Abstract
Crops expressing genes from Bacillus thuringiensis (Bt crops) are among the most successful technologies developed for the control of pests but the evolution of resistance to them remains a challenge. Insect resistant cotton and maize expressing the Bt Vip3Aa protein were recently commercialized, though not yet in Australia. We found that, although relatively high, the frequency of alleles for resistance to Vip3Aa in field populations of H. armigera in Australia did not increase over the past four seasons until 2014/15. Three new isofemale lines were determined to be allelic with previously isolated lines, suggesting that they belong to one common gene and this mechanism is relatively frequent. Vip3Aa-resistance does not confer cross-resistance to Cry1Ac or Cry2Ab. Vip3Aa was labeled with 125I and used to show specific binding to H. armigera brush-border membrane vesicles (BBMV). Binding was of high affinity (Kd = 25 and 19 nM for susceptible and resistant insects, respectively) and the concentration of binding sites was high (Rt = 140 pmol/mg for both). Despite the narrow-spectrum resistance, binding of 125I-labeled Vip3Aa to BBMV of resistant and susceptible insects was not significantly different. Proteolytic conversion of Vip3Aa protoxin into the activated toxin rendered the same products, though it was significantly slower in resistant insects.
Helicoverpa armigera Hübner (Lepidoptera: Noctuidae) is a polyphagous pest feeding on more than 180 species of plants of which the most economically important are grain sorghum, corn and cotton1,2. It has one of the widest distributions of any agricultural pest, until recently being confined to Asia, Europe, Africa and Australia. It was considered a quarantine agricultural pest with no successfully established population in the American continent until 2013 when its intrusion to at least twelve Brazilian states caused economic losses up to 4 billion US dollars2 and in 2015 was detected in pheromone traps in Florida, USA3. Helicoverpa armigera is known for its high level, rapid development of resistance to synthetic insecticides4,5,6. For better control of this pest, Bt crops expressing insecticidal proteins from Bacillus thuringiensis have been commercialized globally. China grows first generation Bt-cotton expressing Cry1Ac7, while Australia, India and the US introduced dual toxin plants expressing Cry1Ac and Cry2Ab8,9,10. Even before the release of dual toxin cotton in Australia, research showed higher than expected levels of resistance alleles in H. armigera and Helicoverpa punctigera to one of the Bt proteins (Cry 2Ab)11,12, and resistance risk remains a critical concern. In addition, various field populations of major lepidopteran pests have now been reported to have developed resistance to Bt crops10,13,14.
The Vip (vegetative insecticidal proteins) proteins discovered in 1996 from Bacillus cereus and B. thuringiensis are compatible with Cry proteins for insect control because they do not share sequence homology and presumably have different modes of action15. Several members of the Vip3A family have high activity against lepidopteran pests16. The vip3Aa gene has been introduced in plants and was first expressed as a single insecticidal protein in cotton17, and later used as a pyramided protein in combination with other cry genes, in cotton and in corn, to confer higher protection and delay insect resistance (http://www.epa.gov/oppbppd1/biopesticides/pips/pip_list.htm). However, a significant threat to Bt-based insect control is the potential development of insect resistance that could jeopardize their-long term success. Resistance to Vip3Aa has already been selected in laboratory colonies of at least three heliothine species. Selection for Vip3Aa resistance in Heliothis virescens over 13 generations resulted in insects with a resistance ratio of 2040-fold relative to susceptible insects18. Of more concern, F2 screens detected frequencies of alleles conferring resistance to Vip3Aa as high as 0.03 and 0.01, respectively, in field populations of H. armigera and H. punctigera from Australia that had not been exposed to plants expressing the toxin19.
Vip3A is an intestine specific virulence factor; after being ingested, the proteins are processed by the insect midgut proteases20,21,22,23. The major proteolytic products of Vip3Aa are approximately 62 and 20 kDa fragments that are inseparable in size-exclusion chromatography24. The processed protein binds to its specific receptors in the midgut epithelial brush border membrane and forms pores20,21,24. Although both Vip3 and Cry proteins need to be activated and bind to membrane receptors to exert their toxic action, the two proteins display different levels of stability and processing rates in the insect midgut23 and bind to different specific receptors in the BBMV of the susceptible insects24,25,26. These differences in the mode of action are thought to be responsible for the absence of cross-resistance to Vip3 proteins observed in Cry-resistant insects from all insect species tested27,28,29,30,31,32,33,34. However, studies on cross-resistance to Cry proteins in Vip3-resistant insects are lacking.
The mechanisms underlying resistance to the Cry proteins have been studied in some field resistant populations and laboratory selected populations. In many cases the gene and mutation responsible has been identified. For Cry1Ac, a number of mutations in resistant individuals have been identified as responsible for phenotypic resistance35 and recently genes involved in Cry1Ca resistance in Spodoptera exigua and in Cry2Ab resistance in H. armigera were also isolated36,37. In addition, gene expression alterations have also been characterized in some other cases38,39. Most cases of insect resistance to Cry proteins reported to date belong to one of the sequential steps proposed for their mode of action: impaired proteolysis activation40,41 or decreased binding to midgut receptors42,43. However, there are a few cases of resistance to Cry proteins that could not be associated with either impaired activation or decreased binding44,45,46.
The first resistance alleles to Vip3A in field populations of Australian H. armigera were isolated in 2009. Pooling F2 screen data across 2009/10 and 2010/11 yielded an r frequency for H. armigera of 0.027 (28 positive lines, 273 tested lines) with a 95% CI between 0.019 and 0.038. Complementation tests involving crosses of the first two isolates (SP85 and SP477) demonstrated that the F1 progeny were also resistant to Vip3A, implying that the resistance in both isolates is due to alleles at a common locus. Characterization of these early isolations showed that the resistance to Vip3Aa is recessive and maps to a locus different from that conferring resistance to Cry2Ab19,33. Herein, in addition to providing up-to-date information on frequencies of resistance alleles in H. armigera field populations and further information on allelism among different resistant families, we provide one of the first demonstrations of a lack of cross-resistance to Cry1Ac and Cry2Ab in a Vip3Aa resistant colony. These studies provide important context to a further investigation reported herein which examines the possible role of Vip3Aa processing and binding to midgut receptors as mechanisms of resistance.
Results
Characteristics of Vip3Aa resistance
Complementation tests involve crossing a standard resistant colony with the new isolate and then testing the offspring by exposing them to the discriminating dose of the toxin. If the offspring survive the discriminating dose of Vip3Aa, then the resistance in each colony is due to the same mutation or variants (alleles) at the same gene which implies a common mechanism. If a complementation test performed on a new resistant isolate is negative (the offspring of a cross between it and the standard colony fail to survive), it is likely that different genes are involved in conferring resistance.
Previously we reported data for two isolations of H. armigera which were allelic with SP85 – here we test an additional four isolations from two seasons of monitoring (2011/2012 and 2012/2013) using F2 tests. Three of these Vip3Aa isolations were found to be clearly allelic to the resistant laboratory line SP85 (11–1112, 12–2602, 12–2998). In one line (11–2201) there was substantial mortality from Vip3Aa (~60%) in the offspring from the crosses to SP85 when compared to the control. This could be explained if another gene is involved in conferring the resistance or the tested individuals were heterozygous which would produce approximately 50% mortality. Unfortunately it was not possible to maintain the 11–2201 line to investigate this issue further. However, in all cases the survival rates are greater than would be expected if no resistance allele was present (p = <0.001 χ2 = 171432). This result supports the notion of a relatively common mechanism for Vip3Aa resistance in field populations of H. armigera in Australia, and justifies using F1 screens to estimate SP85-like Vip3Aa resistance frequencies (Table 1). However, it would be prudent to continue to perform some F2 screens to track whether resistance involving other potential mutations increases in frequency after the deployment of plants expressingVip3Aa.
Table 1. Complementation testing of four field isolated Vip3Aa resistant H armigera colonies with the SP85 type colony.
| Year isolated | Colony crossed to SP85 | Exposure | Dead | Alive | Total | % Dead |
|---|---|---|---|---|---|---|
| 2012 | 11–1112 | buffer | 0 | 20 | 20 | 0.0 |
| Vip3Aa | 5 | 20 | 25 | 20.0 | ||
| 2012 | 11–2201 | buffer | 0 | 45 | 45 | 0.0 |
| Vip3Aa | 27 | 18 | 45 | 60.0 | ||
| 2013 | 12–2602 | buffer | 5 | 40 | 45 | 11.1 |
| Vip3Aa | 7 | 38 | 45 | 15.6 | ||
| 2013 | 12–2998 | buffer | 0 | 45 | 45 | 0.0 |
| Vip3Aa | 0 | 45 | 45 | 0.0 | ||
| Various | Resistant (SP85) | buffer | 0 | 45 | 45 | 0.0 |
| Vip3Aa | 0 | 45 | 45 | 0.0 | ||
| Various | Susceptible (GR) | buffer | 0 | 45 | 45 | 0.0 |
| Vip3Aa | 45 | 0 | 45 | 100.0 |
All crosses were tested with the discriminating dose of Vip3Aa (10 μg/cm2) and scored for survival.
Mahon et al.19 reports frequencies of Vip3Aa resistance alleles for 2009/10 and 2010/11 based on F2 screens. Here we report F2 screen data from 2011/12 to 2012/13, and F1 screen data from 2013/14 and 2014/15. This reflects a shift in the approach used for resistance monitoring. Since the allelism data show one common form of Vip3Aa resistance, in 2013/14 we shifted our focus to the common resistance using the more efficient F1 screen (this shift is outlined in more detail in Walsh et al.33).
F2 screens in 2011/12 and 2012/13 estimated an r frequency for Vip3Aa in H. armigera of 0.025 with a 95% CI between 0.017 and 0.036 (14 positive lines, 284 tested lines) and 0.025 with a 95% CI between 0.016 and 0.037 (12 positive lines, 242 tested lines) respectively. There is no statistically significant difference (Fisher’s Exact test, P = 0.05) between these estimates and those obtained in 2009/10 (0.029, 11 positive lines, 108 tested lines) and 2010/11 (0.028, 17 positive lines, 165 tested lines). Summed across the four years the estimated r frequency for Vip3Aa in H. armigera based on F2 screens is 0.034 with a 95% CI between 0.022 and 0.037 (54 positive lines, 799 tested lines) and has not changed between 2009–2013.
F1 screens in 2013/14 and 2014/15 estimated an r frequency for Vip3Aa in H. armigera of 0.009 with a 95% CI between 0.002 and 0.018 (6 positive lines, 321 tested lines) and 0.016 with a 95% CI between 0.007 and 0.025 (10 positive lines, 313 tested lines) respectively; there is no statistically significant difference between these approximations (Fisher’s Exact test, P < 0.05). Summed across both years the estimated r frequency for Vip3Aa in H. armigera based on F1 screens is 0.013 with a 95% CI between 0.006 and 0.019 (16 positive lines, 634 tested lines).
As part of F2 screens performed during the monitoring program from 2009/10 to 2012/13 we examined cross-resistance against Cry1Ac and Cry2Ab by screening isofemale families of H. armigera that scored positive for carrying a resistance allele for Vip3Aa. Table 2 summarizes these data and shows that none of the randomly selected 16 families examined showed a greater propensity for survival against Cry1Ac and Cry2Ab toxin than did a Vip3Aa susceptible laboratory colony. The sample is representative of the 54 families of H. armigera that scored positive for carrying a resistance allele for Vip3Aa. We therefore conclude that larvae resistant to Vip3Aa are not cross-resistant to Cry1Ac of Cry2Ab.
Table 2. A sample of isofemale lines generated from F2 screens that were confirmed to be homozygous resistant for Vip3Aa resistance, and their responses to Cry1Ac and Cry2Ab toxin in the F3 generation.
| Isofemaleline | Cry1Ac |
Cry2Ab |
||||
|---|---|---|---|---|---|---|
| Dead | Tested | % 3rd | Dead | Tested | % 3rd | |
| 9.1809 | 90 | 90 | 0.00 | 85 | 88 | 0.00 |
| 9.2542 | 90 | 90 | 0.00 | 85 | 90 | 0.00 |
| 10.1634 | 90 | 90 | 0.00 | 90 | 90 | 0.00 |
| 10.1743 | 90 | 90 | 0.00 | 90 | 90 | 0.00 |
| 10.173 | 90 | 90 | 0.00 | 89 | 89 | 0.00 |
| 11.1013 | 82 | 86 | 0.00 | 82 | 88 | 0.00 |
| 11.1112 | 89 | 90 | 0.00 | 0 | 90 | 0.00 |
| 11.2731 | 90 | 90 | 0.00 | 90 | 90 | 0.00 |
| 11.3132 | 87 | 90 | 0.00 | 90 | 90 | 0.00 |
| 12.2169 | 90 | 90 | 0.00 | 78 | 88 | 0.00 |
| 12.2602 | 89 | 89 | 0.00 | 88 | 89 | 0.00 |
| 12.2696 | 90 | 90 | 0.00 | 90 | 90 | 0.00 |
| 12.3256 | 90 | 90 | 0.00 | 90 | 90 | 0.00 |
Assays were performed on neonates. After 7 days they were scored as being alive and at least 3rd instar, or dead or not at 3rd instar.
Vip3Aa processing with midgut juice of susceptible and resistant H. armigera
Since Vip3Aa is found in the protoxin form in cotton leaves47, we searched for differences in its conversion to the activate form between the susceptible and resistant insects. When midgut juice of GR and SP85 was incubated with Vip3Aa protoxin, many proteolytic products were obtained but no difference in the band profile between the two colonies was observed (Fig. 1a,b). The major proteolysis products were the 62 and the 20 kDa fragments in both cases. The kinetic analysis of the 89 kDa activation and the 62 kDa fragment formation showed a difference in the processing rate between the susceptible and the resistant H. armigera colonies (Fig. 1c). The processing of the 89 kDa protoxin was faster in the susceptible colony. After 15 min the protoxin completely disappeared with the midgut juice from the susceptible insects, however, with SP85 there was 31% residual protoxin which was completely activated after 60 min incubation.
Figure 1. Kinetics of the proteolytic processing of Vip3Aa incubated with midgut juice from H. armigera larvae.
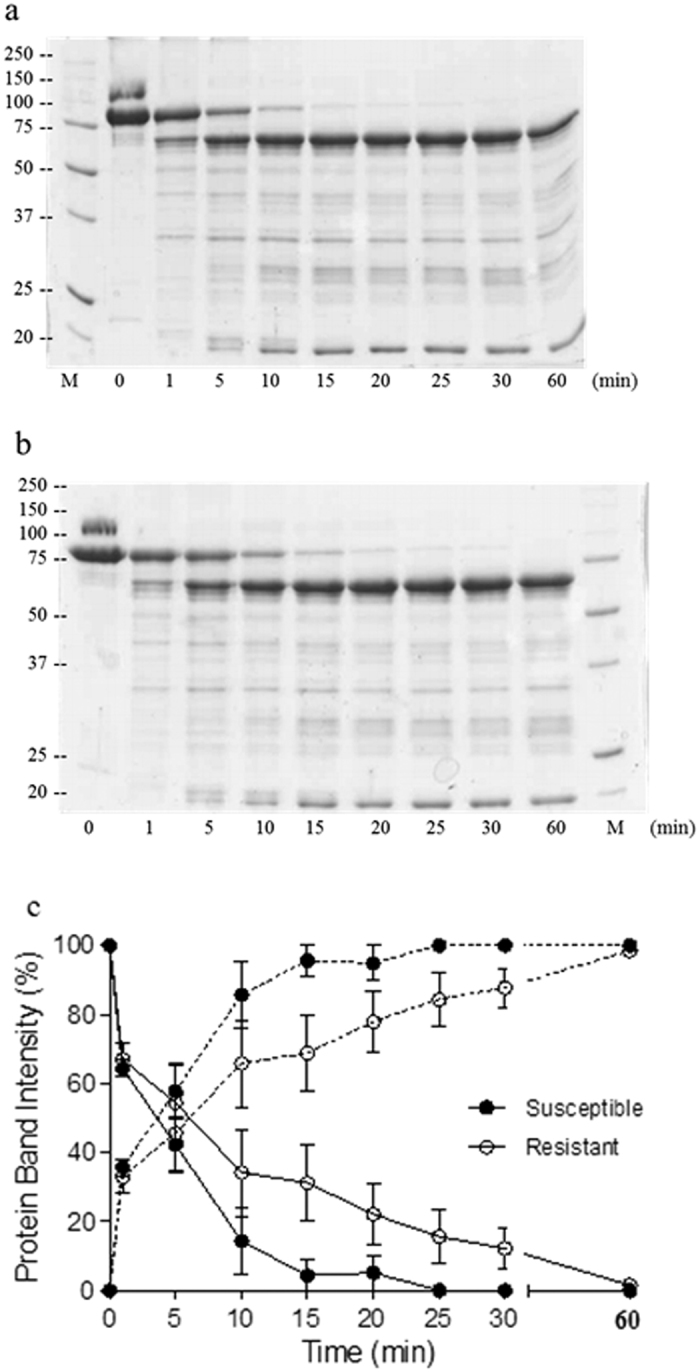
Incubations were performed at 30 °C and 0.1% of midgut juice total protein referred to Vip3Aa protein. Samples from susceptible (a) and resistant (b) insects were subjected to SDS-PAGE at different time intervals and the bands of protoxin (89 kDa, solid lines) and activated protein (62 kDa, broken lines) were quantified by densitometry (c). GR (susceptible) (●) and SP85 (resistant) (○) colonies. Data points represent the mean of three replicates with the standard error indicated by error bars.
125I-Vip3Aa binding to the BBMV of susceptible and resistant H. armigera
Specific binding of 125I-Vip3Aa to H. armigera BBMV was shown by incubating the labeled toxin in the absence and the presence of an excess of unlabeled Vip3Aa (Fig. 2). The difference between total binding and the binding in the presence of competitor is a measure of the specific binding. BBMV from the two colonies showed specific binding, indicating that the resistance was not due to an absence of binding to the epithelial membrane. To determine whether quantitative binding parameters were significantly different between the two colonies, competition binding assays were performed (Fig. 3). Incubation of a fixed amount of 125I-Vip3Aa with increasing concentrations of unlabeled protein showed that Vip3Aa fitted a one-site curve in both the resistant and the susceptible colony. Additionally, there were no significant differences in the equilibrium dissociation constants (Kd) and the concentration of binding sites (Rt) between the two colonies (Table 3). It is worth mentioning that no specific binding could be obtained with BBMV prepared from lyophilized tissue and, therefore, this type of preparation method does not preserve the binding sites involved in Vip3Aa binding.
Figure 2. Binding of 125I-Vip3Aa to BBMV from GR (susceptible) (●, ○, solid lines) and SP85 (resistant) (◼, ◻, broken lines) colonies at increasing concentrations of BBMV.
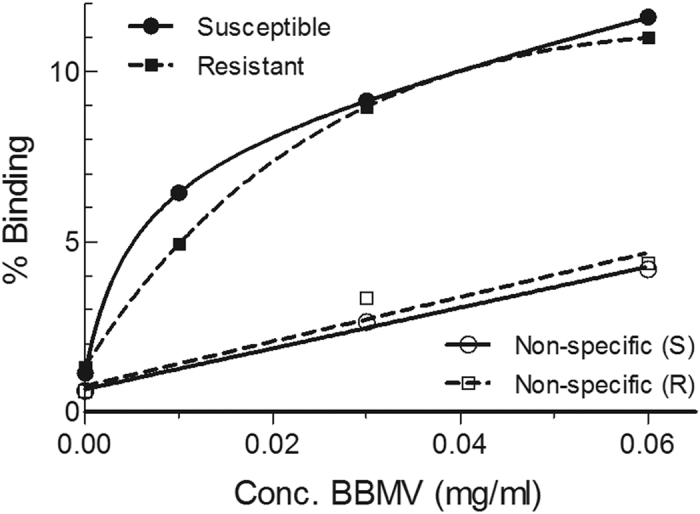
The figure is representative of two independent experiments with different batches of labeled toxin.
Figure 3. Binding of 125I-Vip3Aa to BBMV from resistant and susceptible H. armigera at increasing concentrations of unlabeled Vip3Aa.
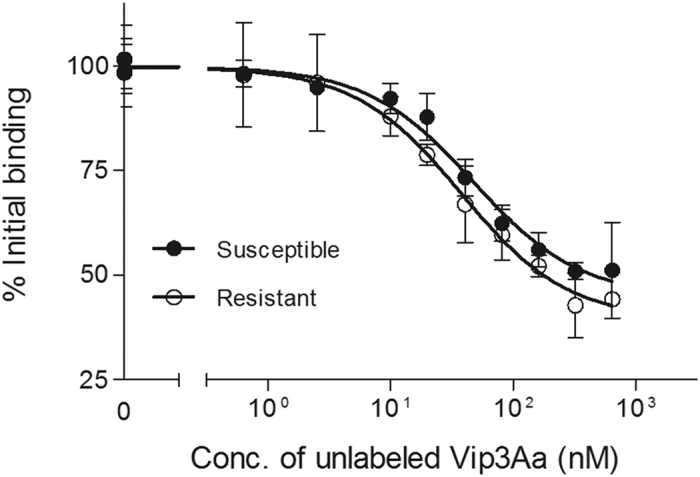
Each data point represents the mean of at least two independent replicates.
Table 3. Equilibrium dissociation constant (Kd) and concentration of binding sites (Rt) of Vip3Aa with BBMV from susceptible and resistant H. armigera.
| Protein | Mean ± SEM | Rt (pmol/mg) |
|---|---|---|
| Kd (nM) | ||
| Susceptible | 25 ± 4 | 139 ± 33 |
| Resistant | 19 ± 5 | 141 ± 25 |
Discussion
In Australia monitoring for resistance to Vip3Aa in field populations of H. armigera has been ongoing since 2009 which enabled isolation of resistant alleles and development of colonies with these genes in the laboratory. These colonies will help with the understanding of Vip3Aa resistance in this global pest. This is timely because despite our ability to detect resistance alleles, until recently, H. armigera was not exposed to significant selection pressure by Vip3Aa. However, with the recent incursion of H. armigera into the New World2 there is enormous potential selection for resistance primarily due to the large areas of corn expressing Vip3Aa proteins to control the closely related Helicoverpa zea and other lepidopteran corn pests. The bioassays performed herein to characterize Vip3Aa resistance support previous research which demonstrates that resistance alleles can readily be detected in Australian field populations despite no obvious selection (for more detail see Mahon et al.19). This relatively high baseline level of resistance may reflect selection at a low level from naturally occurring Vip3 toxins and/or direct or indirect (e.g. linkage) selection to something other than Vip3Aa. Our results suggest that natural variation exists in insect populations that could drive resistance once crops expressing Vip3Aa are introduced unless appropriate resistance management measures are implemented (mean expression levels of Vip3Aa in cotton plants is 25 μg/g dry weight, not substantially different from the discriminant dose used in our study)47.
The frequency of Vip3Aa resistance alleles in Australian populations of H. armigera has not increased significantly in the six seasons that monitoring has taken place, the last four of which are presented herein. F2 screens were performed from 2009/10 until 2012/13 and F1 screens were performed in 2013/14 and 2014/15. This is not surprising given that products expressing Vip3A have yet to be commercialized. Interestingly, the estimates obtained using F2 screens, which can yield false negatives at least for Cry2Ab (S. Downes, unpublished data), are substantially higher than those obtained using F1 screens. It is possible that the different r frequencies from these methods reflect actual changes in frequencies over time but unlikely given the absence of Bt-crops that could impact on selection for Vip3A resistance (although see above for alternative explanations). Regardless of the reason(s), our data verify that Vip3A resistance alleles exist at relatively high frequencies, and are not rising. This is in contrast to Cry2Ab which has been more variable. In 2010–2012 in particular, Cry2Ab resistance in H. armigera doubled compared to the baseline which could have signaled the beginning of a significant resistance problem in response to cotton expressing this toxin48 (see also Downes et al.11 for a similar response in the closely related H. punctigera). In the 2014/15 season, the frequency for Cry2Ab declined to baseline levels, which reflects the variability of the presence of this resistance allele, and Cry1Ac resistance remains rare (Downes, unpublished data). For the Vip3Aa alleles detected to date using F2 screens, cross-resistance to Cry2Ab and Cry1Ac was assessed and not identified. This is one of the first demonstrations of a lack of cross-resistance to Cry proteins in a Vip3Aa resistant colony, in contrast to previous studies in which Cry-resistant colonies were tested for cross-resistance to Vip3Aa.
A number of the different H. armigera Vip3Aa lines were further characterized by performing complementation tests with the first isolated Vip3Aa resistant line (SP85). In the complementation tests, three of them were found to be allelic with the lines previously identified and isolated while the results for another line were less convincing. This suggests that the mechanism present in SP85 is relatively frequent but raises the possibility of other mechanisms/genes being involved in the other F2 isolated Vip3A resistant detections. In the case of Cry1Ac and Cry2Ab resistance in H. armigera, multiple alleles were identified in the same gene which had the same phenotypic effect37,49. The fact that the majority of resistant lines tested were allelic allows us to characterize the mechanism of Vip3Aa resistance in a single line and gives us more confidence in extrapolating the findings to the whole population.
The Vip3Aa protein is produced in planta as a full length protein of ca. 89 kDa and its purification from cotton leaves indicates that the protein is stored in its protoxin form47. Upon ingested by the insect larva, the Vip3Aa protoxin is cleaved by serine proteases to several fragments, with two main products of around 62 kDa and 20 kDa when the incubation is performed under mild conditions (reviewed by Chakroun et al.50). We chose conditions that yielded the 62 kDa and 20 kDa bands as the main products of the Vip3Aa incubation with H. armigera midgut juice to search for differences between the two colonies. Although no differences were observed in the band pattern, the conversion of protoxin (89 kDa) into active toxin (the 62 kDa fragment) was faster, under the same experimental conditions, with midgut juice from the susceptible insects than from their resistant counterparts (Fig. 1). It is difficult to evaluate how this difference in the activation rate may contribute to the resistance to Vip3Aa. In some cases, the kinetics of the protoxin processing to the active toxin has been proposed to be one of the factors determining the potency of Vip3A proteins22,23,51. However, given the narrow spectrum of resistance observed, it is unlikely that a protease-based mechanism is the only factor contributing to the resistance to Vip3Aa42,43. On the contrary, binding site alteration is a well-documented mechanism of resistance to Cry1A and Cry2A toxins42,43,52. This type of alteration is very specific and cross-resistance is found only in those toxins that bind to the altered binding site.
Specific binding of Vip3A proteins to lepidopteran BBMV has been shown in several insect species using biotin-labeled Vip3Aa competed by unlabeled toxin (reviewed in Chakroun et al.50), in particular, in H. armigera53. The use of 125I-labeled ligands allows to increase sensitivity and to obtain quantitative results out of the binding assays. Conditions to successfully label Vip3Aa with 125I, to perform binding analyses, were set up with S. frugiperda BBMV24. We have used these conditions and shown specific binding of 125I-Vip3Aa to H. armigera BBMV (Fig. 2). Equilibrium binding parameters did not show any significant difference between insects from the two colonies (Table 3), indicating that alteration of the binding to the epithelial membrane does not seem to be the reason for the difference in susceptibility of the two insect colonies to Vip3Aa. This result is somewhat unexpected, since binding site alteration confers high levels of resistance to a very small set of structurally related toxins. Several studies have shown that Cry1A and Cry2A toxins do not share binding sites with Vip3A toxins24,25,26,53,54,55. The fact that Vip3Aa-resistant SP85 insects are not cross-resistant to Cry1Ac or Cry2Ab, suggests a highly specific change in the resistant insects but, similarly to other cases of resistance to Cry toxins, this change does not seem to affect binding to the epithelial membrane of the midgut29,56,57.
In conclusion, alleles for Vip3Aa resistance occur at a relatively high frequency in the field in Australian populations of H. armigera, despite the fact that Bt crops expressing this toxin are not yet prevalent in the agroecosystem. Complementation tests with the various alleles isolated by the F2 test in isofemale lines indicate that all alleles identified so far are alleles of the same gene. Biochemical analyses of resistant and susceptible insects have shown no differences at the level of binding and minor differences in the activation rate of the Vip3Aa protoxin, which may or may not contribute to resistance. Since the mode of action of Vip3 proteins is not yet well understood, further study with resistant insects may shed light on specific targets of Vip3A proteins which so far are not known.
Methods
Insect colonies
The H. armigera Vip3Aa resistant colony used in this experiment (SP85) is described in detail elsewhere19. Briefly, it is a laboratory colony which was isolated using an F2 screen during the summer of 2009–10 from individuals collected as eggs on non-Bt cotton from St. George, Queensland, Australia. This resistant colony was outcrossed five times to the susceptible laboratory colony (GR) to maintain fitness and to produce a colony that was 96.2% isogenic with the susceptible colony. Following each outcross, the colony was reselected with levels of toxin that killed all genotypes except those that were homozygous resistant to Vip3A. All subsequent generations were selected at this dose. The assays reported here were performed with individuals from the 3rd to the 5th generation. However, most of the analyses were conducted with assays on the near-isogenic 5th outcross in order to reduce the potentially misleading effects of hybrid vigor that may be evident when crossing colonies of H. armigera.
Individuals from the SP85 colonies survive the maximum concentration of Vip3Aa toxin that can be practically delivered in a surface treatment assay (220 μg/cm2) and larvae develop at the same rate as siblings reared on non-treated diet. Resistance for this colony is essentially recessive, with heterozygotes exhibiting concentration-response characteristics that are similar to those of susceptible insects19. Reciprocal backcrosses of heterozygotes to resistant colonies produced results for concentration-response assays which confirmed that resistance is essentially recessive –that is, 50% of offspring are homozygous resistant while the remainder are heterozygous and thus phenotypically susceptible19. These data are also consistent with the hypothesis that resistance is conferred by a single gene.
The GR colony used in our assays is susceptible to Vip3Aa, Cry1Ac and Cry2Ab toxins. This susceptibility is monitored regularly. The susceptible colony was employed during every screen to verify that a correctly administered discriminating concentration of toxin-containing material was applied. It has been in culture since the mid-1980s and is derived from material collected from cotton fields in the Namoi Valley, northern NSW Australia. On occasions it has been supplemented with additional collections from the same area that were screened for resistance and found to be susceptible.
Source of toxins
Bioassays to characterize Vip3Aa resistance
A Vip3Aa clone in E. coli was used as a source of toxin. Production and calibration of the Vip3Aa toxin was described elsewhere19.
Cry1Ac toxin was produced by the HD-73 strain of B. thuringiensis var. kurstaki (producing only the Cry1Ac toxin and spores). Mass production via fermentation of HD-73 was performed by Genesearch (Brisbane, Australia) with a resulting spore/crystal mix. The pellets produced were resuspended and washed three times before use. The extract was used without activating the toxin by trypsin treatment.
Dried and ground corn leaf material was used as a source of Cry2Ab toxin. This corn powder was provided by Monsanto (US) as a lyophilized Zea mays leaf powder containing transgenically expressed B. thuringiensis crystal protein, Cry2Ab2 at a concentration of 6 mg/g of powder.
Biochemical tests
The E. coli BL21 expressing Vip3Aa1658 used for the biochemical tests was kindly supplied by Dr. Slim Tounsi, CBS (Sfax, Tunisia).
Bioassays to characterize Vip3Aa resistance
Whole organism bioassays were conducted in 45 well (2.7 cm2) trays which contained approximately 2 ml of rearing diet that was overlaid with an aqueous solution of toxin and allowed to air dry. Concentrations were calculated as μg of toxin per cm2 of diet surface. After the addition of one neonate larvae per well, trays were heat sealed and maintained at 25 °C and 45–55% RH. Each bioassay consisted of a control (diet with no toxin), plus one toxin concentration. The concentration used was 10 μg of toxin per cm2. After 7 days, the larvae were scored as “alive” (exhibiting normal movement) or “dead” (dead, moribund, uncoordinated movement). The mortality of neonates in controls was minimal for all assays (mean mortality 4.1 ± 5%, range 0–11%, n = 242 neonate larvae in 6 control assays).
Allelism of different isolations of Vip3Aa resistance
Complementation tests were performed after spending 2 to 5 generations (include the two-generation F2 tests) in the laboratory. They involved setting up reciprocal crosses between new Vip3Aa-resistant colonies and the SP85 colony. To determine if the characteristics of the captured alleles were similar to those of SP85, the response to a discriminating concentration of toxin in bioassays was determined for the progeny from the above cross and from the parental colonies (SP85, the new resistant colony, and GR). Forty five insects were normally tested in the control per test and the same number exposed to 10 μg/cm2 Vip3Aa in 45 well trays (control = 242, tested = 270 in 6 toxin bioassays).
Current frequencies of Vip3Aa resistance
Assays to identify resistant insects included F2 and F1 screens that were conducted using published protocols12. We aimed to expose 90 neonate larvae to Vip3Aa toxin for each line.
(i) F2 method. Eggs collected from field hosts of H. armigera were reared to pupae. On emergence, single male and female moths were placed in individual 850 ml plastic containers with a dilute honey solution. Eggs laid on the gauze opening of the container were collected every 1–2 days. If they were fertile, around 135 hatchings were reared to establish isofemale lines. On pupation, individuals were sexed and equivalent numbers of males and females were placed in a 5 litre container and allowed to mate.
F2 offspring generated from these parents were challenged with a discriminating dose. If either field-collected insect carried a ‘resistant allele’, we would expect at least 6.25% of the toxin-exposed larvae to be homozygous for that allele and thus survive and grow to at least 3rd instar by day 759.
(ii) F1 method. This technique makes use of colonies of resistant insects in a similar fashion to that used by Gould et al.60 to determine the frequency of resistance in H. virescens. Field-collected eggs were reared to pupae and male and female pupae were placed in groups in separate cages. As moths emerged, a male was placed in an 850 ml container with two virgin SP85 females. Similarly, a female was placed in an 850 ml container with two SP85 males.
If fertile eggs were obtained from such crosses, F1 offspring were exposed to a discriminating dose. If the field-derived individual tested in this process was heterozygous for resistance, we would expect approximately 50% of the larvae to be homozygous for resistance and therefore to thrive. In the unlikely event that we collected and tested homozygotes from the field, the frequency of survivors would be close to 100%.
Cross-resistance to Cry1Ac and Cry2Ab in Vip3A resistant colonies
We present data for a sub-set of isofemale lines that were confirmed to be homozygous for alleles conferring resistance to Vip3A toxin and were challenged in the F3 generation as neonates against Cry1Ac and Cry2Ab. The discriminating concentration for Cry1Ac was 0.25 μg/cm2 of Cry1Ac delivered in a 50 μl/well solution. After 7 days this concentration killed 95.7 ± 1.8% of a susceptible general rearing colony (n = 628 larvae in 10 assays conducted over 7 days) and no surviving larvae grew beyond 2nd instar. The discriminating concentration for Cry2Ab was 1 μg/cm2 of Cry2Ab delivered in a 50 μl/ well solution. After 7 days this concentration killed 99.6 ± 0.4% of a susceptible general rearing colony (n = 286 larvae in 6 assays conducted over 7 days) and no surviving larvae grew beyond 3rd instar.
Vip3Aa purification for biochemical analyses
Conditions for bacterial culture and expression of the Vip3Aa16 protein was described previously22. For proteolysis assays the expressed Vip3Aa was purified from an E. coli cell lysate using a HisTrap FF affinity purification column (GE Healthcare) following the manufacturer instructions. Fractions of 1 ml were eluted from the column and collected in tubes containing 50 μl of 0.1 M EDTA. The most concentrated fractions were pooled and dialyzed against 20 mM Tris, 150 mM NaCl, pH 9, before storage at −20 °C.
For binding assays, Vip3Aa protein was purified as described previously24. In brief, the Vip3Aa in the E. coli cell lysate was precipitated adjusting the pH to its isoelectric point using acetic acid. The precipitated protein was recovered in the pellet after centrifugation, dissolved in 20 mM Tris-HCl, 150 mM NaCl, pH 9, and treated with 1% trypsin for 2 h at 37 °C. The protein that was used for labeling was further purified by anion-exchange chromatography in an AKTA explorer 100 system (GE Healthcare, UK).
Midgut juice preparation
Midguts from ten 5th instar larvae of Vip3Aa resistant (SP85) and susceptible (GR) H. armigera colonies reared on standard diet were dissected and the peritrophic membrane extracted with its bolus content, which was then homogenized and centrifuged for 10 min at 16000 g. The supernatant was collected and distributed in small aliquots, flash frozen in liquid nitrogen and stored in −80 °C. Total protein concentration in the midgut juice was quantified with Bradford reagent using BSA as standard.
Proteolytic processing of Vip3Aa
Proteolytic processing of Vip3A protoxin by the midgut juice of the susceptible (GR colony) and resistant (SP85 colony) H. armigera was first performed with different midgut juice dilutions to select the optimal dilution to perform the kinetic study. To compare the kinetics of Vip3Aa activation by the midgut juice of the susceptible and resistant H. armigera, 50 μg of affinity-purified protoxin was incubated with midgut juice at 1/250 (w/w, midgut juice: Vip3Aa) in 70 μl final volume of 20 mM Tris, 150 mM NaCl, pH 9, and incubated for 5, 10, 15, 20, 25, 30 and 60 min at 30 °C. The reaction was stopped by adding the SDS-PAGE loading buffer and heating for 5 min at 99 °C, after which the samples were loaded in 12% polyacrylamide gel. For a quantitative comparison of the processing rate, the amount of Vip3Aa protoxin (89 kDa) and activated toxin (62 kDa) at the different incubation times was quantified densitometrically using the TotalLab 1D v 13.01 software. The densitometry values from the 89 kDa and 62 kDa bands were relativized to the input values in each gel, and the background was corrected. Graphical representation was performed using the software GraphPad Prism v 5.00.
Vip3Aa radiolabeling
Trypsin-activated Vip3Aa was labeled using the chloramine-T method as previously described61,62. The labeled protein was separated from the excess of iodine by size-exclusion chromatography in a PD10 (GE Healthcare) column. The purity of the labeled protein was checked by analyzing the elution fractions by SDS-PAGE with further exposure of the dried gel to an X-Ray film at −20 °C. The calculated specific activity of the protein was 0.38 mCi/mg.
BBMV preparation
Fifth-instar larvae of H. armigera from both the susceptible (GR) and the resistant (SP85) colony reared on standard diet were dissected and the midguts (without the bolus content) were washed in MET buffer (300 mM mannitol, 5 mM EGTA, 17 mM Tris, pH 7.5) and frozen in liquid nitrogen and preserved at −80 °C until required. Alternatively, midguts in MET buffer were lyophilized and kept at 4 °C63. Brush border membrane vesicles (BBMV) were prepared from the frozen or the lyophilized midguts by the differential magnesium precipitation method63,64, and then frozen in liquid nitrogen, and stored at −80 °C until use. The protein concentration in the BBMV preparations was determined by Bradford65 using bovine serum albumin (BSA) as standard.
Binding assays with 125I-labeled Vip3Aa
Prior to use, the buffer of the BBMV was changed to binding buffer (20 mM Tris, 150 mM NaCl, 1 mM MnCl2, pH 7.4) supplemented with 0.1% BSA. To determine the appropriate concentration of BBMV to be used for the binding assays, 125I-Vip3Aa (1.2 nM) was incubated with increasing amounts of BBMV. An excess of unlabeled Vip3Aa was used to calculate the non-specific binding. The reaction was stopped by centrifuging the tubes at 16,000 g for 10 min at 4 °C and the pellet was washed once with 500 μl of cold binding buffer. The radioactivity retained in the pellet was measured in a model 2480 WIZARD2 gamma counter.
Competition experiments were performed by incubating 20 μg/ml of BBMV, from both the susceptible and resistant colonies, with 1.2 nM 125I-Vip3Aa in 0.1 ml final volume of binding buffer for 90 min at 25 °C in the presence of an increasing amount of unlabeled Vip3Aa protein. The reaction was stopped and the remaining radioactivity measured as described above. The dissociation constant (Kd) and the concentration of binding sites (Rt) were calculated using the LIGAND program66.
Additional Information
How to cite this article: Chakroun, M. et al. Characterization of the resistance to Vip3Aa in Helicoverpa armigera from Australia and the role of midgut processing and receptor binding. Sci. Rep. 6, 24311; doi: 10.1038/srep24311 (2016).
Acknowledgments
Research at University of Valencia was supported by the Spanish Ministry of Science and Innovation (grants ref. AGL2009-13340-C02-01 and AGL2012-39946-C02-01), by grants from the Generalitat Valenciana (ACOMP/2011/094, PROMETEO 2011/044 and GVPROMETEOII-2015-001), and by European FEDER funds. MC was supported by a Santiago Grisolía fellowship from the Generalitat Valenciana. NB was recipient of a PhD grant from the Spanish Ministry of Science and Innovation (grant ref. BES-2010-039487). Research at CSIRO was supported by the Cotton Research and Development Corporation (grants no. CSE0002, CSE1103, CSE1201) and by CSIRO Land and Water and CSIRO Agriculture.
Footnotes
Author Contributions J.F., S.D. and T.W. designed the study. M.C., N.B. and B.J. performed the experiments. M.C., S.D., T.W. and J.F. analyzed the data. M.C., N.B., J.F., S.D. and T.W. wrote the manuscript. All authors reviewed the manuscript.
References
- Pogue M. G. A new synonym of Helicoverpa zea (Boddie) and differentiation of adult males of H. zea and H. armigera (Hübner) (Lepidoptera: Noctuidae: Heliothinae). Ann. Entomol. Soc. Am. 97, 1222–1226 (2004). [Google Scholar]
- Tay W. T. et al. A brave New World for an Old World pest: Helicoverpa armigera (Lepidoptera: Noctuidae) in Brazil. Plos ONE 8(11), e80134, doi: 10.1371/journal.pone.0080134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden J. E. & Brambila J. Helicoverpa armigera (Lepidoptera: Noctuidae), the Old World Bollworm (2015) Available at: http://freshfromflorida.s3.amazonaws.com/Media%2FFiles%2FPlant-Industry-Files%2FPest-Alerts%2FPEST + ALERT + Helicoverpa + armigera.pdf (Accessed on 3rd December, 2015).
- Fitt G. P. The ecology of Heliothis species in relation to agroecosystems. Annu. Rev. Entomol. 34, 17–52 (1989). [Google Scholar]
- Fitt G. P. Cotton pest-management: Part 3. an Australian perspective. Annu. Rev. Entomol. 39, 543–562 (1994). [Google Scholar]
- Forrester N. W., Cahill M., Bird L. J. & Layland J. K. Management of pyrethroid and endosulfan resistance in Helicoverpa armigera (Lepidoptera, Noctuidae) in Australia. Bull. Entomol. Res. R1–132 (1993). [Google Scholar]
- Jin L. et al. Dominant resistance to Bt cotton and minor cross-resistance to Bt toxin Cry2Ab in cotton bollworm from China. Evol. Appl. 6, 1222–1235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes S. et al. Adaptive management of pest resistance by Helicoverpa species (Noctuidae) in Australia to the Cry2Ab Bt toxin in Bollgard II® cotton. Evol. Appl. 3, 574–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrick J. A. et al. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. Plos ONE 9(5), e97900, doi: 10.1371/journal.pone.0097900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabashnik B. E., Brévault T. & Carrière Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat. Biotechnol. 31, 510–521 (2013). [DOI] [PubMed] [Google Scholar]
- Downes S., Parker T. & Mahon R. Incipient resistance of Helicoverpa punctigera to the Cry2Ab Bt toxin in Bollgard II cotton. Plos ONE 5(9), e12567 doi: 10.1371/journal.pone.0012567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon R. J., Olsen K. M., Downes S. & Addison S. Frequency of alleles conferring resistance to the Bt toxins Cry1Ac and Cry2Ab in Australian populations of Helicoverpa armigera (Lepidoptera: Noctuidae). J. Econ. Entomol. 100, 1844–1853 (2007). [DOI] [PubMed] [Google Scholar]
- Farias J. R. et al. Field-evolved resistance to Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot. 64, 150–158 (2014). [Google Scholar]
- Gassmann A. J. et al. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc. Natl. Acad. Sci. USA 111, 5141–5146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch J. J. et al. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. USA 93, 5389–5394 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Escudero I. et al. A screening of five Bacillus thuringiensis Vip3A proteins for their activity against lepidopteran pests. J. Invertebr. Pathol. 113, 78–81 (2014). [DOI] [PubMed] [Google Scholar]
- Kurtz R. W., McCaffery A. & O’Reilly D. Insect resistance management for Syngenta’s VipCot (TM) transgenic cotton. J. Invertebr. Pathol. 95, 227–230 (2007). [DOI] [PubMed] [Google Scholar]
- Pickett B. R. Studies on resistance to vegetative (Vip3A) and crystal (Cry1A) insecticidal toxins of Bacillus thuringiensis. In Heliothis virescens (Fabricius). PhD thesis, Imperial College, London, UK (2009).
- Mahon R. J., Downes S. J. & James B. Vip3A resistance alleles exist at high levels in Australian targets before release of cotton expressing this toxin Plos ONE 7(6), e39192, doi: 10.1371/journal.pone.0039192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. G., Mullins M. A., Warren G. W., Koziel M. G. & Estruch J. J. The Bacillus thuringiensis vegetative insecticidal protein Vip3Aa lyses midgut epithelium cells of susceptible insects. Appl. Environ. Microbiol. 63, 532–536 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. K., Walters F. S., Hart H., Palekar N. & Chen J. S. The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3Aa differs from that of Cry1Ab delta-endotoxin. Appl. Environ. Microbiol. 69, 4648–4657 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakroun M., Bel Y., Caccia S., Abdelkefi-Mesrati L., Escriche B. & Ferré J. Susceptibility of Spodoptera frugiperda and S. exigua to Bacillus thuringiensis Vip3Aa insecticidal protein. J. Invertebr. Pathol. 110, 334–339 (2012). [DOI] [PubMed] [Google Scholar]
- Caccia S., Chakroun M., Vinokurov K. & Ferré J. Proteolytic processing of Bacillus thuringiensis Vip3A proteins by two Spodoptera species. J. Insect. Physiol. 67, 76–84 (2014). [DOI] [PubMed] [Google Scholar]
- Chakroun M. & Ferré J. In vivo and in vitro binding of Vip3Aa to Spodoptera frugiperda midgut and characterization of binding sites by 125I radiolabeling. Appl. Environ. Microbiol. 80, 6258–6265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. K., Miles P. & Chen J. S. Brush border membrane binding properties of Bacillus thuringiensis Vip3A toxin to Heliothis virescens and Helicoverpa zea midguts. Biochem. Biophys. Res. Commun. 339, 1043–1047 (2006). [DOI] [PubMed] [Google Scholar]
- Gouffon C., Van Rie J., Jansens S. & Jurat-Fuentes J. L. Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl. Environ. Microbiol. 77, 3182–3188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. E., Marcus M. A., Gould F., Bradley J. R. Jr. & Van Duyn J. W. Cross-resistance responses of Cry1Ac-selected Heliothis virescens (Lepidoptera: Noctuidae) to the Bacillus thuringiensis protein Vip3A. J. Econ. Entomol. 100, 180–186 (2007). [DOI] [PubMed] [Google Scholar]
- Fang J. et al. Characterization of chimeric Bacillus thuringiensis Vip3 toxins. Appl. Environ. Microbiol. 73, 956–996 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilkumar K. J. et al. Production and characterization of Bacillus thuringiensis Cry1Ac-resistant cotton bollworm Helicoverpa zea (Boddie). Appl. Environ. Microbiol. 74, 462–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. et al. Vip3Aa tolerance response of Helicoverpa armigera populations from a Cry1Ac cotton planting region. J. Econ. Entomol. 103, 2169–2173 (2010). [DOI] [PubMed] [Google Scholar]
- Vélez A. M. et al. Inheritance of Cry1F resistance, cross-resistance and frequency of resistant alleles in Spodoptera frugiperda (Lepidoptera: Noctuidae). Bull. Entomol. Res. 103, 700–713 (2013). [DOI] [PubMed] [Google Scholar]
- Huang F. et al. Cry1F resistance in fall armyworm Spodoptera frugiperda: single gene versus pyramided Bt maize. Plos One 9, e112958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. K. et al. Dual Cry2Ab and Vip3A resistant strains of Helicoverpa armigera and Helicoverpa punctigera (Lepidoptera: Noctuidae); testing linkage between loci and monitoring of allele frequencies. J. Econ. Entomol. 107, 1610–1617 (2014). [DOI] [PubMed] [Google Scholar]
- Carrière Y., Crickmore N. & Tabashnik B. E. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat. Biotech. 33, 161–168 (2015). [DOI] [PubMed] [Google Scholar]
- Wu Y. Detection and mechanisms of resistance evolved in insects to Cry toxins from Bacillus thuringiensis. Adv. Insect Physiol. 47, 297–342 (2014). [Google Scholar]
- Park Y. et al. ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biology 12, 46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay W. T. et al. Insect pest resistance to the Bacillus thuringiensis toxin Cry2Ab is conferred by multiple independent mutations in an ABC transporter subfamily A protein. Plos Genet 11(11), e1005534, doi: 10.1371/journal.pgen.1005534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiewsiri K. & Wang P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. USA 108, 14037–14042 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. et al., MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. Plos Genetics 11(4), e1005124, doi: 10.1371/journal.pgen.1005124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppert B., Kramer K. J., Beeman R. W., Johnson D. & McGaughey W. H. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J. Biol. Chem. 272, 23473–23476 (1997). [DOI] [PubMed] [Google Scholar]
- Li et al. Comparative analysis of proteinase activities of Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 34, 753–762 (2004). [DOI] [PubMed] [Google Scholar]
- Ferré J. & Van Rie J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47, 501–533 (2002). [DOI] [PubMed] [Google Scholar]
- Ferré J., Van Rie J. & Macintosh S. C. Insecticidal genetically modified crops and insect resistance management (IRM). (ed. Romeis J., Shelton A. M. & Kennedy G. G.). Integration of Insect Resistant Genetically Modified Crops within IPM Programs Ch. 3, 41–85 (Springer, Netherlands, 2008). [Google Scholar]
- Gould F. et al. Broad-spectrum resistance to Bacillus thuringiensis toxins in Heliothis virescens. Proc. Nat. Acad. Sci., USA 89, 7986–7990 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Tabashnik B. E. & Adang M. J. Binding of Bacillus thuringiensis Cry1Ac toxin to aminopeptidase in susceptible and resistant diamondback moths (Plutella xylostella). Appl. Environ. Microbiol. 63, 1024–27 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero S., Oppert B. & Ferré J. Different mechanisms of resistance to Bacillus thuringiensis toxins in the Indianmeal moth. Appl. Environ. Microbiol. 67, 1085–89 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- U. S. Environmental Protection Agency. Bacillus thuringiensis modified Cry1Ab (SYN-IR67B-1) and Vip3Aa19 (SYN-IR102-7) insecticidal proteins and the genetic material necessary for their production in COT102 X COT67B cotton. (2008) Available at: http://www3.epa.gov/pesticides/chem_search/reg_actions/registration/decision_PC-006529_12-Aug-08.pdf (Accessed: 29th February 2016).
- Downes S. & Mahon R. Successes and challenges of managing resistance in Helicoverpa armigera to Bt cotton in Australia. GM Crops Food 3, 228–234 (2012). [DOI] [PubMed] [Google Scholar]
- Wu Y. D., Zhao J., Jin L. & Yang Y. H. Diverse cadherin mutations conferring resistance to Bacillus thuringiensis toxin Cry1Ac in Helicoverpa armigera. Insect Biochem. Mol. Biol. 40, 113–118 (2010). [DOI] [PubMed] [Google Scholar]
- Chakroun M., Banyuls N., Bel Y., Escriche B. & Ferré J. Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 80, 329–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelkefi-Mesrati L. et al. Investigation of the steps involved in the difference of susceptibility of Ephestia kuehniella and Spodoptera littoralis to the Bacillus thuringiensis Vip3Aa16 toxin. J. Invertebr. Pathol. 107, 198–201 (2011). [DOI] [PubMed] [Google Scholar]
- Caccia S. et al. Binding site alteration is responsible for field-isolated resistance to Bacillus thuringiensis Cry2A insecticidal proteins in two Helicoverpa species. Plos ONE 5(4), e9975, doi: 10.1371/journal.pone.0009975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yang A., Shen X., Hua B. & Shi G. Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J. Invertebr. Pathol. 108, 92–97 (2011). [DOI] [PubMed] [Google Scholar]
- Sena J. A., Hernández-Rodríguez C. S. & Ferré J. Interaction of Bacillus thuringiensis Cry1 and Vip3Aa proteins with Spodoptera frugiperda midgut binding sites. Appl. Environ. Microbiol. 75, 2236–2237 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Hamadou-Charfi D., Boukedi H., Abdelkefi-Mesrati L., Tounsi S. & Jaoua S. Agrotis segetum midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 differs from that of Cry1Ac toxin. J. Invertebr. Pathol. 114, 139–143 (2013). [DOI] [PubMed] [Google Scholar]
- Liu Y. B., Tabashnik B. E., Masson L., Escriche B. & Ferré J. Binding and toxicity of Bacillus thuringiensis protein Cry1C to susceptible and resistant diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 93, 1–6 (2000). [DOI] [PubMed] [Google Scholar]
- Zhao J. Z. et al. Development and characterization of diamondback moth resistance to transgenic broccoli expressing high levels of Cry1C. Appl. Environ. Microbiol. 66, 3784–3789 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelkefi-Mesrati L., Tounsi S. & Jaoua S. Characterization of a novel vip3-type gene from Bacillus thuringiensis and evidence of its presence on a large plasmid. FEMS Microbiol. Lett. 244, 353–358 (2005). [DOI] [PubMed] [Google Scholar]
- Andow D. A. & Alstad D. N. F2 screen for rare resistance alleles. J. Econ. Entomol. 91, 572–578 (1998). [Google Scholar]
- Gould F. et al. Initial frequency of alleles for resistance to Bacillus thuringiensis toxins in field populations of Heliothis virescens. Proc. Natl. Acad. Sci. USA 94, 3519–3523 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Rodríguez C. S., Van Vliet A., Bautsoens N., Van Rie J. & Ferré J. Specific binding of Bacillus thuringiensis Cry2A insecticidal proteins to a common site in the midgut of Helicoverpa species. Appl. Environ. Microbiol. 74, 7654–7659 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rie J., Jansens S., Höfte H., Degheele D. & Van Mellaert H. Specificity of Bacillus thuringiensis delta-endotoxins. Importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur. J. Biochem. 186, 239–47 (1989). [DOI] [PubMed] [Google Scholar]
- Hernández C. S., Rodrigo A. & Ferré J. Lyophilization of lepidopteran midguts: a preserving method for Bacillus thuringiensis toxin binding studies. J. Invertebr. Pathol. 85, 182–187 (2004). [DOI] [PubMed] [Google Scholar]
- Wolfersberger M. G. et al. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86, 301–308 (1987). [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. J. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Munson P. & Rodbard D. LIGAND: A versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 107, 220–239 (1980). [DOI] [PubMed] [Google Scholar]


