Abstract
The rationale of this study is to compare and integrate two heterologous datasets intended to unravel the spatiotemporal specificities of gene expression in a rapidly growing and complex organ. We implemented medium-throughput RNA in situ hybridization (ISH) for 39 genes mainly corresponding to cell wall proteins for which we have particular interest, selected (i) on their sequence identity (24 class III peroxidase multigenic family members and 15 additional genes used as positive controls) and (ii) on their expression levels in a publicly available Arabidopsis thaliana seed tissue-specific transcriptomics study. The specificity of the hybridization signals was carefully studied, and ISH results obtained for the 39 selected genes were systematically compared with tissue-specific transcriptomics for 5 seed developmental stages. Integration of results illustrates the complementarity of both datasets. The tissue-specific transcriptomics provides high-throughput possibilities whereas ISH provides high spatial resolution. Moreover, depending on the tissues and the developmental stages considered, one or the other technique appears more sensitive than the other. For each tissue/developmental stage, we finally determined tissue-specific transcriptomic threshold values compatible with the spatiotemporally-specific detection limits of ISH for lists of hundreds to tens-of-thousands of genes.
The post-genomics era leads to the generation of increasing amount of publicly available large transcriptomic datasets. In October 2015, 3,848 array- and sequence-based datasets covering 1,604,299 samples from all living kingdoms were deposited on the gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo/). Transcriptomics can reach cellular/tissular levels using accurate sampling methods. These include for example fluorescence activated cell sorting (FACS) of animal stem cells1 or plant protoplast populations2,3, isolation of nuclei tagged in specific cell types4, or laser capture microdissection (LCM) of single prokaryotic cell5, functionally distinct human neurons6 or plant tissue serial sections7,8,9,10. Spatiotemporally-specific transcriptomes are accessible through raw data files or user-friendly web-based tools6,11,12,13. These tissue-specific transcriptomic studies need validation and refinement of the expression profiles through qRT-PCR, promoter::reporter genes or RNA in situ hybridization (ISH). The most popular ISH protocols use whole mount permeabilized samples14,15 or paraplast-embedded sections16,17. ISH is a powerful approach often underestimated probably due to its perception as being difficult to perform, its relative sensitivity and its moderate throughput. Rare reports of elegant medium-to-high throughput ISH studies exist3,18 but with relatively poor spatiotemporal resolution.
A biological question that particularly needs spatiotemporally-resolved gene expression data concerns the dynamics of plant cell walls. Their study is of high interest since they constitute an active zone of molecular dialogue and signalling during plant growth and plant microorganism interactions19, and they harbour stocks of renewable carbon of high interest for pulp and paper and biofuel production20,21. However, plant cell wall study is complex since they form diverse, complex and dynamic networks with highly cell-specific patterns as illustrated by numerous immunocytochemical studies of cell wall polymers such as polysaccharides and lignins22,23,24,25,26,27,28. The knowledge of plant cell wall proteomes has increased to about 2,000 proteins in a given organism in the last ten years and these proteomic studies argue for species- and organ-specific profiles29,30,31. High-throughput plant cell-specific profiles remain so far unachievable and these proteomic studies have to be completed by more spatially resolutive approaches such as immunolabelling dedicated to a subset of proteins30.
The present study uses the fast and complex development kinetics of the ~400 μm-long seeds from the model plant Arabidopsis thaliana (Supplementary Fig. S1) well adapted to the study of cell wall dynamics25. The rationale of the present study was to evaluate and demonstrate the complementarity between the results obtained from a publicly available high-throughput tissue-specific transcriptomic study7 (Supplementary Table S1) and our medium-throughput ISH analysis implemented for 39 selected genes and 6 seed developmental stages. We primarily selected 24 genes of the multigenic family of Class III peroxidases (73 members) mainly corresponding to cell wall proteins32. Recent studies demonstrated that these oxidoreductases that may accommodate a wide range of substrate in vitro became highly specific in vivo due to their accurate spatiotemporal expression and positioning in cell wall microdomains where they encounter specific partners and substrates33,34. Studying members of such a multigenic family also allowed to clearly assess the specificity of ISH signals given the high sequence identities. To reinforce our gene sampling, we added 15 genes mostly corresponding to cell wall proteins primarily selected based on their tissue-specific transcriptomic expression values.
Tissue-specific transcriptomic high-throughput was not challenged by ISH medium-throughput, however, ISH allowed to obtain specific signals for genes with high sequence identity that were not distinguished by tissue-specific transcriptomics. We also proved that ISH was more accurate than tissue-specific transcriptomics regarding the localisation of the transcripts in precise cell types. Our results were discussed and compared with the literature describing microphenotypes or cell-specific expression data for some of these genes. For all other genes, our results constitute a basis for future functional studies. To widen the interest of our method, we finally estimated, for each tissue/developmental stage, thresholds of tissue-specific transcriptomic expression values for which a specific ISH signal could be reasonably detected for lists of hundreds to tens-of-thousands of genes.
Methods
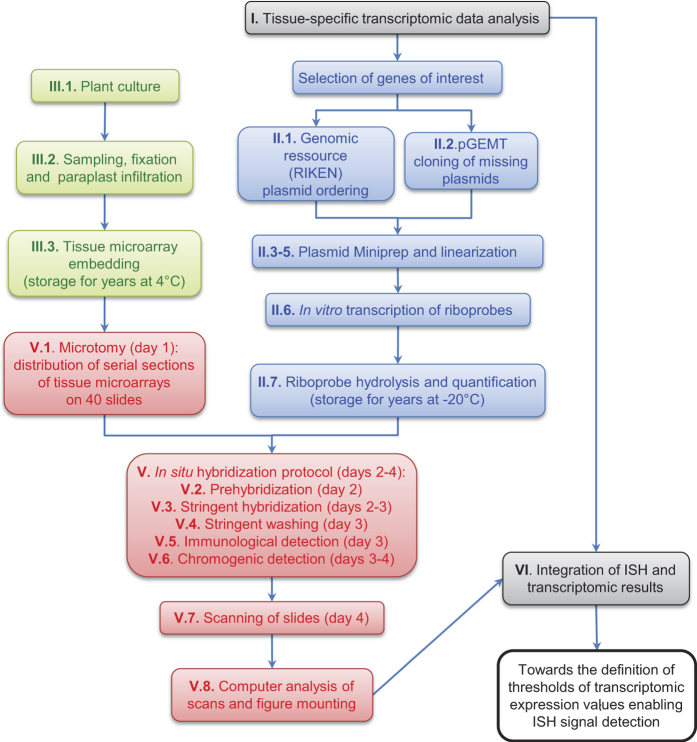
The overall flowchart of the method is presented in Fig. 1. The following sections explain the principle of the 6 main steps of the method displayed in Fig. 1. Extended details of the methods and step-by-step protocol are provided in the Supplementary Method in the same order.
Figure 1. Flowchart of the method used for medium throughput in situ hybridization based on/compared to published tissue-specific transcriptomic data.
This flowchart is labelled according to the paragraph numbers appearing in the Material and Methods and Supplementary Methods sections.
Tissue-specific transcriptomic data analysis (step I)
Gene Expression Omnibus accession series GSE124047 was downloaded as series matrix files at www.ncbi.nlm.nih.gov/geo. The “.txt” file was converted to “.xls” file which was further annotated and edited (Supplementary Table S1). The expression data of the 73 Class III peroxidases (AtPRXs)35,36 (http://peroxibase.toulouse.inra.fr/) was filtered (Supplementary Table S2). We first selected the 16 most highly expressed AtPRXs spot IDs from this resource and from two additional resources (seed eFP browser at http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Seed12, and the Supplementary data 2 provided in the original publication7). We performed a Pearson correlation coefficient analysis showing that, despite different data treatment, the three resources corresponded to the same original data (Supplementary Table S3). The circa 3 ratio of signal intensities between the GSE12404/eFP browser datasets and the Belmonte et al. dataset (Supplementary Table S3) allowed to convert the detection limit value of 15 defined in the original publication7 to a detection limit value of 45 for the GSE12404/eFP browser datasets. In the following, we used the most complete GSE12404 dataset (Supplementary Table S1). For the ISH study, we selected 24 AtPRXs (about 1/3 of the family) corresponding to the 16 most responsive spot IDs (14 unique genes and 2 ambiguous duplicated gene pairs), to 6 AtPRXs whose maximum transcriptomic expression value was below a 300 arbitrary cut-off and 2 AtPRXs for which no spot ID was available (Supplementary Table S2). We added 15 non-peroxidase genes, that corresponded to two peroxidase partners (RBOHE, RBOHF), one housekeeping gene (TUB4), and 12 putative cell wall proteins in which we have particular interest29,31. We paid attention to select candidates presenting maximum tissue-specific transcriptomic expression values ranging from very high to medium levels (Supplementary Table S5).
The most recent release of the Arabidopsis cDNA sequences dataset (TAIR10) was downloaded from The Arabidopsis Information Resource FTP server (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR10_blastsets/). Each sequence of this dataset was searched locally using Blastall 2.2.24 (“blastn” command with the default parameters37) against the TAIR10 cDNA database. The second best hit (defined as the best hit beside the query itself) for each of the 33,602 cDNA sequences of TAIR10 is indicated in Supplementary Table S6. The Supplementary Table S7 corresponds to the extraction from the Supplementary Table S6 of the second best hit of the 15,866 cDNAs with a nucleotide number above 300. The Supplementary Table S8 corresponds to the extraction from Supplementary Table S7 of the second best hit among the 73 AtPRXs and the 15 additional non peroxidase genes selected for our in situ hybridization study. More detailed protocols are provided in Supplementary Methods.
Plasmid resources and riboprobe in vitro transcription (step II)
The full length cDNA (pda clones) ordered at the RIKEN bioresource center [ http://www.brc.riken.jp/lab/epd/catalog/cdnaclone.html;38,39] are listed in the Supplementary Table S9. These cDNA clones are provided within pBluescript-derived vector series allowing their direct utilization for in vitro transcription of riboprobes since the multiple cloning sites are framed by the T3 and T7 RNA polymerase promoters, respectively (Supplementary Fig. S2). Five cDNAs not available at RIKEN were amplified using a pool of retro-transcribed mRNAs from various seed developmental stages, and cloned in pGEM-T Easy (Promega) (Supplementary Table S10). These clones are included in the Supplementary Table S9, and the multiple cloning site (MCS) including SP6 and T7 RNA polymerase promoter position is shown in Supplementary Fig. S2.
Plasmids (5–7 μg) were linearized with excess (20 units) of single cut 5′ overhang or blunt restriction enzymes (Promega, Roche) (Supplementary Table S9) for 4 h at 37 °C. The digestion occurred either at the 5′ or the 3′ end of the cDNA, in order to produce the template for in vitro transcription of the antisense or the sense probes, respectively. Sense probes were used as classical negative control and the significance/limits of this type of control and the use of additional negative controls will be discussed hereafter. Whenever possible, the linearization was performed using enzymes single-cutting in the multiple cloning site (MCS) and not cutting the cDNA (Supplementary Fig. S2). In some cases, we used enzymes single-cutting at the cDNA ends and not cutting the vector (Supplementary Fig. S3).
In vitro transcription of Digoxigenin (Dig)-labelled riboprobes for ISH or unlabelled antisense riboprobes for competitive inhibition was performed during 2 h at 37 °C, using Dig RNA labelling mix (Roche) and unlabelled NTP mix (Promega), respectively, and T3, T7 or SP6 RNA polymerase (Promega) (Supplementary Table S9). Riboprobes were further chopped to an average size of 400 b by alkaline hydrolysis (Supplementary Table S9). Step-by-step detailed protocol is provided in Supplementary Methods.
Plant material and tissue microarray preparation (step III)
The same wild-type Arabidopsis thaliana ecotype (Wassilewskija, Ws) and similar culture conditions (115 μmol.m−2.sec−1 continuous light using 25% Osram 58 W Fluora and 75% lumilux cool daylight 58 W tubular fluorescent lamps, respectively; 22 °C; 75% relative humidity) as used for the tissue-specific transcriptomic reference study7 was primarily used. For specificity studies, atprx36 T-DNA knock out insertion line [per36-1; SAIL_194_G0333;] and its corresponding A. thaliana wild type background (Columbia 0, Col-0) were cultivated in the same conditions. RNase-free conditions were strictly observed for all steps. In order to cover the whole kinetics of seed development, 39, 40, 43 and 45-day old plants were sampled each in three batches (siliques from the top, medium and bottom of the floral stem, the youngest siliques being at the top). In each case, dozens of whole siliques were harvested by cutting the pedicel with a razor blade and rapidly vacuum infiltrated in FAA (10% Formalin (37% formaldehyde solution, Sigma-Aldrich); 50% ethyl alcohol; 5% acetic acid; 35% DEPC-treated water) and fixed for 16 h at 4 °C. The dehydration and paraplast infiltration protocol was adapted from40. Siliques were concentrated in embedding molds constituting tissue microarrays emcompassing up to 1,000 developing seeds (Supplementary Fig. S1b). Step-by-step detailed protocol is provided in Supplementary Methods.
Medium-throughput in situ hybridization conditions (step IV)
The respect of RNase-free conditions is crucial for the success of ISH. The detail of material, chemicals and solutions including preparation guide is provided in Supplementary Methods.
Medium-throughput in situ hybridization (step V)
The basis of this protocol comes from41 following several adaptations40,42,43 and final simplification using a unique buffer for most of the steps and substituting the acetic anhydre/triethanolamine tedious charge equilibration step by a more simple diethylpyrocarbonate (DEPC)-mediated carbethoxylation step, inspired by16. 10 μm-serial sections of tissue microarrays were disposed on precoated microscopy slides. 40 slides, corresponding to 40 riboprobes were processed in one ISH experiment. Hybridization was performed overnight at 50 °C, and following stringent washing steps, hybridized probes were immunodetected using anti-Dig-alkaline phosphatase (AP) antibody (Roche) and colour development of AP reaction was performed overnight at room temperature. Step-by-step detailed protocol is provided with timing in Supplementary Methods. Slides were mounted in Eukitt and scanned at high-throughput using a nanozoomer HT slide scanner (Hamamatsu). We routinely scanned all slides at ×20 focus (=0.46 μm per pixel on a single z plan) and these scans were viewed and analysed using the NDP view freeware (Hamamatsu) and directly used to prepare Figures (Fig. 2; Supplementary Figs. S4–S45).
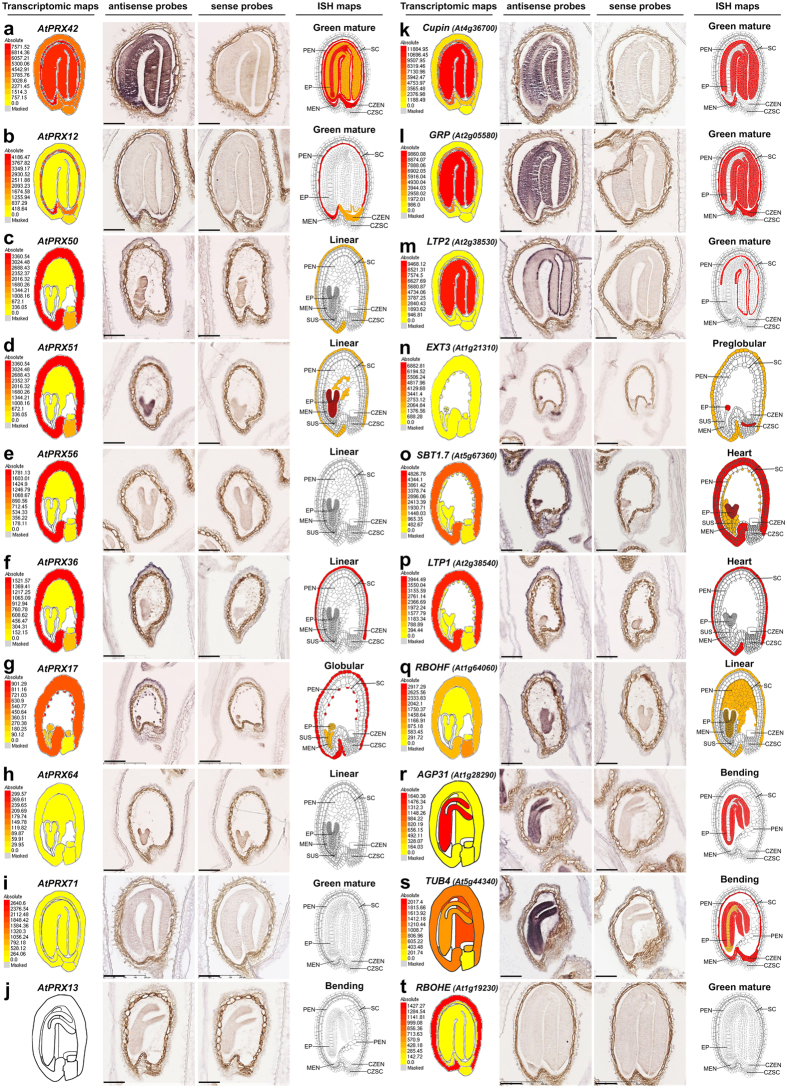
Figure 2. Illustration of the in situ hybridization results obtained for a selection of genes and developmental stages.
Results for 10 AtPRXs (a–j) and for 10 non peroxidase genes (k–t) are displayed in the decreasing order of their maximal individual tissue-specific transcriptomic expression values7. For each gene and each developmental stage, a screen copy of the seed eFP browser tissue-specific transcriptomic map including the individual absolute heatmap scale (red-to-yellow colour codes correspond to high-to-low expression values) that is different for each gene12 is compared with the corresponding ISH results (for both the antisense and sense probes). The final corresponding new ISH map (re-coloured after original drawings from ref. 7 available at Seedgenenetwork http://estdb.biology.ucla.edu/seed/) gave increased cellular resolution with a unique colour code for all genes (red corresponding to visually detected strong signals, orange corresponding to moderate signals and white corresponding to the absence of detected ISH signal). Note that the naturally brown colour of seed coat does not correspond to ISH signal. The full ISH results corresponding to a selection of 5 developmental stages for all 39 genes can be found in Supplementary Figs. S4–S42. Scale bars: 100 μm. Abbreviations: AS, anti-sense; CZEN, chalazal endosperm; CZSC, chalazal seed coat; EP, embryo proper; MEN, micropylar endosperm; PEN, peripheral endosperm; S, sense; SC, seed coat; SUS, suspensor.
Integration of ISH and tissue-specific transcriptomic results (step VI)
One Figure was assembled for each gene studied by ISH. In each Figure, we compared the screen copy of the individual tissue-specific transcriptomic maps and relative heatmap scale available through seed eFP browser at http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Seed12 for 5 selected developmental stages, with corresponding ISH antisense and sense images directly extracted from NDP view. Finally, we constructed using Corel Photopaint a new detailed expression map for each gene based on our ISH results, with a red/orange/white colour code (red, strong ISH signals; orange, moderate ISH signals; white, no ISH signal), and using the original detailed cartoon from ref. 7 available at Seedgenenetwork (http://estdb.biology.ucla.edu/seed/) as a basis for the drawing except for the bending cotyledons stage for which we drew our own cartoon. Note that the new ISH maps corresponded to the observation of numerous seed sections from at least 3 experimental repeats and not only to the individual displayed ISH images.
The Supplementary Table S1 was used to extract tissue-specific transcriptomic expression profiles from all the AtPRXs and the 39 genes studied in ISH. This data was gathered in single Microsoft Excel files (Supplementary Table S4 for peroxidases and Supplementary Table S5 for the 39 genes studied by ISH), keeping the original information about the maximum expression value and the ranking position of each gene. The individual tissue-specific transcriptomic values were framed in bold when either strong or moderate ISH signal was observed in the corresponding tissues according to Supplementary Figs. S4–S42. The gene annotation was framed in red (strong ISH signal in at least one tissue), orange (moderate-to-low ISH signal in at least one tissue) or yellow (no ISH signal detected) following the similar colour code as in Supplementary Figs. S4–S42. Demonstration of ISH specificity is provided in Supplementary Figs. S43–S45.
The Supplementary Table S1 was sequentially used to distribute, for each of 36 tissue-specific transcriptomic samples (all samples except the 6 whole seed samples), the 23,933 genes within 10 groups according to their transcriptomic expression value. The groups corresponded to expression values of 0–44 (below the detection limit of the transcriptomic study; 55–65% of the genes depending on the sample), 45–299 (below the arbitrary 300 cut-off used for our first selection of candidate genes), 1000–1999, 2000–2999, 3000–3999, 4000–4999, 5000–9999, 10000–19999, 20000-max. In order to fully integrate our ISH results with the whole tissue-specific transcriptomic data, the abbreviation of the 37 genes of the 39 genes studied by ISH that were present on the array (all 39 genes except AtPRX13 and AtPRX32) was positioned on the individual histograms on the top of the range corresponding to their tissue-specific transcriptomic value (Fig. 3; Supplementary Figs. S46–S51). In order to summarize the ISH results, we kept the same red/orange/white colour code described above. Less genes were studied in ISH for the preglobular stage which was under-represented on the tissue arrays. We finally calculated within colour-coded double arrows positioned on the top of the histograms, the sum of genes for which ISH could be sensitive enough to various degrees for each tissue/developmental stage.
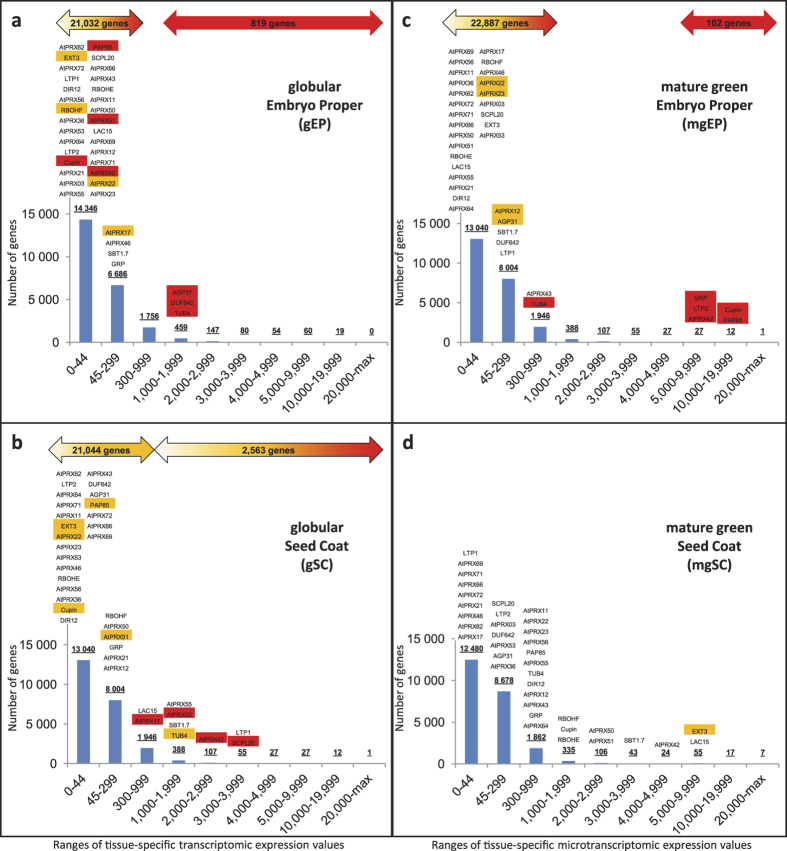
Figure 3. Thresholds of tissue-specific transcriptomic values compatible with ISH are specific to each developmental stage/tissue.
For each developmental stage/tissue tissue-specific transcriptomic sample ((a,b) globular developmental stage; (c,d) mature green developmental stage; (a,c) embryo proper tissue; (b,d) seed coat tissue), the 23,933 genes present on the microarray were classified according to their tissue-specific transcriptomic expression values. The range 0–44 corresponded to the expression values below the 45 detection limit of the transcriptomic study (55–65% of the genes); the range 45–299 corresponded to the expression values between the 45 detection limit of the transcriptomics and the 300 arbitrary cut-off that we initially defined for our ISH study; the other ranges were arbitrarily set to allow the distribution of the genes in various expression value groups. The resulting number of genes within each range was plotted on individual histograms. All the genes analysed by ISH in this study were positioned above the histograms according to their individual tissue-specific transcriptomic expression value. The ISH results were colour-coded in red/orange/white as in Fig. 2, according to Supplementary Figs. S4–S42. The deducted total number of genes compatible, to various extend, with ISH was posted on the top of each graph within double arrows using shades of the same colour coding. This clearly illustrates that the sensitivity of ISH is dependent on spatiotemporal parameters and the complementarity of transcriptomics and ISH. The corresponding histograms for all developmental stages and tissues are available in Supplementary Figs. S46–S51.
Results and Discussion
Comparison and integration of in situ hybridization and tissue-specific transcriptomic results
We systematically compared, for each of the 39 genes and 5 selected developmental stages, a screen copy of an user-friendly web-based tissue-specific transcriptomic map12 including the individual absolute heatmap scale that is different for each gene, with the corresponding ISH results (Fig. 2a–t; Supplementary Figs. S4–S42). All together, the analysis of ISH results for tens of thousands of seed sections allowed drawing the corresponding new ISH maps giving increased cellular resolution, using a unique colour code for all genes; red, orange and white corresponding to strong, moderate and no visually detected ISH signal, respectively (Fig. 2a–t; Supplementary Figs. S4–S42). Since preglobular stage was underrepresented on the tissue microarray, only a subset of the 39 genes could be studied at this developmental stage. Additionally, in some circumstances, small developing embryo could not be recovered on all serial section series hindering their presence in some pictures. In a first attempt to integrate ISH and tissue-specific transcriptomic results, the initial selection of genes belonging to the Class III peroxidase multigenic family (AtPRXs; Supplementary Table S2) were organized within Fig. 2 and among Supplementary Figs. S4–S27, following decreasing maximum individual tissue-specific transcriptomic expression values. In a first screening with the 16 most highly expressed AtPRXs, ISH signals overall confirmed tissue-specific transcriptomic data for 8 AtPRXs (AtPRX42, 12, 50, 51, 03, 36, 46, 17; Fig. 2a–d,f,g; Supplementary Figs. S4–S7 and S11–S14). For AtPRX42, 51, 03, ISH provided new signals not detected by tissue-specific transcriptomics in young embryo and peripheral endosperm tissues (Fig. 2d; Supplementary Figs. S4, S7 and S11). For 3 other AtPRXs (AtPRX55, 22, 23), ISH was fully complementary to tissue-specific transcriptomics since all ISH signals were not detected by tissue-specific transcriptomic data (Supplementary Figs. S10, S16 and S17). AtPRX12 and AtPRX36 highly specific spatiotemporal ISH profiles were fully consistent with tissue-specific transcriptomic data, and interestingly, the resolution reached by ISH allowed to demonstrate the AtPRX12 expression in all endosperm cells and the restriction of AtPRX36 expression to the outermost cell layer of the 5-layered teguments used for laser capture microdissection (Fig. 2b,f; Supplementary Figs. S5 and S12). We acknowledged that tissue-specific transcriptomics is more sensitive than ISH in chalazal tissues and mature seed coat (Fig. 2a,e,g,h; Supplementary Figs. S4–S11 and S13–S19). Neither the 6 lowly expressed AtPRXs nor the 2 AtPRXs without spot ID gave positive ISH signal (Fig. 2h–j; Supplementary Figs. S20–S27). To better understand these detection limits, we integrated ISH and tissue-specific transcriptomic results for the 24 AtPRXs studied in a single table (Supplementary Table S4). An overall gradient of ISH-positive genes towards ISH-negative genes followed the ranking of decreasing maximum transcriptomic expression values. However, no clear single cut-off between undetected and detected AtPRX gene expression could be highlighted, indicating that the ISH sensitivity was dependent on spatiotemporal parameters.
To decipher how spatiotemporality influences sensitivity, we analysed 15 additional non-peroxidase genes with medium to very-high maximum tissue-specific transcriptomic expression values (Supplementary Table S5). We detected ISH signal in agreement with transcriptomic data for the 14 most expressed genes (Fig. 2k–t; Supplementary Figs. S28–S42; Supplementary Table S5). All together, this illustrated how both techniques were complementary since ISH gave new signals not detected through tissue-specific transcriptomics, mostly illustrated in the younger developmental stages for AtPRX51, AtPRX22, Cupin, PAP85, EXT3, SBT1.7, RBOHF, AGP31 and DUF642; Fig. 2d,n,o,q; Supplementary Figs. S7, S16, S28, S29, S32, S34, S38, S39 and S41; Supplementary Table S5); whereas tissue-specific transcriptomics was more powerful than ISH for 14 AtPRXs, Cupin, EXT3, LAC15, SBT1.7, DIR12, SCPL20, LTP1, RBOHF, TUB4, mostly illustrated in chalazal tissues at young stages and in seed coat at mature green stages (Fig. 2a,c,d,k; Supplementary Figs. S4, S6, S7, S9–S11, S14–S21, S28, S32–S38 and S40; Supplementary Table S5). The scarcity of ISH signal for preglobular stage could be explained by its underrepresentation on the tissue-arrays.
To propose final simplified integrated models taking into account the high-throughput tissue-specific transcriptomic and medium-throughput ISH data, we sorted, for each tissue/developmental stage, the 23,933 genes available in Supplementary Table S1 according to several ranges of their individual expression value and plotted the summarized colour-coded ISH results for the 39 genes on the top of the resulting histograms (Fig. 3; Supplementary Figs. S46–S51). This highlighted how ISH signal detection thresholds primarily relied on spatiotemporal parameters and allowed to estimate that the number of genes putatively detectable with ISH ranged from hundreds to putatively the whole genome depending on the tissue/developmental stage considered. For example, from the 23,933 genes present on the array, 55–65% were below the tissue-specific transcriptomic detection limit (value of 45), depending on the tissue/developmental stage (Fig. 3; Supplementary Figs. S46–S51). Interestingly, ISH allowed signal detection for several of those genes in tissues such as the embryo at all developmental stages (e.g. 7 out of 30 genes in the embryo at the globular stage) giving a chance of ISH success for the whole genome in these tissues/developmental stages (Fig. 3a,c; Supplementary Figs. S46–S51). On the other hand, ISH sensitivity within the seed coat tissues decreased along the seed development kinetics and therefore tissue-specific transcriptomics became more sensitive (Fig. 3b,d; Supplementary Figs. S46–S51). Increased ISH sensitivity could be theoretically obtained for recalcitrant tissues with branched DNA ISH44 but these techniques remain expensive prohibiting their application to medium-throughput.
Medium-throughput in situ hybridization spatial resolution and specificity
Beyond the primary interest of our study which is to demonstrate the complementarity of both datasets, we particularly want to emphasise additional advantages of this medium-throughput ISH protocol not achievable through tissue-specific transcriptomics. The first one is the possibility to extend the study to several serial sections for a given gene. For example, the spatiotemporal specific expression of AtPRX36 in the outermost cell layer of the 5-layered teguments (Fig. 2f; Supplementary Fig. S12; Supplementary Tables S4 and S5) can be refined with a tomographical study allowing some spatial modelization (Supplementary Fig. S43). The second point concerns the commonly underestimated specificity of ISH. AtPRX50/51, AtPRX22/23 and LTP1/LTP2, three pairs of tandem duplicated genes with high identity level (82–92% nucleotide identity all along the sequence; Supplementary Table S8) corresponded to ambiguous tissue-specific transcriptomic spot IDs, but gave different -and thus specific- ISH signals (Fig. 2c,d; Supplementary Figs. S6, S7, S16, S17,S31, S37 and S44; Supplementary Tables S4 and S5). The fact that AtPRX50/51 presented the highest nucleotide identity among the 39 studied genes (92.4%; Supplementary Table S8) contributes demonstrating the ISH specificity for the 37 other genes studied. This also indicates that signal specificity in future ISH studies would have to be demonstrated for the 4,176 genes whose second best hit is above this value, and would be secured for all the other genes (Supplementary Table S7). In this study we used sense riboprobe as traditional negative controls. In most cases, we observed no significant background reinforcing corresponding labelling with antisense probes whenever present (Supplementary Figs. S4–S42). In some circumstances, we observed faint signals with both antisense and sense riboprobes mostly in young embryo epidermis (Supplementary Figs. S11, S13, S14, S21, S26 and S38). We arbitrarily chose to classify these signals as background and to not consider corresponding labelling with antisense probes as specific signals. However, it has to be noticed that some residual in situ labelling with sense probes could also have some physiological significance since natural antisense RNA, mostly corresponding to lowly expressed genes, are growingly being discovered, e.g. in plant or human tissues45,46. More dedicated and careful studies would be necessary to fully consider the significance of the observed faint labelling with these few sense probes. Two other ways to further demonstrate the signal specificity are illustrated in the case of AtPRX36 by testing both the loss of ISH signal in a knockout mutant previously characterized and by applying competitive inhibition between AtPRX36 labelled antisense probes and AtPRX36 or AtPRX72 (82.2% nucleotide identity; Supplementary Table S8) unlabelled antisense probes (Supplementary Fig. S45).
Biological significance of ISH results
Literature also supports the specificity and validity of ISH results. The ISH expression profiles and previously published localization and/or phenotyping are in good agreement for some of the studied genes. AtPRX36 specific expression in the outermost cell layer of seed coat [also called mucilage secretory cells (MSCs)47] at linear cotyledon stage (Supplementary Figs. S12, S43 and S45) is in good agreement with recently published GFP imaging studies under the control of AtPRX36 promoter and the corresponding phenotype of delayed mucilage release33). SBT1.7 expression in the MSCs from heart to bending cotyledon stages (Supplementary Fig. S34) can be also related to promoter-reporter gene and immunolabelling profiles in this cell layer and the related phenotype of delayed mucilage release48. Expression of SBT1.7 in additional tissues such as the embryo argues for additional roles to be uncovered for this protein. DIR12/DP1 expression in the MSCs detected by ISH (Supplementary Fig. S35) is correlated to the promoter activity49. Contrary to AtPRX36 and SBT1.7, the DIR12/DP1 expression profile does not correspond to an obvious mucilage release phenotype50 and is rather to be linked to a secondary metabolism phenotype51. LAC15 expression was detected in the second cell layer underneath MSC (Supplementary Fig. S33) in agreement with LAC15 promoter activity and a role in oxidative polymerization of flavonoids in this cell layer52. The strong ISH signal detected in young developing embryo for EXT3, (Supplementary Fig. S32) and not detected by transcriptomics can be directly related to the role of EXT3 in structuring the cell plate during initial cell divisions of the embryo leading to a lethal phenotype in a KO mutant53,54. For all other genes, ISH results constitute a refined spatiotemporal information that is crucial for future functional study of their role.
Concluding Remarks
Finally, our study allowed
To provide a highly detailed ISH protocol explaining the means allowing to reach medium-throughput, to acknowledge its relative sensitivity and to demonstrate its high specificity.
To confirm numerous expression profiles obtained by the tissue-specific transcriptomic approach validating the ISH protocol.
To complement the transcriptomic data by studying genes having no probe set IDs, corresponding to ambiguous probe set IDs or by detecting ISH signals not revealed by tissue-specific transcriptomics.
To draw new expression maps with spatially -and temporally- refined expression profiles that will allow to write new hypotheses for functional studies of the cell wall protein candidate genes.
To integrate both datasets to estimate spatiotemporally-specific thresholds of tissue-specific transcriptomic signal intensity putatively compatible with the ISH protocol for hundreds to tens-of-thousands genes.
We anticipate that this method should be more widely applied in parallel to any tissue-specific transcriptomic studies, contributing to the functional study of numerous genes along the development of various plant or animal models. However, this method is not restricted to model organisms with available transcriptomic facilities since the complementarity of RNAseq large datasets with ISH approach have been recently illustrated for dozens of genes involved in the anticancer alkaloid biosynthetic pathway from the Madagascar periwinkle non model plant55,56,57.
Additional Information
How to cite this article: Francoz, E. et al. Complementarity of medium-throughput in situ RNA hybridization and tissue-specific transcriptomics: case study of Arabidopsis seed development kinetics. Sci. Rep. 6, 24644; doi: 10.1038/srep24644 (2016).
Supplementary Material
Acknowledgments
Université Paul Sabatier Toulouse 3 and Centre National de la Recherche Scientifique (CNRS) granted this work. We thank Alain Jauneau Jean-Philippe Galaud and Elisabeth Jamet for discussions, and Cécile Pouzet and Yves Martinez for technical supports. This work has been done at LRSV, part of the “Laboratoire d’Excellence” (LABEX) entitled TULIP (ANR -10-LABX-41).
Footnotes
Author Contributions E.F. designed and performed most experiments, analysed images and built all ISH figures. P.R. performed part of cloning and second best hit analysis (Supplementary Tables 5–7). C.P. and V.P. performed cloning, probe synthesis and assisted in set up of ISH protocol. A.L.R. assisted with slide scanning. C.D. and V.B. directed research. V.B. designed experiments, set up the method, analysed the results and wrote the manuscript.
References
- Munoz J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 31, 3079–3091, doi: 10.1038/emboj.2012.166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960, doi: 10.1126/science.1090022 (2003). [DOI] [PubMed] [Google Scholar]
- Yadav R. K., Girke T., Pasala S., Xie M. T. & Reddy V. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106, 4941–4946, doi: 10.1073/pnas.0900843106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R. B. & Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 6, 56–68, doi: 10.1038/nprot.2010.175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., McMillan I., Norris M. H. & Hoang T. T. Single prokaryotic cell isolation and total transcript amplification protocol for transcriptomic analysis. Nat. Protoc. 10, 974–984, doi: 10.1038/nprot.2015.058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W. S. et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol. Genomics 28, 311–322, doi: 10.1152/physiolgenomics.00208.2006 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte M. F. et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 110, E435–E444, doi: 10.1073/pnas.1222061110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B. J. W. et al. Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol. 163, 205–215, doi: 10.1104/pp.113.223511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan D. et al. Transcriptome atlas of the Arabidopsis funiculus - a study of maternal seed subregions. Plant J. 82, 41–53, doi: 10.1111/tpj.12790 (2015). [DOI] [PubMed] [Google Scholar]
- Le B. H. et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107, 8063–8070, doi: 10.1073/pnas.1003530107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R. et al. Expression Atlas update-a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 42, D926–D932, doi: 10.1093/nar/gkt1270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D. et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLos One 2, e718, doi: 10.1371/journal.pone.0000718 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L. & Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632, doi: 10.1104/pp.104.046367 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejatko J. et al. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1, 1939–1946, doi: 10.1038/nprot.2006.333 (2006). [DOI] [PubMed] [Google Scholar]
- Thisse C. & Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69, doi: 10.1038/nprot.2007.514 (2008). [DOI] [PubMed] [Google Scholar]
- Brewer P. B., Heisler M. G., Hejatko J., Friml J. & Benkova E. In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat. Protoc. 1, 1462–1467, doi: 10.1038/nprot.2006.226 (2006). [DOI] [PubMed] [Google Scholar]
- Capodieci P. et al. Gene expression profiling in single cells within tissue. Nat. Methods 2, 663–665, doi: 10.1038/nmeth786 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski C. C. et al. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 95, 9973–9978, doi: 10.1073/pnas.95.17.9973 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Hematy K. & Hofte H. In Annual Review of Plant Biology (ed Merchant S. S.) Vol. 63, 381–407 (Annual Reviews, 2012). [DOI] [PubMed] [Google Scholar]
- Carroll A. & Somerville C. In Annu. Rev. Plant Biol. Vol. 60,165–182 (Annual Reviews, 2009). [DOI] [PubMed] [Google Scholar]
- Mansfield S. D. Solutions for dissolution-engineering cell walls for deconstruction. Curr. Opin. Biotechnol. 20, 286–294, doi: 10.1016/j.copbio.2009.05.001 (2009). [DOI] [PubMed] [Google Scholar]
- Burlat V., Ambert K., Ruel K. & Joseleau J. P. Relationship between the nature of lignin and the morphology of degradation performed by white-rot fungi. Plant Physiol. Biochem. 35, 645–654 (1997). [Google Scholar]
- Hall H. C., Cheung J. & Ellis B. E. Immunoprofiling reveals unique cell-specific patterns of wall epitopes in the expanding Arabidopsis stem. Plant J. 74, 134–147, doi: 10.1111/tpj.12111 (2013). [DOI] [PubMed] [Google Scholar]
- Joseleau J. P. & Ruel K. Study of lignification by noninvasive techniques in growing maize internodes - An investigation by Fourier transform infrared cross-polarization magic angle spinning C-13-nuclear magnetic resonance spectroscopy and immunocytochemical transmission electron microscopy. Plant Physiol. 114, 1123–1133, doi: 10.1104/pp.114.3.1123 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J. D. et al. Distinct cell wall architectures in seed endosperms in representatives of the Brassicaceae and Solanaceae. Plant Physiol. 160, 1551–1566, doi: 10.1104/pp.112.203661 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J. D., Marcus S. E. & Knox J. P. Cell wall biology: Perspectives from cell wall imaging. Mol. Plant. 4, 212–219, doi: 10.1093/mp/ssq075 (2011). [DOI] [PubMed] [Google Scholar]
- Moller I. et al. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 50, 1118–1128, doi: 10.1111/j.1365-313X.2007.03114.x (2007). [DOI] [PubMed] [Google Scholar]
- Musel G. et al. Structure and distribution of lignin in primary and secondary cell walls of maize coleoptiles analyzed by chemical and immunological probes. Planta 201, 146–159, doi: 10.1007/bf01007699 (1997). [DOI] [Google Scholar]
- Albenne C., Canut H. & Jamet E. Plant cell wall proteomics: the leadership of Arabidopsis thaliana. Front. Plant Sci. 4, 111, doi: 10.3389/fpls.2013.00111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douche T. et al. Brachypodium distachyon as a model plant toward improved biofuel crops: Search for secreted proteins involved in biogenesis and disassembly of cell wall polymers. Proteomics 13, 2438–2454, doi: 10.1002/pmic.201200507 (2013). [DOI] [PubMed] [Google Scholar]
- Irshad M., Canut H., Borderies G., Pont-Lezica R. & Jamet E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 8, 94, doi: 10.1186/1471-2229-8-94 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz E. et al. Roles of cell wall peroxidases in plant development. Phytochemistry 112, 15–21, doi: 10.1016/j.phytochem.2014.07.020 (2015). [DOI] [PubMed] [Google Scholar]
- Kunieda T. et al. Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 25, 1355–1367, doi: 10.1105/tpc.113.110072 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rubio M. C., Alassimone J. & Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412, doi: 10.1016/j.cell.2013.02.045 (2013). [DOI] [PubMed] [Google Scholar]
- Fawal N. et al. PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 41, D441–D444, doi: 10.1093/nar/gks1083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koua D. et al. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 37, D261–D266, doi: 10.1093/nar/gkn680 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402, doi: 10.1093/nar/25.17.3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Carninci P., Nishiyama Y., Hayashizaki Y. & Shinozaki K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15, 707–720, doi: 10.1046/j.1365-313x.1998.00237.x (1998). [DOI] [PubMed] [Google Scholar]
- Seki M. et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145, doi: 10.1126/science.1071006 (2002). [DOI] [PubMed] [Google Scholar]
- Burlat V., Oudin A., Courtois M., Rideau M. & St-Pierre B. Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 38, 131–141, doi: 10.1111/j.1365-313X.2004.02030.x (2004). [DOI] [PubMed] [Google Scholar]
- Jackson D. In Molecular Plant Pathology: a Practical Approach (eds Gurr S. J., McPherson M. J. & Bowles D. J.) 163–174 (IRL Press at Oxford University, 1992). [Google Scholar]
- Mahroug S., Courdavault V., Thiersault M., St-Pierre B. & Burlat V. Epidermis is a pivotal site of at least four secondary metabolic pathways in Catharanthus roseus aerial organs. Planta 223, 1191–1200, doi: 10.1007/s00425-005-0167-y (2006). [DOI] [PubMed] [Google Scholar]
- St-Pierre B., Vazquez-Flota F. A. & De Luca V. Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11, 887–900, doi: 10.1105/tpc.11.5.887 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. RNA imaging in situ. Nat. Methods 9, 787–790 (2012). [Google Scholar]
- Wang H. et al. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Research 24, 444–453, doi: 10.1101/gr.165555.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554, doi: 10.1016/j.cell.2015.03.010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz E., Ranocha P., Burlat V. & Dunand C. Arabidopsis seed mucilage secretory cells: regulation and dynamics. Trends Plant Sci. 20, 515–524, doi: 10.1016/j.tplants.2015.04.008 (2015). [DOI] [PubMed] [Google Scholar]
- Rautengarten C. et al. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 54, 466–480, doi: 10.1111/j.1365-313X.2008.03437.x (2008). [DOI] [PubMed] [Google Scholar]
- Esfandiari E. et al. Identification and analysis of an outer-seed-coat-specific promoter from Arabidopsis thaliana. Plant Mol. Biol. 81, 93–104, doi: 10.1007/s11103-012-9984-0 (2013). [DOI] [PubMed] [Google Scholar]
- Ranocha P., Francoz E., Burlat V. & Dunand C. Expression of PRX36, PMEI6 and SBT1.7 is controlled by complex transcription factor regulatory networks for proper seed coat mucilage extrusion. Plant signaling & behavior 9, e977734, doi: 10.4161/15592324.2014.977734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F. et al. AtMetExpress Development: A Phytochemical Atlas of Arabidopsis Development. Plant Physiol. 152, 566–578, doi: 10.1104/pp.109.148031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L. et al. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17, 2966–2980, doi: 10.1105/tpc.105.035154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Q. & Cannon M. C. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14, 1161–1172, doi: 10.1105/tpc.010477 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M. C. et al. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 105, 2226–2231, doi: 10.1073/pnas.0711980105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K. et al. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 5, 3606, doi: 10.1038/ncomms4606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courdavault V. et al. A look inside an alkaloid multisite plant: the Catharanthus logistics. Curr. Opin. Plant Biol. 19, 43–50, doi: 10.1016/j.pbi.2014.03.010 (2014). [DOI] [PubMed] [Google Scholar]
- Geu-Flores F. et al. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492, 138–142, doi: 10.1038/nature11692 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.