Abstract
Achromobacter xylosoxidans, an opportunistic pathogen, is responsible for various nosocomial and community-acquired infections. We isolated phiAxp-3, an N4-like bacteriophage that infects A. xylosoxidans, from hospital waste and studied its genomic and biological properties. Transmission electron microscopy revealed that, with a 67-nm diameter icosahedral head and a 20-nm non-contractile tail, phiAxp-3 has features characteristic of Podoviridae bacteriophages (order Caudovirales). With a burst size of 9000 plaque-forming units and a latent period of 80 min, phiAxp-3 had a host range limited to only four A. xylosoxidans strains of the 35 strains that were tested. The 72,825 bp phiAxp-3 DNA genome, with 416-bp terminal redundant ends, contains 80 predicted open reading frames, none of which are related to virulence or drug resistance. Genome sequence comparisons place phiAxp-3 more closely with JWAlpha and JWDelta Achromobacter phages than with other N4 viruses. Using proteomics, we identified 25 viral proteins from purified phiAxp-3 particles. Notably, investigation of the phage phiAxp-3 receptor on the surface of the host cell revealed that lipopolysaccharide serves as the receptor for the adsorption of phage phiAxp-3. Our findings advance current knowledge about A. xylosoxidans phages in an age where alternative therapies to combat antibiotic-resistant bacteria are urgently needed.
Achromobacter xylosoxidans is a medically important opportunistic pathogen frequently associated with various nosocomial and community-acquired infections1. Infections caused by A. xylosoxidans result in significant morbidity and mortality in debilitated individuals2. A. xylosoxidans, an emerging and major pathogen that attacks people with cystic fibrosis3, also has the potential to cause serious infections in premature babies4. One of the main threats from A. xylosoxidans is the high rate of resistance it has to antibiotics. This species exhibits innate resistance to many antibiotic types, including cephalosporins (except ceftazidime), aztreonam, and aminoglycosides5. In the last few years, numerous cases of multidrug-resistant A. xylosoxidans infections have been documented in immunocompromised and cystic fibrosis patients6, and this has complicated the treatment of such infections. Little is known about the optimal therapy for A. xylosoxidans. In addition to the known intrinsic antibiotic resistance patterns in A. xylosoxidans, acquired resistance is also widely reported for this bacterium7. One potential option to combat A. xylosoxidans is use of bacteriophages8. Biocontrol using phages can be applied through food, agriculture, and medical fields9. Phages have higher bacterial specificity than antibiotics and have the advantage of minimal impact on commensal bacteria in the host10. Accordingly, phages that specifically target A. xylosoxidans may be a good choice for the control of A. xylosoxidans infections, especially for antibiotic-resistant A. xylosoxidans since avoiding an antibiotic treatment would avoid the spread of multiresistant bacteria11. Additionally, phages play an important role in bacterial evolution and microbial ecology12. The genes and activities of phages are suggested to be a driving force in maintaining genetic diversity of the bacterial community13. To date, however, only a few A. xylosoxidans-specific phages have been studied in detail14, there are only three reported fully sequenced A. xylosoxidans phages, including phiAxp-1 (GenBank accession number KP313532)15, JWAlpha (KF787095)14 and JWDelta (KF787094)14. Thus, isolating and characterizing new A. xylosoxidans phages is an essential prerequisite for developing efficient biocontrol agents against A. xylosoxidans. Herein, we isolated and genome sequenced a virulent A. xylosoxidans bacteriophage (phiAxp-3) of the Podoviridae family and identified its receptor. We also investigated the effect of various physicochemical treatments on phage stability.
Results and Discussion
Morphology and host range
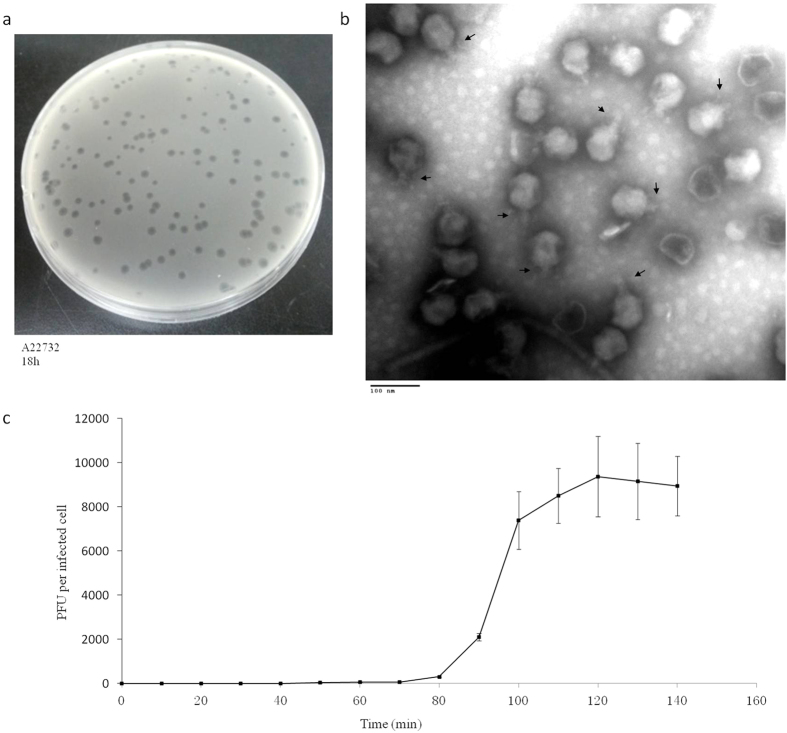
Phage phiAxp-3 was isolated from raw hospital sewage in China, using the A. xylosoxidans A22732 strain as the host; this bacterium produces OXA-114e and IMP-1 carbapenemases, which confer resistance to multiple β-lactam antibiotics including carbapenems16. Phage phiAxp-3 formed round plaques with transparent centres on double-layer plates (Fig. 1a). Transmission electron microscopy of the phiAxp-3 particles showed that phiAxp-3 possesses an isometric head with a diameter of about 67 nm and a short tail with an approximate length of 20 nm (Fig. 1b), thereby matching the typical morphological features of Podoviridae family viruses. Host range testing suggested that phiAxp-3 was able to successfully infect all A. xylosoxidans strains tested, unlike other species that were tested (Table 1). Besides the A22732 strain, which is reported to be multidrug-resistant16, all three of the other clinical A. xylosoxidans strains investigated here have been shown to be resistant to aztreonam and tobramycin15.
Figure 1. Isolated Achromobacter phage phiAxp-3.
(a) Plaque morphology of phage phiAxp-3. (b) Transmission electron micrographs of phiAxp-3. Arrows indicate the short noncontractile tails. Phage particles were negatively stained with 2% phosphotungstic acid. Scale bar, 100 nm. (c) One-step growth curves for phiAxp-3 with A. xylosoxidans strain A22732. Plaque-forming units per ml of A22732 culture at different time points. Each time point represents the mean value of three experiments.
Table 1. Host range infection of the phage phiAxp-3. −absent; +present.
| Species | ID | Infection |
|---|---|---|
| Achromobacter xylosoxidans | A22732 | + |
| A. xylosoxidans | 5271 | + |
| A. xylosoxidans | 844 | + |
| A. xylosoxidans | 6065 | + |
| Enterobacter aerogenes | 3-SP | − |
| E. aerogenes | 201316724 | − |
| E. aerogenes | 2015-301 | − |
| E. aerogenes | 13208 | − |
| E. aerogenes | A29864 | − |
| E. aerogenes | A36179 | − |
| Escherichia coli | ATCC 25922 | − |
| E. coli | DH10B | − |
| E. coli | EC600 | − |
| Klebsiella pneumoniae | ATCC BAA-1706 | − |
| K. pneumoniae | ATCC BAA-2146 | − |
| K. pneumoniae | ATCC BAA-1705 | − |
| K. pneumoniae | K2044 | − |
| K. pneumoniae | 511 | − |
| Serratia marcescens | wk2050 | − |
| S. marcescens | 201315732 | − |
| S. marcescens | wj-1 | − |
| S. marcescens | wj-2 | − |
| S. marcescens | wj-3 | − |
| E. cloacae | T5282 | − |
| E. cloacae | TI3 | − |
| E. sakazakii | 45401 | − |
| E. sakazakii | 45402 | − |
| Leclercia adcarboxglata | P10164 | − |
| Raoultella ornithinolytica | YNKP001 | − |
| Stenotrophomonas maltophilia | 9665 | − |
| Citrobacter freundii | P10159 | − |
| Vibrio parahaemolyticus | J5421 | − |
| Pseudomonas aeruginosa | PA01 | − |
| Acinetobacter baumannii | N1 | − |
| Shigella sonnei | #1083 | − |
One-step growth curve
We performed a one-step growth curve experiment for phiAxp-3 to determine its latent time period and phage burst size. Burst size and latent period in phages are influenced by the host, the composition of the growth medium, the incubation temperature and the specific growth rate17. The latent period for phiAxp-3 was about 80 min, after which there was a gradual increase in the number of viral particles released (Fig. 1c). It took about 40 min for the phages to reach the growth plateau phase and this resulted in burst sizes of ca. 9000 plaque-forming units (PFU) per infected cell.
Phage stability
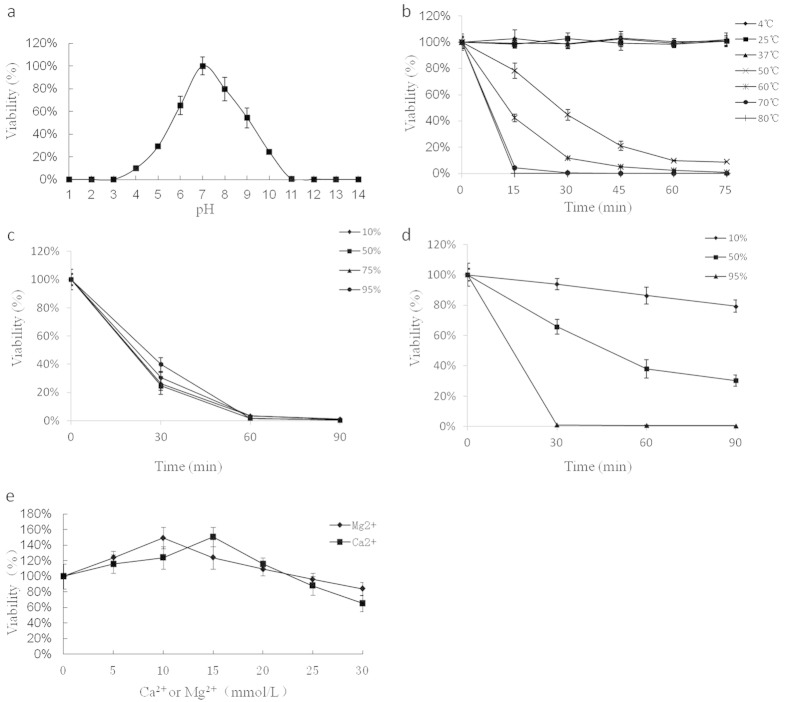
Figure 2a shows the pH sensitivity of phage phiAxp-3. The phage titres decreased to different extents when the pH was above or below 7. At pH 4 and pH 10, reductions of 90.25% and 75.76% in phage particle counts were observed, respectively. Almost no viral particles were detected at pH 1 and pH 14. The viability loss when phiAxp-3 was subjected to temperatures of 25 °C, 37 °C, 50 °C, 60 °C, 70 °C and 80 °C is shown in Fig. 2b. A control at temperature of 4 °C was also included. The phage titres reduced dramatically at 50 °C, 60 °C, 70 °C and 80 °C. After 75 min at 50 °C, the phage titre reduced by 91.2%. At 80 °C, a 99.86% reduction in viral particles was recorded after 15 min, and compared with the control, after 75 min only 0.0002% of the viral particles were detected. Scarcely any reduction in the phage titres were observed at 4 °C, 25 °C and 37 °C after 75 min of treatment. The survivor curves for phiAxp-3 in different biocides are shown in Fig. 2c,d. The results show that the presence of ethanol at low (10%) and high (95%) concentrations reduced the phage titres (Fig. 2c). The phage titres reduced by 20.75%, 69.76% and 99.62% after 75 min of treatment with isopropanol at 10%, 50% and 95%, respectively (Fig. 2d). Divalent ions such as Ca2+ or Mg2+ are necessary for phage attachment and intracellular growth18. phiAxp-3 showed divalent cation dependency for plaque development, but the concentration of Ca2+ or Mg2+ had to be less than or equal to 20 mM (Fig. 2e).
Figure 2. Resistance of phage phiAxp-3 to physical and chemical agents.
(a) The effect of pH on the adsorption of phage phiAxp-3 to A. xylosoxidans A22732 in LB broth. (b) Inactivation kinetics of phage phiAxp-3 at 4 °C, 25 °C, 37 °C, 50 °C, 60 °C, 70 °C and 80 °C. (c) Inactivation kinetics of phage phiAxp-3 in the presence of 10%, 50%, 75% and 95% ethanol. (d) Inactivation kinetics of phage phiAxp-3 in the presence of 10%, 50% and 95% isopropanol. (e) Effect on phage phiAxp-3 titre of incubation in LB broth with and without CaCl2 or MgCl2 (0, 5, 10, 15, 20, 25 and 30 mmol/l) at 37 °C. For all the graphs, the values represent the mean of three determinations.
Genomic features of bacteriophage phiAxp-3
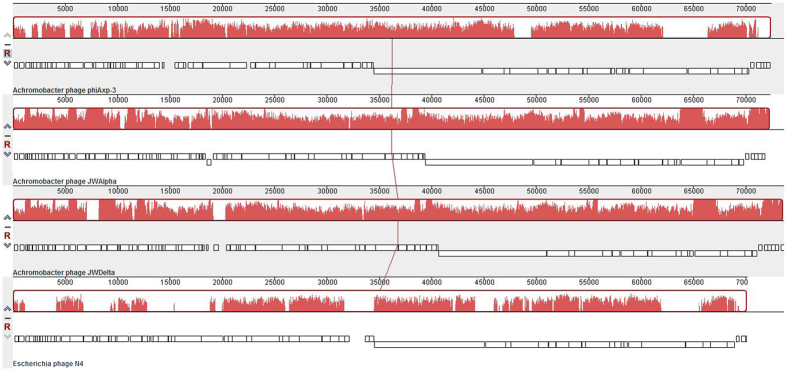
Analysis of a bacteriophage’s genome is an important preliminary step towards the development of phage therapy19. Whole-genome sequencing and assembling of the phiAxp-3 genome generated a circular molecule of 72,409 bp in size. The assembly was terminally permuted but not redundant after the original sequencing was completed. An initial whole genome Basic Local Alignment Search Tool (BLAST) analysis of phiAxp-3 against the National Center for Biotechnology Information database and multiple genome alignments showed that phiAxp-3 is related to two N4-like viruses (i.e., JWAlpha and JWDelta), indicating that phiAxp-3 is an N4-like phage (Fig. 3). It is well-known that N4-like phages have linear genomes and terminal repeats, but the terminal repeats are usually not identical20. Additionally, it is important to verify experimentally the ends of the phage genome rather than relying on genome assembly programs21. Therefore, to determine whether the phiAxp-3 genome is linear or circularly permuted and whether the ends are fixed or variable, restriction enzyme analyses were performed. BlpI restriction enzyme digestion of phiAxp-3 DNA produced two distinct fragments, thereby indicating the presence of a single recognition site in the viral DNA, and four distinct fragments when cut with EagI, thereby indicating the presence of three sites in the viral DNA (Supplementary Figure 1a). These findings clearly indicate that phiAxp-3 has a fixed linear genome structure without circular permutation. Terminal restriction enzyme fragments and primer walking experiments were used to determine the sequence of the phiAxp-3 genomic ends. The 5′ genomic end was predicted to be contained within a 3.5 kb BlpI fragment (Supplementary Figure 1b); hence, the gel-purified BlpI fragment was used as a template for sequencing reactions. Primer (P1), which was designed to read off the 5′ end of the genome, exhibited a detectable drop in signal intensity, indicating that the likely 5′ genome end had been reached. Sequencing from the predicted 3′ end, using the 3′ 5.4 kb EagI fragment and primer P2 (Supplementary Figure 1b), produced the repeat region at the 3′ end. Therefore, the phiAxp-3 genome has direct terminal repeats of 416 bp and possesses no cohesive ends; this result is consistent with those described previously for JWAlpha and JWDelta phages14. An alignment of the direct terminal repeats of phiAxp-3 is very similar to those for JWAlpha and JWDelta (Supplementary Figure 2). The terminal repeats in phiAxp-3 are 51 bp longer than that of JWAlpha (365 bp) and 4 bp shorter than that of JWDelta (420 bp). When the non-consecutive indels were removed, we found that they shared about 77% identity with those of JWAlpha and JWDelta. The additional 416-bp repeat means that the genome is 72,825-bp-long (GC content, 55.2%), rather than 72,409-bp-long (Supplementary Figure 1c).
Figure 3. Multiple genome alignment generated by Mauve software (http://asap.ahabs.wisc.edu/mauve/), and the chromosomes of Achromobacter phages phiAxp-3, JWAlpha and JWDelta and the Enterobacter phage, N4.
Genome similarity is represented by the height of the bars, which correspond to the average level of conservation in that region of the genome sequence. Completely white regions represent fragments that were not aligned or contained sequence elements specific to a particular genome.
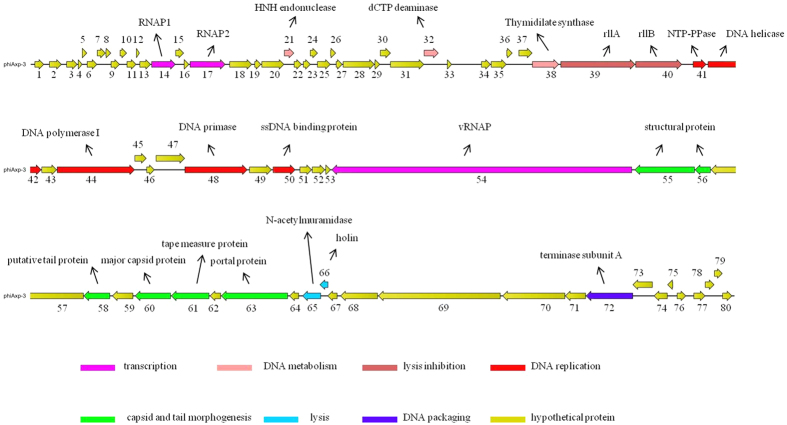
The order and arrangement of the open reading frames of the revised genome are the same as the previously sequenced version and were not affected by the reorganisation of the terminal regions of the genome. A total of 80 protein coding genes were predicted in the genome and ranged from 120 to 10,287 bp, 22 of which are leftward oriented while the others are rightward oriented (Fig. 4). N4-like phages are a class of virulent Podoviridae phages and members of this group are lytic against their hosts22. The phiAxp-3 genome sequence shares 51.6% and 50.4% nucleotide identity with JWAlpha and JWDelta, respectively. The three phages were isolated from samples obtained from two locations that are geographically far apart (phiAxp-3 was isolated in China, JWAlpha and JWDelta were isolated in Germany)14. For comparison, phiAxp-3 shares 40.8% nucleotide identity with phage N4. JWAlpha and JWDelta share 96.6% nucleotide sequence identity. The overall architecture of N4 is shared among all phages of this group. Based on our analysis, the annotated proteins of phiAxp-3 can be categorised into the following functional groups: Transcription (RNA polymerase; RNAP1, RNAP2, vRNAP), DNA metabolism (HNH endonuclease, dCTP deaminase, thymidylate synthase), lysis inhibition (rllA, rllB), DNA replication (NTP-PPase, DNA helicase, DNA polymerase, DNA primase, ssDNA-binding protein), virion morphogenesis (structural proteins, tail protein, major capsid protein, tape measure protein, portal protein), host lysis (N-acetylmuramidase, holin) and DNA packaging (large terminase subunit) (Fig. 4). No tRNA was identified in the phiAxp-3 genome; this indicates that upon entry into the host, the phage is completely reliant on the host tRNA for its protein synthesis. Table 2 shows a detailed comparison of phiAxp-3, N4, and JWAlpha and JWDelta proteins. In our analyses, 25 phage proteins were detected using LC/ESI/MS/MS, of which 10 had annotated functions (Table 3).
Figure 4. Genome map of phage phiAxp-3.
Representation of the open reading frame (ORF) (ORF 1 to 80) organisation of phage phiAxp-3. The predicted genes are indicated as arrows. Blue arrows, DNA regulation module; purple arrows, packaging module; yellow arrows, phage structural proteins; red arrows, host lysis proteins; green arrows, lysis/lysogeny module; black arrows, hypothetical proteins.
Table 2. phiAxp-3 gene annotations.
| ORFs | Start | End | Strand | Length (aa)a | Vs |
Function | Conserved Protein Domain Family | ||
|---|---|---|---|---|---|---|---|---|---|
| JWAlpha | JWDelta | N4 | |||||||
| ORF01 | 141 | 461 | + | 106 | 73 | 73 | 51 | ||
| ORF02 | 640 | 1065 | + | 141 | 67 | 67 | 43 | ||
| ORF03 | 1229 | 1606 | + | 125 | |||||
| ORF04 | 1628 | 1774 | + | 48 | |||||
| ORF05 | 1767 | 1937 | + | 56 | |||||
| ORF06 | 1934 | 2284 | + | 116 | 41 | 41 | |||
| ORF07 | 2281 | 2571 | + | 96 | 28 | 28 | |||
| ORF08 | 2574 | 2759 | + | 61 | |||||
| ORF09 | 2752 | 3057 | + | 101 | 72 | ||||
| ORF10 | 3061 | 3300 | + | 79 | 50 | 50 | |||
| ORF11 | 3297 | 3623 | + | 108 | 55 | ||||
| ORF12 | 3620 | 3739 | + | 39 | |||||
| ORF13 | 3736 | 4119 | + | 127 | 71 | 73 | 55 | ||
| ORF14 | 4142 | 4969 | + | 275 | 80 | 80 | 62 | RNA polymerase 1 | PHA00452; COG5108 |
| ORF15 | 4966 | 5253 | + | 95 | |||||
| ORF16 | 5256 | 5435 | + | 59 | |||||
| ORF17 | 5462 | 6676 | + | 404 | 83 | 55 | RNA polymerase 2 | pfam00940; PHA00452 | |
| ORF18 | 6814 | 7587 | + | 257 | 49 | 41 | |||
| ORF19 | 7674 | 7901 | + | 75 | 54 | 45 | |||
| ORF20 | 7916 | 8689 | + | 257 | 42 | 44 | |||
| ORF21 | 8689 | 9027 | + | 112 | 79 | 80 | 64 | HNH endonuclease | pfam13392 |
| ORF22 | 9024 | 9290 | + | 88 | |||||
| ORF23 | 9340 | 9594 | + | 84 | 44 | 44 | |||
| ORF24 | 9578 | 9841 | + | 87 | 74 | 76 | |||
| ORF25 | 9823 | 10278 | + | 151 | 66 | 77 | |||
| ORF26 | 10275 | 10463 | + | 62 | 64 | ||||
| ORF27 | 10465 | 10686 | + | 73 | 39 | 40 | |||
| ORF28 | 10704 | 11786 | + | 360 | 73 | 73 | 51 | ||
| ORF29 | 11789 | 11974 | + | 61 | |||||
| ORF30 | 11978 | 12340 | + | 120 | 46 | 46 | |||
| ORF31 | 12325 | 13482 | + | 385 | 80 | 80 | 57 | ||
| ORF32 | 13479 | 13976 | + | 165 | 74 | 75 | 58 | dCTP deaminase | cd07557; COG0717; PRK00416; TIGR02274; PHA01707 |
| ORF33 | 14274 | 14435 | + | 53 | 53 | 53 | |||
| ORF34 | 15442 | 15765 | + | 107 | 70 | 69 | |||
| ORF35 | 15776 | 16309 | + | 177 | 24 | 27 | |||
| ORF36 | 16309 | 16509 | + | 66 | 47 | ||||
| ORF37 | 16724 | 17191 | + | 155 | 67 | ||||
| ORF38 | 17188 | 18099 | + | 303 | 78 | 54 | Thymidilate synthase | pfam02511; TIGR02170; COG1351; PRK00847 | |
| ORF39 | 18162 | 20717 | + | 851 | 61 | 61 | 28 | rIIAlike protein | cd00075; pfam13589; smart00387; COG1389; PRK04184; TIGR01052 |
| ORF40 | 20727 | 22319 | + | 530 | 69 | 68 | 33 | rIIBlike protein | |
| ORF41 | 22695 | 23153 | + | 152 | 87 | 86 | 45 | NTP pyrophosphohydrolase | cd11530; COG4696; pfam01503 |
| ORF42 | 23208 | 24515 | + | 435 | 79 | 80 | 56 | DNA helicase | pfam13604; pfam13538; pfam13086; COG0507; TIGR01448; PRK13826; PRK10875 |
| ORF43 | 24525 | 25046 | + | 173 | 65 | 65 | 40 | ||
| ORF44 | 25055 | 27715 | + | 886 | 81 | 81 | 63 | DNA polymerase I | pfam00476; cd08637; smart00482; smart00474; COG0749; TIGR00593; PRK05755 |
| ORF45 | 27712 | 28116 | + | 134 | 66 | 66 | 49 | ||
| ORF46 | 28119 | 28394 | + | 91 | 71 | 75 | 59 | ||
| ORF47 | 28440 | 29432 | + | 330 | 82 | 82 | 58 | ||
| ORF48 | 29429 | 31576 | + | 715 | 91 | 91 | 76 | DNA primase | pfam08708; smart00942 |
| ORF49 | 31640 | 32407 | + | 255 | 85 | 85 | 67 | ||
| ORF50 | 32450 | 33217 | + | 255 | 64 | 64 | 49 | ssDNA binding protein | |
| ORF51 | 33368 | 33781 | + | 137 | 83 | 52 | |||
| ORF52 | 33792 | 34235 | + | 147 | 63 | 58 | |||
| ORF53 | 34252 | 34431 | + | 59 | 58 | ||||
| ORF54 | 34464 | 44750 | 3428 | 82 | 82 | 55 | RNA polymerase | PRK10811; COG0810 | |
| ORF55 | 44849 | 46900 | 683 | 70 | 70 | 34 | structural protein | ||
| ORF56 | 46913 | 47446 | 177 | 69 | 68 | structural protein | |||
| ORF57 | 47459 | 50110 | 883 | 54 | 54 | 46 | |||
| ORF58 | 50114 | 51013 | 299 | 77 | 65 | putative tail protein | |||
| ORF59 | 51090 | 51806 | 238 | 68 | 68 | 55 | |||
| ORF60 | 51873 | 53096 | 407 | 94 | 95 | 84 | major capsid protein | TIGR04387 | |
| ORF61 | 53109 | 54413 | 434 | 79 | 53 | tape measure protein | |||
| ORF62 | 54439 | 54813 | 124 | 54 | 39 | ||||
| ORF63 | 54824 | 57106 | 760 | 86 | 87 | 66 | portal protein | ||
| ORF64 | 57162 | 57485 | 107 | ||||||
| ORF65 | 57608 | 58240 | 210 | 90 | 93 | 62 | Nacetylmuramidase | pfam05838; pfam09374; COG3926 | |
| ORF66 | 58203 | 58490 | 95 | 85 | 76 | 31 | putative holin protein | ||
| ORF67 | 58469 | 58804 | 111 | 68 | 63 | 55 | |||
| ORF68 | 58909 | 60186 | 425 | 69 | |||||
| ORF69 | 60206 | 64393 | 1395 | 54 | 160 kDa protein | ||||
| ORF70 | 64462 | 66606 | 714 | 40 | 40 | 39 | |||
| ORF71 | 66610 | 67314 | 234 | 61 | 61 | 54 | |||
| ORF72 | 67322 | 68932 | 536 | 87 | 87 | 76 | terminase subunit A | TIGR01630; pfam03237 | |
| ORF73 | 68925 | 69605 | 226 | 90 | 88 | 65 | |||
| ORF74 | 69649 | 70116 | 155 | 65 | 66 | ||||
| ORF75 | 70113 | 70304 | 63 | ||||||
| ORF76 | 70440 | 70751 | + | 103 | 29 | 29 | 30 | ||
| ORF77 | 71010 | 71411 | + | 133 | |||||
| ORF78 | 71401 | 71718 | + | 105 | |||||
| ORF79 | 71715 | 71999 | + | 94 | 46 | ||||
| ORF80 | 71996 | 72328 | + | 110 | 42 | ||||
aamino acids.
Table 3. Virion proteins detected by LC/ESI/MS/MS.
| Protein ID | Theoretical avg. mass (Da) | Score | Matches | Sequences | Annotated Function |
|---|---|---|---|---|---|
| GI:921956017 | 15700 | 5256 | 107 (103) | 13 (12) | Hypothetical protein |
| GI:921956033 | 26713 | 19132 | 296 (290) | 19 (18) | Hypothetical protein |
| GI:921956035 | 29169 | 1309 | 35 (31) | 14 (14) | Hypothetical protein |
| GI:921956040 | 17465 | 169 | 3 (3) | 3 (3) | Hypothetical protein |
| GI:921956043 | 40735 | 25 | 1 (1) | 1 (1) | Hypothetical protein |
| GI:921956046 | 44368 | 30 | 2 (2) | 2 (2) | Hypothetical protein |
| GI:921956053 | 34122 | 152 | 3 (3) | 3 (3) | Thymidilate synthase |
| GI:921956054 | 96533 | 198 | 7 (5) | 6 (5) | Hypothetical protein |
| GI:921956055 | 57942 | 105 | 4 (3) | 4 (3) | rIIB like protein |
| GI:921956062 | 38520 | 41 | 2 (1) | 2 (1) | Hypothetical protein |
| GI:921956064 | 29341 | 185 | 6 (4) | 5 (3) | Hypothetical protein |
| GI:921956065 | 27758 | 136 | 4 (4) | 4 (4) | ssDNA-binding protein |
| GI:921956069 | 369998 | 3409 | 70 (61) | 47 (41) | virion RNA polymerase |
| GI:921956070 | 72167 | 16474 | 268 (258) | 44 (43) | Structural protein |
| GI:921956071 | 17648 | 1468 | 38 (38) | 6 (6) | Structural protein |
| GI:921956072 | 98259 | 68 | 2 (2) | 2 (2) | Hypothetical protein |
| GI:921956045 | 32808 | 2670 | 53 (53) | 15 (15) | Putative tail protein |
| GI:921956074 | 26168 | 750 | 15 (15) | 9 (9) | Hypothetical protein |
| GI:921956075 | 44531 | 18929 | 285 (274) | 32 (32) | Major capsid protein |
| GI:921956077 | 14174 | 149 | 4 (3) | 4 (3) | Hypothetical protein |
| GI:921956078 | 84941 | 732 | 20 (19) | 5 (5) | Portal protein |
| GI:921956083 | 49043 | 592 | 13 (11) | 10 (8) | Hypothetical protein |
| GI:921956084 | 157480 | 293 | 5 (5) | 5 (5) | 160 kDa protein |
| GI:921956085 | 76176 | 1632 | 33 (30) | 25 (23) | Hypothetical protein |
| GI:921956086 | 26536 | 4608 | 103 (92) | 15 (14) | Hypothetical protein |
Transcription module
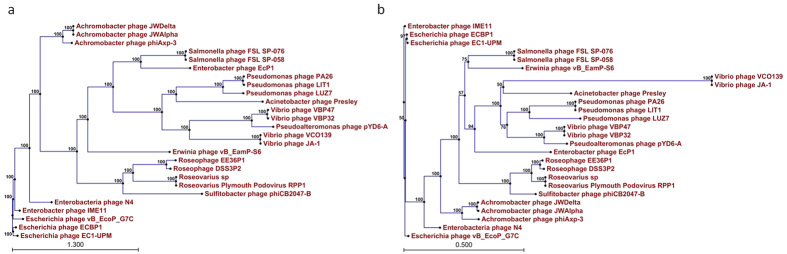
Phage N4 employs at least three genes encoding RNAPs for the transcription of genes in different stages of its life cycle23. The most remarkable and highly conserved signature is the virion RNA polymerase (vRNAP), which is by far the largest protein described among all known phages23. vRNAP is packed into the capsid and is injected into the host cell together with phage DNA, which makes N4 the only known phage that does not depend on host RNAP for transcription of its early genes24. As an N4 like virus, phiAxp-3 also harbours three different RNAPs, suggesting the same transcription strategy as that used by N4. The large 3428 amino acid vRNAP (ORF54), which represents approximately 14% of the whole genome length of phiAxp-3, contains no cysteine residues. The vRNAP of phiAxp-3 shares amino acid 82% sequence identity with JWAlpha and JWDelta. Phylogenetic analysis of vRNAP from different N4 viruses revealed that phiAxp-3, JWAlpha, and JWDelta formed a separate clade from the other N4-like viruses (Fig. 5a). Besides vRNAP, phiAxp-3 possesses two different RNA polymerase subunits for transcription of phage middle genes: RNAP1 (ORF14) and RNAP2 (ORF17). RNAP1 and RNAP2 are transcribed in the opposite direction to vRNAP. In the N4 genome the RNAP1 gene is followed directly by RNAP2, but in the phiAxp-3 genome insertions of two small genes (ORF15 and ORF16) occur between RNAP1 and RNAP2. This situation differs from JWAlpha and JWDelta as they encode two RNAP2 in their genomes14.
Figure 5.
Phylogenetic tree based on the virion RNA polymerase (a) and large terminase subunits (b) of N4-like bacteriophages for which genome sequences are available. The 26 virion RNA polymerase and large terminase subunits were compared using the ClustalW program, and the phylogenetic tree was generated with the neighbour-joining method and 1000 bootstrap replicates (CLC Genomics Workbench 6).
DNA metabolism
phiAxp-3 has three genes encoding proteins involved in nucleotide metabolism: an HNH endonuclease (ORF21), a deoxycytidine triphosphate (dCTP) deaminase (ORF32) and a thymidylate synthase (TS) protein (ORF38). These proteins each play a role in regulating some of the enzymes involved in DNA metabolism or replication14 and are similar to homologous proteins from JWAlpha, JWDelta and N4. Homing endonucleases (HEs) are able to transfer genetic elements from an HE-encoding genome to an HE-lacking recipient to promote gene recombination in phages25. One HE family is the HNH endonucleases, which are small DNA binding and digestion proteins characterised by two histidine residues and an asparagine residue26. The phiAxp-3 HNH endonuclease contains an HNH_3 domain (pfam13392) predicted to possess HNH endonuclease activity. phiAxp-3 also possesses a dCTP deaminase with a conserved dcd (PRK00416) domain. Thymidylate synthase (TS) is essential for production of dTMP and is a key enzyme involved in DNA synthesis and transcriptional regulation in organisms27. phiAxp-3 TS contains a Thy1 (pfam02511) domain and appears to be flavin-dependent. A thioredoxin gene that has been reported in the genomes of JWAlpha and JWDelta is absent from the phiAxp-3 genome.
DNA replication
DNA replication genes are concentrated in a region that stretches from ORF41 to ORF50 in phiAxp-3. They are separated from the structural module by a particularly large vRNAP gene. The DNA helicase (ORF42) present in phiAxp-3 possesses greater similarity to helicases from JWAlpha and JWDelta than the helicase from N4. The same situation also exists for DNA polymerase I (ORF44), DNA primase (ORF48) and the ssDNA binding protein, ssb (ORF50). The phiAxp-3 DNA helicase contains an AAA_30 (pfam13604) domain at the N terminal and a UvrD_C_2 (pfam13538) domain at the C terminal. This DNA helicase can probably be classified within the RecD-like helicase superfamily. The DNA polymerase I present in phiAxp-3 shares 81% similarity with those present in JWAlpha and JWDelta, while Ssb is involved in DNA replication/recombination and host RNA polymerase activation in late N4 transcription23. It has been reported that Ssb from most N4-like viruses is located next to the DNA primase18. However, in the phiAxp-3, JWAlpha, JWDelta and N4 genomes, there is a gene next to the DNA primase that encodes a protein with a similar size to that of the Ssb protein (200–250 amino acids), but this protein shares no amino acid similarity with the Ssb protein.
Virion morphogenesis
Sequence-based predictions identified the following six ORFs involved in virion morphogenesis: two phage structural proteins (ORF55 and 56), phage tail protein (ORF58), major capsid protein (ORF60), tail tape measure protein (ORF61) and portal protein (ORF63). The morphogenesis-related proteins are similar to those found in JWAlpha, JWDelta and N4. Portal proteins, which have molecular masses between 40 and 90 kDa, are not well conserved28. Accordingly, the phiAxp-3 portal protein is 760 amino acid residues in length, which corresponds to an 85 kDa molecular mass. Although tape measure proteins act as scaffolds for assembly of the phage-tail in Myoviridae and Siphoviridae members23, the presence of tape measure proteins in Podoviridae phages is not unusual.
Lysis and lysis inhibition
In the phiAxp-3 genome downstream of the structurally clustered genes involved in cell lysis, we identified two ORFs located contiguously that encode a predicted N-acetylmuramoyl-L-alanine amidase (ORF65) and a putative phage holin (ORF66). These two proteins are required for host cell lysis and the release of new virions at the end of the lytic cycle12. The presence of a lysis gene but no lysogeny-related gene indicates that bacteriophage phiAxp-3 is a lytic bacteriophage. The putative amidase, predicted to be a 210-amino-acid protein, is presumably involved in cleaving the amide bond between N-acetylmuramoyl and the L-amino acid in peptidoglycan12. The predicted holin protein gene encodes a 95-amino acid molecule responsible for controlling the timing of lysis. It was assigned as a class II holin with two transmembrane domains. phiAxp-3 rIIA-like (ORF39) and rIIB-like (ORF40) proteins, which might play roles in lysis inhibition, are located upstream of the replication cluster. These types of protein were first described in phage T4, where the rI gene was found to somehow be able to detect superinfection at any point until just before the normal time of lysis and was also able to delay lysis for several hours29.
DNA packaging
We were only able to identify the large subunit of the terminase (ORF72) used for DNA packaging in phiAxp-3. The large terminase subunit shares high amino acid sequence similarity with JWAlpha and JWDelta, and probably uses the same mechanism for packaging as other N4-like phages. Large terminase protein sequences have been used to construct phylogenies and decipher evolutionary relationships among phages belonging to different families30. Clustering of the amino acid sequences of the large terminase proteins encoded by phiAxp-3 with the other N4-like bacteriophages for which genome sequences are available31, clearly placed phiAxp-3 within the branch of JWAlpha and JWDelta (Fig. 5b).
Host receptor identification
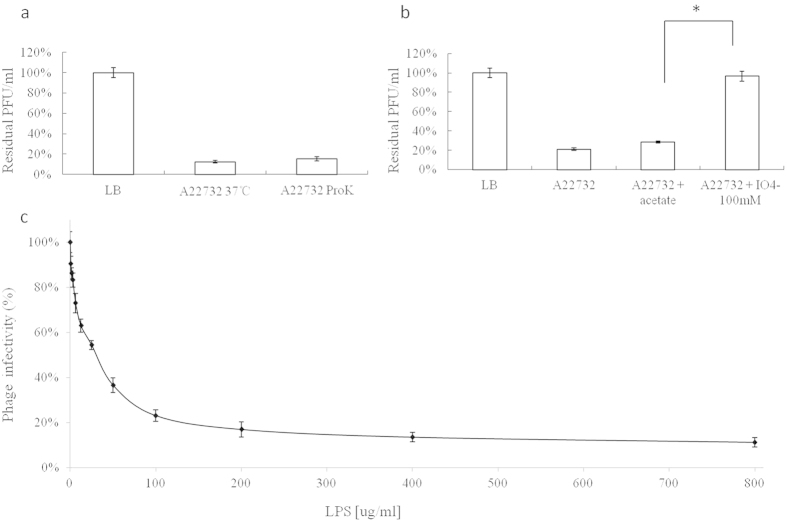
Phage infection is dependent on the presence of an attachment site on the host cell surface and any exposed component of the cell surface can potentially act as a receptor32. As a Gram-negative bacterium, the exposed surface of A. xylosoxidans consists essentially of a complex of lipopolysaccharide (LPS) and proteins32. Thus it is important to determine whether LPS and proteins are recognisable by phages during infection. To identify the host receptor for phiAxp-3, the outer membrane proteins and the carbohydrate structure of the A. xylosoxidans cell surface were destroyed by proteinase K and periodate, respectively (Fig. 6a,b). The results revealed that the absence of carbohydrate structure inhibits phage propagation, suggesting that phiAxp-3 uses the bacterial LPS layer as its specific receptor. The results were confirmed by the phage inactivation assay performed with pure LPS isolated from strain A22732. The experiments revealed a direct correlation between LPS concentration and inhibition of viral particle infectivity (Fig. 6c). LPS at 25 μg per ml was needed to inhibit the activity of 3.2 × 104 pfu phiAxp-3 by 50%, while LPS at 800 μg per ml resulted in 89% inactivation of phiAxp-3.
Figure 6. The effects of various bacterial treatments on phiAxp-3 adsorption to host cells, as determined by residual plaque-forming unit percentages.
(a) Effect of proteinase K treatment on the adsorption of phiAxp-3 to A. xylosoxidans strain A22732. (b) Effect of periodate treatment on the adsorption of phiAxp-3 to A. xylosoxidans strain A22732. The control (LB and “A22732 + acetate”), untreated strain (A22732), and treatment (“A22732 + ProtK” for proteinase K treatment and “A22732+IO4−” for periodate treatment) groups were tested for adsorption as indicated by the x axes. Error bars denote statistical variations. Statistical significance was determined by a Student t test for comparison between the treated and untreated groups. *P 0.05. (c) Inactivation of phage phiAxp-3 by lipopolysaccharide derived from A. xylosoxidans A22732. The percentage infectivity was determined after 1 h of incubation at 37 °C. Error bars denote statistical variations.
Concluding remarks
In this study, we have presented the characteristics of phiAxp-3, a lytic phage that was found to infect clinical isolates of A. xylosoxidans. We propose that phiAxp-3 is assigned to the Caudovirales order (Podoviridae phage family) based on its morphological features and genomic characteristics. Characterisation and analysis of genome structure and gene function are necessary steps before bacteriophages can be approved as therapeutic agents. According to the overall genomic organisation and sequence similarities revealed herein, we suggest that phiAxp-3 is classified as the N4-like phage group. In its 72,825 bp linear DNA genome, phiAxp-3 has fixed ends with direct terminal repeats of 416 bp. Phage phiAxp-3 is genetically related to the N4-like phages JWAlpha and JWDelta, and phylogenetic analysis of its RNAPs and large terminase subunits supports this assignment.
The phage infection process begins with the adsorption of the phage to the bacterial receptor, which is present on the cell surface33. Exploration of the receptors used by phages is essential for understanding the processes underlying phage lysis and for research on phage therapy. Analysis of the phiAxp-3 putative cell wall receptor revealed that phiAxp-3 recognises LPS as its primary receptor for adsorption, thereby accounting for the specificity of its interactions with its host bacterium. Although bacteria can develop resistance to their viral predators, finding new phages that can kill drug-resistant bacteria is not difficult, because phages continually evolve alongside mutated bacteria10. LPS acts as an important virulence factor for A. xylosoxidans34, and receptor mutated strains will be avirulent or attenuated. Furthermore, phage cocktails, containing different types of phages, can effectively prevent bacteria from developing resistance to phages10. Facing the emerging threat from multi-drug resistance A. xylosoxidans, the lytic power of phiAxp-3 combined with its specificity for A. xylosoxidans makes phiAxp-3 an appealing agent for therapeutic or disinfection applications.
Methods
Bacterial strains and growth conditions
All bacterial strains (including the phage indicator strain and the strains used for host range identification) were grown at 37 °C in Luria-Bertani (LB) broth. To isolate and purify the phages, the A. xylosoxidans strain A22732 was used as an indicator strain to reveal the presence of phages in the hospital sewage collected from the Second Artillery General Hospital of Chinese People’s Liberation Army (Beijing, China), using the double agar overlay plaque assay described previously for the isolation of lytic phages35. Plaques picked from agar plates were placed in 5 ml of LB broth and incubated with 0.3 ml of an overnight culture of the host strain. Incubation at 37 °C was performed until lysis of the culture was complete. The host range of the phages were examined using 35 clinical strains of different bacterial species stored at our microorganism centre using standard spot tests36.
Transmission electron microscopy
One drop of purified phiAxp-3 particles was adsorbed to a 230-mesh Formvar/carbon-coated copper grid for several minutes, followed by staining with 2% (wt/vol) phosphotungstic acid (pH 7). Samples were examined with a Philips EM 300 electron microscope operated at 80 kV.
One-step growth curve
Mid-exponential growing cultures of A. xylosoxidans A22732 cells were harvested and suspended in LB broth. Phages were added at a multiplicity of infection of 0.1. At 10-min intervals over 140 min, aliquots from each dilution were collected for phage counts37. Latent period, burst time and burst size were calculated from the one-step growth curve, as described previously38. Measurement of the duration of a phage’s latent-period was accomplished by detecting the delay between phage adsorption of a bacterium and the liberation of phage virions39,40,41. We calculated the burst size from the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells during the latent period.
Stability studies
We assayed phage stability in LB broth at pH values ranging from 1 to 14, after incubation for 60 min at 37 °C, and the phages that survived were diluted and counted immediately. Seven temperatures (4, 25, 37, 50, 60, 70 and 80 °C) were selected to study the thermal tolerance of phiAxp-3 in LB broth at 15-min sampling intervals. Biocide resistance was determined using the common biocides ethanol (10%, 50%, 75% and 95% v/v) and isopropanol (10%, 50% and 95%) at 30-min intervals for sampling. The influence of Ca2+ and Mg2+ on phage lysis was investigated by incubation (37 °C) of infected A. xylosoxidans A22732 in LB agar with and without CaCl2 or MgCl2 (0, 5, 10, 15, 20, 25 and 30 mmol/l). Plaque formation was investigated using the double-layer plate technique. We expressed the results as a percentage of the initial viral counts.
DNA isolation and genome sequencing
Genomic DNA was extracted from purified phage particles with phenol-chloroform (24:1, vol/vol) method described previously15. Whole-genome sequencing of the phiAxp-3 phage was performed with an Illumina HiSeq2500 sequencer. The reads were assembled using the CLC genomics Workbench de novo assembly algorithm (CLC bio, Cambridge, MA). The BLASTP program was used to search putative homologies and proteins sharing similarities with predicted phage proteins (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignment and phylogenetic analysis were performed using ClustalW (Slow/Accurate, IUB) and Mauve software (http://asap.ahabs.wisc.edu/mauve/). LC/ESI/MS/MS spectra (Q-TOF Ultima API, Micromass UK Ltd.) were used to identify phage proteins, as described previously42.
Structure determination of phiAxp-3 genome ends
The physical genome structure (linear vs circular) of phiAxp-3 was assessed by analysing its restriction digestion profiles using the conditions recommended by the manufacturer (New England Biolabs, Ipswich, MA). Lambda DNA/HindIII Markers (Thermo Scientific, Waltham, MA) were used for estimating DNA fragment sizes. Phage phiAxp-3 genome-end fragments resulting from digestion with BlpI or EagI restriction enzymes were excised from agarose gels and extracted using a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). The DNA fragments isolated were checked for purity by electrophoresis and used as templates for sequencing.
Receptor identification
The receptor properties of phiAxp-3 were determined as described previously43. Briefly, A. xylosoxidans A22732 cultures were treated with proteinase K (0.2 mg/ml; Promega) at 37 °C for 3 h and sodium acetate (50 mM, pH 5.2) containing 100 mM IO4− at room temperature for 2 h (protected from light) to determine whether proteinase K or periodate could destroy the phage receptor. Next, a phage adsorption assay was performed as described previously44. LPS extraction from A. xylosoxidans cultures was performed using an LPS extraction kit from Intron Biotechnology (17144; Boca Scientific, Boca Raton, FL) according to the manufacturer’s instructions. Phage inactivation by LPS was performed as described previously45.
Nucleotide sequence accession number
The annotated genome sequence for the phage phiAxp-3 was deposited in the NCBI nucleotide database under the accession number KT321317.
Additional Information
How to cite this article: Ma, Y. et al. Isolation and molecular characterisation of Achromobacter phage phiAxp-3, an N4-like bacteriophage. Sci. Rep. 6, 24776; doi: 10.1038/srep24776 (2016).
Supplementary Material
Acknowledgments
This work received support from National Natural Science Foundation of China Grant (31200137 and 31360034).
Footnotes
Author Contributions Y.M., E.L. and Z.Q. did the experiments and contributed equally to this study as joint first authors. H.L., X.W. and W.L. analyzed the data. R.Z., A.J., H.Y., Z.Y. and J.Y. provided the bacterial strains. Z.Q. managed the project (31360034). X.Z. managed the project (31200137), designed the experiments and wrote the article.
References
- Roy P. Pulmonary Infection Caused by Achromobacter xylosoxidans in a Patient with Carcinoma of Epiglottis: A Rare Case. J Clin Diagn Res 8, DD01–02 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Jasser A. M. & Al-Anazi K. A. Complicated septic shock caused by Achromobacter xylosoxidans bacteremia in a patient with acute lymphoblastic leukaemia. Libyan J Med 2, 218–219 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca Otero L. et al. Achromobacter xylosoxidans infection in an adult cystic fibrosis unit in Madrid. Enferm Infecc Microbiol Clin doi: 10.1016/j.eimc.2015.05.006 (2015). [DOI] [PubMed] [Google Scholar]
- Turel O. et al. Bacteremia due to Achromobacter xylosoxidans in neonates: clinical features and outcome. Braz J Infect Dis 17, 450–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bador J., Amoureux L., Blanc E. & Neuwirth C. Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob Agents Chemother 57, 603–605 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traglia G. M. et al. Achromobacter xylosoxidans: an emerging pathogen carrying different elements involved in horizontal genetic transfer. Curr Microbiol 65, 673–678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott I. J. & Peleg A. Y. Stenotrophomonas, Achromobacter, and nonmelioid Burkholderia species: antimicrobial resistance and therapeutic strategies. Semin Respir Crit Care Med 36, 99–110 (2015). [DOI] [PubMed] [Google Scholar]
- Oliveira H. et al. Unexploited opportunities for phage therapy. Front Pharmacol 6, 180, doi: 10.3389/fphar.2015.00180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. S. et al. Therapeutic effects of bacteriophages against Salmonella gallinarum infection in chickens. J Microbiol Biotechnol 23, 1478–1483 (2013). [DOI] [PubMed] [Google Scholar]
- Kong M. & Ryu S. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl Environ Microbiol 81, 2274–2283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F. et al. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Biomed Res Int 2015, 752930, doi: 10.1155/2015/752930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal B. S. et al. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: genomic organization and proteomic analysis of the virions. Arch Virol 156, 25–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. C. et al. Genomic analysis of bacteriophage varphiAB1, a varphiKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Genomics 97, 249–255 (2011). [DOI] [PubMed] [Google Scholar]
- Wittmann J. et al. First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol J 11, 14, doi: 10.1186/1743-422X-11-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E. et al. Characterization of a novel Achromobacter xylosoxidans specific siphoviruse: phiAxp-1. Sci Rep 6, 21943, doi: 10.1038/srep21943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. IMP-1 encoded by a novel Tn402-like class 1 integron in clinical Achromobacter xylosoxidans, China. Sci Rep 4, 7212, doi: 10.1038/srep07212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey-Smith G. V. et al. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol Lett 258, 182–186 (2006). [DOI] [PubMed] [Google Scholar]
- Haberer K. & Maniloff J. Adsorption of the tailed mycoplasma virus L3 to cell membranes. J Virol 41, 501–507 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K. H., Stothard P. & Dennis J. J. Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genomics 13, 223, doi: 10.1186/1471-2164-13-223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D. E. et al. Whole genome sequencing and comparative genomic analyses of two Vibrio cholerae O139 Bengal-specific Podoviruses to other N4-like phages reveal extensive genetic diversity. Virol J 10, 165, doi: 10.1186/1743-422X-10-165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehi G. & Lindell D. The P-SSP7 cyanophage has a linear genome with direct terminal repeats. PLos One 7, e36710, doi: 10.1371/journal.pone.0036710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H. M. et al. The complete genome sequence of EC1-UPM, a novel N4-like bacteriophage that infects Escherichia coli O78:K80. Virol J 10, 308, doi: 10.1186/1743-422X-10-308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Heredia I., Rodriguez-Valera F. & Martin-Cuadrado A.-B. Novel group of podovirus infecting the marine bacterium Alteromonas macleodii. Bacteriophage 3, e24766, doi: 10.4161/bact.24766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S., Zehring W., Rothman-Denes L. & DNA-dependent R. N. A. polymerase from bacteriophage N4 virions. Purification and characterization. J Biol Chem 255, 4339–4347 (1980). [PubMed] [Google Scholar]
- Wang N., Dykhuizen D. E. & Slobodkin L. B. The evolution of phage lysis timing. Ecol Evol 10, 545–558 (1996). [Google Scholar]
- Moodley S., Maxwell K. L. & Kanelis V. The protein gp74 from the bacteriophage HK97 functions as a HNH endonuclease. Protein Sci 21, 809–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. & Zhang X. Identification and characterization of a novel thymidylate synthase from deep-sea thermophilic bacteriophage Geobacillus virus E2. Virus genes 37, 218–224 (2008). [DOI] [PubMed] [Google Scholar]
- Wittmann J., Gartemann K.-H., Eichenlaub R. & Dreiseikelmann B. Genomic and molecular analysis of phage CMP1 from Clavibacter michiganensis subspecies michiganensis. Bacteriophage 1, 6–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E. M. et al. Characterization of a ViI-like phage specific to Escherichia coli O157: H7. Virol J 8, 430, doi: 10.1186/1743-422X-8-430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. R. et al. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J Bacteriol 187, 1091–1104 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. Z., Millard A. D., Mann N. H. & Schafer H. Comparative genomics defines the core genome of the growing N4-like phage genus and identifies N4-like Roseophage specific genes. Front Microbiol 5, 506, doi: 10.3389/fmicb.2014.00506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin A. & Manning P. A. Bacteriophage CP-T1 of Vibrio cholerae. Eur J Biochem 153, 89–94 (1985). [DOI] [PubMed] [Google Scholar]
- Kot W., Hammer K., Neve H. & Vogensen F. K. Identification of the receptor-binding protein in lytic Leuconostoc pseudomesenteroides bacteriophages. Appl Environ Microbiol 79, 3311–3314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. P., Levy C. E. & Yano T. A heat-stable cytotoxic factor produced by Achromobacter xylosoxidans isolated from Brazilian patients with CF is associated with in vitro increased proinflammatory cytokines. J Cyst Fibros 11, 305–311 (2012). [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang K., Jiao N. & Chen F. Genome sequences of two novel phages infecting marine roseobacters. Environ Microbiol 11, 2055–2064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E. Phage host range and efficiency of plating. Methods Mol Biol 501, 141–149 (2009). [DOI] [PubMed] [Google Scholar]
- Chow J. J., Batt C. A. & Sinskey A. J. Characterization of Lactobacillus bulgaricus Bacteriophage ch2. Appl Environ Microbiol 54, 1138–1142 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Isolation and characterization of a novel virulent phage (phiLdb) of Lactobacillus delbrueckii. Int J Food Microbiol 137, 22–27 (2010). [DOI] [PubMed] [Google Scholar]
- Abedon S. T. Lysis of lysis-inhibited bacteriophage T4-infected cells. J Bacteriol 174, 8073–8080 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbruck M. The Growth of Bacteriophage and Lysis of the Host. J Gen Physiol 23, 643–660 (1940). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A., Manson M. D. & Young R. Holins kill without warning. Proc Natl Acad Sci USA 98, 9348–9352 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar W. M., Blackburn R. K., Krise J. M. & Moseley M. A. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. J Am Soc Mass Spectrom 14, 971–979 (2003). [DOI] [PubMed] [Google Scholar]
- Kiljunen S. et al. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage phiA1122. J Bacteriol 193, 4963–4972 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. et al. Outer membrane proteins ail and OmpF of Yersinia pestis are involved in the adsorption of T7-related bacteriophage Yep-phi. J Virol 87, 12260–12269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis-Wlodarczyk K. et al. Characterization of the Newly Isolated Lytic Bacteriophages KTN6 and KT28 and Their Efficacy against Pseudomonas aeruginosa Biofilm. PLos One 10, e0127603, doi: 10.1371/journal.pone.0127603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.