Abstract
It has been proposed that livestock production effluents such as wastewater, airborne dust and manure increase the density of antimicrobial resistant bacteria and genes in the environment. The public health risk posed by this proposed outcome has been difficult to quantify using traditional microbiological approaches. We utilized shotgun metagenomics to provide a first description of the resistome of North American dairy and beef production effluents, and identify factors that significantly impact this resistome. We identified 34 mechanisms of antimicrobial drug resistance within 34 soil, manure and wastewater samples from feedlot, ranch and dairy operations. The majority of resistance-associated sequences found in all samples belonged to tetracycline resistance mechanisms. We found that the ranch samples contained significantly fewer resistance mechanisms than dairy and feedlot samples, and that the resistome of dairy operations differed significantly from that of feedlots. The resistome in soil, manure and wastewater differed, suggesting that management of these effluents should be tailored appropriately. By providing a baseline of the cattle production waste resistome, this study represents a solid foundation for future efforts to characterize and quantify the public health risk posed by livestock effluents.
Livestock production effluent has been implicated in the transmission of antimicrobial resistant bacteria into aquatic, terrestrial and atmospheric ecosystems1,2,3. Regulation of livestock manure and wastewater is currently designed to limit levels of organic nutrients and coliform bacteria within ground and surface waters, and manured soils. Such regulations may not adequately control transfer of antimicrobial resistance genes (ARGs) from livestock facilities into the surrounding environment, particularly when antimicrobial drug residues are present in effluents4,5,6. However, significant increases in soil ARG levels have been observed after application of antimicrobial-free soil7,8, or when manure application rates are high or manure is improperly stored9. Furthermore, non-manured or “pristine” soil contains a diverse repertoire of ARGs, making it difficult to distinguish between natural and anthropogenically impacted ARG content10.
In North American cattle production systems, antimicrobials are used to treat, prevent and control disease, as well as to improve production efficiency. Antimicrobial use in extensive management settings (i.e., cow-calf production settings on pasture) is low, with 1.9% of US and 2.7% of Canadian beef cows reportedly treated with antimicrobials11,12. In contrast, >20% of US feedlot cattle receive antimicrobials for prevention of respiratory disease, >13% for treatment of active respiratory disease, and >70% for the prevention of liver abscesses13. Ionophores are the most common antimicrobial administered to cattle in North American feedlots, but they are not classified as medically important14,15. Macrolides, which are classified as critically important, are the second most commonly administered antimicrobial13,15. Most North American dairy operations (>94%) administer antimicrobials to prevent mastitis during the dry period, with >97% of this use attributed to beta-lactam antimicrobials, including first- and third-generation cephalosporins16. Pre-weaned dairy calves are most commonly treated for respiratory disease and diarrhea, with phenicols, cephalosporins, macrolides, sulfonamides, tetracyclines and aminoglycosides all used in approximately the same proportions16.
Given these use practices and evidence that antimicrobial drug residues in livestock effluent can contribute to elevated antimicrobial resistance (AMR) levels, it is important to improve our understanding of how current management systems may impact AMR transmission to the public. Culture-independent methods can contribute to this understanding, as the genetic dynamics of AMR are complex and pan-microbial. For instance, ARGs often move together in a networked fashion17,18,19, and can be exchanged between distantly-related bacteria20,21,22,23. Transfer rates among bacteria increase under conditions that induce stress, including antibiotic exposure24,25. Pan-microbial approaches enable access to this microbial resistance ecology and provide information on how livestock production practices influence the density and composition of ARGs (i.e., the resistome26). With this insight, practical mitigation strategies can be proposed to minimize the flow of ARGs into aquatic, terrestrial and atmospheric ecosystems.
However, very little is currently known about the cattle production resistome. The few published studies are descriptive in nature, utilizing samples taken from a small number of non-commercial animals27,28. A lack of basic study design information is a major impediment to execution of larger, inferential studies. For instance, the sample size needed to detect resistome differences between conventional and organic operations is currently not estimable because we do not have a baseline of normal resistome variability among different cattle. In addition, many livestock production factors may confound the relationship between antimicrobial use and the resistome, e.g., stocking density or ration formulation. However, to-date these potential factors have not been identified, making it difficult to control for confounders in study design.
Therefore, the goal of this study was to provide a description of the dairy and beef production resistome in raw feces, soil and wastewater, and to identify factors that commonly influence this resistome. A conventional and an organic dairy were chosen to investigate the impact of antimicrobial use practices on the resistome; a US and a Canadian feedlot were chosen to explore the impact of diet (US feedlots typically feed corn-based diets, while Canadian feedlots utilize barley); and a cow-calf ranch was selected to compare resistome differences between intensive (i.e., feedlot) and extensive (i.e., pastured) production systems.
Results and Discussion
Sequencing produced over 1.05 billion sequencing reads across all samples, at an average of 30.8 million reads per sample (range 14.9–48.9 M, Supplementary Table S1). Trimming resulted in removal of 20.9% of reads (range 15–29%) and filtering of Bos taurus removed a further 0.38% (range 0.003–5.4%) of reads (Supplementary Table S1).
Across all samples, 212 128 reads aligned to 34 resistance mechanisms that confer resistance to 15 classes of antimicrobial drugs (Supplementary Table S2). The majority of reads aligned to genes that confer tetracycline resistance (70.0%), and within this class, 98% of reads aligned to ribosomal protection proteins with the largest representation in TetQ and TetW (45.6% and 23.2% of all reads aligning to tetracycline ribosomal protection proteins, respectively). Of the 30 samples that contained reads that aligned to ARGs within the resistance database, 27 contained TetQ and 22 contained TetW. Interestingly, these resistance groups were also the most prevalent in fecal samples collected from >1 000 humans as part of two metagenomic studies29,30, suggesting that they may be common in both agricultural and human populations. Studies across diverse agricultural ecosystems also document the ubiquity of tetracycline resistance genes31,32.
Reads aligning to the macrolide-lincosamide-streptogramin (MLS) class of antimicrobials comprised 10.9% of all resistance-aligned reads. Among MLS-aligned reads, 61.1% aligned to macrolide efflux pumps, 22.1% to Erm 23s rRNA methyltransferases, and 16.7% aligned to lincosamide nucleotidyltransferases. The mefA efflux pump was the most frequently identified mechanism within the macrolide class.
Few resistance mechanisms were identified in samples collected from the cow-calf ranch
None of the sequencing reads obtained from the soil and wastewater samples collected from the cow-calf ranch aligned to the resistance database (n = 4). This finding is unexpected given previous reports of a diverse and abundant resistome in soils and water not impacted by human activity33,34,35. However, these studies utilized functional metagenomics, PCR and/or very deep sequencing to detect ARGs, which may be better able to detect very low-abundance ARGs compared to shotgun metagenomics. In addition, the use of functional metagenomics offers the potential for novel resistance gene discovery36. While deeper sequencing may have revealed the presence of resistance gene sequences in the pasture soil and water samples, sequencing depth (as well as DNA quantity and quality) was not significantly different when comparing feedlot, dairy and ranch samples, suggesting that extensive rearing methods may result in lower resistance gene levels compared to more intensive rearing methods such as feedlots. Alternatively, soil and wastewater ARGs on ranches may not be as geographically concentrated and therefore more samples may be needed to gain a representative portrait of the ranch environmental resistome. However, soil samples were collected from areas where cattle commonly congregated, and water was collected from a dugout, and therefore these samples represent the resistome of locations where ARGs were likely to be most concentrated. In addition, the two fecal samples collected from the pasture contained 2 resistance mechanisms in total, as compared to 4, 11, 7 and 21 in the conventional dairy, organic dairy, US and Canadian feedlot samples, respectively. This suggests that extensive rearing may promote fewer mechanisms of resistance overall. Previous studies have documented a significantly lower level of integrons (which commonly carry ARGs) in grass-fed compared to lot-fed cattle, although the content of the integrons did not differ substantially between the two groups37. Culture-based studies also have documented lower levels of resistance in generic Escherichia coli isolated from beef cows on pasture compared to feedlot cattle38,39,40,41. However, the feedlot and pastured cattle populations differ in many other aspects, including genetics, sex and age of the cattle, and therefore more controlled studies are needed to determine whether differences in AMR are driven by antimicrobial use. Such studies should also take into account the “dilution effect” of cattle on pasture, and the likelihood that environmental samples from such settings are comprised largely of soil or water that has not been impacted by cattle activity.
Resistome composition and diversity differ between calves and adult cattle
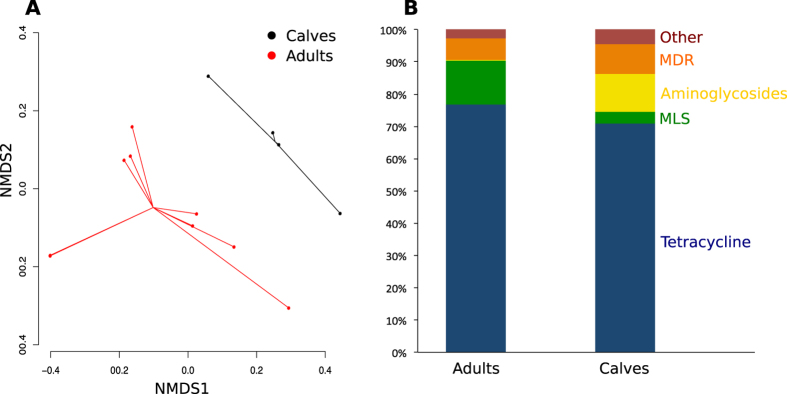
The resistome of fecal samples collected from preweaned dairy calves was different from that of mature cattle of all types (Stress = 0.11 and 0.03, ANOSIM R = 0.28 and 0.41, ANOSIM P = 0.05 and 0.02 at the mechanism and class levels, respectively, Fig. 1). Shannon’s diversity and richness at the mechanism and class levels were both lower in adult cattle compared to calf feces (Kruskal-Wallis P = 0.02 and 0.05 for richness and P = 0.09 and 0.09 for Shannon’s diversity, respectively). A descriptive comparison of dairy calf and adult dairy cow feces reflects this same pattern (Fig. 2), suggesting that production system (i.e., beef versus dairy) was not confounding this comparison and that the resistome of adult cattle feces is less diverse than that of calves. In addition, sequencing depth and DNA quantity and quality were not significantly different between samples collected from calves and samples collected from adult cattle, providing further evidence that resistome differences between these samples were not artifactual. Previous studies in calves have documented significant changes in fecal microbial diversity and composition as calves moved from milk-fed to weaning42,43 and these changes could be driving resistome composition.
Figure 1. A comparison of fecal samples collected from calves and adult cattle.
(A) NMDS ordination of fecal sample resistomes from calf vs. adult cattle. (B) Proportion of all aligned reads that aligned to ARGs within different resistance classes, in adult cattle versus calf feces. MDR = Multi-drug resistant mechanisms; MLS = Macrolide-lincosamide-streptogramin.
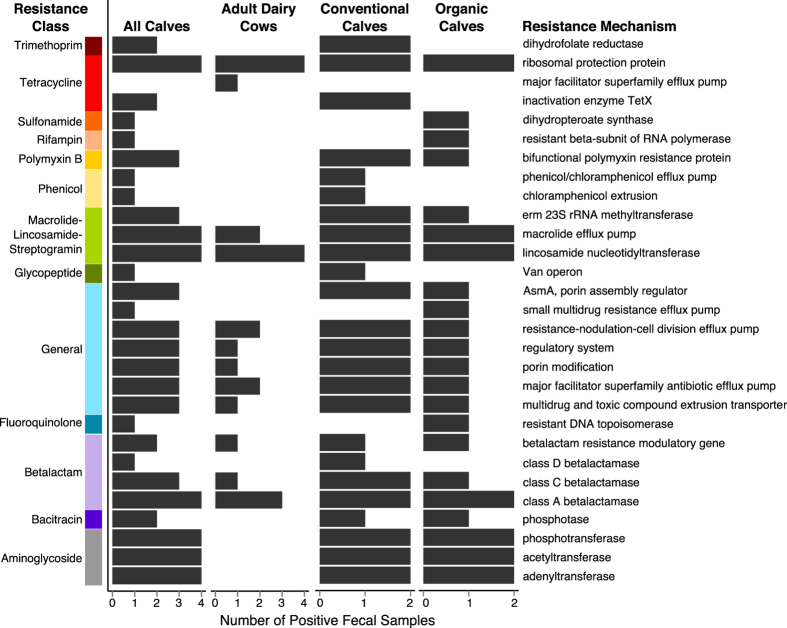
Figure 2. The calf fecal resistome is significantly more diverse and rich than the adult fecal resistome.
Number of fecal samples containing alignments within each mechanism and class of resistance, separated by calves vs. adults, and conventionally raised vs. organically raised calves.
Reads aligning to trimethoprim and aminoglycoside ARGs were identified in calf feces, but not in mature cattle feces (Fig. 2). This finding could reflect antimicrobial use practices in the conventional dairy, which administered trimethoprim-sulfadiazine and sulfamethazine to calves for treatment of scours, but did not administer these classes of antimicrobials to adult dairy cattle. Culture-based studies in generic E. coli report higher prevalence of resistance to trimethoprim-sulfamethoxazole and aminoglycosides (i.e., streptomycin, kanamycin and gentamycin) in beef calves compared to their corresponding dams38. However, aminoglycosides were not used in either dairy operation in this study, and therefore antimicrobial use practices do not directly explain these findings. A similar pattern regarding aminoglycoside resistance and use has been previously described in swine production, suggesting that aminoglycoside resistance may be co-selected by use of other antimicrobials4. We did not identify any tetracycline major facilitator superfamily (MFS) alignments in calf feces, but did in adult cattle feces, and reads aligning to macrolide efflux pumps and lincosamide nucleotidyltransferases were significantly more abundant in adult compared to calf feces. Lincosamides were used to treat mastitis on the conventional dairy farm, which could account for the increased lincosamide resistance in relation to preweaned calves. Alternatively, differences in resistome composition between calves and adult cattle could be driven by taxonomic differences in their respective fecal microbiomes. Studies in swine have shown significant shifts in the fecal microbiome as a function of age and diet, suggesting that the same patterns could be driving the differences observed in this study44,45. However, the degree to which microbiome drives resistome composition can vary dramatically20,46, and further research is needed to tease apart these potential confounders.
Comparison of the resistomes of conventionally- and organically-reared calves may shed light on the role of antimicrobial use practices in driving resistome differences. In this study population, organically raised calves did not receive any antimicrobial drugs, while the conventional dairy operation utilized beta-lactams (including cephalosporins), florfenicols, tetracyclines, macrolides, trimethoprim-sulfadiazine, sulfamethazine and macrolides. The 2 fecal samples collected from conventionally raised dairy calves contained slightly more unique resistance mechanisms (24/29) than those from organically raised dairy calves (22/29, Fig. 2). Among the 24 resistance mechanisms identified in feces from conventionally raised calves, 18 were identified in both conventional samples, while 7 of the 22 identified in feces from organically raised calves were identified in both organic samples (Fig. 2). These findings suggest that the resistome of organically raised calves may contain fewer unique resistance mechanisms and at lower frequency than the resistome of conventionally raised calves. However, the fecal resistome of organically raised calves contained a diverse set of resistance mechanisms, despite the naiveté of these calves to antimicrobial drugs. Similar findings have been reported in 6-month old infants that were unexposed to antimicrobials47, and culture-based studies of generic E. coli isolated from preweaned calf feces support the difference in AMR in conventional and organic calf feces48,49,50,51. Due to the stark difference in calf feces versus mature cattle feces, calf samples were removed from further analyses to enable a direct comparison of adult cattle feces.
Beef feedlot resistome differs from the dairy resistome
Resistome composition at the mechanism and class levels differed between beef (feedlot and pasture) and dairy operations, for all sample matrices (NMDS Stress = 0.11 and 0.06, ANOSIM R = 0.15 and 0.11, and ANOSIM P = 0.01 and 0.03, respectively, Fig. 3A). However, Shannon’s diversity and richness indices did not differ between beef and dairy cattle (data not shown). Sequencing depth and DNA quality and quantity did not differ significantly between beef and dairy samples. Within fecal samples, ordination at the mechanism level showed clear separation of beef and dairy resistomes (NMDS Stress = 0.006, ANOSIM R = 0.38 and ANOSIM P = 0.03, Fig. 3B); soil and wastewater resistomes of beef versus dairy could not be compared due to data sparseness. Of the 13 classes of resistance identified in all samples collected from adult cattle, 5 were more abundant in beef samples versus dairy, 2 were more abundant in dairy samples, 1 was nearly equally abundant in dairy and beef samples, and 5 did not pass filtering criteria due to low prevalence across samples. Even among classes that did pass filtering, several were present in very low abundance, and therefore estimates of log-fold differences in abundance may not be reliable. Consequently, the discussion is restricted to resistance classes present in at least 10 of the 30 soil, wastewater and adult fecal samples, i.e., tetracyclines, MLS, aminoglycosides, beta-lactams, and general-purpose mechanisms. The MLS resistance class was nearly equally abundant between dairy and beef samples. Alignments to tetracycline ARGs were more abundant in feedlots than in dairies, while aminoglycoside acetyltransferases and phosphotransferases were significantly more abundant in dairy samples. This pattern could reflect differential antimicrobial use practices in feedlots and dairies. Although the American Association of Bovine Practitioners strongly discourages dairy and beef veterinarians from administering aminoglycosides to cattle, nationwide surveys suggest that they are still being used with some frequency in dairies16, much less so in beef feedlots13. In addition, tetracyclines are used more frequently by US feedlots than US dairies, both in-feed and parenterally13,16. However, many other factors could account for these differences, including frequency of pen cleaning (which could influence the soil resistome), lagoon construction and management (which could impact the lagoon resistome), and feed composition (which could affect the fecal resistome). In addition, alignments to beta-lactam ARGs were more abundant in feedlot samples, even though these antimicrobial drugs are reported to be much more widely used in dairy production13,52. These findings highlight the complexity of dynamics between antimicrobial use and AMR, and suggest that use practices do not solely or directly influence AMR patterns in livestock production.
Figure 3. Beef and dairy systems have different resistomes.

NMDS ordination at the mechanism level of (A) (adult) fecal, soil and wastewater samples and (B) only adult fecal samples were both significantly different based on system, e.g., beef vs. dairy (ANOSIM P < 0.05).
Resistomes of feces, soil and wastewater are distinct
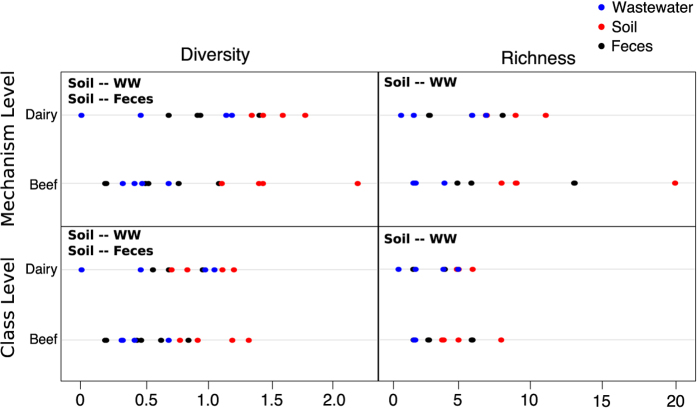
The resistome composition of feces, soil and wastewater were significantly different at the mechanism and class levels (NMDS Stress = 0.11 and 0.06, ANOSIM R = 0.34 and 0.30, and ANOSIM P = 0.001 and 0.001, respectively). Shannon’s diversity and richness were significantly higher in soil versus wastewater at the mechanism and class levels; however, soil samples received significantly more sequencing reads than water samples, and this could account for such differences53. Shannon’s diversity but not richness was significantly higher in soil versus feces at the mechanism and class levels (Fig. 4), and sequencing depth was comparable between the two matrices. The difference between soil and fecal diversity is especially interesting given that pen floors in dairies and feedlots are often a mixture of dried, compacted feces and underlying soil, rather than undisturbed soil matrix. In this context, the differentiation of “soil” (i.e., compacted feces) from fresh feces could stem from simple mixing of the two components, or from changes that occur within the soil-fecal matrix over time and under varying environmental exposures, as has been shown in E. coli54. Indeed, the fecal collection in this study focused on collection of recently voided fecal pats, in which exposure to aerobic conditions was of relatively short duration. Therefore, the environmental milieu to which the fecal and soil samples were exposed was likely very different.
Figure 4. Soil samples are significantly more diverse and rich than wastewater.
Dotplots showing Shannon’s diversity and richness at the mechanism and class levels, separated by system (beef vs. dairy) and colored by sample matrix, i.e., feces (black), soil (red) and wastewater (blue). Bolded text within each panel indicates which matrices differed based on Nemenyi post-hoc pairwise comparisons (WW = wastewater). Diversity and richness were not significantly different between beef and dairy at any level.
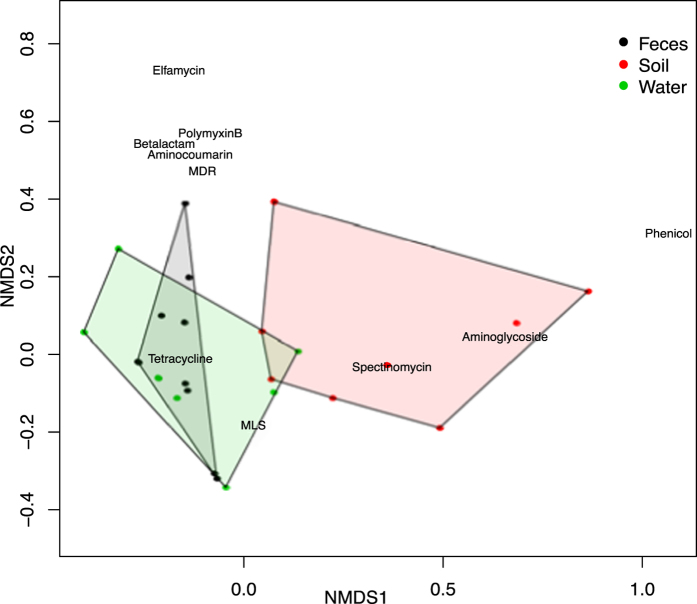
Ordination biplot results indicated that aminoglycoside, spectinomycin, phenicol and tetracycline resistance classes strongly influenced separation of soil samples from wastewater and feces (Fig. 5). Phenicol and spectinomycin resistance classes were not identified in adult cattle fecal samples, and reads aligning to aminoglycoside ARGs were less abundant in adult cattle fecal samples compared to soil samples (log-fold change = −4.6, adjusted P < 0.0001). Reads aligning to genes that confer tetracycline resistance–which were highly abundant in all samples–were more abundant in adult feces than in soil (log-fold difference = 1.50, adjusted P < 0.0001). This difference was driven overwhelmingly by the abundance of ribosomal protection proteins in feces, as major facilitator superfamily (MFS) efflux pumps were less abundant in feces compared to soil (log-fold difference = −2.1, adjusted P < 0.001). This pattern of tetracycline resistance mechanism differentiation between soil and feces has been observed in a comparison of human gut samples and agricultural soils55. Beta-lactam resistance alignments were also more abundant in feces and soil than in wastewater. Beta-lactams have been shown to degrade relatively quickly in the environment, and this could account for significantly lower levels in wastewater if antimicrobial drug residues influence the density of beta-lactam ARGs in this environment56.
Figure 5. Specific resistance classes drive separation of soil from fecal and wastewater resistomes.
NMDS ordination of fecal (black), soil (red) and wastewater (green) samples based on normalized counts of alignments aggregated at the resistance class level. Biplot coordinates of resistance classes are labeled with the class name, and show that aminoglycoside, phenicol and spectinomycin resistances differentiate the soil from the fecal and wastewater resistomes.
The potentially confounding effect of sample matrix on resistome composition should guide future sampling efforts by providing a logical rubric on which to base future study designs. Fortuitously, wastewater, manure and soils are often managed separately in large livestock production systems, and therefore research efforts can focus within one matrix without forfeiting real-world applicability.
Antimicrobial exposures on dairies did not significantly impact resistome composition
The resistomes of conventional and organic dairy samples (excluding calf fecal samples) were not different upon ordination, and there were no significant differences in Shannon’s diversity, richness or sequencing depth (P > 0.05). Abundance of resistance classes and mechanisms also did not differ, with the exception of macrolide resistance efflux pumps, which were more abundant in conventional dairy samples (log-fold difference = 9.4, adjusted P < 0.0001). The lack of differences may stem from insufficient study power, as descriptive analysis suggests major structural differences, particularly in the wastewater samples. For example, we identified only 2 resistance mechanisms in the organic dairy lagoon samples: lincosamide nucleotidyltransferases and tetracycline ribosomal protection proteins (specifically, TetQ and TetW). These samples clearly differed from the conventional dairy lagoon samples, which contained 9 resistance mechanisms. Previous studies have described significantly higher concentrations of TetO, TetW and Sul1 in lagoons at conventional dairies compared to organic dairies57, as well as higher levels of TetO, TetQ, TetW and TetM in feedlot lagoons draining pens exposed to moderate and high levels of antimicrobials as compared to lagoons draining antimicrobial-free pens58. Taken together, these findings suggest that antimicrobial use may substantially impact the resistome of stored wastewater, and therefore regulations regarding lagoon construction, wastewater use and management should be tailored to parameters of livestock operations, i.e., housing density or specific antimicrobial use protocols59. However, the influence of taxonomic composition on these resistome differences cannot be discounted, and such findings highlight the need for high-throughput methods that can definitively link microbiome composition with resistance genes. This is particularly important given recent studies reporting varying degrees of correlation between microbiome and resistome content, as well as varying rates of inferred horizontal gene transfer within microbial communities20,46. A shotgun metagenomic approach with short read sequence data can only provide a measure of microbiome-resistome correlation; such data do not enable definitive linking of resistance genes with the bacteria that harbor them. Long-read technology may represent a solution to this knowledge gap60, but relatively low throughput remains an obstacle for metagenomic applications.
Across soil, water and fecal samples, we identified 15 resistance mechanisms in non-calf conventional dairy samples and 18 in non-calf organic dairy samples. Reads aligning to class D beta-lactamases (bla-OXA), erm 23S rRNA methyltransferases, macrolide resistance efflux pumps, trimethoprim-resistant dihydrofolate reductases and the TetX inactivation enzyme were all identified in conventional but not organic samples. Conversely, organic samples contained several general-purpose resistance mechanisms not found in conventional samples (e.g., MATE, MFS efflux pumps and RND efflux pumps, porin modification genes, and regulators of resistance mechanisms). Resistance mechanisms with broad substrate specificity have been identified on organically-grown vegetables61, and our own research suggests that these types of mechanisms may be more abundant in cattle populations not exposed to antimicrobials. Under this theory, general purpose resistance mechanisms are favored in the absence of antimicrobial selective pressure as a means for bacteria to defend against numerous and varied environmental selective pressures, while resistance mechanisms with a more targeted purpose are favored in the presence of antimicrobial drugs. Further work is needed to test this hypothesis under more controlled settings.
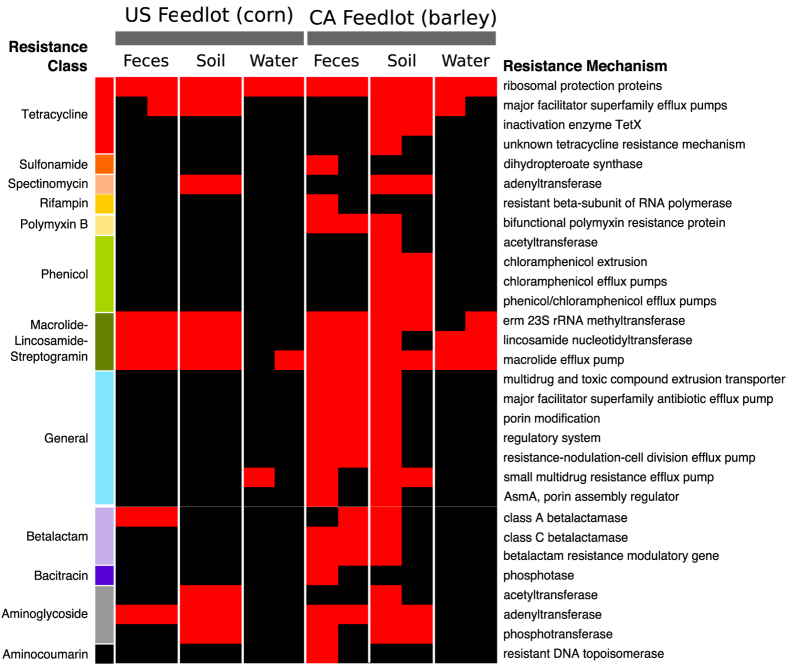
Resistome composition did not differ between US and Canadian feedlot samples
No significant resistome differences were observed between US and Canadian feedlot samples, which may indicate that diet did not have a major impact (NMDS Stress = 0.11 and 0.04, ANOSIM R = 0.02 and 0.05, and ANOSIM P = 0.31 and 0.26 for the mechanism and class levels, respectively). Diversity, richness and sequencing depth also did not differ significantly. However, descriptive results show differences, indicating that lack of statistical difference could stem from low study power (Fig. 6). Canadian samples contained reads that aligned to 12 classes and 30 mechanisms of resistance. In comparison, reads aligning to 6 classes and 11 mechanisms of resistance were identified in US feedlot samples. Previous studies have shown that diet significantly impacts the cattle fecal microbiome, which could impact the resistome62,63. While the US feedlot fed a corn-based and the Canadian feedlot a barley-based ration, antimicrobial use in these feedlots was largely similar (both feedlots administered in-feed tylosin) and this uniformity may have overpowered the influence of diet and microbial composition, leading to a lack of difference between samples.
Figure 6. Binary heatmap of resistance mechanisms and classes identified in fecal, soil and wastewater samples collected from a US and a Canadian (CA) feedlot.
Black = absent, red = present.
Utility of resistome analysis and implications for future studies
This pilot study compared the fecal, soil and wastewater resistomes of beef and dairy production. Using shotgun metagenomics, we revealed the diversity of the beef and dairy resistome through identification of 34 resistance mechanisms within 15 resistance classes. Wastewater, soil and pre-composted manure from intensive production operations contained varying types and levels of resistance mechanisms, and resistome composition was influenced by sample matrix, stage of animal development, and production system. These findings suggest that future research, regulations and decisions regarding livestock waste management should be tailored to effluent as well as to production system. Importantly, this study also highlights areas in which the shotgun metagenomics approach could be improved; specifically, longer reads could help to definitively identify the origin of resistance genes in metagenomic data and to link taxonomic differences with resistome differences. Finally, reference-based identification methods are ipso facto reliant on the accuracy and completeness of available databases. As with organisms in the microbiome, it is highly likely that many resistance mechanisms remain undiscovered, producing varying rates of false negatives and misclassification using available databases and bioinformatics tools64,65; our use of nucleotide homology for ARG identification may have also resulted in under-detection if the ARGs in the sample contained high rates of synonymous substitutions. Conversely, false positive identification can occur if sequences show high homology with known ARGs, but contain mutations that render them non-resistant. The necessary trade-off between sensitivity and specificity should be decided on a case-by-case basis, with deference to the goals of the study and full acknowledgement of limitations and potential misclassification. Regardless, as databases and classification methods continue to improve, so too should our ability to interpret results from metagenomic data. As this study demonstrates, resistome analysis can provide actionable guidance on complex agricultural AMR issues, and therefore improvement in associated methods and databases should be prioritized66,67. The results presented here provide a foundation for further research in this area, while also highlighting areas in which methodological improvements should be aggressively pursued.
Methods
Study population and sampling sites
Both dairies and the US feedlot were located in northeastern Colorado, while the Canadian feedlot and ranch were located in southern Alberta. The conventional dairy milked 990 cows on one location. Cows and calves were treated with a variety of antimicrobials for clinical illness and dry-off. The organic dairy milked ~16 500 mature cows daily and managed ~23 000 animals across 5 locations. No antimicrobials were used on the organic dairy. The US feedlot had a one-time holding capacity of ~90 000 head and fed cattle a steam-flaked corn-based diet. The Canadian feedlot had a one-time holding capacity of 17 000 cattle and fed a barley-based diet. Both the US and Canadian feedlots administered in-feed macrolides and ionophores to all cattle. The cow-calf ranch was located at the Onefour Agriculture Canada research station, which consisted of 17 000 hectare and carried ~600 cow-calf pairs. Less than one percent of the herd received antimicrobials, with treatment limited to a proportion of those individuals that exhibited clinical illness.
Sample collection and processing
Sampling procedures were approved by the Colorado State University Institutional Animal Care and Use Committee, and were carried out in accordance with the Committee’s approved guidelines.
Feces
At each of the 5 operations described above, 2 composite fecal samples were collected from adult cattle. Additionally, 2 composite fecal samples from preweaned calves were collected at each of the 2 dairies, resulting in 14 total fecal samples for the study (10 adult and 4 calf). At the ranch, fecal samples were collected from an area where animals congregated at the time of sampling. Fecal samples at the feedlots and dairies were collected from 2 purposively selected pens. Feedlot pens identified as ready for slaughter were chosen such that samples represented cattle that had been >165 days-on-feed. At dairies, cows were grouped by level of milk production, and pens with highest production were chosen. To collect composite samples, personnel walked through pens or pasture area in a prescribed pattern, collecting ~1–2 grams (g) of freshly voided feces from 20 fecal pats. These 20 individual samples were placed in a sterile bag and thoroughly mixed by hand for 1 min, after which enough fecal material was removed to fill a sterile 50 milliliter (ml) Falcon tube. Dairy calves were housed in individual hutches, and therefore fecal samples were collected from the floor of 20 randomly selected individual hutches and mixed together as described above.
Soil
The same feedlot pens, dairy pens and pasture areas sampled for feces were also sampled for soil, using methods described for compositing of feces. Soil was not collected from dairy calf hutches, and therefore a total of 10 composite soil samples were collected (2 at each operation).
Wastewater
At each operation, 2 wastewater samples were collected. In the feedlots and dairies, wastewater was collected from the holding lagoon closest to the pens sampled during fecal collection. If lagoons were separated into “high-solids” and “low-solids”, a single sample was collected from each, and they were collected near the bank from opposite sides. Winter weather at the US feedlot prevented access to the entire lagoon, and therefore both samples were collected from the same side. At the cow-calf ranch, samples were taken from opposite sides of a dugout that cattle could fully access for drinking water.
Sample processing and sequencing
Samples were placed on ice for transport to the laboratory. Fecal and soil samples were stored at −80 °Celsius (C) until processed. Water samples were stored at 4 °C for a maximum of 24 hours (h), and then centrifuged at 15 000 × g for 20 minutes (min). The resulting supernatant was decanted and each pellet was stored in a 15 ml Falcon tube at −80 °C.
After thawing, fecal and soil samples were pre-sedimented, allowing for processing of a greater sample volume (up to 10 g) while simultaneously removing PCR inhibitors known to be present in soil and feces. Briefly, 30 ml of buffered peptone water (BPW) was added to 10 g of soil or feces in a 50 ml conical tube and samples were shaken vigorously before allowing to sediment for 10 min. Supernatants, including limited soil/fecal debris, were transferred to a new 50 ml conical tube and centrifuged for 10 min at 4 300 × g. The BPW was removed and the resulting sample pellet was rinsed with 5 ml of molecular grade sterile PBS, and centrifuged again at 4 300 × g for 10 min. The supernatant was removed and the resulting pellet was resuspended in 15 ml of PowerBead solution before being processed with the Mo Bio PowerMax Soil DNA Isolation Kit (Mo Bio Laboratories, Solana Beach, CA, USA) according to manufacturer’s instructions. Water samples (250 milligrams [mg]) were processed using the Mo Bio PowerFecal DNA Isolation Kit according to manufacturer’s instructions. Extracted DNA from fecal, soil and wastewater samples was eluted in 400 microliters (μl), 200 μl and 50 μl of elution buffer, respectively. After extraction, DNA concentration was measured at 260 nanometers (nm) using a NanoDropTM spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Samples that were not sufficiently concentrated (defined as <20 nanograms/μl [ng/μl]) were subjected to ethanol precipitation to achieve a concentration of >50 ng/μl. Briefly, 1/10 volume of 3 molar (M) sodium acetate, pH 5.2, and 2 volumes of cold 100% molecular grade ethanol were added to the sample, which was then incubated at −20 °C for 1 h and centrifuged at 11 000 × g for 20 min at 4 °C. Supernatant was discarded and 150 μl of 70% cold ethanol was added; samples were centrifuged a final time at 11 000 × g for 10 min at 4 °C. Supernatant was again discarded and the DNA pellets allowed to air dry before resuspending in ¼ the original volume.
After DNA extraction and ethanol precipitation, samples were checked for purity using a NanoDropTM spectrophotometer (Thermo Fisher Scientific); all samples attained a 260:280 ratio of >1.4. Finally, ≥10 μl of DNA from each sample was delivered for library preparation (according to manufacturer’s protocol) and single-end sequencing on the Ion ProtonTM platform using the P1 chip (Thermo Fisher Scientific).
Bioinformatics
Reads were trimmed and filtered for quality68: the leading 3 and trailing 3 nucleotides were removed, then nucleotides were removed from the 3′ end until the average Phred score across the first 4 nucleotides was at least 15. Sequences with <36 nucleotides were discarded. Adaptors were removed using a maximum of 2 mismatches in the initial seed, and adapter clipping occurred if a match score of 30 was reached. Upon clipping, both reads of a pair were retained to supply more reads for downstream applications. Reads classified as host (i.e., bovine) were removed by alignment to the UMD 3.1 Bos taurus genome with BWA using default parameters69. Non-host reads were then aligned to a custom non-redundant database of 3 111 ARG sequences compiled from the actively curated ARG databases ARG-ANNOT70, Resfinder71 and CARD (all downloaded on August 12th, 2014)72 using BWA with default settings. These databases were chosen because they are specific to antimicrobial resistance genes, they contain nucleotide sequences (as opposed to amino acid sequences), and they are actively curated and frequently updated. Redundant sequences between ARG-ANNOT and Resfinder were identified using CD-HIT-EST-2D73 with local alignment (-G 0) and the following parameters: -c 1.0 -AS 0 -AL 0 -aL 1.0 -aS 1.0. A single representative sequence was selected from each resulting cluster (n = 1,427), and these sequences were appended to the list of unique gene sequences in ARG-ANNOT (n = 261) and Resfinder (n = 715). This process was then repeated for the CARD database using the combined ARG-ANNOT/Resfinder non-redundant database. Seven hundred and eight sequences were unique to CARD, resulting in a final non-redundant database containing 3,111 unique ARG sequences. ARGs with >80% gene fraction (i.e., >80% of the nucleotides in the ARG sequence were covered by at least one read) were considered to be positively identified in a sample. Reads that aligned to ARGs that confer resistance via substitutions, insertions and/or deletions within antibiotic drug target genes were manually verified to match the reference ARG with 100% amino acid homology across 100% of the middle 95% of the ARG sequence (the very ends of the sequence were not included as reads rarely align to these “overhang” regions); synonymous substitutions were permitted. This list comprised the gene groups gyrA and parC (fluoroquinolone resistance), parE (aminocoumarin resistance), rpoB (rifampin resistance), folP (sulfonamide resistance), dfr (trimethoprim resistance) and EF-Tu (elfamycin resistance). This post-processing step was necessary in order to prevent potential false positives due to the presence of “wild-type” genes (i.e., genes not containing the resistance-conferring mutation) within the metagenomic data. Such reads could align to the reference ARG sequence with near-perfect identity, thus resulting in false identification of ARGs at the >80% gene fraction cutoff. Manual verification was conducted by visually inspecting alignment results using Tablet74. Alignments that did not pass post-processing verification were not included in downstream analyses.
Data Analysis
Samples without identified ARGs (n = 4) were removed from further analysis due to an inability to normalize and ordinate. Alignments to each positively identified ARG were summed within sample and normalized using a data-driven approach based on shifts in count distributions75,76. Because conventions for naming ARGs based on sequence homology vary between resistance classes, normalized counts were aggregated to the mechanism and class level to generate a less biased comparison among samples.
Primary comparisons included resistome differences between beef and dairy systems; conventional and organic dairy samples; calves and adult cattle; pasture and intensively managed operations; and between Canadian and US feedlots. In addition, we investigated differences in sample matrix (e.g., feces vs. soil vs. wastewater).
Non-metric multidimensional scaling (NMDS) using Euclidean distances between Hellinger-transformed normalized counts was used to ordinate samples based on resistome composition77. Significant dissimilarity of ordination between groups was assessed using the Analysis of Similarities statistic78.
To compare Shannon’s diversity and richness across groups, as well as to assess potential sequencing bias, a Kruskal-Wallis test statistic with Nemenyi post-hoc comparisons adjusted for rank ties was used. Separate comparisons were made between system (beef vs. dairy vs. pasture), sample matrix (feces vs. soil vs. water), dairy production type (organic vs. conventional) and feedlot diet type (corn vs. barley).
To assess differences in abundance of resistance alignments at the mechanism and class levels while accounting for potential uneven sequencing depth, zero-inflated Gaussian mixture models were used75. Prior to modeling, features (i.e., classes or mechanisms) present in fewer than 3 samples were removed from the dataset, as sparse features create model instability and do not provide reliable model estimates. When applicable, sample matrix and/or system (e.g., beef versus dairy) were added to models as potential confounders. Pairwise comparisons of log-fold change in abundance between groups were conducted using limma79, adjusting for multiple comparisons using the Benjamini-Hochberg procedure and a critical α of 0.0580.
Additional Information
Accession codes: The sequence information in this paper has been submitted to the GenBank Sequence Read Archive database under BioProject accession number SRP067931.
How to cite this article: Noyes, N. R. et al. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci. Rep. 6, 24645; doi: 10.1038/srep24645 (2016).
Supplementary Material
Acknowledgments
The authors thank all of the livestock operations that allowed us access to their facilities to obtain samples. The authors also acknowledge Erin Petrilli and the Colorado State University Next Generation Sequencing Core for their assistance. This project was supported by a grant from the Colorado State University Infectious Disease Supercluster. N.N. was supported by NIH T32OD012201.
Footnotes
Author Contributions N.R.N., H.Y., D.R.W., I.G., S.P.G., T.A.M., K.E.B. and P.S.M. conceptualized the project and study design. N.R.N., X.Y., S.R.C., R.Z. and J.A.M. designed and executed sample collection. X.Y., L.M.L., R.J.M. and S.R.C. processed samples. N.R.N., K.L.J., C.A.B. and J.R. performed bioinformatics and data analyses. All the authors contributed to discussions and comments on the manuscript.
References
- Sura S. et al. Transport of three veterinary antimicrobials from feedlot pens via simulated rainfall runoff. Sci. Total Environ. 521, 191–199 (2015). [DOI] [PubMed] [Google Scholar]
- McNab Walt W., Singleton M. J., Moran J. E. & Esser B. K. Assessing the Impact of Animal Waste Lagoon Seepage on the Geochemistry of an Underlying Shallow Aquifer. Environ. Sci. Technol. 41, 753–758 (2007). [DOI] [PubMed] [Google Scholar]
- McEachran A. D. et al. Research. Antibiotics, Bacteria, and Antibiotic Resistance Genes: Aerial Transport from Cattle Feed Yards via Particulate Matter. Environ. Health Perspect. 123, 337–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.-G. et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. 110, 3435–3440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Sun Y., Ding X., Wang M. & Zeng Z. Selective pressure of antibiotics on ARGs and bacterial communities in manure-polluted freshwater-sediment microcosms. Front. Microbiol. 6, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Occurrence of antimicrobials and antimicrobial resistance genes in beef cattle storage ponds and swine treatment lagoons. Sci. Total Environ. 463–464, 631–638 (2013). [DOI] [PubMed] [Google Scholar]
- Udikovic-Kolic N., Wichmann F., Broderick N. A. & Handelsman J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. 111, 15202–15207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyselková M. et al. Cow excrements enhance the occurrence of tetracycline resistance genes in soil regardless of their oxytetracycline content. Chemosphere 93, 2413–2418 (2013). [DOI] [PubMed] [Google Scholar]
- Ghosh S. & LaPara T. M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1, 191–203 (2007). [DOI] [PubMed] [Google Scholar]
- Cytryn E. The soil resistome: the anthropogenic, the native, and the unknown. Soil Biol. Biochem. 63, 18–23 (2013). [Google Scholar]
- USDA-APHIS-VS-CEAH-NAHMS. USDA. 201. Beef 2007–08, Antimicrobial Drug Use and Antimicrobial Resistance on US Cow-calf Operations, 2007–08 (2012).
- Gow S. P. & Waldner C. L. Antimicrobial drug use and reason for treatment in 203 western Canadian cow–calf herds during calving season. Prev. Vet. Med. 90, 55–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA–APHIS–VS–CEAH–NAHMS. Feedlot 2011. Part IV: Health and Health Management on US Feedlots with a Capacity of 1,000 or More Head. (2013).
- FDA. Guidance for Industry #213: New Animal Drugs and New Animal Drug Combination Products Administered in or on Medicated Feed or Drinking Water of Food- Producing Animals: Recommendations for Drug Sponsors for Voluntarily Aligning Product Use Conditions with GFI #209. (2013).
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 3rd revision. (2011).
- USDA–APHIS–VS–CEAH–NAHMS. Dairy 2007, Part V: Changes in Dairy Cattle Health and Management Practices in the United States, 1996–2007 (2009).
- Herrick J. B., Haynes R., Heringa S., Brooks J. M. & Sobota L. T. Coselection for resistance to multiple late-generation human therapeutic antibiotics encoded on tetracycline resistance plasmids captured from uncultivated stream and soil bacteria. J. Appl. Microbiol. 117, 380–389 (2014). [DOI] [PubMed] [Google Scholar]
- Tremblay C.-L., Letellier A., Quessy S., Daignault D. & Archambault M. Antibiotic-resistant Enterococcus faecalis in abattoir pigs and plasmid colocalization and cotransfer of tet(M) and erm(B) genes. J. Food Prot. 75, 1595–1602 (2012). [DOI] [PubMed] [Google Scholar]
- Gow S. P., Waldner C. L., Harel J. & Boerlin P. Associations between Antimicrobial Resistance Genes in Fecal Generic Escherichia coli Isolates from Cow-Calf Herds in Western Canada. Appl. Environ. Microbiol. 74, 3658–3666 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K. J. et al. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 337, 1107–1111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smorawinska M. et al. Mobilizable narrow host range plasmids as natural suicide vectors enabling horizontal gene transfer among distantly related bacterial species. FEMS Microbiol. Lett. 326, 76–82 (2012). [DOI] [PubMed] [Google Scholar]
- Stecher B. et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 109, 1269–1274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Populaire F., Trieu-Cuot P., Dosbaa I., Andremont A. & Courvalin P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 35, 185–187 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber J. W., Hochhut B. & Waldor M. K. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74 (2004). [DOI] [PubMed] [Google Scholar]
- Guerin E. et al. The SOS response controls integron recombination. Science 324, 1034 (2009). [DOI] [PubMed] [Google Scholar]
- Noyes N.R., Yang X., Linke L.M., Magnuson R.J., Dettenwanger A., Cook S., Geornaras I., Woerner D.R., Gow S.P., McAllister T.A., Yang, H., Ruiz, J., Jones K.L., Boucher C.A., Morley P.S., Belk K.E. Resistome diversity in cattle and the environment decreases during beef production. eLife. 2016 Mar 8;5. pii: e13195. doi: 10.7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso L. M., Harhay G. P., Bono J. L. & Smith T. P. L. Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J. Microbiol. Methods 84, 278–282 (2011). [DOI] [PubMed] [Google Scholar]
- Wichmann F., Udikovic-Kolic N., Andrew S. & Handelsman J. Diverse Antibiotic Resistance Genes in Dairy Cow Manure. mBio 5, e01017–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K. et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23, 1163–1169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4, 2151 (2013). [DOI] [PubMed] [Google Scholar]
- Schmitt H., Stoob K., Hamscher G., Smit E. & Seinen W. Tetracyclines and tetracycline resistance in agricultural soils: microcosm and field studies. Microb. Ecol. 51, 267–276 (2006). [DOI] [PubMed] [Google Scholar]
- Tian B., Fadhil N. H., Powell J. E., Kwong W. K. & Moran N. A. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3, e00377–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BaoWei et al. Metagenomic profiles of antibiotic resistance genes (ARGs) between human impacted estuary and deep ocean sediments. Environ. Sci. Technol. 47, 12753–12760 (2013). [DOI] [PubMed] [Google Scholar]
- Walsh F. & Duffy B. The Culturable Soil Antibiotic Resistome: A Community of Multi-Drug Resistant Bacteria. PLos ONE 8, e65567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-X., Zhang T. & Fang H. H. P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 82, 397–414 (2009). [DOI] [PubMed] [Google Scholar]
- Pehrsson E. C., Forsberg K. J., Gibson M. K., Ahmadi S. & Dantas G. Novel resistance functions uncovered using functional metagenomic investigations of resistance reservoirs. Front. Microbiol. 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow R. S., Fegan N. & Gobius K. S. Integron-containing bacteria in faeces of cattle from different production systems at slaughter. J. Appl. Microbiol. 107, 540–545 (2009). [DOI] [PubMed] [Google Scholar]
- Gow S. P., Waldner C. L., Rajić A., McFall M. E. & Reid-Smith R. Prevalence of antimicrobial resistance in fecal generic Escherichia coil isolated in western Canadian beef herds. Part II–cows and cow-calf pairs. Can. J. Vet. Res. Rev. Can. Rech. Vét. 72, 91–100 (2008). [PMC free article] [PubMed] [Google Scholar]
- Gow S. P., Waldner C. L., Rajić A., McFall M. E. & Reid-Smith R. Prevalence of antimicrobial resistance in fecal generic Escherichia coil isolated in western Canadian cow-calf herds. Part I–beef calves. Can. J. Vet. Res. Rev. Can. Rech. Vét. 72, 82–90 (2008). [PMC free article] [PubMed] [Google Scholar]
- Carson C. A., Reid-Smith R., Irwin R. J., Martin W. S. & McEwen S. A. Antimicrobial resistance in generic fecal Escherichia coli from 29 beef farms in Ontario. Can. J. Vet. Res. 72, 119–128 (2008). [PMC free article] [PubMed] [Google Scholar]
- Berge A. C., Hancock D. D., Sischo W. M. & Besser T. E. Geographic, farm, and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the western United States. J. Am. Vet. Med. Assoc. 236, 1338–1344 (2010). [DOI] [PubMed] [Google Scholar]
- Oikonomou G. et al. Fecal Microbial Diversity in Pre-Weaned Dairy Calves as Described by Pyrosequencing of Metagenomic 16S rDNA. Associations of Faecalibacterium Species with Health and Growth. PLos ONE 8, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Jöbstl D. et al. Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front. Microbiol. 5, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umu Ö. C. O. et al. Resistant starch diet induces change in the swine microbiome and a predominance of beneficial bacterial populations. Microbiome 3, 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N. et al. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 7, 554–569 (2015). [DOI] [PubMed] [Google Scholar]
- Forsberg K. J. et al. Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy F. et al. Identification of Aminoglycoside and β-Lactam Resistance Genes from within an Infant Gut Functional Metagenomic Library. PLos ONE 9, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge A. C., Atwill E. R. & Sischo W. M. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69, 25–38 (2005). [DOI] [PubMed] [Google Scholar]
- Pereira R. V. et al. Effect of on-farm use of antimicrobial drugs on resistance in fecal Escherichia coli of preweaned dairy calves. J. Dairy Sci. 97, 7644–7654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R. V. V. et al. Antimicrobial resistance and prevalence of virulence factor genes in fecal Escherichia coli of Holstein calves fed milk with and without antimicrobials. J. Dairy Sci. 94, 4556–4565 (2011). [DOI] [PubMed] [Google Scholar]
- B. Berge A. C., Moore D. A. & Sischo W. M. Field Trial Evaluating the Influence of Prophylactic and Therapeutic Antimicrobial Administration on Antimicrobial Resistance of Fecal Escherichia coli in Dairy Calves. Appl. Environ. Microbiol. 72, 3872–3878 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA. Dairy 2007, Part V: Changes in Dairy Cattle Health and Management Practices in the United States, 1996–2007 (2009).
- Smith D. P. & Peay K. G. Sequence Depth, Not PCR Replication, Improves Ecological Inference from Next Generation DNA Sequencing. PLos ONE 9, e90234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander T. W. et al. Longitudinal Characterization of Resistant Escherichia coli in Fecal Deposits from Cattle Fed Subtherapeutic Levels of Antimicrobials. Appl. Environ. Microbiol. 75, 7125–7134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. K., Forsberg K. J. & Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. M., Ullman J. L., Teel A. L. & Watts R. J. pH and temperature effects on the hydrolysis of three β-lactam antibiotics: Ampicillin, cefalotin and cefoxitin. Sci. Total Environ. 466–467, 547–555 (2014). [DOI] [PubMed] [Google Scholar]
- McKinney C. W., Loftin K. A., Meyer M. T., Davis J. G. & Pruden A. tet and sul Antibiotic Resistance Genes in Livestock Lagoons of Various Operation Type, Configuration, and Antibiotic Occurrence. Environ. Sci. Technol. 44, 6102–6109 (2010). [DOI] [PubMed] [Google Scholar]
- Peak N. et al. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 9, 143–151 (2007). [DOI] [PubMed] [Google Scholar]
- Ham J. M. & DeSutter T. M. Toward site-specific design standards for animal-waste lagoons: protecting ground water quality. J. Environ. Qual. 29, 1721–1732 (2000). [Google Scholar]
- Conlan S. et al. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci. Transl. Med. 6, 254ra126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Fuentes M. Á., Ortega Morente E., Abriouel H., Pérez Pulido R. & Gálvez A. Antimicrobial resistance determinants in antibiotic and biocide-resistant gram-negative bacteria from organic foods. Food Control 37, 9–14 (2014). [DOI] [PubMed] [Google Scholar]
- Durso L. M. et al. Comparison of bacterial communities in faeces of beef cattle fed diets containing corn and wet distillers’ grain with solubles. Lett. Appl. Microbiol. 55, 109–114 (2012). [DOI] [PubMed] [Google Scholar]
- Shanks O. C. et al. Community Structures of Fecal Bacteria in Cattle from Different Animal Feeding Operations. Appl. Environ. Microbiol. 77, 2992–3001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody M. A., Van Rossum T., Lo R. & Brinkman F. S. L. Evaluation of shotgun metagenomics sequence classification methods using in silico and in vitro simulated communities. BMC Bioinformatics 16, 363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Noyes, N.R., Doster, E., Martin, J.N., Linke, L.M., Magnuson, R.J., Yang, H., Geornaras, I., Woerner, D., Jones, K.L., Ruiz, J., Boucher, C.A., Morley, P.S., Belk, K.E. Use of shotgun metagenomic sequencing technology to detect foodborne pathogens within their microbiome in beef production chain. Applied and Environmental Microbiology. 2016; doi:10.1128/AEM.00078-16 [DOI] [PMC free article] [PubMed]
- Martínez J. L., Coque T. M. & Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 13, 116–123 (2015). [DOI] [PubMed] [Google Scholar]
- Hall R. M. & Schwarz S. Resistance gene naming and numbering: is it a new gene or not? J. Antimicrob. Chemother. 71, 569, doi: 10.1093/jac/dkv351 (2016) [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. & Usadel B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics. 30, 2114, doi: 10.1093/bioinformatics/btu170 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K. et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A. G. et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 57, 3348–3357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S. & Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinforma. Oxf. Engl. 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I. et al. Tablet–next generation sequence assembly visualization. Bioinforma. Oxf. Engl. 26, 401–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J. N., Stine O. C., Bravo H. C. & Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P. J. & Holmes S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLos Comput Biol 10, e1003531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P. & Gallagher E. D. Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280 (2001). [DOI] [PubMed] [Google Scholar]
- Clarke K. R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143 (1993). [Google Scholar]
- Ritchie M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.