined by Western blotting with anti-Myc and anti-FLAG antibodies, respectively.
FLC Sumoylation Is Stimulated by HPY2
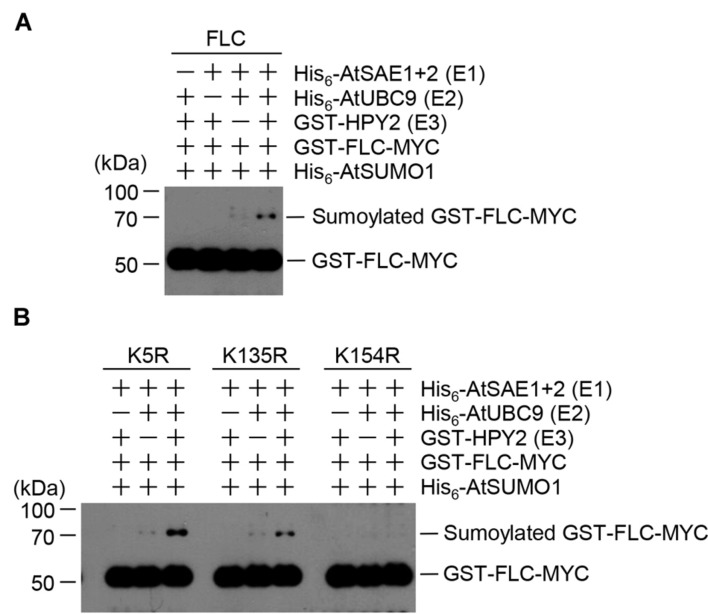
The strong interaction between HPY2 and FLC (Figures 1B,C) suggested that FLC could be modified by SUMO through E3 SUMO ligase activity of HPY2. Therefore, we next tested whether HPY2 had E3 SUMO ligase activity for FLC. In vitro sumoylation reactions were performed using His6-AtSAE1b, His6-AtSAE2, His6-AtSCE1, His6-AtSUMO1, GST-HPY2, and GST-FLC-Myc. The amount of sumoylated GST-FLC-Myc was increased by HPY2 in an E1- and E2-dependent manner (Figure 2A).
FIGURE 2.
Flowering Locus C (FLC) is sumoylated by HPY2 in vitro. Arabidopsis His6-AtSAE1b, His6-AtSAE2, His6-AtSCE1, GST-HPY2, His6-AtSUMO1, and GST-FLC-Myc were overexpressed in E. coli and purified with Ni2+-NTA or glutathione affinity columns as appropriate. (A) Sumoylation of GST-FLC-Myc was assayed in the presence or absence of E1 (His6-AtSAE1b and His6-AtSAE2), E2 (His6-AtSCE1), E3 (GST-HPY2), and His6-AtSUMO1. FLC sumoylation was detected by Western blotting with anti-Myc antibody. (B) To identify the sumoylation site on FLC, GST-FLCm1-Myc (K54R), GST-FLCm2-Myc (K135R), and GST-FLCm3-Myc (K154R) were overexpressed in E. coli and purified using a glutathione affinity column. The reaction mixture contained E1 (His6-AtSAE1b and His6-AtSAE2), E2 (His6-AtSCE1), E3 (GST-HPY2), and His6-AtSUMO1 without (-) or with (+) a mutant protein instead of GST-FLC-Myc. FLC sumoylation was detected by Western blotting with anti-Myc antibody.
Our previous study showed that FLC contained three possible sumoylation sites (Son et al., 2014). Single- or double-mutant derivatives with the mutations K154R, K5R/K135R, K5R/K154R, or K135R/K154R were produced, and their sumoylation was examined by in vitro sumoylation assays (Son et al., 2014). The results showed K154 to be the principal site of SUMO conjugation on FLC (Son et al., 2014). In the current study, HPY2-mediated sumoylation was examined in the mutant FLC proteins GST-FLCm1(K5R)-Myc, GST-FLCm2(K135)-Myc, and GST-FLCm3(K154R)-Myc. Consistent with the previous study, GST-FLCm3(K154R)-Myc was not sumoylated, even in the presence of HPY2, confirming that the lysine residue at position 154 was a true sumoylation site (Figure 2B).
HPY2 Stabilizes FLC
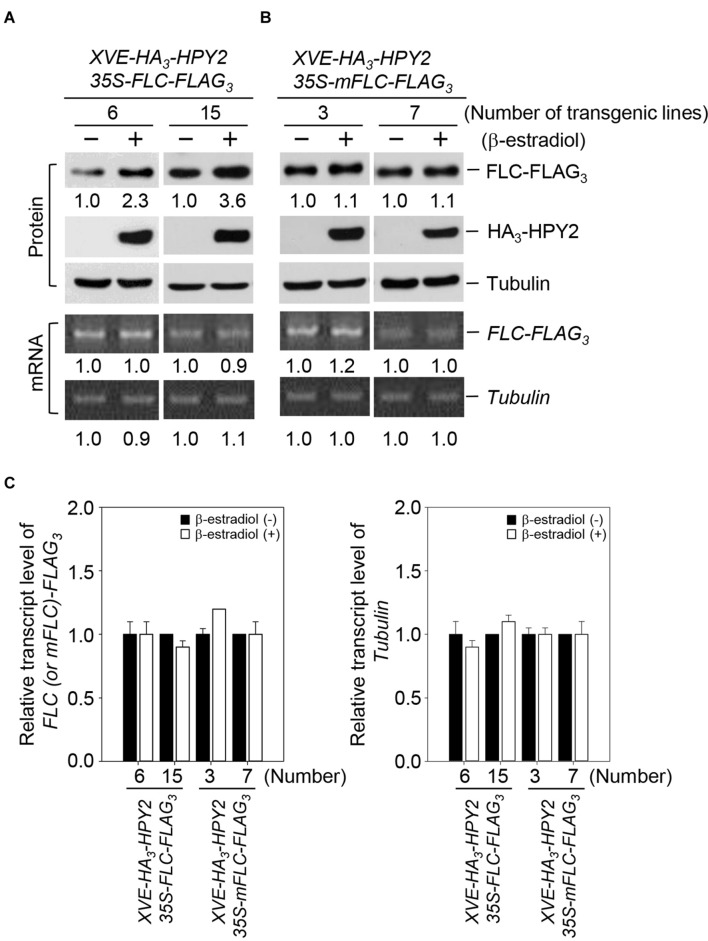
Based on the strong interaction of HPY2 with FLC and the increase in FLC sumoylation as a result of HPY2 activity, we inferred that FLC stability could be modulated by HPY2. We therefore measured the effect of HPY2 on FLC levels using double transgenic plants with XVE-HA3-HPY2 and 35S-FLC-FLAG3 or with XVE-HA3-HPY2 and 35S-mFLC-FLAG3. In mFLC, the lysine at position 154 (sumoylation site) was mutated to arginine (K154R). HPY2 induction increased FLC levels by up to 2.3- and 3.6-fold in two independent transgenic plants (Figures 3A,C). However, no increase in mFLC level was seen after induction of HPY2 in two independent transgenic plants (Figures 3B,C). FLC and mFLC transcript levels were similar after induction of HPY2 expression, as determined using real-time qRT-PCR (Figures 3A,B). These data indicate that the FLC protein is stabilized by the E3 SUMO ligase activity of HPY2.
FIGURE 3.
Flowering Locus C is stabilized by HPY2 in vivo. Double transgenic plants containing 35S-FLC-FLAG3 and XVE-HA3-HPY2 (A) or 35S-mFLC (K154R)-FLAG3 and XVE-HA3-AtSIZ1 (B) were incubated in liquid medium with β-estradiol to induce HPY2 expression. After incubation for 15 h, HA3-HPY2, FLC-FLAG3, and mFLC-FLAG3 levels were assessed by Western blotting with anti-HA or anti-FLAG antibodies. Tubulin was used as a loading control. Numbers under lanes indicate relative intensities. Protein levels were normalized to a value of 1.00 for FLC or mFLC levels without inducer (“–” in both panels). RNA concentrations of FLC-FLAG3 and mFLC-FLAG3 were determined by real-time qRT-PCR using a FLAG primer and a gene-specific primer. Tubulin RNA was used as a loading control. (C) Graphical expression of Tubulin, FLC-FLAG3, and mFLC-FLAG3 transcript levels from (B). Bars indicate standard errors (n = 3).
A hpy2 Mutant Exhibits Early Flowering
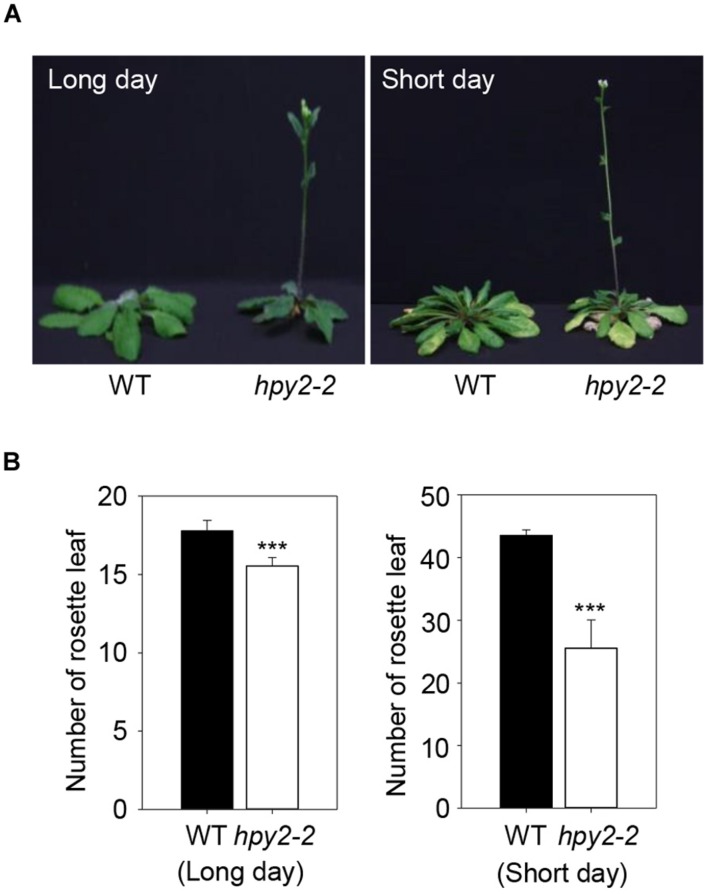
Flowering Locus C is stabilized by HPY2-mediated sumoylation, suggesting that FLC functions may be compromised in a hpy2 mutant background and that flowering may be affected. Two independent T-DNA insertion mutant lines, hpy2-1 and hpy2-2, were characterized previously (Ishida et al., 2009, 2012). The hpy2-2 mutant exhibits a relatively weak phenotype, but the hpy2-1 mutant shows a severe growth defect phenotype and growth frequently stops prior to bolting. The hpy2-2 mutant was therefore chosen to examine flowering time. Rosette leaves were counted just after bolting in WT and hpy2-2 plants. Compared to WT, flowering was delayed in hpy2-2 mutants under both long- and short-day conditions (Figures 4A,B). Flowering delay in hpy2-2 was significant under short-day conditions but was less pronounced under long-day conditions (Figures 4A,B).
FIGURE 4.
Flowering time of hpy2-2 mutants. Early flowering was observed in the hpy2-2 mutant grown under both long- and short-day conditions (A). Flowering time was investigated by counting the number of rosette leaves present when inflorescences appeared (B). The number of rosettes in WT and hpy2-2 plants differed significantly (∗∗∗P < 0.0001, t-test, n = 20). Bars indicate standard errors.
Expression of Flowering-related Genes Is Affected by HPY2
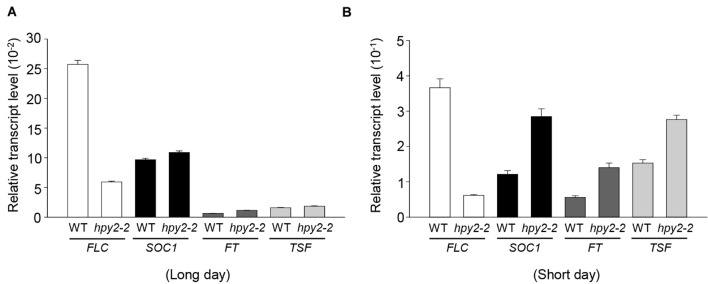
HPY2 was identified as an E3 SUMO ligase for FLC, suggesting that the early flowering phenotype seen in hpy2-2 mutants might be partly caused by low levels of FLC and decreased FLC activity. However, it was also possible that early flowering in hpy2-2 mutants occurred as a consequence of altered gene expression. To investigate this, real-time qRT-PCR was used to measure transcript levels of FLC and the flowering-related genes SOC1, FT, and TSF in WT and hpy2-2 mutants grown in long- or short-day conditions (Figures 5A,B). SOC1, FT, and TSF were slightly upregulated in hpy2-2 mutants compared with their expression in WT plants under long-day conditions (Figure 5A). More substantial differences in the expression of SOC1, FT, and TSF were apparent under short-day conditions, with approximately twofold higher transcript levels observed in hpy2-2 plants compared to WT plants (Figure 5B).
FIGURE 5.
Transcript levels of flowering-related genes in hpy2-2 mutants. Total RNA was isolated from the leaves of WT and hpy2-2 mutant plants grown in soil under long (A)-or short-day (B) conditions. Transcript levels were examined using real-time qRT-PCR with gene-specific primers. Results are expressed as the mean ± SD (n = 3). FLC, FLOWERING LOCUS C; SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1; FT, FLOWERING LOCUS T; TSF, TWIN SISTER OF FT.
Discussion
In this study, HPY2 was shown to be a functional E3 SUMO ligase that was able to directly interact with FLC to catalyze its sumoylation.
To date, two SIZ-type E3 SUMO ligases have been identified in plants, namely, AtSIZ1 and HPY2. The functional roles and phenotypic features of AtSIZ1 are well-characterized. However, the functions of HPY2 are poorly understood. A recent study showed that HPY2 had self-sumoylation activity in vitro, and that mutation of HPY2 reduced SUMO-conjugate levels in Arabidopsis (Ishida et al., 2009). In addition, loss of HPY2 function resulted in a premature mitotic-to-endocytic transition that led to severe dwarfism (Ishida et al., 2009). Whereas the phenotype of hpy2-1 mutants did not depend on the accumulation of salicylic acid (SA), the siz1-2 mutant phenotype was caused by SA accumulation, indicating that the two proteins functioned through different pathways and that their roles did not overlap (Ishida et al., 2012).
Sumoylation of target proteins by AtSIZ1 is well-characterized (Elrouby and Coupland, 2010; Miller et al., 2010; Miura and Hasegawa, 2010; Park et al., 2011; Zheng et al., 2012; Conti et al., 2014; Son et al., 2014; Kim D.Y. et al., 2015; Kim S.I. et al., 2015). However, the targets of HPY2 remain unidentified, and E3 ligase activity of HPY2 against target proteins has not been reported to date. In a previous study, we showed that AtSIZ1 interacts directly with FLC and stabilizes the protein (Kim S.I. et al., 2015). However, our results showed that AtSIZ1 inhibited FLC sumoylation, which led us to speculate that HPY2 may act as a specific E3 SUMO ligase for FLC. An in vitro sumoylation assay showed that HPY2 had E3 SUMO ligase activity against FLC through direct interaction (Figures 1B,C and 2A). This is the first report demonstrating HPY2 E3 SUMO ligase activity for a target protein.
Modification of target lysines by SUMO occurs by two different mechanisms. In the first mechanism, target proteins are directly modified by conjugating enzyme E2 (Bernier-Villamor et al., 2002; Meulmeester et al., 2008; Zhu et al., 2008). The second mechanism involves E3 ligase-mediated sumoylation (Park and Yun, 2013). Our previous study showed that FLC could be sumoylated without the help of an E3 SUMO ligase, and that FLC sumoylation was inhibited by the E3 SUMO ligase AtSIZ1. In the present study, we found that the level of sumoylated FLC was increased by the addition of HPY2 (Figure 2A), indicating that FLC sumoylation was stimulated by HPY2 via its E3 SUMO ligase activity.
Our previous study showed that AtSIZ1 inhibited FLC sumoylation but stabilized the FLC protein (Son et al., 2014). In the current study, HPY2 overexpression led to an increase in levels of FLC but not in levels of mFLC, which lacked the critical lysine residue for sumoylation (Figures 3A–C), indicating that FLC sumoylation by HPY2 stabilized FLC. In yeast, animal, and plant systems, modification by SUMO affects the stability and activity of target proteins (Park and Yun, 2013). Consistent with this, our results showed that sumoylation had a positive effect on FLC function.
Since FLC is a central regulator of flowering and HPY2 has E3 SUMO ligase activity against FLC, we next examined flowering time in the hpy2-2 mutant. Mutant plants flowered earlier than WT plants (Figure 4). Previously, we reported that flowering time was significantly delayed by the overexpression of WT FLC but that flowering time was unaffected by overexpression of mFLC (Son et al., 2014), indicating that sumoylation was critical for the floral repressor function of FLC. This suggests that early flowering in hpy2-2 mutants is caused by the reduced stability and lower activity of FLC resulting from the loss of HPY2 E3 SUMO ligase activity.
Our data suggest that early flowering in the hpy2-2 mutant results from the loss of HPY2 activity, which leads to low FLC levels or low FLC activity. However, it was possible that early flowering of the hpy2-2 mutant might be a consequence of changes in gene expression. FLC transcript levels were decreased in the hpy2-2 mutant (Figure 5) indicating that HPY2 regulated FLC-mediated flowering at the transcriptional level as well as at the post-translational level.
Conclusion
HPY2 controls FLC-mediated floral transition through its E3 SUMO ligase activity as well as through direct binding to FLC. Taken together with previous findings, our data indicate that two E3 SUMO ligases, AtSIZ1 and HPY2, are involved in the regulation of FLC stability, and that sumoylation is a critical modification for the regulation of FLC function. Elucidation of the mechanisms underlying the regulation of FLC function by AtSIZ1 and HPY2, alone or in combination, will enhance our understanding of the modulation of flowering time by the SUMO system.
Author Contributions
HS designed the project. JK, GS, and S-IK carried out experiments. JK, GS, S-IK, JS, and HS analyzed and interpreted the data. JK and HS wrote the manuscript. All authors commented on the results and the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center no. PJ01108701), Rural Development Administration, Republic of Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00530
References
- Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002). Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108 345–356. 10.1016/S0092-8674(02)00630-X [DOI] [PubMed] [Google Scholar]
- Castro P. H., Tavares R. M., Bejarano E. R., Azevedo H. (2012). SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci. 69 3269–3283. 10.1007/s00018-012-1094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I. A., Hu Y., Seo H., Zhang X., et al. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19 2952–2966. 10.1105/tpc.106.049981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong M. S., Park H. C., Hong M. J., Lee J., Choi W., Jin J. B., et al. (2009). Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. 151 1930–1942. 10.1104/pp.109.143719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Colby T., Matthai A., Boeckelmann A., Stuible H. P. (2006). SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 142 318–332. 10.1104/pp.106.085415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Nelis S., Zhang C., Woodcock A., Swarup R., Galbiati M., et al. (2014). Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 28 102–110. 10.1016/j.devcel.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Conti L., Price G., O’Donnell E., Schwessinger B., Dominy P., Sadanandom A. (2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20 2894–2908. 10.1105/tpc.108.058669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N., Coupland G. (2010). Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. U.S.A. 107 17415–17420. 10.1073/pnas.1005452107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez M., March-Diaz R., Reyes J. C. (2008). The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J. Biol. Chem. 283 21469–21477. 10.1074/jbc.M708176200 [DOI] [PubMed] [Google Scholar]
- Greb T., Mylne J. S., Crevillen P., Geraldo N., An H., Gendall A. R., et al. (2007). The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 17 73–78. 10.1016/j.cub.2006.11.052 [DOI] [PubMed] [Google Scholar]
- He Y., Amasino R. M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10 30–35. 10.1016/j.tplants.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 76–79. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Hotson A., Chosed R., Shu H., Orth K., Mudgett M. B. (2003). Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50 377–389. 10.1046/j.1365-2958.2003.03730.x [DOI] [PubMed] [Google Scholar]
- Huang L., Yang S., Zhang S., Liu M., Lai J., Qi Y., et al. (2009). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 60 666–678. [DOI] [PubMed] [Google Scholar]
- Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., et al. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21 2284–2297. 10.1105/tpc.109.068072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Yoshimura M., Miura K., Sugimoto K. (2012). MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS ONE 7:e46897 10.1371/journal.pone.0046897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. B., Jin Y. H., Lee J., Miura K., Yoo C. Y., Kim W. Y., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S., Gupta A. A. (2001). An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106 735–744. 10.1016/S0092-8674(01)00491-3 [DOI] [PubMed] [Google Scholar]
- Kagey M. H., Melhuish T. A., Wotton D. (2003). The polycomb protein Pc2 is a SUMO E3. Cell 113 127–137. 10.1016/S0092-8674(03)00159-4 [DOI] [PubMed] [Google Scholar]
- Kahyo T., Nishida T., Yasuda H. (2001). Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8 713–718. 10.1016/S1097-2765(01)00349-5 [DOI] [PubMed] [Google Scholar]
- Kim D. Y., Han Y. J., Kim S., Song J. T., Seo H. S. (2015). Arabidopsis CMT3 activity is positively regulated by AtSIZ1-mediated sumoylation. Plant Sci. 239 209–215. 10.1016/j.plantsci.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Kim S. I., Park B. S., Kim D. Y., Yeu S. Y., Song S. I., Song J. T., et al. (2015). E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signaling and plant development. Biochem. J. 469 299–314. 10.1042/BJ20141302 [DOI] [PubMed] [Google Scholar]
- Krichevsky A., Gutgarts H., Kozlovsky S. V., Tzfira T., Sutton A., Sternglanz R., et al. (2006). C2H2 zinc finger-SET histone methyltransferase is a plant-specific chromatin modifier. Dev. Biol. 303 259–269. 10.1016/j.ydbio.2006.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Walker J. M., Smalle J., Gosink M. M., Davis S. J., Durham T. L., et al. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278 6862–6872. 10.1074/jbc.M209694200 [DOI] [PubMed] [Google Scholar]
- Lee J., Nam J., Park H. C., Na G., Miura K., Jin J. B., et al. (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49 79–90. 10.1111/j.1365-313X.2006.02947.x [DOI] [PubMed] [Google Scholar]
- Lois L. M., Lima C. D., Chua N. H. (2003). Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15 1347–1359. 10.1105/tpc.009902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008). Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30 610–669. 10.1016/j.molcel.2008.03.021 [DOI] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. J., Barrett-Wilt G. A., Hua Z., Vierstra R. D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107 16512–16517. 10.1073/pnas.1004181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Hasegawa P. M. (2010). Sumoylation and other ubiquitin-like posttranslational modifications in plants. Trends Cell Biol. 20 223–232. 10.1016/j.tcb.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J. B., Hasegawa P. M. (2007a). Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10 495–502. 10.1016/j.pbi.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J. B., Lee J., Yoo C. Y., Stirm V., Miura T., et al. (2007b). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 9 1403–1414. 10.1105/tpc.106.048397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Lee J., Miura T., Hasegawa P. M. (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 51 103–113. 10.1093/pcp/pcp171 [DOI] [PubMed] [Google Scholar]
- Miura K., Ohta M. (2010). SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J. Plant Physiol. 167 555–560. 10.1016/j.jplph.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A. S., Raghothama K. G., et al. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U.S.A. 102 7760–7765. 10.1073/pnas.0500778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas G., Reeves P. H., Fu Y. F., Bancroft I., Dean C., Coupland G. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15 2308–2319. 10.1105/tpc.015487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Shimizu K., Ishida T., Sugimoto K., Umeda M. (2014). Differential regulation of B2-type CDK accumulation in Arabidopsis roots. Plant Cell Rep. 33 1033–1040. 10.1007/s00299-014-1581-z [DOI] [PubMed] [Google Scholar]
- Park B. S., Sang W. G., Yeu S. Y., Choi Y. D., Paek N. C., Kim M. C., et al. (2007). Post-translational regulation of FLC is mediated by an E3 ubiquitin ligase activity of SINAT5 in Arabidopsis. Plant Sci. 173 269–275. 10.1016/j.plantsci.2007.06.001 [DOI] [Google Scholar]
- Park B. S., Song J. T., Seo H. S. (2011). Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. 2 400 10.1038/natcomms1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. J., Yun D. J. (2013). New insights into the role of the small ubiquitin-like modifier (SUMO) in plants. Int. Rev. Cell Mol. Biol. 300 161–209. 10.1016/B978-0-12-405210-9.00005-9 [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108 109–120. 10.1016/S0092-8674(01)00633-X [DOI] [PubMed] [Google Scholar]
- Rose A., Meier I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. U.S.A. 98 15377–15382. 10.1073/pnas.261459698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S. E., Ditta Z. S., Schwarz-Sommer Z., Yanofsky M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. 10.1126/science.288.5471.1613 [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D. (2006). PIAS proteins and transcriptional regulation-more than just SUMO E3 ligases? Genes Dev. 20 754–758. 10.1101/gad.1421006 [DOI] [PubMed] [Google Scholar]
- Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., Peacock W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458. 10.1105/tpc.11.3.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G., Dean C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296 285–289. 10.1126/science.296.5566.285 [DOI] [PubMed] [Google Scholar]
- Son G. H., Park B. S., Song J. T., Seo H. S. (2014). FLC-mediated flowering repression is positively regulated by sumoylation. J. Exp. Bot. 65 339–351. 10.1093/jxb/ert383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 799–802. 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- Verger A., Perdomo J., Crossley M. (2003). Modification with SUMO. EMBO Rep. 4 137–142. 10.1038/sj.embor.embor738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K. A., Henley J. M. (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428 133–145. 10.1042/BJ20100158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C. Y., Miura K., Jin J. B., Lee J., Park H. C., Salt D. E., et al. (2006). SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 42 1548–1558. 10.1104/pp.106.088831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K. S., Guo Y. (2012). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109 12822–12827. 10.1073/pnas.1202630109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu S., Guzzo C. M., Ellis N. A., Sung K. S., Choi C. Y., et al. (2008). Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 283 29405–29415. 10.1074/jbc.M803632200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.