Abstract
Mitochondrial organization is often altered to accommodate cellular bioenergetic and biosynthetic demands. Changes in metabolism are a hallmark of a number of diseases, including cancer; however, the interdependence between mitochondrial metabolic function and organization is not well understood. Here, we present a noninvasive, automated and quantitative method to assess mitochondrial organization in three-dimensional (3D) tissues using exclusively endogenous two-photon excited fluorescence (TPEF) and show that mitochondrial organization reflects alterations in metabolic activities. Specifically, we examine the organization of mitochondria within live, engineered epithelial tissue equivalents that mimic normal and precancerous human squamous epithelial tissues. We identify unique patterns of mitochondrial organization in the different tissue models we examine, and we attribute these to differences in the metabolic profiles of these tissues. We find that mitochondria are clustered in tissues with high levels of glycolysis and are more highly networked in tissues where oxidative phosphorylation is more dominant. The most highly networked organization is observed within cells with high levels of glutamine consumption. Furthermore, we demonstrate that mitochondrial organization provides complementary information to traditional morphological hallmarks of cancer development, including variations in nuclear size. Finally, we present evidence that this automated quantitative analysis of endogenous TPEF images can identify differences in the mitochondrial organization of freshly excised normal and pre-cancerous human cervical tissue specimens. Thus, this method could be a promising new modality to assess the role of mitochondrial organization in the metabolic activity of 3D tissues and could be further developed to serve as an early cancer clinical diagnostic biomarker.
Keywords: mitochondrial organization, cancer bioenergetics, auto-fluorescence, human papilloma virus, optical biomarkers
Mitochondria are key energy producing organelles, which are involved in processes leading to the production of important biosynthetic substrates and signaling molecules. In addition, mitochondria play an important role in the integration of death signals during apoptosis.1 Therefore, alterations in mitochondrial function are implicated in the development of numerous diseases, with the Warburg effect during cancer development as a prominent example.2 In fact, fundamental differences in metabolic preferences between healthy and cancerous tissues have been observed, leading to the recognition that metabolic alterations are important cancer hallmarks (as reviewed in Refs. 3 and 4). These metabolic modifications confer advantages to cancer cells by activating pathways that support cell proliferation and promote survival and often engage mitochondria.2–4
Changes in mitochondrial function have been associated with changes in mitochondrial structure and organization.5–7 Mitochondria are typically organized as tubular networks that have the ability to change dynamically through fusion and fission.7,8 The factors that affect mitochondrial organization and their impact on cell function are diverse and not thoroughly understood. However, studies performed with cancer and stem cells have indicated the formation of more extensive mitochondrial networks when oxidative phosphorylation levels increase relative to glycolysis, as a result of substrate changes or during differentiation.9–14 To date, the majority of studies examining mitochondrial morphology employ electron microscopy or mitochondria-specific fluorescent dyes.9–12,15,16 These methods can give rise to artifacts, are not well suited for measurements in three-dimensional (3D) tissues and cannot be translated to a clinical setting. Furthermore, most analytical approaches to assess mitochondrial organization rely on qualitative visual inspection or manual tracing of mitochondrial networks.5,6,9,10,15,16
To overcome these limitations, we considered the automated analysis of endogenous two-photon excited fluorescence (TPEF) images as a means to quantify depth-resolved mitochondrial organization in 3D tissue specimens. We have previously used such images to assess the redox state of epithelial tissues, relying on the natural fluorescence detected from two coenzymes, nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD).17 NADH fluorescence images are sensitive predominantly to the bound NADH form residing within mitochondria.18,19 In fact, the organization of the bright autofluorescent features in NADH images is correlated with the organization of traditional mitochondrial dyes.20 Thus NADH autofluorescence is essentially localized within mitochondria, while prominent dark features in these images are formed by cell borders and nuclei that have little or no mitochondria. Therefore, these images contain morphological information regarding key organelles (nuclei and mitochondria) that serve important cellular functions, and it may be valuable to consider their combined use as diagnostic biomarkers.
Here, we demonstrate that it is possible to identify differences in the mitochondrial organization of 3D normal and precancerous tissues using noninvasive imaging that relies on endogenous NADH fluorescence and an analysis method that is fully automated and quantitative. Specifically, we characterize mitochondrial features by identifying the intrinsic NADH TPEF signal and using a digital object cloning and Fourier-based analysis approach that provides a quantitative measure of mitochondrial clustering. We detect significant differences in mitochondrial clustering between normal and precancerous engineered epithelial tissue equivalents (EETEs) and we observe similar differences in freshly excised human cervical epithelial specimens. We attribute these differences to changes in the relative levels of glycolysis and oxidative phosphorylation, with higher levels of glycolysis yielding more clustered mitochondria. Interestingly, cells fueling oxidative phosphorylation via enhanced glutamine consumption appear to have the least clustered (i.e., most highly networked) mitochondria. Thus, we illustrate how this approach may enable studies that examine the interdependence of mitochondrial function and organization. These studies in turn will be important for the diagnosis and treatment monitoring of not only cancer, but also potentially a number of metabolic diseases.
Material and Methods
Engineered epithelial tissue equivalents
To model normal and precancerous epithelial tissues we constructed EETEs based on previously established protocols21 using five different types of epithelial cells: primary human foreskin keratinocytes serving as normal controls (C HFKs), HFKs with stable expression of HPV16 E7, HPV16 E6 and HPV16 E6/E7 and HPV 16 immortalized HKc/DR cells. Further details are provided in Supporting Information Methods.
Glucose and oxygen starvation studies
HFKs were cultured on 50 mm glass bottom dishes (Mattek) until reaching confluence. Cell cultures were imaged after exposure for at least 2–4 hr to: (i) commercially available KSFM, (ii) media prepared in-house with the same glucose and glutamine concentrations as KSFM (Low Glu), or (iii) media prepared in-house with the same glutamine concentration as KSFM, but with no glucose (No Glu; See Supporting Information methods for detailed composition). In addition, images were acquired from cells during the first 30 min of exposure to KSFM media that had been nitrogen bubbled for 6 hr and contained very low concentrations of O2.
Data were acquired from at least two different dishes and several fields from each group for a total of 40, 19 and 20 fields for the KSFM, Low Glu and No Glu groups, respectively. For the low-O2 experiment, 31 and 35 fields were imaged from the KSFM and low-O2 groups, respectively.
Cervical specimens
All human tissue investigations were approved by the Institutional Review Board of Tufts Medical Center and Tufts University and informed consent was obtained from each subject. Four normal ectocervical specimens were obtained from the discarded cervical tissue acquired immediately following hysterectomy of four patients with no known cervical tissue abnormalities. Abnormal cervical specimens were excised from the transformation zone of the uterine cervix of four patients undergoing colposcopy before a loop electrosurgical excision procedure. Upon tissue excision, each specimen was placed in Ringer’s solution and imaged within two hours. One to four regions of each specimen were imaged yielding a total of 11 and 12 stacks from the normal and CIN specimens, respectively. Histology was performed following imaging and an experienced pathologist (M. G-M) classified two abnormal specimens as cervical intraepithelial neoplasia (CIN) 1, one as focal CIN 2, and one as CIN 2 of the surface epithelium.
TPEF data acquisition
TPEF images were acquired on a Leica TCS SP2 confocal microscope (Wetzlar, Germany) equipped with a Ti:sapphire laser (Spectra Physics, Mountain View, CA). Samples were placed on glass coverslips, excited with 755, 800 and 860 nm light and imaged using a 63×, 1.2 NA water immersion objective. The microscope was equipped with a microincubation system that kept samples in a humid chamber maintained at 37°C and 5% CO2 (Okolabs, Ottaviano, Italy). TPEF images were acquired using emitter bandpass filters centered at 460 and 525 nm. Image stacks from one to four regions from 43 independent EETE tissues (nC_HFK = 10, nHKc/DR = 4, n16E6_HFK = 10, n16E7_HFK = 6 and n16E6E7_HFK = 13) were acquired. TPEF 3D images were rendered in ImageJ (v10.2) and OsiriX (v3.0.2).
Mitochondrial feature isolation in EETEs and cervical tissues
To assess mitochondrial organization, we analyzed endogenous TPEF images acquired at 755 nm excitation and 460 nm emission dominated by mitochondrial bound NADH autofluorescence. A series of image preprocessing steps was necessary to remove keratin-associated fluorescence and prominent image features associated with the nonfluorescent nuclear and extracellular features22 (Supporting Information Methods and Supporting Information Fig. S1). Thus, the processed images contained intensity fluctuation patterns that primarily reflect mitochondrial morphology. To acquire quantitative information regarding the organizational characteristics of these features, we performed a two-dimensional (2D) Fourier transform. This standard mathematical operation describes, in this case, the relative prevalence of intensity fluctuations at different characteristic spatial frequencies. As spatial frequency, k, is inversely proportional to length scale, the squared amplitude of the Fourier transform of an image, referred to as the power spectral density (PSD) or R (k), is a metric that can be used to quantify the occurrence of morphological features of different length scales within the image. The morphology of biological cells and tissue often exhibits PSDs with an inverse power law dependence on spatial frequency [i.e., R (k) ∝ k−β]. Increasing values of the power law exponent, β, correlate with an increased degree of clustering in the image features22 (Supporting Information Fig. S1c). Thus, to assess clustering, we fit to the PSD an inverse power law equation of the form R (k) = Ak−β. The power law exponent, β, was computed for each mitochondrial image and referred to as the level of mitochondrial clustering.
Quantification of tissue organization in autofluorescence images
To assess depth-dependent changes in TPEF image features with sizes on the order of individual cells and cell nuclei, we also used a Fourier-based analysis approach, which has been described previously23 and is detailed in Supporting Information Methods. This method relies on the fact that nuclei and extracellular spaces that lack mitochondria appear dark in NADH TPEF images that have not been pre-processed. Because the size of individual cells and nuclei have characteristic length scales, their presence has a significant effect on the PSD magnitude of the images at the spatial frequencies corresponding to those length scales. Therefore, features with sizes in the 8–12 µm range, attributed primarily to cell nuclei, affect the magnitude of the PSD in the 0.125–0.083 µm−1 range. If the size and prevalence of features in this frequency range does not change as a function of depth within an epithelial tissue, we expect that the magnitude of the PSD values will remain fairly constant and the variation of the PSD signal will be small (Supporting Information Fig. S1a). If the size and prevalence of such morphological features (i.e., cell nuclei) changes as a function of depth, the magnitude of the PSD within the corresponding spatial frequency range will change accordingly (Supporting Information Fig. S1a). Thus, we averaged PSD values within the spatial frequency range corresponding to nuclear sizes and then computed the variance of these average PSD values from the image slices at each depth. This metric of PSD variation within each EETE stack that we imaged provided an estimate of nucleus-to-cytoplasm ratio variation as a function of depth.
Statistics
To assess the significance of the differences in either β or the normalized PSD variation between the five EETE groups, an ANOVA was performed with post-hoc Tukey Honestly Significant Different tests in Jmp 8 SAS (Cary, NC). For each ANOVA, a nested design considered multiple regions from individual samples nested within each tissue group.
Results
Distinct depth-dependent variations in morphological and differentiation features are detected in endogenous TPEF images of normal and precancerous epithelia
We have employed five different types of EETEs to model normal squamous epithelia and pre-cancerous epithelial lesions. EETEs corresponding to normal, fully differentiated squamous epithelia were generated from primary HFKs (C HFKs) seeded on top of a fibroblast containing collagen matrix (Fig. 1). EETEs were also constructed using HFKs that are immortalized following expression of the full-length “high-risk” human papillomavirus type 16 (HPV16) genome (HKc/DR cells) (Fig. 1). HKc/DR EETEs are considered good in vitro models for high-grade premalignant squamous intraepithelial lesions, since these cells show an mRNA expression profile that is similar to that of HPV16 positive cervical lesions and cancers.24 We also examined EETEs consisting of HFKs expressing HPV16 E6 and/or E7. It is well established that E6 and E7 inhibit the p53 and retinoblastoma (pRB) tumor suppressor pathways, respectively, that can affect cell apoptosis, differentiation and proliferation.25 Since expression of viral proteins, such as E6 and E7, cause cancer by interfering with cellular pathways that are also often perturbed as a result of carcinogenic genomic variations,26 the HPV16 E6 and/or E7 expressing HFK EETEs may serve as useful models for a broad range of cancers.
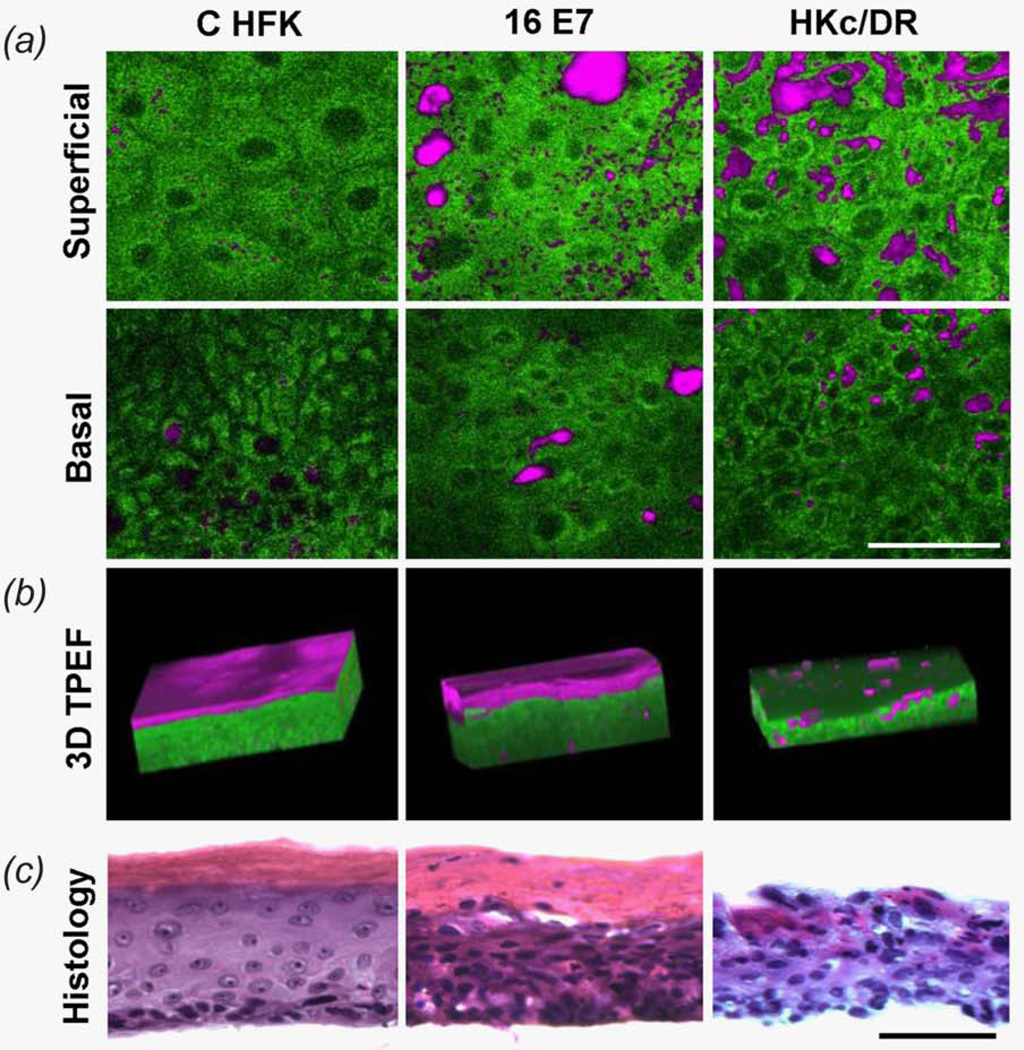
Figure 1.
(a) Representative endogenous TPEF images from distinct depths of CHFK, 16E7 HFK and HKc/DR EETEs. NADH TPEF is pseudo-colored green, while keratin positive pixels are shown in magenta. (b) Corresponding 3D projections of optical stacks. (c) Corresponding histological sections following hematoxylin and eosin staining. Scale bars are 50 µm. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The intrinsic fluorescence excited at 755 nm and detected from these epithelial tissues in the 460 nm region is dominated by mitochondrial bound NADH autofluorescence and reveals substantial differences in tissue organization and morphology between the different EETEs (Figs. 1a and 1b and Supporting Information Fig. S2). For C HFK EETEs, a gradient of morphological changes is observed from the basal to the superficial layers, as demonstrated by a gradually decreasing nuclear to cytoplasmic ratio, increased cell size (Fig. 1a) and a high level of keratin fluorescence in the topmost, cornified layer (Figs. 1b and 1c). Co-staining of DAPI and PCNA localized to the basal layer confirms that, as expected, proliferation is confined to the basal layer in C HFK EETEs (Supporting Information Fig. S2d). Generally, 16E6 HFK EETEs exhibit similar, but overall less pronounced morphological changes with respect to depth compared with the C HFK EETEs (Supporting Information Fig. S2). This is consistent with the corresponding transverse histological cross sections and PCNA staining that remains primarily confined to the basal layer (Supporting Information Fig. S2c and S2d). In contrast, 16E7 and 16E6E7 HFK EETEs consist of irregularly shaped and arranged cells throughout the depth of the epithelium (Fig. 1a and Supporting Information Fig. S2). Despite an absence of clear morphological changes in cell size and shape as a function of depth, these EETEs maintain a cornified layer, as shown by the high level of keratin fluorescence that is particularly prominent within the top-most tissue layers (Fig. 1b and Supporting Information Fig. S2) and confirmed by hematoxylin and eosin staining (Fig. 1c and Supporting Information Fig. S2). Furthermore, 16E7 and 16E6E7 HFK EETEs stain positive for PCNA throughout the basal and parabasal epithelial layers, indicating delocalized proliferation in differentiated layers of the epithelium (Supporting Information Fig. S2d). In HKc/DR EETEs, low levels of cellular differentiation (Fig. 1a) are apparent along with proliferation in upper epithelial layers (Supporting Information Fig. S2d), as well as abnormal and spotty keratinization (Fig. 1b). Rotating 3D-renderings and optical section stacks of the intrinsic fluorescence images from representative EETEs are included as Supporting Information videos and provide more detailed representations of these features. The optical sections and 3D reconstructions demonstrate that the EETEs we have examined provide a robust platform for assessing a range of morphological and differentiation alterations that can potentially occur at the onset of cancer. In addition, it is evident that these differences are readily observable within high-resolution TPEF images that rely entirely on endogenous fluorescence contrast.
Mitochondrial clustering is altered in precancerous epithelial tissues
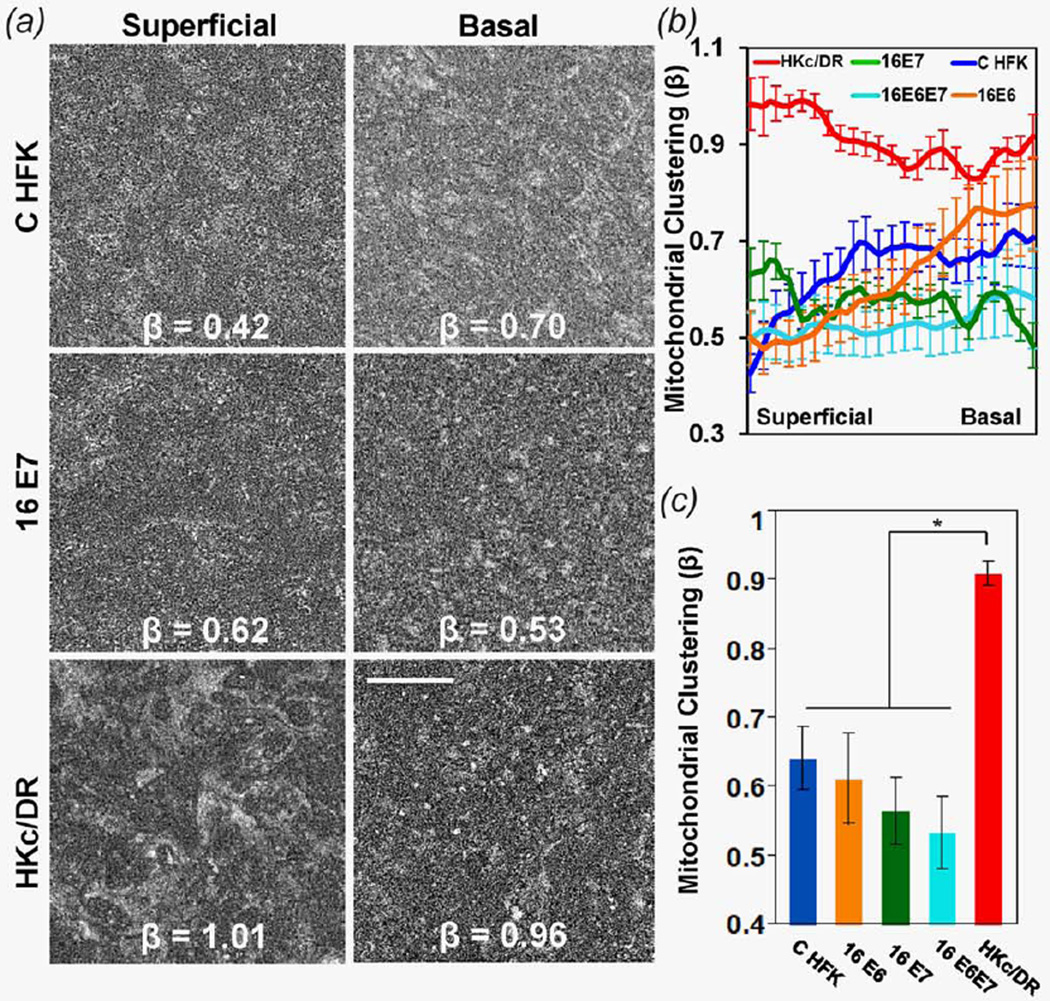
The features that dominate the variations we observe readily between tissues and/or as a function of depth are mainly associated with the borders of cells and nuclei, which appear dark. To characterize intensity fluctuations that are influenced primarily by the organization of the mitochondria themselves, it is important to pre-process the images in order to remove keratin fluorescence and the larger scale (low spatial frequency) intensity variations associated with nuclear and cell features (Methods and Supporting Information Fig. S1). Representative resulting images are shown in Figure 2a and Supporting Information Figure S3 from different layers for each EETE group we examined. The characteristic intensity fluctuations in these images are described by the PSD, R (k), as a function of spatial frequency, k. We fit an equation of the form R (k) = Ak−β to the PSD for a range of spatial frequencies higher than 0.1 µm−1 (corresponding to features that are smaller than 10 µm) to extract the value of the parameter β (Supporting Information Fig. S1c). Since the intensity fluctuations in these images emanate to a great extent from mitochondrial bound NADH fluorescence, β describes the level of characteristic mitochondrial organization (i.e., clustering) and its value is shown for each one of the panels (Fig. 2a and Supporting Information Fig. S3). The detailed depth-dependence of this clustering parameter exhibits distinct patterns for the different types of EETEs we examined (Fig. 2b). Specifically, EETEs constructed with C HFKs exhibit a gradual increase in mitochondrial clustering from the superficial to the basal layer (Fig. 2b). Less variable clustering values, similar in magnitude to those found in the basal layer of the C HFK EETEs, are observed within HKc/DR EETEs (Fig. 2b). This trend results in a significant (p < 0.05) increase in the overall mitochondrial clustering features observed in the HKc/DR EETEs compared with all other EETEs (Fig. 2c).
Figure 2.
(a) Representative processed images containing predominantly mitochondrial organization features from distinct epithelial layers of CHFK, 16E7 and HKc/DR EETE with corresponding mitochondrial clustering values, β. Scale bar is 50 µm for all images. (b) Mean mitochondrial clustering as a function of normalized depth, from the basal to the upper-most superficial layer for each EETE type we examined. (c) Mean overall mitochondrial clustering for each EETE group. Error bars indicate standard error for each group. Statistical significance is indicated for p <0.05. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Expression of E6 or E7 within the cells of the 16E6 and/or 16E7 HFK EETEs resulted in distinct patterns of mitochondrial organization. Specifically, similar to C HFK EETEs, 16E6 HFK EETEs exhibit a gradual increase in mitochondrial clustering from the superficial to the basal layer (Fig. 2b). The overall clustering is also similar in magnitude to that of C HFK EETEs and significantly lower (p < 0.05) than that of HKc/DR EETEs (Fig. 2c). 16E7 HFK EETEs and 16E6E7 HFK EETEs also have overall lower mitochondrial clustering than what is observed for the HKc/DR EETEs (p < 0.05; Fig. 2c). Interestingly, the 16E7 and 16E6E7 HFK EETEs are characterized by the lowest overall levels of clustering (Fig. 2c). Taken together, these data demonstrate that mitochondrial clustering reveals depth-dependent and overall differences not only between normal and precancerous tissues, but also among different types of precancerous epithelia.
Changes in mitochondrial clustering reflect alterations in mitochondrial function
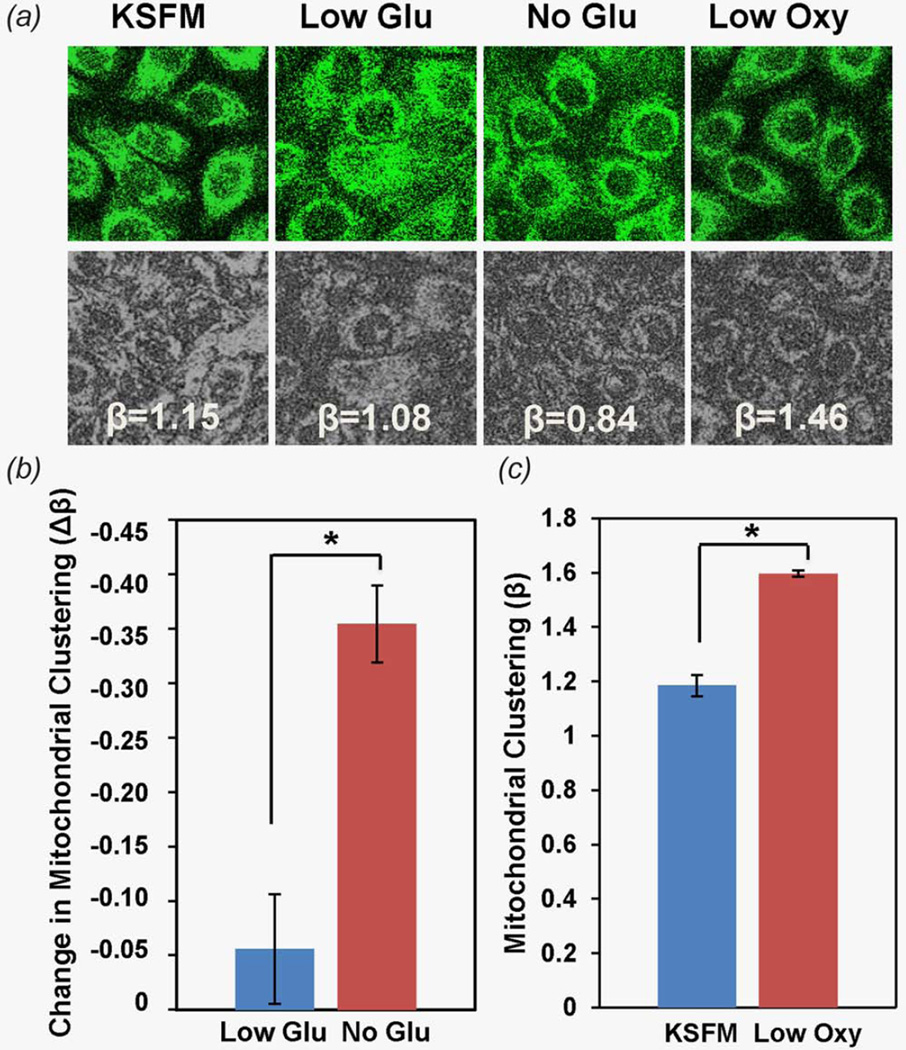
To investigate the mechanisms that may drive these distinct clustering alterations in HKc/DR, 16E7 and 16E6E7 HFK EETEs we performed dynamic measurements over time using 2D HFK cultures exposed to different media intended to enhance the levels of either glutaminolysis or glycolysis (Fig. 3). When cells are forced to use glutamine as a carbon source for energy production by depriving them of glucose and pyruvate, we detect a significant decrease in the level of mitochondrial clustering (Figs. 3a and 3b, p < 0.05). When cells are forced to produce energy predominantly via glycolysis due to limited availability of oxygen, levels of mitochondrial clustering increase significantly (Figs. 3a and 3c, p < 0.05). These results support the hypothesis that enhanced mitochondrial clustering in HKc/DR EETEs represents enhanced levels of glycolysis, while the decreased levels of clustering in 16E7 and 16E6E7 HFK EETEs may signal enhanced levels of glutamine consumption.
Figure 3.
(a) Representative endogenous TPEF images detected at 460 nm following 755 nm excitation from cells exposed to KSFM, Low Glu (low glucose), No Glu (no glucose) and Low Oxy (low O2) media (top row). Corresponding images following processing to isolate mitochondrial features are shown in the bottom row. (b) Changes in mitochondrial clustering (Δβ) observed after 2 hr exposure of HFK cells either to Low glucose or No glucose media. (c) Mitochondrial clustering observed before and within 30 min of the exposure of cells to low O2 media. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Mitochondrial organization provides complementary information to cell and nuclear morphological features
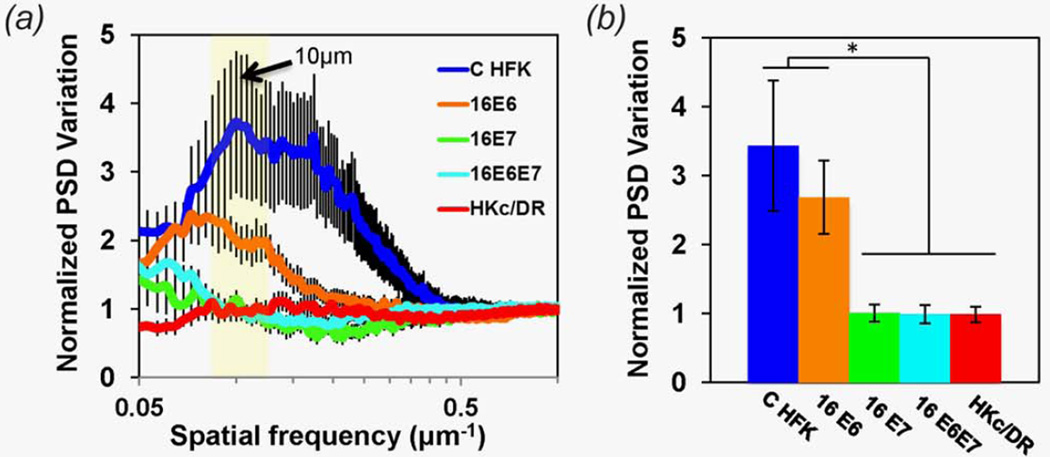
To examine further the potential utility of mitochondrial organization as a diagnostic biomarker, we assessed its relationship with another parameter that is often used in early cancer diagnosis, namely nuclear size variation or pleomorphism. The PSD variation over a wide range of spatial frequencies (1–0.05 µm−1) corresponding to features with characteristic lengths of 1 to 20 µm is shown for C HFK, 16E6 HFK, 16E7 HFK, 16E6E7 HFK and HKc/DR EETEs in Figure 4a. We find that PSD variation is significantly higher for the C HFK and 16E6 HFK EETEs compared with 16E7, 16E6E7 HFK and HKc/DR EETEs. This is especially true in the 0.125–0.083 µm−1 range corresponding to image features ~8–12 µm in size, which is consistent with the size range of nuclei (Fig. 4a, yellow shaded area, Fig. 4b, p ≤ 0.05). The high PSD variation values indicate that the relative prevalence of nuclei vary significantly as a function of depth in the C HFK and 16E6 HFK EETEs compared to the other EETEs. This difference is highly consistent with our qualitative observations of the depth-dependent variations in nuclei and cell size in the endogenous TPEF image stacks, which is high for the C HFK and 16E6 HFK EETEs and relatively small for the 16E7, 16E6E7 HFK and the HKc/DR EETEs (Fig. 1, Supporting Information Fig. S2 and Supporting Information videos).
Figure 4.
(a) Mean normalized PSD depth-dependent variation as a function of spatial frequency for each tissue type. Error bars represent standard error (SE) values. The yellow region corresponds to spatial frequencies in the 10−0.983–10−1.081 µm−1 or 0.125–0.083 µm−1 region, relevant to features over the 8–12 µm length scale. (b) Mean (+/−SE) normalized PSD variation of features on the order of 8–12 µm. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
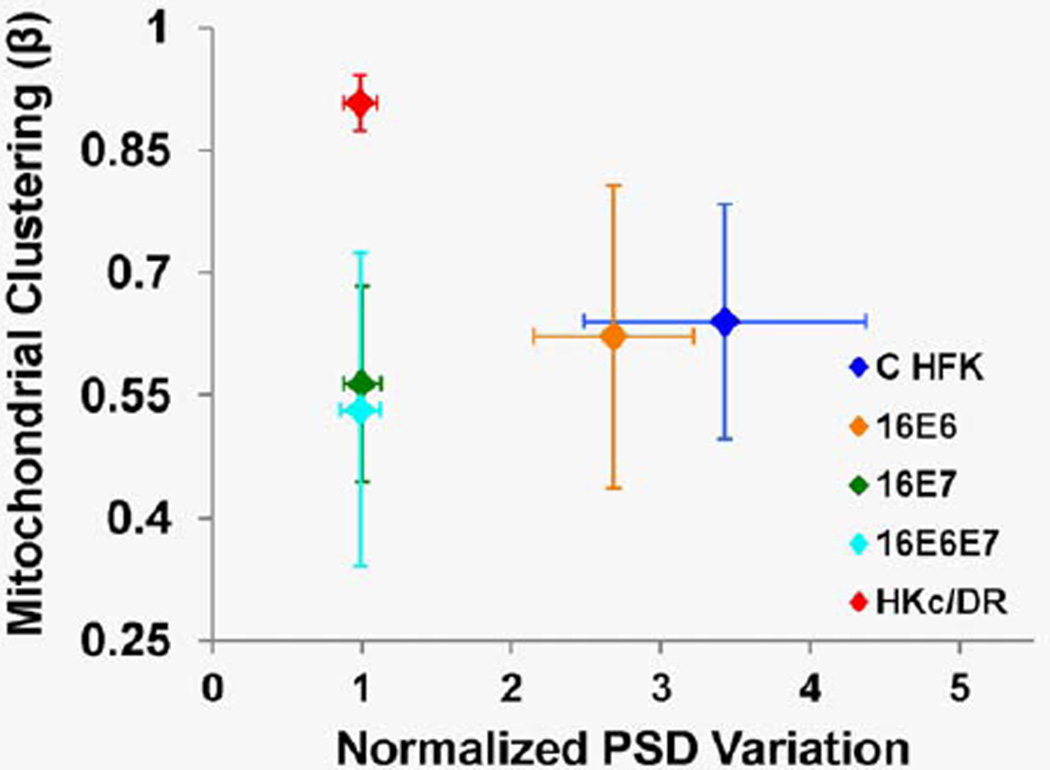
Interestingly, differences in the PSD variation among different EETE types are not well correlated with corresponding differences in mitochondrial clustering (Fig. 5). While mitochondrial clustering exhibits greater differences between the EETEs constructed with HKc/DR and those constructed with 16 E6E7 and 16E7 HFKs, PSD variation distinguishes the C and 16E6 HFK EETEs from the remaining groups. These differences suggest that mitochondrial clustering and nuclear pleomorphism (as assessed by PSD variation) may be sensitive to different cellular and tissue features that can change at the onset of cancer development and may serve as complementary cancer biomarkers.
Figure 5.
Mean overall mitochondrial clustering as a function of the mean normalized depth-dependent PSD variation for features on the order of 8–12 µm, for each EETE group. Error bars indicate standard deviation for each group. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Mitochondrial organization within human cervical tissues
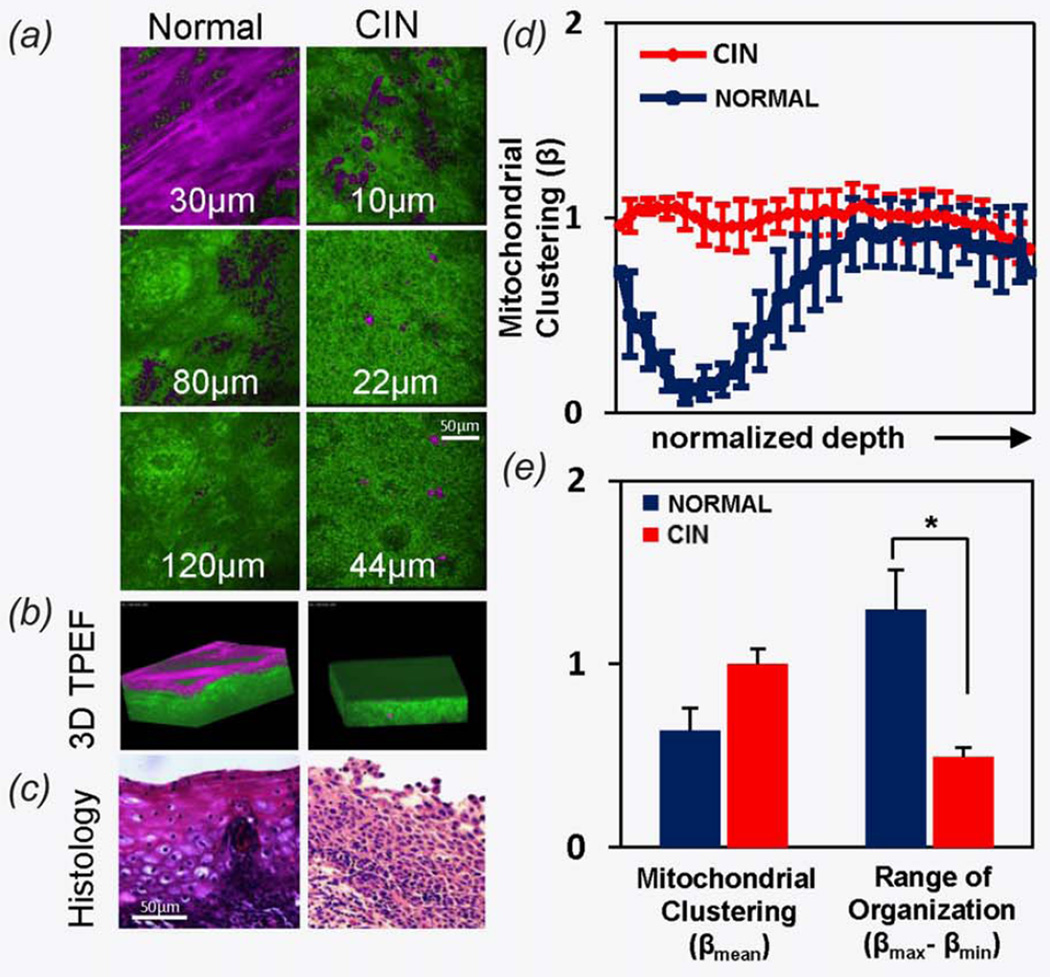
To determine whether the mitochondrial organization biomarker we developed based on analyses of EETEs may be relevant to the characterization of human tissues, we performed a proof-of-principle study on a small cohort of normal (n = 4) and pre-cancerous human cervical tissue specimens (n = 4) using image acquisition and analysis approaches that we developed for the analysis of the EETEs (Fig. 6). Although EETEs recapitulate the basic architecture of human ectocervical tissues, the cervical stroma extends intermittently toward the surface of the epithelium such that basal cells may reside adjacent to invaginating stromal tissue (Figs. 6a and 6b and Supporting Information videos). Thus, in the normal ectocervix specimens, differentiation advances as a function of axial and lateral distance from the dermal papillae. The epithelium of the CIN specimens lacks fully differentiated superficial cells and contains round basal-like cells in the lower and middle tissue layers (Figs. 6a and 6b). Histological staining confirms these findings (Fig. 6c). Despite the more complex architecture of human tissue specimens compared with that of the EETEs, depth-dependent mitochondrial organization differences can be detected in endogenous TPEF images when we apply the same algorithm developed for the analysis of the EETEs (Fig. 6d). Similar to C HFK EETEs, mitochondria become increasingly clustered within the normal cervical tissues as a function of depth from the surface, while the mitochondrial clustering in the CIN specimens remains constant throughout the full thickness of the epithelium. This is readily observed by the highly significant decrease in the range of clustering, that is, simply the difference between the maximum and minimum clustering values detected within the epithelium, between the CIN and normal tissues (p < 0.03, Fig. 6e).
Figure 6.
(a) Depth-resolved endogenous TPEF images and (b) 3D rendered optical stacks of representative fields from healthy and CIN freshly excised human cervical tissue specimens. TPEF dominated by NADH signal is shown in green, while keratin pixels are identified as magenta. (c) Hematoxylin and eosin stained sections from the corresponding tissue specimens. (d) Mean mitochondrial clustering as a function of depth and (e) mean overall mitochondrial clustering and mitochondrial clustering range for normal (n = 4) and CIN (n = 4) cervical tissue specimens. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
However, the more complex architecture of the human ectocervical tissue specimens in combination with the complexity of arranging the tissue so that each optical section that is acquired corresponds to a distinct cell layer of the epithelium, currently limits to some extent the application of the approach we developed with EETEs, where cells of uniform differentiation and morphological features are present within each optical section. This was particularly true for the assessment of nuclear pleomorphism through PSD variation, since due to the papillar architecture of the tissue, many of the optical sections through the depth of the normal epithelia contain cells representing basal, parabasal and in some cases even superficial cell layers with a wide range of nuclear sizes. Thus, the magnitude of the PSD does not change significantly as a function of depth and PSD variation is not significantly higher for the normal than the CIN tissues. We are currently developing more sophisticated algorithms that take into account the papillar architecture of the tissue and which may allow assessment of nuclear pleomorphism in human tissues through PSD analysis.
Discussion
Changes in mitochondrial function play a significant role during normal tissue development and are implicated in the establishment and progression of a number of diseases, including cancer.1,3 As such, they are considered important diagnostic biomarkers. The factors that affect mitochondrial function and their relationship with mitochondrial structure and organization are not fully understood. However, several studies have suggested that changes in mitochondrial structure and organization may be linked to alterations in bioenergetic pathways that are observed during pathogenic processes, including cancer.9–16 Typically, such mitochondrial morphological features are characterized using exogenous chromophores and described with qualitative assessments of organization. This limits their use as biomarkers, particularly in live, 3D specimens, where permeability of the tissue to a stain and stain toxicity are potential problems. To overcome these limitations, we developed a robust, noninvasive and quantitative method for assessing mitochondrial organization in live normal and precancerous EETEs, as well as freshly excised human cervical tissue specimens. Using only endogenous sources of fluorescence (namely NADH TPEF) and automated analytical techniques, we show that distinct depth-resolved mitochondrial organization patterns are detected in epithelial tissues upon the onset of precancerous changes (Fig. 2). Importantly, these changes in mitochondrial organization reflect the underlying metabolic pathway that is disturbed, with more clustered mitochondria representing enhanced levels of glycolysis and less clustered (more highly networked) mitochondria evolving in cells that rely more on glutamine for energy production (Fig. 3). Finally, we demonstrate that the mitochondrial clustering patterns provide complementary information relative to more traditional cellular and tissue morphological metrics used routinely as histopathological biomarkers associated with nuclear and cell shape features (Fig. 5).
A number of interesting patterns are observed in the depth-dependence and overall values of mitochondrial clustering features of C HFK, 16E6 HFK, 16E7 HFK, 16E6E7 HFK and HKc/DR EETEs that suggest some consistent relationships between mitochondrial organization and metabolic function. For example, our results are consistent with the model that the observed high levels of mitochondrial clustering are associated with increased levels of glycolysis. The most direct evidence for this hypothesis is provided by the increase in mitochondrial clustering observed in HFK cultures exposed to media with no or minimal oxygen (Fig. 3). In addition, we observe significantly increased mitochondrial clustering throughout the depth of HKc/DR EETEs compared with all other EETE types. This increased level of mitochondrial clustering could be attributed to enhanced levels of glycolysis that are expected to support the high biosynthetic demands imposed upon the pre-cancerous cells within the HKc/DR EETEs. In fact, we have recently reported that HKc/DR EETEs exhibit higher levels of glycolysis than all of the other EETE types we examined using a standard biochemical assay that quantifies the amount of lactate produced relative to the amount of consumed glucose.27 Furthermore, optical studies of the redox ratio of C HFK and HKc/DR EETEs have also reported changes consistent with enhanced glycolytic activity relative to oxidative phosphorylation in the HKc/DR EETEs.17,27 Finally, others have reported that cells undergoing glycolysis exhibit more clustered and vesicular mitochondrial organization.9,13,14 Thus, the increased mitochondrial clustering that we identify non-invasively within the basal layers of C HFK EETEs and throughout the depth of the HKc/DR EETEs may serve as a biomarker of increased glycolysis relative to oxidative phosphorylation.
A shift to oxidative phosphorylation-based energy production along with the establishment of more extensive mitochondrial networks has been observed within differentiated stem cells and cardiomyocytes.10,11,16 This shift may explain the decreased mitochondrial clustering, which indicates the presence of thinner, more elongated mitochondrial networks, in the well-differentiated superficial layers of the C HFK EETEs (Figs. 1, 2a and 2b and Supporting Information Figs. S2 and S3). In addition, it is possible that this type of organization is more optimally suited to support energy demands that are distributed more uniformly throughout the cell.
The organization of mitochondria within 16E7 HFK and 16E6E7 HFK EETEs is characterized by low clustering levels throughout the epithelium (Fig. 2 and Supporting Information Fig. S3). These patterns may also be associated with enhanced rates of oxidative phosphorylation relative to glycolysis. However, it is curious that clustering levels decrease even further within the basal layers of these tissues, suggesting that additional metabolic modifications may be involved. It has been shown that E7 expression catalyzes the formation of the dimeric form of pyruvate kinase type M2,28 which prevents pyruvate from entering the mitochondria, requiring the use of glutamine as a substrate for the Krebs cycle via a process typically referred to as glutaminolysis.29 Additionally, high levels of myc expression can induce glutamine-addiction and have been observed in the presence of both HPV E6 and E7 expression.30,31 Therefore, higher levels of glutamine consumption are expected in the HPV16 E7 and HPV16 E6E7 expressing EETEs, which exhibit the lowest levels of mitochondrial clustering. In addition, our studies with standard tissue culture cells provide direct evidence that energy production through enhanced glutamine consumption results in more highly networked mitochondria (Fig. 3). This hypothesis is also supported by our recent mass spectrometry-based findings that HPV16 E7 and HPV16 E6E7 EETEs have significantly higher levels of glutamine consumption than all other EETE types we examined.27 Taken together, these studies suggest that mitochondrial clustering may serve as a sensitive biomarker for changes in the balance between glycolytic and oxidative phosphorylation-based energy production, and it may be further enhanced when the primary carbon source for the latter switches from glucose (pyruvate) to glutamine. More detailed studies will be needed to assess the sensitivity of mitochondrial clustering as a biomarker for such specific metabolic changes.
Our initial examination of human cervical tissue specimens reveals promising results in terms of the potential for using mitochondrial organization as a biomarker to detect differences between normal and pre-cancerous lesions (Fig. 6). Specifically, we find that the CIN specimens are characterized by elevated, depth-invariant mitochondrial clustering levels, consistent with enhanced glycolytic energy production relative to oxidative phosphorylation and a Warburg-like phenotype, as discussed above. In contrast, the normal epithelium exhibits significant depth-dependent variations in organization, suggesting a gradient in the balance between glycolysis and oxidative phosphorylation activity. While the current experiments provide a proof-of-principle that this method may be used for assessment of clinical specimens, a significantly larger number of clinical specimens that represent the full spectrum of changes associated with HPV-associated precancerous cervical lesions will need to be analyzed in the future. We anticipate that algorithm modifications that take into account the more complex architecture of tissue due to the presence of papillae will be necessary to fully exploit the diagnostic capabilities of the approach developed in this study. However, these promising preliminary results suggest that assessment of mitochondrial organization may represent a clinically useful diagnostic biomarker, especially with the advent of compact TPEF-capable endoscopes,32–34 and the use of TPEF imaging clinically.35,36 This is further supported by the complementary nature of the information provided by mitochondrial organization and traditional cancer hallmarks focusing on nuclear morphological features (Fig. 5).
It is interesting to note that E7 appears to be the major factor in the metabolic alterations as well as the changes in nuclear morphology and mitochondrial organization that are detected in HPV16 E6/E7 expressing EETEs. This is not entirely unexpected since, as previously described, E7 can induce the Warburg effect28 and drive genomic destabilization,37 which presumably contribute to the metabolic alterations and nuclear changes observed in Figures 2 and 5, respectively. Our finding is also consistent with experiments using transgenic mouse models that have shown that HPV16 E7 is a more potent oncogene in mucosal tissues than E6.38,39 Nonetheless, E6 and E7 are uniformly found coexpressed in HPV-associated cancers, and HPV positive cervical carcinoma cells are addicted to both E6 and E7 expression.40,41
Mitochondrial clustering reported in this study as a biomarker of mitochondrial organization is assessed automatically, in near real-time and without applying exogenous fluorescent probes or contrast agents. The intrinsic high-resolution 3D imaging capabilities of TPEF enables us to extract mitochondrial clustering information in 3D specimens dynamically, in an unperturbed state. Therefore, this approach is applicable to in vitro, ex vivo and in vivo specimens. The weak NADH-based TPEF signal compared to exogenous chromophores, and the need for high resolution imaging using visible-NIR light, limits use of this approach to tissue characterization within the upper 200–300 µm tissue layers, which should be sufficient for many epithelial cancers. The differences that we detect in C HFK, 16E6 and/or 16E7 HFK and HKc/DR EETEs suggest that mitochondrial clustering is sensitive to subtle changes in the relative utilization of major metabolic pathways, such as glycolysis, oxidative phosphorylation and potentially glutaminolysis. Therefore, this study establishes the presence of a relationship between mitochondrial function and organization in 3D tissues and provides a platform to examine these relationships in more detail in the context of the diagnosis or treatment monitoring of cancer.
Supplementary Material
What’s new?
Mitochondrial organization is based on dynamic tubular networks and is related to mitochondrial function, such that functional changes that occur at the onset of cancer may be reflected in mitochondrial organization. This concept has not been rigorously tested, however, especially in three-dimensional tissues. Here, automated analysis of endogenous two-photon excited fluorescence (TPEF) images was used to quantify mitochondrial organization in three-dimensional specimens, revealing differences between normal and pre-cancerous tissues. The differences were associated with changes in major bioenergetic pathways. The findings suggest that mitochondrial organization may serve as a useful noninvasive diagnostic biomarker for early cancer.
Acknowledgments
Grant sponsor: American Cancer Society Research Scholar; Grant number: RSG-09–174-01-CCE; Grant sponsor: NIBIB/NIH; Grant number: R01 EB007542; Grant sponsor: NCI/NIH; Grant number: R01 CA066980 and K01 CA143010; Grant sponsor: NIAMS/NIH; Grant number: F32 AR061933
Abbreviations
- CIN
cervical epithelial neoplasia
- EETE
engineered epithelial tissue equivalent
- FAD
flavin adenine dinucleotide
- HFK
human foreskin keratinocyte
- HPV
human papilloma virus
- KSFM
keratinocyte serum free media
- NADH
nicotinamide adenine dinucleotide
- PSD
power spectral density
- TPEF
two-photon excited fluorescence
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Lyamzaev KG, Izyumov DS, Avetisyan AV, et al. Inhibition of mitochondrial bioenergetics: the effects on structure of mitochondria in the cell and on apoptosis. Acta Biochim Pol. 2004;51:553–562. [PubMed] [Google Scholar]
- 6.Hackenbrock CR, Rehn TG, Weinbach EC, et al. Oxidative phosphorylation and ultrastructural transformation in mitochondria in the intact ascites tumor cell. J Cell Biol. 1971;51:123–137. doi: 10.1083/jcb.51.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 8.Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 9.Rossignol R, Gilkerson R, Aggeler R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 10.Varum S, Rodrigues AS, Moura MB, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Blerkom J. Mitochondria in early mammalian development. Semin Cell Dev Biol. 2009;20:354–364. doi: 10.1016/j.semcdb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Benard G, Bellance N, James D, et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 13.Folmes CD, Dzeja PP, Nelson TJ, et al. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilkerson RW, Margineantu DH, Capaldi RA, et al. Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS Lett. 2000;474:1–4. doi: 10.1016/s0014-5793(00)01527-1. [DOI] [PubMed] [Google Scholar]
- 15.Rambold AS, Kostelecky B, Elia N, et al. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hom JR, Quintanilla RA, Hoffman DL, et al. The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev Cell. 2011;21:469–478. doi: 10.1016/j.devcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitt JM, McLaughlin-Drubin ME, Munger K, et al. Automated biochemical, morphological, and organizational assessment of precancerous changes from endogenous two-photon fluorescence images. PLoS One. 2011;6:e24765. doi: 10.1371/journal.pone.0024765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng J, Lynch RM, Balaban RS. Nicotinamide adenine dinucleotide fluorescence spectroscopy and imaging of isolated cardiac myocytes. Biophys J. 1989;55:621–630. doi: 10.1016/S0006-3495(89)82859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chance B, Thorell B. Localization and kinetics of reduced pyridine nucleotide in living cells by microfluorometry. J Biol Chem. 1959;234:3044–3050. [PubMed] [Google Scholar]
- 20.Quinn KP, Sridharan GV, Hayden RS, et al. Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci Rep. 2013;3:3432. doi: 10.1038/srep03432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers C, Frattini M, Hudson J, et al. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257 doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 22.Xylas J, Quinn KP, Hunter M, et al. Improved Fourier-based characterization of intracellular fractal features. Opt Express. 2012;20:23442–23455. doi: 10.1364/OE.20.023442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levitt JM, Hunter M, Mujat C, et al. Diagnostic cellular organization features extracted from autofluorescence images. Opt Lett. 2007;32:3305–3307. doi: 10.1364/ol.32.003305. [DOI] [PubMed] [Google Scholar]
- 24.Wan F, Miao X, Quraishi I, et al. Gene expression changes during HPV-mediated carcinogenesis: a comparison between an in vitro cell model and cervical cancer. Int J Cancer. 2008;123:32–40. doi: 10.1002/ijc.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 26.Rozenblatt-Rosen O, Deo RC, Padi M, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varone A, Xylas J, Quinn KP, et al. Endogenous two-photon fluorescence imaging elucidates metabolic changes related to enhanced glycolysis and glutamine consumption in pre-cancerous epithelial tissues. Cancer Res. 2014;74:3067–3075. doi: 10.1158/0008-5472.CAN-13-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zwerschke W, Mazurek S, Massimi P, et al. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1999;96:1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Munger K. The role of human papillomaviruses in human cancers. Front Biosci. 2002;7:d641–d649. doi: 10.2741/a800. [DOI] [PubMed] [Google Scholar]
- 31.McLaughlin-Drubin ME, Munger K. Oncogenic activities of human papillomaviruses. Virus Res. 2009;143:195–208. doi: 10.1016/j.virusres.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera DR, Brown CM, Ouzounov DG, et al. Compact and flexible raster scanning multiphoton endoscope capable of imaging unstained tissue. Proc Natl Acad Sci USA. 2011;108:17598–17603. doi: 10.1073/pnas.1114746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murari K, Zhang Y, Li S, et al. Compensation-free, all-fiber-optic, two-photon endomicroscopy at 1.55 mum. Opt Lett. 2011;36:1299–1301. doi: 10.1364/OL.36.001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Akins ML, Murari K, et al. A compact fiber-optic SHG scanning endomicroscope and its application to visualize cervical remodeling during pregnancy. Proc Natl Acad Sci USA. 2012;109:12878–12883. doi: 10.1073/pnas.1121495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konig K, Ehlers A, Riemann I, et al. Clinical two-photon microendoscopy. Microsc Res Tech. 2007;70:398–402. doi: 10.1002/jemt.20445. [DOI] [PubMed] [Google Scholar]
- 36.Balu M, Mazhar A, Hayakawa CK, et al. In vivo multiphoton NADH fluorescence reveals depth-dependent keratinocyte metabolism in human skin. Biophys J. 2013;104:258–267. doi: 10.1016/j.bpj.2012.11.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munger K, Hayakawa H, Nguyen CL, et al. Viral carcinogenesis and genomic instability. Exs. 2006:179–199. doi: 10.1007/3-7643-7378-4_8. [DOI] [PubMed] [Google Scholar]
- 38.Jabbar SF, Park S, Schweizer J, et al. Cervical cancers require the continuous expression of the human papillomavirus type 16 E7 oncoprotein even in the presence of the viral E6 oncoprotein. Cancer Res. 2012;72:4008–4016. doi: 10.1158/0008-5472.CAN-11-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas MK, Pitot HC, Liem A, et al. Dominant role of HPV16 E7 in anal carcinogenesis. Virology. 2011;421:114–118. doi: 10.1016/j.virol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeFilippis RA, Goodwin EC, Wu L, et al. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J Virol. 2003;77:1551–1563. doi: 10.1128/JVI.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magaldi TG, Almstead LL, Bellone S, et al. Primary human cervical carcinoma cells require human papillomavirus E6 and E7 expression for ongoing proliferation. Virology. 2012;422:114–124. doi: 10.1016/j.virol.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.