Abstract
Background
Drought is a widespread limiting factor in coffee plants. It affects plant development, fruit production, bean development and consequently beverage quality. Genetic diversity for drought tolerance exists within the coffee genus. However, the molecular mechanisms underlying the adaptation of coffee plants to drought are largely unknown. In this study, we compared the molecular responses to drought in two commercial cultivars (IAPAR59, drought-tolerant and Rubi, drought-susceptible) of Coffea arabica grown in the field under control (irrigation) and drought conditions using the pyrosequencing of RNA extracted from shoot apices and analysing the expression of 38 candidate genes.
Results
Pyrosequencing from shoot apices generated a total of 34.7 Mbp and 535,544 reads enabling the identification of 43,087 clusters (41,512 contigs and 1,575 singletons). These data included 17,719 clusters (16,238 contigs and 1,575 singletons) exclusively from 454 sequencing reads, along with 25,368 hybrid clusters assembled with 454 sequences. The comparison of DNA libraries identified new candidate genes (n = 20) presenting differential expression between IAPAR59 and Rubi and/or drought conditions. Their expression was monitored in plagiotropic buds, together with those of other (n = 18) candidates genes. Under drought conditions, up-regulated expression was observed in IAPAR59 but not in Rubi for CaSTK1 (protein kinase), CaSAMT1 (SAM-dependent methyltransferase), CaSLP1 (plant development) and CaMAS1 (ABA biosynthesis). Interestingly, the expression of lipid-transfer protein (nsLTP) genes was also highly up-regulated under drought conditions in IAPAR59. This may have been related to the thicker cuticle observed on the abaxial leaf surface in IAPAR59 compared to Rubi.
Conclusions
The full transcriptome assembly of C. arabica, followed by functional annotation, enabled us to identify differentially expressed genes related to drought conditions. Using these data, candidate genes were selected and their differential expression profiles were confirmed by qPCR experiments in plagiotropic buds of IAPAR59 and Rubi under drought conditions. As regards the genes up-regulated under drought conditions, specifically in the drought-tolerant IAPAR59, several corresponded to orphan genes but also to genes coding proteins involved in signal transduction pathways, as well as ABA and lipid metabolism, for example. The identification of these genes should help advance our understanding of the genetic determinism of drought tolerance in coffee.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0777-5) contains supplementary material, which is available to authorized users.
Keywords: Candidate gene, Coffee, Drought, Differential gene expression, RNA-Seq, Real-time PCR (RT-qPCR)
Background
Coffee is the single most important tropical commodity traded worldwide and is a source of income for many developing countries in Tropics [1]. In the coffee genus, Coffea arabica accounts for approximately 70 % of total production worldwide, estimated at 8.5 million tons in 2015 [2]. Coffee production is subject to regular fluctuations mainly due to the natural biennial cycle but also caused by adverse climatic effects. Among them, drought is a widespread limiting factor and affects flowering and bean development, hence coffee yield [3]. Marked variations in rainfall also increase bean defects and modify the biochemical composition of beans, hence the final quality of the beverage [4]. Periods of drought may become more pronounced as a consequence of global climate change and geographical coffee growing regions may shift considerably, leading to environmental, economic and social problems [5]. In such a context, the creation of drought-tolerant coffee varieties has now become a priority for coffee research.
Genetic variability for drought tolerance exits in the coffee genus, particularly in Coffea canephora [6, 7] but also in C. arabica [8]. Although molecular mechanisms of drought tolerance have been widely studied in model plants [9], they are less well understood in Coffee sp. In a previous study analysing the effects of drought on gene expression, we recently identified a set of 30 genes differentially expressed in the leaves of drought-tolerant and drought-susceptible clones of C. canephora grown in the greenhouse under control (unstressed) and drought conditions [10, 11]. In that case, the expression of genes encoding glycine-rich proteins, heat shock proteins, dehydrins, ascorbate peroxidase, as well as trans-acting factors (such as DREB1D), for example, increased under drought conditions.
In Coffea sp., EST resources have been developed for various species and tissues including roots, leaves, and fruits [12–16]. However, no genomic resources are available for shoot apices, which are considered as key organs for plant development by integrating several signals, such as environmental stimuli as well as hormones (abscisic acid [ABA], auxins, cytokinins) and transcription [17]. On the other hand, next-generation sequencing (NGS) provides new opportunities to study transcriptomic responses and to combine high-throughput sequencing with the functional annotation capacity of generated ESTs [18].
In order to identify candidate genes involved in drought tolerance in coffee plants, we collected the shoot apices from drought-tolerant IAPAR59 and drought-susceptible Rubi cultivars of C. arabica under control and drought conditions to generate libraries that were sequenced using the GS-FLX Titanium strategy. A reference full transcriptome was annotated and compared to pre-identify genes differentially expressed between cultivars and drought conditions. The transcription profiles of these genes were further analysed by qPCR in the plagiotropic buds of these plants.
Methods
Plant material
We compared two cultivars of Coffea arabica, the drought-susceptible (DS) Rubi MG1192 (also referred to hereafter as RUB) and the drought-tolerant (DT) IAPAR59 (also referred to hereafter as I59). Rubi did not undergo recent introgression with C. canephora genomic DNA, while IAPAR59 is the result of a cross between the Timor hybrid HT832/2 and the Villa Sarchi cultivar [19].
Field experiment
Seeds of these two commercial cultivars came from fruits harvested in May 2007 in the coffee experimental fields of the Institute for Research and Rural Assistance (Incaper, Vitoria, Espirito Santo, Brazil) and germinated (September 2007) in greenhouse of this institute. Five-month-old plantlets of the Rubi and IAPAR59 were then planted (January 2008) in a field experiment (0.7 m spacing between plants and 3 m spacing between rows) at the Cerrado Agricultural Research Center (Planaltina-DF, Brazil 15°35’44”S - 47°43’52”W) under full-sunlight conditions in two blocks of 30 plants for each cultivar. Under the conditions of the Cerrado climate [20], the rainfall pattern is divided into a dry season (from May to September) followed by a wet season (from October to April) that concentrates more than 80 % of annual precipitations. For each cultivar, one control (C) block was irrigated while the drought (D) block was not irrigated during the dry seasons. For the control condition, irrigation was supplied by sprinklers (1.5 m in height) set up in the field in such a way that irrigation was uniform. Soil water content was monitored using PR2 profile probes (Delta-T Devices Ltd), and irrigation was applied regularly so as to maintain a moisture content above 0.27 cm3 H2O.cm-1.
Sampling
For both cultivars and experiments, leaf predawn water potentials (Ψpd) were measured once a week during the 2009 dry season (from May to October) of (23-month-old plants) and only once in 2011 (at the end of the dry season) (47-month-old plants) using a Scholander-type pressure chamber (Plant Water Status Console, Model 3000 F01, Soil Moisture Equipment Corp, Santa Barbara, CA USA) in fully expanded leaves (8–15 cm long) from the third pair from the apex of plagiotropic branches located in the upper third of the plant canopy. For 454 sequencing, between 30 and 50 shoot apices were collected (between 10:00 and 11:00 am) from three different plants at the end of the dry season from Rubi and IAPAR59 under the control and drought conditions, and further dissected to isolate the shoot apex (Fig. 1b). For microscopic analyses, leaves identical to those used for Ψpd measurements were also collected from the same plants. At the end of the 2011 dry season, Ψpd were measured once for Rubi and IAPAR59 plants under control and drought treatments, and shoot apices were collected (Fig. 1a) for gene expression analyses (qPCR).
Fig. 1.
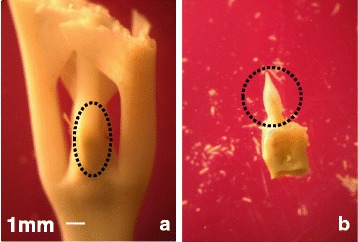
Tissue dissection of plagiotropic buds. a The plagiotropic buds (including small leaves) were collected from plants during the 2011 dry season and used to extract RNA for qPCR expression analysis. b Meristem and leaf primordium dissected from plagiotropic buds harvested during the 2009 dry season and used to extract RNA for pyrosequencing. The dotted circles show the position of meristem and leaf primordium. The same scale (white bar = 1 mm) is used for both documents
RNA isolation, DNA synthesis and 454-sequencing
The plagiotropic buds were incubated for 5 min in the washing buffer (66 % chloroform, 33 % methanol, 1 % HCl) [21] and further incubated twice for 30 min under a vacuum in the fixation buffer (25 % acetic acid, 75 % ethanol RNAse-free) then cooled to 4 °C. Samples were stored in 75 % RNAse-free ethanol. For the control and drought conditions, shoot apices (meristems and primordium leaves) of three different plants were separated from plagiotropic buds under a binocular microscope by dissection and then ground to powder in liquid nitrogen using a pestle and mortar. Total RNA was extracted using the Nucleospin RNA Plant kit (Macherey-Nagel), including a DNAse-I treatment. The quality and quantity of RNA were checked with a Bioanalyzer (2100, RNA Nano 6000 Agilent). The 1st strand cDNA synthesis was performed using 1 μg total RNA and the SMARTer™ PCR cDNA Synthesis Kit (Clontech). Double-stranded DNA was then produced for each library (I59-C, I59-D, RUB-C and RUB-D). For each sample, DNA (around 5 μg) was nebulized to a mean fragment size of 650 bp, ligated to an adapter using standard procedures [22] and then sequenced by performing two runs (1 library per DNA sample x 2) using GS-FLX Titanium (Beckman Coulter Genomics SA, Grenoble, France) which generated one million reads corresponding to more than 255 Mb.
Transcriptome assembly and automatic annotation
All 454-sequencing reads were inspected for low quality reads and 454 adapters that were identified by SSAHA2 software [23]. A reference full transcriptome was then built using C. arabica reads originating from the present project and from the Brazilian Coffee Genome Project (BCGP) available in the GenBank public database [14, 24]. The Sanger and 454 reads were submitted for a trimming pipeline using bdtrimmer software [25] that was used to exclude ribosomal, vector, low quality (regions with a PHRED score less than 20) and short sequences (less than 100 bp). All sequences (454 and Sanger reads) were assembled using MIRA software [26]. The contigs formed by only Sanger reads were discarded from the full transcriptome assembly. The reference full transcriptome was annotated by Blast2GO software version 2.8 [27] using Non-Redundant protein (NCBI/NR), InterPro and Gene Ontology (GO) databases. The same program was also used to group datasets in GO according to the biological process. Further details on the automatic annotation of all contigs are provided in Additional file 1: Table S1. The complete bioinformatic pipeline used for this work is described in Additional file 2: Figure S1.
Digital gene expression analysis
The reference full transcriptome was also used to count all 454 reads/libraries individually by parsing the ACE file generated by MIRA software. The number of sequences anchored in each contig (read counts) was subjected to differential expression analysis between the libraries using DEseq [28] and EdgeR [29] software in the R/Bioconductor package. A unigene was considered as differentially expressed when it was identified in at least one software considering fold-change ≥ 2 (or fold-change ≤ -2) and p-value ≤ 0.05. The libraries were compared based on (1) differentially expressed genes in IAPAR59 between C (control) and D (drought) conditions (with the calculation of fold-change based on the I59-D/I59-C ratio), (2) differentially expressed genes in Rubi between C and D conditions (RUB-D/RUB-C), (3) differentially expressed genes in the control library between Rubi and IAPAR59 (RUB-C/I59-C) and (4) differentially expressed genes in the drought library between Rubi and IAPAR59 (RUB-D/I59-D). Further information about differentially expressed genes in all the libraries is given in Additional file 3: Table S2.
Functional annotation of differentially expressed genes
The lists of differentially expressed genes in each analysis were separated into UP and DOWN regulated and subjected to GO enrichment analysis to identify significantly enriched GO slim terms (Plant GO slim) using Blast2GO software and a p-value ≤ 0.05.
Selection of candidate genes
The comparison of DNA libraries led to the identification of 80 (20 for each library) candidate genes (CGs) that were up- and down-regulated (see Additional file 3: Table S2). For each CG, primer pairs were designed using Primer Express software (Applied Biosystems) and tested of their specificity and efficiency against a mix of ss-DNAs of plagiotropic buds (data not shown). The best primer pairs (n = 20) were used to monitor the expression of corresponding CGs in plagiotropic buds of Rubi and IAPAR59 under control and drought conditions. These genes corresponded to CaAEP1, CaCAB2, CaCHI1, CaCHI2, CaCHI3, CaDLP1, CaELIP3, CaGAS2, CaGRP2, CaH2A, CaHSP3, CaIPS1, CaJAMT1, CaMAS1, CaPP2, CaPSBB, CaSAMT1, CaSDC1, CaSLP1 and CaSTK1 (Table 1). This list of CGs was increased by adding other genes such as 14 orphan genes (CaUNK2-CaUNK7, CaUNK9 and CaUNK11-CaUNK17 already described to present differential gene expression profiles in different organs of C. canephora [30]. This list was finally completed by including the CaUNK1, CaUNK8 and CaUNK10 orphan genes, and LTP genes that were already studied in C. canephora [10, 11, 31] and C. arabica [32], respectively.
Table 1.
Candidate genes and corresponding primers used for qPCR experiments
| Gene | Protein name | C. canephora | GB | ATP | SGN | Primer | Primer sequences | bp |
|---|---|---|---|---|---|---|---|---|
| CaUBQ10 | Ubiquitin | Cc02_g31600 | GW488515 | 32782 | U637098 | BUBI-F BUBI-R |
5’ AAGACAGCTTCAACAGAGTACAGCAT 3’ 5’ GGCAGGACCTTGGCTGACTATA 3’ |
104 |
| CaAEP1 a | Putative aldose 1-epimerase | Cc07_g03170 | GT005185 | 716 | U637659 | 716-1 F 716-1R |
5' CGGTGATGTCCTCTCTGATGAG 3’ 5’ GTTGGGATGAGCTGGTTGTTC 3’ |
75 |
| CaCAB2 a | Chlorophyll a/b-binding protein | Cc09_g09030 | GT003492 | 33540 | U629601 | 48565-F 48565-R |
5’ GTTCAAGGCTGGATCCCAAA 3’ 5’ GCAAGCCCAGATAGCCAAGA 3’ |
100 |
| CaCHI1 a | Class III chitinase | Cc11_g00410 | GT012279 | 32745 | U637166 | 50103-F 50103-R |
5’ AATCAAGCGACCGTCCATTC 3’ 5’ GTGTTTCCGCTGTGGATGTG 3’ |
70 |
| CaCHI2 a | Putative chitinase | Cc00_g14300 | GT011845 | 32737 | U638035 | 53058-F 53058-R |
5’ CCTGCTCGCGGTTTCTACAC 3’ 5’ TTGTTCCAAAAGCCCCATTG 3’ |
70 |
| CaCHI3 a | Chitinase-like protein | Cc03_g13720 | GW491433 | 32875 | U645893 | 23638-F 23638-R |
5’ AAACGGCCCGTCCAGAA 3’ 5’ GCTTTGTCCTGCTGGTCCAT 3’ |
130 |
| CaDLP1 a | Dirigent-like protein | Cc00_g27410 | GW477731 | 35149 | nf | 39577-F 39577-R |
5’ TTGGTAGTCCGGCGAGAGAA 3’ 5’ GCATATCCCCGAGCAAACCT 3’ |
70 |
| CaELIP3 a | Early light-induced protein (ELIP) | Cc03_g04300 | GR985685 | 32771 | U631550 | 32771-F 32771-R |
5’ TCGGTTGCCATGCAATCTT 3’ 5’ GCAGATGAAGCCCACAGCTT 3’ |
100 |
| CaGAS2 a | Glucosyltransferase arbutin synthase | Cc02_g39100 | GT697284 | 3945 | U632419 | 632419-F 632419-R |
5’ GCTGACGACGTTAGGATTGAGA 3’ 5’ AACTTGGCGGTGTCAACCAA 3’ |
101 |
| CaGRP2 a | Glycin-rich protein | Cc00_g16260 | GW430980 | 32799 | U635030 | 53139-1 F 53139-1R |
5’ CACATATGCTGGTGAGCCAAA 3’ 5’ AGGCATTTAAGCGCCATGAT 3’ |
100 |
| CaH2A a | Putative histone H2A | Cc01_g12440 | GT723387 | 33557 | U630412 | 53417-F 53417-R |
5’ GCACTGGAGCTCCGGTCTAC 3’ 5’ AGCAGCATTTCCAGCCAATT 3’ |
80 |
| CaHSP3 a | Heat schock protein (HSP) 70 kDa | Cc02_g08040 | GR982512 | 33197 | U636531 | 33197-1 F 33197-1R |
5’ GGCGTCTGGCAACACGAT 3’ 5’ CGATGAGACGCTCGGTGTCT 3’ |
100 |
| CaIPS1 a | Myo-inositol 1-phosphate synthase | Cc07_g15530 | GT003538 | 10496 | U632517 | 10496-1 F 10496-1R |
5’ AAGCAACCTGAATTTGGCTGAT 3’ 5’ GAGAGGGACCATGGATTCCA 3’ |
100 |
| CaJAMT1 a | Jasmonate O-methyltransferase | Cc03_g07330 | GR989151 | 33008 | U631389 | 47327-F 47327-R |
5’ CTGTGGCTGAACCCTTGCTT 3’ 5’ TCTTTGGACATGCGATCAGAAA 3’ |
100 |
| CaMAS1 a | Momilactone-A synthase | Cc00_g13640 | GW479615 | 33413 | nf | 33413-F 33413-R |
5’ GGGCAGAGGCACGAAAAA 3’ 5’ GGTACCCTGCCGCAACTATG 3’ |
60 |
| CaPP2 a | Putative phloem protein 2 (PP2) | Cc03_g13000 | GR995691 | 33207 | U633544 | 33207-F 33207-R |
5’ GGTGTTGGCGATGTCGAGAT 3’ 5’ TTCCTTGGGTCGAAGCTCAA 3’ |
90 |
| CaPSBB a | Photosystem II CP47 (psbB)-like protein | nf | GW447378 | 22102 | U630312 | 55586-F 55586-R |
5’ ATCGGAAATAATCCGGCAAA 3’ 5’ AACCATCCAATCGCTATTCCA 3’ |
80 |
| CaSAMT1 a | S-adenosyl-methionine-methyltransferase | Cc03_g05630 | DV672716 | 754 | U629783 | 34318-F 34318-R |
5’ AACGTTTGGGTGATGAATGTTG 3’ 5’ GTGCCAATAAGCCCTCTATCGT 3’ |
80 |
| CaSDC1 a | S-adenosyl-L-methionine decarboxylase | Cc11_g11130 | GT002431 | 8508 | U629687 | 8508-1 F 8508-1R |
5’ CTCGATTCCTCCCATCCTGAA 3’ 5’ TGACTGTGCCCCAGGGAATA 3’ |
100 |
| CaSLP1 a | Subtilisin-like protein | Cc00_g19100 | GW430663 | 1620 | nf | 7961-F 7961-R |
5’ CCATCGTTCTCGGTGGTCTT 3’ 5’ GCATTGCTCCCCACATTCTT 3’ |
80 |
| CaSTK1 a | Hypothetical S/T protein kinase | Cc00_g18670 | GT687049 | 6301 | U631794 | 6301-1 F 6301-1R |
5’ CCACCCACAAGCTGTATTCTCA 3’ 5’ GACCCAATGGGATGTCATCAC 3’ |
80 |
| CaUNK1 c | Unknown protein 1 | Cc03_g08880 | DV689820 | 33062 | U614843 | 182052-F 182052-R |
5’ TATAGTGTTTATGGTGTGGCTTTCAGT 3’ 5’ GTACCACCGTAGGGAGACGTATG 3’ |
79 |
| CaUNK2 b | Unknown protein 2 | Cc07_g01940 | DV708962 | 31492 | U637447 | 33353-F 33353-R |
5’ GAACTTACAAACGCGCGTAACC 3’ 5’ CATGGTCGAATCCAGATTTCATT 3’ |
80 |
| CaUNK3 b | Unknown protein 3 | nf | nf | 22823 | nf | 22823-F 22823-R |
5’ GGAAGCATGCACACAGAAAATAGA 3’ 5’ TTCCTGTTTACGTCTTTTTCAATTGA 3’ |
80 |
| CaUNK4 b | Unknown protein 4 | Cc06_g11210 | GW465088 | 39984 | nf | 55677-F 55677-R |
5’ GCTGTGGTTTTAAAGTTTTGATGGA 3’ 5’ TGCAAAATTAAGGTCCCAACAGT 3’ |
81 |
| CaUNK5 b | Unknown protein 5 | Cc08_g09510 | GW474926 | 4578 | nf | 4578-F 4578-R |
5’ GGAGTTCCTGTCCGAAGTTGTT 3’ 5’ GGCATGCTGTCACCTGAAAA 3’ |
80 |
| CaUNK6 b | Unknown protein 6 | Cc03_g06850 | GT002178 | 34993 | U632634 | 34993-F 34993-R |
5’ AAGCCAATGCCGATCGATT 3’ 5’ CGCCGCCGAAGATCTCTAG 3’ |
100 |
| CaUNK7 b | Unknown protein 7 | Cc03_g00560 | GW444736 | 33613 | U631416 | 25639-F 25639-R |
5’ CGAGGAAGCTGAAGGAAAGGA 3’ 5’ TCCGACTGGCCTAACAAGGT 3’ |
61 |
| CaUNK8 b | Unknown protein 8 | Cc00_g04970 | DV695331 | 33190 | U640780 | LP18101-F LP18100-R |
5’ CTCGCGTGGCCGAGATC 3’ 5’ CCCTCACATTTCCACGTGAAT 3’ |
100 |
| CaUNK9 b | Unknown protein 9 | Cc03_g08920 | GT649500 | 32762 | U636808 | 30926-F 30926-R |
5’ CGGAGGAGGCCATGGAGGT 3’ 5’ CCGTGTCCATAACCACCATGT 3’ |
123 |
| CaUNK10 c | Unknown protein 10 | nf | GT648004 | 14813 | U645073 |
D18240-F D18240-R |
5’ TAGCCTTGTTCTTTTAGGGAGTCTTATC 3’ 5’ AGAGCTTCGTCCAGGAAGAAGA 3’ |
134 |
| CaUNK11 b | Unknown protein 11 | Cc03_g14330 | GR991912 | 8598 | U637116 | 32792-F 32792-R |
5’ GCTGGGAAAGCTACAGAAACCA 3’ 5’ GAACTCCAACGCCAAGCATT 3’ |
100 |
| CaUNK12 b | Unknown protein 12 | Cc10_g12840 | nf | 53029 | nf | 53029-F 53029-R |
5’ CTTCACACCATTCAGACAATCGA 3’ 5’ GACCGTAATTGGGCGTCAAT 3’ |
100 |
| CaUNK13 b | Unknown protein 13 | Cc00_g17760 | GT673421 | 14198 | U639484 | 33980-F 33980-R |
5’ ATTGCCCTGTTTGCATGCAT 3’ 5’ CTGCATGGTGATTGTCCTCAGT 3’ |
100 |
| CaUNK14 b | Unknown protein 14 | Cc00_g16260 | GT672564 | 48325 | U635030 | 11524-F 11524-R |
5’ GGCGGTTGTCATGGATACG 3’ 5’ TTTGGCTCACCAGCATATGTG 3’ |
119 |
| CaUNK15 b | Unknown protein 15 | Cc00_g04970 | GR983286 | 33190 | U636790 | 05517-F 05517-R |
5’ AAAATTTCACCACGGCAAGCT 3’ 5’ TTGCCTCCCTCACATTTCCA 3’ |
72 |
| CaUNK16 b | Unknown protein 16 | nf | GW464209 | 9761 | U639049 | 18112-F 18112-R |
5’ TGTGAACTGCCATCCCAAGA 3’ 5’ AAGACTACCATGTCCAACAACTTCAG 3’ |
88 |
| CaUNK17 b | Unknown protein 17 | Cc03_g08920 | GT685623 | 32762 | U636800 | 42747-F 42747-R |
5’ AGGTGGCTGCCAAGTCAGTT 3’ 5’ ATGGTACTTGGCTTCTCCTTCCTC 3’ |
71 |
|
CaLTP1
d
CaLTP2 d |
Non-specific lipid transfer protein (nsLTP) | Cc11_g09700 |
HG008739 HG008740 |
46897 | U632702 | LTP-R2 LTP-FT |
5’ CACCATTACATGGGAACGTTGC 3’ 5’ CTGTGGTCTGAAATGGCCAACT 3’ |
120 |
| CaLTP3 d | Non-specific lipid transfer protein (nsLTP) | Cc04_g06890 | HG008741 | 33368 | U632702 | LTP-R1 LTP-FT |
5’ ATTCAACACCATTACTAGTTTTCGAGC 3’ 5’ CTGTGGTCTGAAATGGCCAACT 3’ |
113 |
| LTP d | nf | - | U632702 | LTP-F100 LTP-R100 |
5’ TGCAATTTTATCAAAGATCCAGC 3’ 5’ AGTTGGCCATTTCAGACCACA 3’ |
93 |
Gene names were assigned based on the best BLAST hit obtained by comparing the coffee ESTs with public databases. C. canephora means coffee sequences that aligned with the candidate genes using BLASTx searches against NR/NCBI and filtration (http://coffee-genome.org [59]). GenBank (GB: http://blast.ncbi.nlm.nih.gov/Blast.cgi), ATP (http://www.lge.ibi.unicamp.br/cirad/) and SGN (Sol Genomics Network, http://solgenomics.net/) accession numbers of coffee ESTs are also given, as well as the length of base pairs (bp) of amplicons. nf: no-hits found (SGN: tools/blast/SGN Clusters [current version] / Coffee species Clusters, GB: BLASTn/Nucleotide collection [nr/nt]). The size of amplicons is based on the unigene. (a): candidate genes (n = 20) identified during this study. (b): orphan genes (n = 14) previously described [35] and analysed in this study. (c): orphan genes (n = 3) with expression already been studied in leaves of DT and DS clones of C. canephora conilon [10, 11, 36]. (d): LTP-encoding genes were previously described [37]
Real-time quantitative PCR assays
For qPCR experiments, plagiotropic buds containing shoot apices and small leaves (Fig. 1a) were immediately frozen in liquid nitrogen after collection, and stored at -80 °C before being extracted and converted into single-strand cDNA as previously described [33]. Real-time qPCR assays were carried out using the protocol recommended for the use of 7500 Fast Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA). DNA preparations were diluted (1/50) and tested by qPCR using CG primer pairs (Table 1). RT-qPCR was performed with 1 μl of diluted ss-DNA and 0.2 μM (final concentration) of each primer in a final volume of 10 μl with SYBR green fluorochrome (SYBRGreen qPCR Mix-UDG/ROX, Invitrogen). The reaction mixture was incubated for 2 min at 50 °C (Uracil DNA-Glycosilase treatment), then for 5 min at 95 °C (inactivation of UDGase), followed by 40 amplification cycles of 3 sec at 95 °C and finally for 30 sec at 60 °C. Data were analysed using SDS 2.1 software (Applied Biosystems) to determine cycle threshold (Ct) values. The specificity of the PCR products generated for each set of primers was verified by analysing the Tm (dissociation) of amplified products. PCR efficiency (E) was estimated using absolute fluorescence data captured during the exponential phase of amplification of each reaction with the equation E (in %) = (10(-1/slope) -1) x 100 [34]. Efficiency values were taken into account in all subsequent calculations. Gene expression levels were normalized to expression levels of CaUBQ10 as a constitutive reference. Relative expression was quantified by applying the formula (1 + E)−ΔΔCt where ΔCt target = Ct target gene – Ct reference gene and ΔΔCt = ΔCt target – ΔCt internal calibrator, with the internal reference always being the Rubi-control (RUB-C) sample with relative expression equal to 1.
Leaf histological analysis of cuticle
Mature leaves of the IAPAR59 and Rubi genotypes were fixed for 48 h in 100 mM phosphate buffer at pH 7.2, supplemented with 1 % (v/v) glutaraldehyde, 2 % (v/v) paraformaldehyde, and 1 % (w/v) caffeine, at room temperature [35]. The samples were dehydrated and embedded in Technovit 7100 resin (Heraeus Kulzer) according to the manufacturer’s recommendations. Three-micrometer semi-thin sections were cut with glass knives on a Leica RM2065 Microtome. The resulting sections were double stained according to Buffard-Morel et al. [36]. Briefly, polysaccharides were stained dark pink with periodic acid Schiff (PAS) and soluble proteins were stained blue with naphthol blue-black (NBB) [37]. Sections were then mounted in Mowiol. The slides were observed with a Leica DM6000 microscope (Leica, Germany) under bright field or epifluorescent light (A4 filter). Pictures were taken with a Retiga 2000R camera (QImaging Co.) and the images were processed with Volocity 4.0.1 (Improvision, Lexington, MA, USA). Cuticle thickness was measured with the freeware Image J software (http://imagej.nih.gov/ij/). Experiments were conducted on the “Plate-Forme d’Histocytologie et Imagerie Cellulaire Végétale (PHIV platform)” (http://phiv.cirad.fr/) using microscopes belonging to the Montpellier Rio Imaging platform (www.mri.cnrs.fr). The results are expressed as means (μm) of 11 measured values. The data were statistically processed using (1) an analysis of variance computer program (Statistica, StatSoft, Inc.), and (2) the Student-Newman-Keuls (SNK) mean comparison test [38] when the effect of the factor tested was found to be statistically significant. A probability level of P ≤ 0.05 was considered significant for all the statistical analyses.
Results
Monitoring drought under field conditions
In 2009, leaf predawn water potential (Ψpd) values were similar in the leaves of irrigated Rubi and IAPAR59 plants, ranging from -0.06 to -0.16 MPa (Fig. 2a). This confirmed the unstressed status of these plants which were considered as the control in our experiment. At the same time, the Ψpd values decreased gradually during the dry season in the leaves of Rubi and IAPAR59 under drought conditions reaching the lowest values at the end of the dry season (Fig. 2a). At that time, the less negative Ψpd values in IAPAR59 indicated that it had better access to soil water. The first rains then occurred and the Ψpd values of drought-stressed plants increased almost to those measured in irrigated plants, illustrating the complete recovery of stressed plants. In 2011, Ψpd was measured at the peak of the drought (end of dry season). Under drought conditions, both Rubi and IAPAR59 had similar Ψpd values that were more negative than those measured in 2009, indicating more severe drought stress in 2011 (Fig. 2b).
Fig. 2.
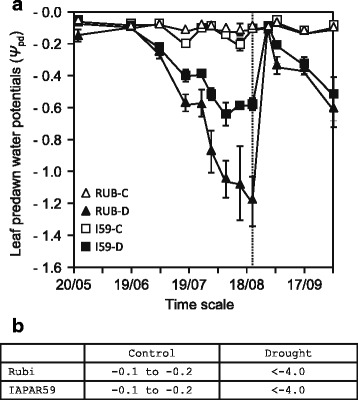
Predawn leaf water potentials (Ψ pd) measured in plants of C. arabica. Rubi (RUB, triangle) and IAPAR59 (I59, square) cultivars were grown under control (C, open symbols) and drought (D, black symbols) conditions. Ψ pd values (expressed in mega-Pascal, MPa) were measured once a week during the 2009 dry season (23-month-old plants) (a). The time scale is in days and months (dd/mm, from 20/05 to 02/10). Vertical bars are standard deviations (n = 9 leaves) and the dashed vertical line (20/08) represents the harvest point of plagiotropic buds for RNA extraction for 454 sequencing and leaves for microscopic analyses. b Ψ pd of Rubi and IAPAR59 plants (47-month-old plants) measured at the end of the 2011dry season. In this case, Ψ pd values ranged from -0.1 to -0.2 MPa for the control conditions, but were below (< -4.0 MPa = severe drought) the range of use of a Scholander-type pressure chamber for drought conditions
Sequencing, assembly and annotation of the Coffee shoot apex transcriptome
The final reference assembly generated a total of 34,743,872 bp (34.7 Mbp) with coverage of 6.5x and 43,087 clusters, corresponding to 41,512 contigs and 1,575 singletons. These data are composed of: (1) 17,719 clusters (16,238 contigs and 1,575 singletons) from 454 sequences, exclusively; and (2) 25,368 hybrid clusters that contain 454 reads, and at least one contig from Sanger sequencing (public database). The contigs formed by only Sanger reads were discarded from the full transcriptome assembly. On average, 22.4 % and 55.6 % of the total raw data were discarded from Sanger and 454, respectively, due to low quality. After removing the adapters, these reads had a size of 379.2 bp (on average). The statistical data for the Sanger and 454 reads are listed in Table 2.
Table 2.
Characteristics of reads used in this work
| Libraries | Total reads | Trimmed reads | Average length of reads |
|---|---|---|---|
| Public Sanger database | 195,110 | 151,403 | 518 |
| I59-C | 135,304 | 66,641 | 325 |
| I59-D | 282,213 | 112,518 | 351 |
| RUB-C | 230,064 | 101,394 | 360 |
| RUB-D | 345,751 | 153,572 | 342 |
| Total | 1,188,442 | 585,528 | 379.2 |
Statistics of all reads used in this work: public Sanger reads and 454 sequenced reads from two cultivars under two conditions. Cultivars (RUB: Rubi and I59: IAPAR59) of C. arabica and treatments (C control and D drought) are indicated. The number of total reads, trimmed reads and average read length (in bp) are indicated
Transcriptome annotation by Blast2GO using Non-Redundant protein (NCBI/NR) and InterPro databases resulted in 36,965 transcriptome clusters (85.8 %) with a known protein function, 1,824 conserved proteins of unknown function (4.2 %), 1,515 proteins identified by InterPro only (3.5 %) and 2,783 unidentified proteins (6.5 % no-hits found).
The results of the digital gene expression analysis (Table 3) showed more differentially expressed genes (DEG) in the cultivars Rubi (RUB) and IAPAR59 (I59) cultivars under drought (D) conditions (RUB-D/I59-D), totalling 490 clusters (1.14 % of the total), with 320 clusters classified as up-regulated. Under the control (C) conditions, a few DEG were found (RUB-C/I59-C), corresponding to 184 clusters (0.43 % of total clusters). The comparison between control and drought conditions showed a prevalence of up-regulated genes (165 clusters) and a total of 226 DEG in IAPAR59 (I59-D/I59-C) with 0.52 % of total clusters, and 343 clusters in Rubi (RUB-D/RUB-C) with 0.80 % of total clusters.
Table 3.
Reads showing differential expression between cultivars and/or treatments
| Libraries | EdgeR DEG (% of total clusters) | DEseq DEG (% of total clusters) | Total DEG (% of total clusters) | Up-regulated clusters (% of total clusters) | Down-regulated clusters (% of total clusters) |
|---|---|---|---|---|---|
| I59-D/I59-C | 209 (0.49 %) | 176 (0.41 %) | 226 (0.52 %) | 165 (0.38 %) | 61 (0.14 %) |
| RUB-D/RUB-C | 323 (0.75 %) | 306 (0.71 %) | 343 (0.80 %) | 251 (0.58 %) | 92 (0.21 %) |
| RUB-C/I59-C | 173 (0.40 %) | 169 (0.39 %) | 184 (0.43 %) | 104 (0.24 %) | 80 (0.19 %) |
| RUB-D/I59-D | 392 (0.91 %) | 433 (1.00 %) | 490 (1.14 %) | 320 (0.74 %) | 170 (0.39 %) |
Differentially expressed genes (DEG) were obtained with the R/Bioconductor packages DEseq and EdgeR. Total DEG values mean the union of DEseq and EdgeR results. The calculation of percentage was based on total of clusters (43,087 clusters). Cultivars (RUB Rubi and I59: IAPAR59) of C. arabica and treatments (C control and D drought) are indicated
The results of the gene ontology (GO) enrichment analysis are shown in Fig. 3 and all GO enrichment data are listed in Additional file 1: Tables S1 and Additional file 3: Table S2. For IAPAR59, the comparison of drought and control conditions (I59-D/I59-C) identified over-represented GO terms characterized by up-regulated genes involved in expression (gALL_c3501) and translation (gALL_c2033, gALL_c4461, gALL_c6492) processes and in the generation of precursor metabolites and energy (gALL_c921, gALL_c4013, gALL_c4540). For Rubi, a comparison of the RUB-D/RUB-C libraries revealed an over-representation of the following GO terms which were up-regulated: protein metabolic process (gALL_c2021, gALL_c3355), response to stress (gALL_rep_c33197/CaHSP3) and response to abiotic stimulus (gALL_rep_c32771/CaELIP3, gALL_c2829, gALL_rep_c32766). When comparing both cultivars under drought conditions (RUB-D/I59-D), GO terms were identified related to increased enrichment of tropism for up-regulated genes (gALL_c1270, gALL_c1524, gALL_c1864) and photosynthesis for down-regulated genes (gALL_c27215, gALL_rep_c34074, gALL_rep_c34746). Under the control conditions (RUB-C/I59-C), proteins of translational machinery were identified for up-regulated genes (gALL_c3061, gALL_c16674, gALL_c19094) and photosynthesis for down-regulated genes (gALL_rep_c34074, gALL_rep_c37283, gALL_rep_c50892).
Fig. 3.
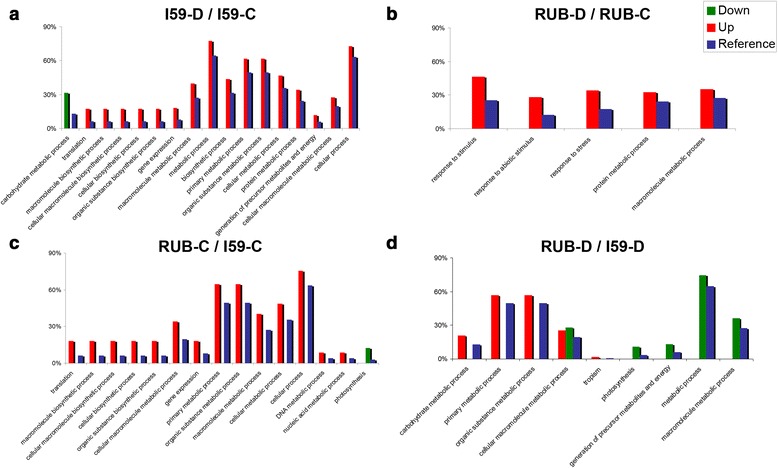
Gene ontology (GO) enrichment analysis on a list of differentially expressed genes up- and down-regulated under four conditions. The calculation of fold change was based on the ratio of: (a) I59-D/I59-C; (b) RUB-D/RUB-C; (c) RUB-C/I59-C; and (d) RUB-D/I59-D. The Y axis indicates the number of genes normalized by the total number of genes used in each comparison from each library. Cultivars (RUB: Rubi and I59: IAPAR59) of C. arabica and treatments (C: control and D: drought) are indicated
Expression profiles of candidate genes
Among the candidate genes (CGs) identified in silico as presenting up- and down-regulation, expression profiles from 20 of them were analysed by qPCR together with the expression of 17 orphan genes (3 of them already studied in C. canephora [10, 11, 30, 31]) and LTP genes [32]. For all these genes, expression profiles were analysed in plagiotropic buds of Rubi and IAPAR59 under control and drought conditions. These results are presented in separate sections below, according to the observed expression patterns.
Genes with induced expression under drought conditions
Twenty-five genes showing up-regulated expression profiles under drought conditions, mainly in IAPAR59 and to a lesser extent in Rubi, were identified (Fig. 4). This was observed for CaSTK1 which encodes a putative oxidative stress response serine/threonine protein kinase with 87 % identity with a predicted protein of Populus trichocarpa (XP_002299433). In that case, expression of this gene was highly induced by drought in the DT cultivar IAPAR59. Similar profiles were also observed for the CaSAMT1 gene encoding a putative S-adenosyl-L-methionine-dependent methyltransferase and the orphan genes CaUNK2 and CaUNK3. The latter gene had no open reading frame but presented high identity (e-value 2E-45) with the SGN-U637447 contig and also with various coffee ESTs mainly found in C. canephora cherries at early developmental stages (data not shown).
Fig. 4.
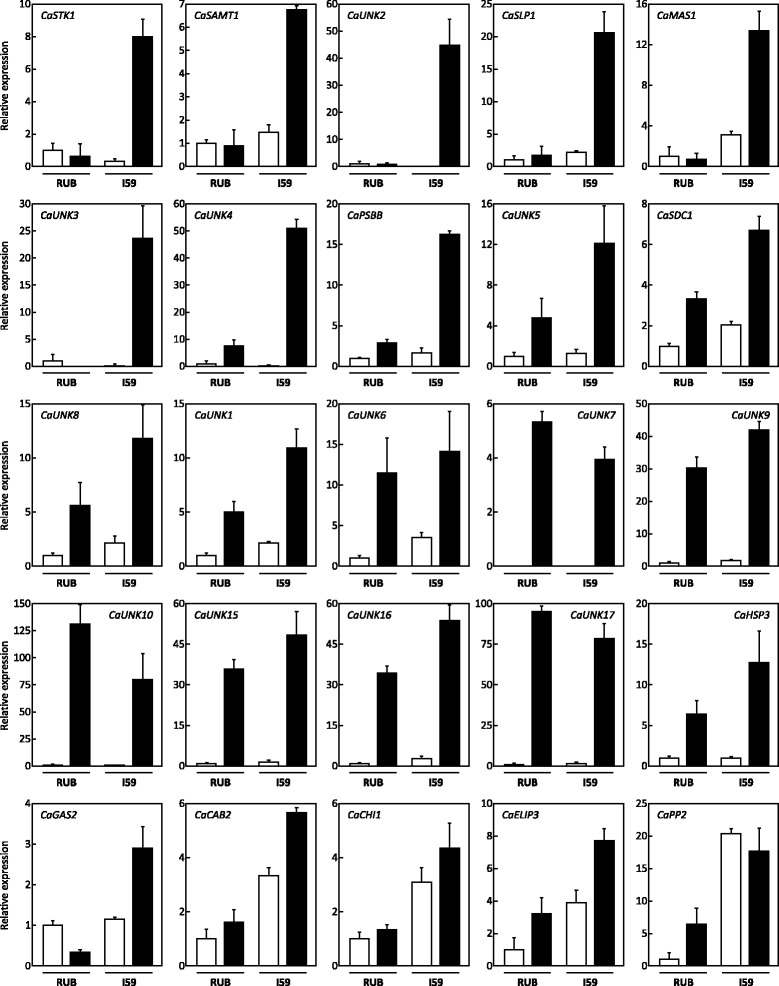
Expression profiles of genes up-regulated under drought conditions. Gene expression was analysed in plagiotropic buds of Rubi (RUB) and IAPAR59 (I59) cultivars of C. arabica grown under control (white isobars) and drought (black isobars) conditions. The gene names are indicated in the histograms. Transcript abundances were normalized using the expression of the CaUBQ10 gene as the endogenous control. Results are expressed using RUB-C as the reference sample (Relative expression = 1). Values of three technical replications are presented as mean ± SD (bar)
Expression of the CaSLP1 gene encoding a putative protein homologous (65 % identity, 74 % similarity) to a protein of Nicotiana benthamiana containing a peptidase S8/subtilisin-related domain, was also higher in IAPAR59 than in Rubi under drought conditions. A similar situation was observed for the CaMAS1gene encoding a protein of 311 amino acid residues sharing similarities (e-value 2E-121, 66 % identity, 82 %, similarity) with momilactone A synthase-like protein from Vitis vinifera (XP_002275768) that contains a secoisolariciresinol dehydrogenase conserved domain.
Similar expression profiles, characterized by high up-regulation under drought conditions particularly in IAPAR59, were observed for the orphan genes CaUNK1, CaUNK4, CaUNK5, CaUNK8, and for CaPSBB (similar to the gene of C. arabica chloroplast genome encoding the photosystem II CP47 chlorophyll apoprotein) and CaSDC1 encoding a putative protein related (81 % identity, 88 %, similarity) to the adenosylmethionine decarboxylase proenzyme of Catharanthus roseus). Expression of the CaUNK6 gene was also induced under drought conditions but without significant difference in expression between the two cultivars.
Interestingly, the expression profiles of orphan genes CaUNK7, CaUNK9, CaUNK10, CaUNK15, CaUNK16 and CaUNK17 were similar to that of HSP-encoding gene CaHSP3 in the sense that gene expression was highly up-regulated under drought conditions in both cultivars. In the case of CaUNK10, it is worth noting that expression increased 145- and 88-fold under drought conditions in Rubi and IAPAR59, respectively.
Under drought conditions, expression of the CaGAS2 gene encoding a putative protein homologous (73 % identity, 86 % similarity) to the arbutin synthase from Rauvolfia serpentina (AJ310148), was slightly increased in IAPAR59 but reduced in Rubi. The CaCAB2, CaCHI1 and CaELIP3 genes encoding a photosystem II light harvesting chlorophyll A/B binding protein of Gardenia jasminoides (ACN41907), a class III chitinase of C. arabica (ADH10372) and an early light-induced protein (ELIP) of Glycine max (NP_001235754), respectively, showed similar profiles but with lower expression in Rubi than in IAPAR59, under control and drought conditions. Lastly, expression of the CaPP2 gene encoding a putative phloem protein 2 (PP2) of Vitis vinifera (XP_002279245) increased under drought conditions in Rubi but was quite stable in IAPAR59 under both conditions.
Expression of type II nsLTP genes
The expression of Type II nsLTP-encoding genes was also monitored using the primer pairs LTP-FT/LTP-R1 (specific to the CaLTP1 and CaLTP2 genes from the C. eugenioides sub-genome of C. arabica, hereafter referred to as CaCe), LTP-FT/LTP-R2 (specific to CaLTP3 genes from the C. canephora of C. arabica, hereafter CaCc) and LTP-F100/LTP-R100 recognizing all homologous genes [32]. No expression of nsLTP genes was detected under the control conditions in both cultivars (Fig. 5). However, expression of nsLTP genes was highly up-regulated in IAPAR59 but not in Rubi under drought conditions. It is worth noting that the CaLTP1-CaLTP2 and CaLTP3 genes were co-expressed in IAPAR59, and that the expression of CaCc genes was slightly higher than that of CaCe genes.
Fig. 5.
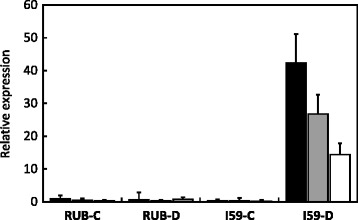
Expression of nsLTP genes. Expression of CaLTP1-CaLTP2 (CaCe: white isobars), CaLTP3 (CaCc: grey isobars) and all (CaLTP1, CaLTP2 and CaLTP3: black isobars) genes was analysed by qPCR in plagiotropic buds of Rubi (RUB) and IAPAR59 (I59) cultivars of C. arabica grown under control (C) and drought (D) conditions, using the LTP-FT/LTP-R2, LTP-FT/LTP-R1 and LTP-F100/LTP-R100 primer pairs, respectively [37]. Expression levels are expressed in arbitrary units (AU) of nsLTP genes using the expression of the CaUBQ10 gene as the endogenous control and RUB-C (with LTP100 primers) as the reference sample (Relative expression = 1). Values of three technical replications are presented as mean ± SD (bar)
Drought influences leaf cuticle thickness
Leaf anatomical analyses were also performed, revealing that the abaxial epidermis of IAPAR59 had a thicker cuticle than Rubi under drought conditions (Fig. 6). There was also a strong interaction between genotype and drought conditions (F1, 40 = 16,2). For example, in the DT cultivar IAPAR59, the abaxial epidermis cuticle thickness greatly increased under drought conditions compared with the control treatment (Table 4). However, no significant variation in abaxial epidermis cuticle thickness could be observed between the control and drought treatments for Rubi leaves.
Fig. 6.
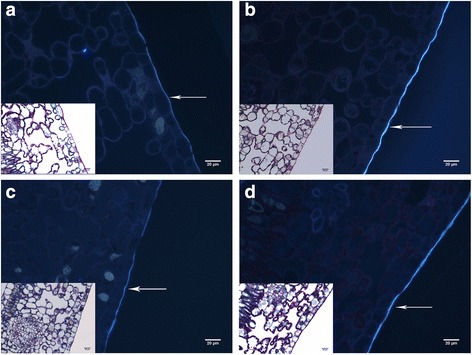
Comparative analysis of leaf histological cross sections of IAPAR59 (a and b) and Rubi (c and d) cultivars of C. arabica under control (irrigation: a and c) and drought (b and d) conditions. Samples were double stained with Schiff and NBB and observed under wide field (at the bottom left of each image) and fluorescent microscopy (A4 filter). LE = Lower (abaxial) epidermis. The white arrows indicate the fluorescent cuticle. Values of leaf cuticle thickness are given in Table 4. Bars = 20 μm
Table 4.
Influence of drought on leaf cuticle thickness
| Cuticle thickness (μm) | ||
|---|---|---|
| Treatment | IAPAR59 | Rubi |
| Control | 1.49 ± 0.19(a) | 1.75 ± 0.15(b) |
| Drought | 1.98 ± 0.19(c) | 1.73 ± 0.28(b) |
Leaves of IAPAR59 and Rubi cultivars of C. arabica grown under control (irrigation) and drought conditions were analysed to measure the cuticle thickness of the abaxial faces. Values (in μm) correspond to the average calculated from 11 independent measurements. Those marked with different letters are significantly different (Student-Newman-Keuls mean comparison test, P < 0.05)
Genes with reduced expression under drought conditions
The qPCR experiments led to the identification of several genes whose expression was reduced under drought conditions (Fig. 7). In both cultivars, expression of the orphan genes CaUNK11 and CaUNK12, and of the CaDLP1 gene encoding a putative protein containing a dirigent-like protein domain homologous to the hypothetical protein (CAN61316) of Vitis vinifera, was greatly reduced under drought conditions. Expression of the CaCHI2 gene encoding a protein homologous to the putative chitinase of Catharanthus roseus (ADK98562), was 5-fold higher in IAPAR59 than in Rubi under the control conditions but decreased under drought conditions. However, the expression level of the CaCHI2 gene was similar in IAPAR59 and Rubi under drought conditions. For the genes CaCHI3 (putative protein related to chitinase-like protein Artemisia annua [ABJ74186]), CaUNK13 and CaJAMT1 (putative protein containing a methyltransferase domain [pfam03492] found in enzymes acting on salicylic acid, jasmonic acid and 7-methylxanthine), similar expression profiles were found. In these cases, drought reduced gene expression in both cultivars but expression levels were always higher in IAPAR59 than in Rubi, particularly for CaJAMT1.
Fig. 7.
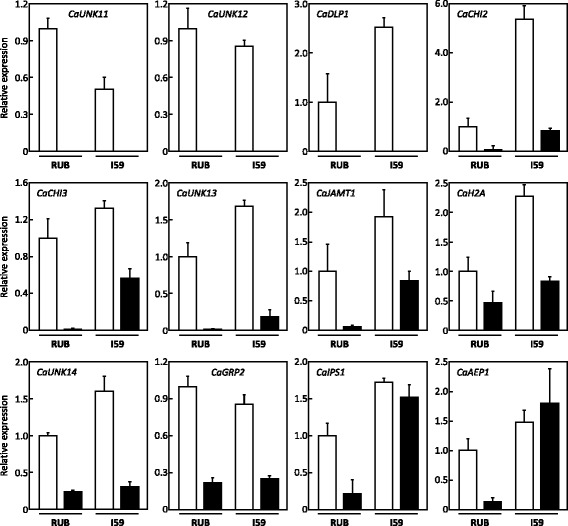
Expression profiles of genes down-regulated under drought conditions. Gene expression was analysed in plagiotropic buds of Rubi (RUB) and IAPAR59 (I59) cultivars of C. arabica grown under control (white isobars) and drought (black isobars) conditions. The gene names are indicated in the histograms. Transcript abundances were normalized using the expression of the CaUBQ10 gene as the endogenous control. Results are expressed using RUB-C as the reference sample (Relative expression = 1). Values of three technical replications are presented as mean ± SD (bar)
Gene expression levels of the CaH2A (H2A histone protein), CaGRP2 (putative glycin-rich protein) and CaUNK14 genes, were similar in Rubi and IAPAR59. For the CaAEP1 (putative aldose 1-epimerase) and CaIPS1 (myo-inositol 1-phosphate synthase) genes, gene expression remained high in IAPAR59 under both control and drought conditions, but decreased drastically in Rubi under drought conditions.
Discussion
In this study, we obtained 34.7 Mbp (coverage 6.5x) of sequences with longer reads (mean of 379.2 bp) from plagiotropic shoot apices enriched in meristems and primordium leaves of two cultivars of C. arabica under control (irrigation) and drought conditions. These sequences were assembled giving 43,087 clusters (17,719 contigs exclusively from 454-sequencing and 25,368 hybrid contigs formed by 454 and Sanger sequences) with a mean size ≥ 300 bp each. These RNAseq data, which complement those already available in public databases for coffee ESTs (407 million ESTs: dbEST release June 2015), can be considered as innovative and relevant in the sense that they were produced from C. arabica tissues (meristems) that have never previously been studied [39].
The transcriptome annotation by Blast2GO provided information based on the nomenclature and organism of origin of genes in the NCBI/NR database, the enzyme family, a functional analysis of proteins from the InterPro database, and metabolic functions, biological processes and cellular location from gene ontology. Our results showed that a large percentage of transcriptome alignment had 36,965 hits with known function (85.8 %), 1,824 genes with unknown function (4.2 %) both in the NCBI/NR database, and only 1,515 hits in the Interpro database (3.5 %), thereby enabling the identification of most genes. With this analysis, we identified 34,857 genes related to Coffea sp. (80.9 % of the total). We also found 1,383 genes from Solanum sp., 573 genes from Populus trichocarpa, 482 genes from Vitis vinifera and 156 genes from Arabidopsis sp. Thus, the transcriptome was aligned with several genes from different plant species and these genes may be conserved among these species, including Coffea sp. On the other hand, our results also included 2,783 “no-hit” genes (6.5 %), perhaps indicating the presence of unannotated or new genes.
The comparisons of DNA libraries undertaken during this work led to the identification of 1,243 genes (Table 3: ∑ Total DEG %) with differential expression profiles in silico between the drought-susceptible (Rubi) and drought-tolerant (IAPAR59) cultivars of C. arabica with drought conditions. The expression profiles of these genes, as well as those of other previously identified genes [10, 11, 30–32], were analysed by qPCR in plagiotropic buds (containing meristems and small leaves) taken from control and drought-stressed plants of Rubi and IAPAR59. For most of the CGs identified during this work, in vivo gene expression profiles confirmed those deduced from in silico comparisons of DNA libraries. For example, this was the case for the CaHSP3 (heat shock protein) gene whose up-regulated expression under drought conditions can be considered as a “molecular control” of stress applied to the plants during this study and confirmed by leaf water potential (Ψpd) measurements. Many ESTs encoding putative HSPs were also found in leaf cDNA libraries of C. arabica (SH2) and C. canephora (SH3) plants grown under drought conditions [31], heat stress [40], leaf infection by Hemileia vastatrix [15, 16] and also during bean development [14].
Our results also identified several genes differentially expressed in plagiotropic buds of IAPAR59 and Rubi, as for the CaSTK1 gene encoding a putative serine/threonine protein kinase containing a conserved domain (cd06610) of mitogen-activated protein kinases (MAPKs). These kinases are known to have a central role in the transduction of extra- and intracellular signals in plants, including cell division and differentiation, as well as in responses to various types of stress [41]. In Pisum sativum, there is evidence that the MAPK cascade is involved in ABA-regulated stomatal activity as well as ABA-induced gene expression in the epidermal peels [42]. In a recent study, Shen et al. [43] showed that the phosphorylation of OsWRKY30 protein by MAPKs is a key step in conferring drought tolerance in transgenic rice. According to our results, higher CaSTK1 expression under drought conditions in IAPAR59 than in Rubi could enhance the MAPK cascade and therefore be involved in the drought tolerance of IAPAR59. In this cultivar, the over-expression of CaSAMT1 under drought conditions is also particularly interesting because this sequence encodes a putative S-adenosyl-L-methionine-dependent methyltransferase related to the TUMOROUS SHOOT DEVELOPMENT2 (TSD2) gene. In Arabidopsis thaliana, tsd2 is a pleiotropic mutation that affects leaf, root and shoot meristem development [44]. Expression of a TSD2:: GUS reporter gene has mainly been detected in meristems where this gene is essential for cell adhesion and coordinated plant development. The weaker expression of CaSAMT1 in Rubi than in IAPAR59 under drought conditions, points to the existence of major developmental differences between these two cultivars. The differential expression in Rubi and IAPAR59 of the CaSLP1 gene encoding a putative subtilisin-like protein is also worth noting. In Arabidopsis, the subtilisin-like serine-protease SDD1 (stomatal density and distribution) gene was shown to be strongly expressed in stomatal precursor cells (meristemoids and guard mother cells) [45]. In addition, sdd1 mutation increased leaf stomatal density (SD) while SDD1 over-expression led to the opposite phenotype with decreased SD. In C. arabica, maximum and minimum average stomatal densities were observed in full sunlight and shaded conditions respectively, providing evidence for the existence of plasticity for this characteristic in this coffee species [46, 47]. Even though no SD were observed between Rubi and IAPAR59 under moderate drought conditions [48], the CaSLP1 expression profiles presented here do not preclude the involvement of this gene in the genetic determinism of drought tolerance in coffee.
Another interesting response concerned the differential expression of the CaMAS1 gene encoding a putative protein containing the conserved domain [cd05326]. This domain is also found in secoisolariciresinol dehydrogenase-like proteins catalyzing the NAD-dependent conversion of (-)-secoisolariciresinol to (-)-matairesinol, like the Arabidopsis ABA2 protein considered to be one of the key regulators of ABA biosynthesis [49]. Based on the CaMAS1 expression profiles presented here, it is possible that ABA synthesis was enhanced by drought in plagiotropic buds of IAPAR59 but not (or to a lesser extent) in those of Rubi. This hypothesis is also reinforced by the fact that higher CaJAMT1 expression was observed in IAPAR59 than in Rubi buds. Indeed, in addition to well-known functions of jasmonates in plant defence mechanisms in response to biotic stress [50], recent studies also demonstrated that methyl jasmonate stimulates ABA biosynthesis under drought conditions in panicles of Oryza sativa [51].
Higher expression of CaSDC1 (encoding a protein sharing 89 % similarity with the S-adenosyl-L-methionine decarboxylase from Catharanthus roseus) under drought conditions in IAPAR59 than in Rubi is also worth noting because this enzyme catalyzes the synthesis of polyamines (e.g. spermine, spermidine and putrescine) involved in stress tolerance in higher plants [52]. In Theobroma cacao, ABA and drought induced the expression of TcSAMDC increasing spermine and spermidine leaf contents correlated with changes in stomatal conductance [53]. More recently, SAMDC over-expression in transgenic rice was also shown to facilitate drought tolerance [54]. Investigation of polyamine levels in plagiotropic buds and leaves of IAPAR59 and Rubi would be of particular interest to see if these compounds are involved in drought tolerance in coffee.
In mature plants, nuclear-encoded early-light inducible proteins (ELIPs) accumulate in response to various stress conditions including ABA or desiccation [55]. These proteins are presumed to protect the chloroplast apparatus from photo-oxidation occurring after stomatal limitation of photosynthesis [56]. In a recent study, transgenic plants of Medicago truncatula over-expressing the Dsp22 gene from Craterostigma plantagineum were shown to be able to recover from water deprivation better than wild type plants, thereby reinforcing the idea of using ELIP-encoding genes to improve abiotic stress resistance in crops [57]. Our results clearly highlight the increased expression of the CaELIP3 (ELIP-like), CaPSBB (CP47-like) and CaCAB2 (PSII Cab proteins) genes, respectively, under drought conditions. Interestingly, the expression levels of all these genes were always higher in IAPAR59 than in Rubi. These results are also in accordance with electronic Northern experiments which showed high accumulation of ELIP and Cab-encoding ESTs in cDNA libraries of C. arabica and C. canephora subjected to drought [58].
Another surprising result concerned the CaPSBB gene that was reverse-transcribed and detected during our qPCR experiments despite the fact that it corresponds to a chloroplast gene [59]. However, preliminary analyses of a whole genome sequence of C. canephora revealed the presence of a CP47/like nuclear gene [60]. Interestingly, photosystem II CP47 chlorophyll apoproteins encoding ESTs have also been reported to be expressed in C. arabica beans [61], leaves infected by Hemileia vastatrix [62] and also in the cDNA libraries (SH2 and SH3) of drought-stressed coffee plants [14, 24, 31], demonstrating increased expression of this gene under biotic and abiotic stress. As CP47 and ELIP proteins are essential for the activity and protection of the photosynthetic apparatus [55], the expression profiles reported here probably reflect a better photosynthetic and physiological status of IAPAR59 compared to Rubi.
Differential expression was also observed for the chitinase-encoding gene CaCHI1, with higher expression in IAPAR59 than in Rubi. An opposite situation was observed with respect to the chitinase-encoding genes CaCHI2 and CaCHI3, whose expression was reduced under drought conditions. It is worth noting that the expression of these genes under drought conditions was always higher in IAPAR59 than in Rubi. These results also show that coffee chitinase-encoding genes responded in different ways to drought. A large number of chitinase-encoding ESTs were identified in the BCGP project [24], mainly in the SH2 cDNA library of drought-stressed plants of C. arabica var. Catuai [58], but also in the leaves of C. arabica infected by leaf rust [62]. Even though chitinases are defence-related enzymes induced by abiotic stress, some evidence also indicates their participation in tolerance to abiotic stress [63]. Even though the roles of pathogenesis-related proteins in abiotic stress are still not fully understood, DT transgenic plants over-expressing chitinase genes have been obtained [64]. In that sense, the high level of expression for CaCHI1 in plagiotropic buds of IAPAR59 under both control and drought conditions could have an important function in drought tolerance.
Arbutin is a phenolic glucoside (4-hydroxyphenyl-β-D-glucopyranoside) abundant in the leaves of many freezing- or desiccation-tolerant plants [65] and also present in coffee fruits [66]. In a previous study, down-regulation of the CcGAS1gene encoding arbutin synthase was reported in leaves of C. canephora under drought conditions [10]. The results presented here clearly demonstrated differential expression profiles for CaGAS2 between the two cultivars of C. arabica. Gene expression increased under drought conditions in IAPAR59 while the opposite was observed in Rubi. Even though the presence of arbutin in coffee leaves has never been demonstrated, further analyses of this metabolite should be performed to investigate the role of this glucoside (and of other phenolic compounds) in preventing cell damage in coffee subject to abiotic stresses.
The CaPP2 gene (encoding a putative phloem protein 2, PP2) also showed differential expression profiles, with higher expression in IAPAR59 than in Rubi. In higher plants, PP2s are sieve elements (SE) very abundant in the phloem sap. These proteins are believed to play an important role in the establishment of phloem-based defence mechanisms induced by insect attacks and feeding stress [67], but also by wounding and oxidative conditions [68]. The functions of PP2 proteins are still not clear but they could act by forming high molecular weight polymers to close (“SE plugging”) the sieve pores caused by external injuries mainly due to biotic stress [69]. When Arabidopsis was treated with HrpNEa (a proteinaceous elicitor of plant defences produced by gram-negative plant pathogenic bacteria), the suppression of phloem-feeding activities by aphids was attributed to over-expression of the PP2-encoding gene AtPP2-A1 [70]. Other studies showed that HrpN activated ABA signalling, thereby inducing drought tolerance in Arabidopsis thaliana [71]. Based on these results, the involvement of PP2 proteins in plant response mechanisms to abiotic stress can be hypothesized, for example by maintaining (or protecting) the integrity of vessels under drought conditions by forming sieve plate filaments upon oxidation [72]. In that case, higher synthesis of CaPP2 which would be expected to occur in IAPAR59 plagiotropic buds under drought conditions could play a role in drought-tolerance by reducing sap-flow in young leaves and consequently increasing the water use efficiency of this cultivar [48].
Other interesting results concerned the gene expression stability of the CaAEP1 (putative aldose 1-epimerase) and CaIPS1 (myo-inositol 1-phosphate synthase) genes observed in IAPAR59 under control and drought conditions, whereas expression of both genes decreased under drought conditions in Rubi. Plant cells use myo-inositol to synthesize a variety of low molecular weight compounds and sugar alcohols such as the galactinol, a key element in the formation of raffinose family oligosaccharides. Nishizawa et al. [73] found that plants with high galactinol and raffinose contents were less susceptible to oxidative stress. In C. arabica, up-regulation of CaGolS genes involved in galactinol biosynthesis was reported in leaves of plants subjected to severe drought [74]. In addition, drought up-regulated the expression of mannose 6-phosphate reductase (involved in mannitol biosynthesis) in leaves of C. canephora [10, 11] and C. arabica [75, 76]. Even though little is known about the biochemical mechanisms of drought tolerance in coffee, the accumulation of carbohydrates expected in leaves of drought-stressed plants as a consequence of the up-regulated expression of these genes, could play an important role in the genetic determinism of this phenotype in coffee [77].
In addition to the previously described genes, our results also identified several orphan genes that presented differential expression profiles between the cultivars and treatments, such as CaUNK2, CaUNK3 and CaUNK4 whose expression was highly induced under drought conditions in IAPAR59 and to a lesser extent in Rubi. Orphan genes are also expected to interact specifically with the environment as a consequence of lineage-specific adaptations to that environment [78].
Interestingly, the expression profiles of the CaUNK2 and CaUNK3 orphan genes were very similar to those of Type II nsLTP-encoding genes, with high expression mainly detected under drought conditions in plagiotropic buds of IAPAR59 but not in those of Rubi. Up-regulation of LTP genes under drought conditions is well documented in higher plants [79–81]. Lipid transfer proteins (LTPs) are thought to be involved in the transfer of lipids through the extracellular matrix for the formation of cuticular wax [82]. In fact, together with the lipophilic cutin polymer matrix, waxes enter in the composition of cuticle, which forms the first barrier between plants and environmental stresses by limiting non-stomatal water loss and gas exchanges, hence mitigating the effects of drought by controlling water loss associated with epidermal conductance [83]. In Nicotiana glauca, LTP genes are predominantly expressed in the guard and epidermal cells and are induced under drought conditions [84], providing evidence that LTP play an important role in the development of drought tolerance. Even though the up-regulation of CaLTP genes observed under drought in plagiotropic buds of IAPAR59 cannot explain directly the greater thickness of leaf cuticle observed in this cultivar than in Rubi, these results strongly suggested that lipid metabolism plays a major role in coffee drought tolerance.
As reported in other higher plants, our study also highlighted the differential expression of many genes encoding proteins known to be over-expressed under biotic stress (e.g. chitinases and PP2), by drought. The fact that our experiment was conducted with drought-stressed plants grown under uncontrolled (field) conditions, could explain such a situation. However, it is also probable that these results reflect a biological reality since it is well known that crosstalk exists in higher plants between signalling pathways for biotic and abiotic stress responses [85].
Conclusions
During this work, we produced some new transcriptomic information for C. arabica with a total of 34.7 Mbp of sequences assembled into 43,087 clusters (41,512 contigs and 1,575 singletons) from genes expressed in plagiotropic shoot apices enriched in meristems and primordium leaves in DT (IAPAR59) and DS (Rubi) cultivars grown under control and drought conditions. Major differences between these plants concerned their phenotypic behaviour (e.g. predawn leaf water potential, Ψpd) and transcriptome expression profiles. Differences between these plants affected genes of specific pathways such as those involved in abscisic acid biosynthesis, perception and transduction of drought stress, plant development and lipid metabolism. In that sense, the present study increased the number of CGs potentially involved in the genetic determinism of drought tolerance firstly identified in C. canephora. Because C. arabica is an amphidiploid species (originating from a natural hybridization event between C. canephora and C. eugenioides), its transcriptome is a mixture of homologous genes expressed from these two sub-genomes in which C. eugenioides is assumed to express genes mainly for proteins involved in basal biological processes (e.g. photosynthesis), while the C. canephora sub-genome is assumed to regulate Arabica gene expression by expressing genes for regulatory proteins and adaptation processes [86]. In this genetic context, it is possible that the characteristics of IAPAR59 that enable it to better withstand drought stress than Rubi, really originated from the specific expression of C. canephora genes recently introgressed (through the Timor hybrid HT832/2 [19]) in this cultivar of C. arabica [33]. Even though this study provides further indications about the way in which different coffee cultivars activate their transcriptomes, additional work is still required to understand how epigenetics and epistasis regulate gene expression in the different coffee sub-genomes (CaCe and CaCc) in C. arabica under drought conditions.
Source of the plant materials and permissions
This work was carried out as part of the scientific cooperation project entitled “Study of genetic determinism of drought tolerance in coffee” (2006–2010) approved between Embrapa and CIRAD. It complied with all institutional, national, or international guidelines. In the frame of this project, field experiments were conducted at the Cerrado Agricultural Research Center (Planaltina-DF, Brazil) with all permissions of partners and in accordance with local legislation.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Availability of supporting data
The reads were submitted to GenBank and to the BioProject/NCBI database under the accession number PRJNA282394.
Acknowledgments
The authors would like to thank Dr Aymbiré Francisco Almeida da Fonseca from the INCAPER Institute for kindly providing seeds of the Rubi and IAPAR59 commercial cultivars of C. arabica. The authors are also very grateful to Drs Antonio Fernando Guerra and Omar Cruz Rocha (Embrapa Cerrados) for their assistance during the field trial experiments. The authors wish also to thank Daphne Goodfellow, as well as Peter Biggins and Cécile Fovet-Rabot (CIRAD-DGDRS) for English revision of the manuscript.
Funding
PM acknowledges financial support from CIRAD (Centre de Coopération Internationale en Recherche Agronomique pour le Développement, Montpellier, France) and the CIRAD ATP project “Analysis of phenotypic plasticity in response to water constraints in perennial plants growing under different field conditions”. ACA acknowledges financial support from the Brazilian Coffee R&D Consortium, FINEP and INCT-café (CNPq/FAPEMIG). MFC and GAGP acknowledge financial support from the Center for Computational Engineering and Sciences - FAPESP/Cepid (2013/08293-7). The authors acknowledge the scholarships from the Brazilian agencies CAPES (KED, NGV), Brazilian Consortium of Coffee Research (FAC, JCA) and CAPES-COFECUB Project Sv738-12 (MGC).
Abbreviations
- EST
expressed sequence tag
- qPCR
quantitative polymerase chain reaction
Additional files
Summary of Blast2GO automatic annotation of the transcriptome clusters. (XLS 15226 kb)
Complete bioinformatics pipeline of the transcriptome assembly and automatic annotation methods used in this work. (TIF 105 kb)
Summary of the DEseq/ EdgeR fold changes and p-values between the cultivars (I59: IAPAR59 and RUB: Rubi) and between control (C) and drought (D) conditions. These tables also contain Blast2GO automatic annotation of the transcriptome clusters. Table lanes coloured in grey are related to clusters aligned to the new candidate genes tested by RT-qPCR (see Table 1). (XLS 51895 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GCR and PM measured predawn leaf water potentials and harvested the samples. FdB and TL performed meristem dissections, RNA extractions and cDNA synthesis. LSM, MFC, GAGP and RV were responsible for the bioinformatic processing of the data. JLV, FLM and ML performed the histology and microscopy analyses. PM, ACA and DP selected the candidate genes qPCR-analysed by FAC, NGV, KED, JCA and MGC. GCR, ACA and PM designed the study, drew up the experimental design and implemented it. RV, MFC, GAGP, ACA and PM drafted the manuscript. All the authors read and approved the final version of the manuscript.
Contributor Information
Luciana Souto Mofatto, Email: luciana@lge.ibi.unicamp.br.
Fernanda de Araújo Carneiro, Email: fearca14@gmail.com.
Natalia Gomes Vieira, Email: ngvieira1@gmail.com.
Karoline Estefani Duarte, Email: karollduarte31@gmail.com.
Ramon Oliveira Vidal, Email: ramon.vidal@gmail.com.
Jean Carlos Alekcevetch, Email: jeancarlosembrapa@gmail.com.
Michelle Guitton Cotta, Email: michellegcotta@gmail.com.
Jean-Luc Verdeil, Email: jean-luc.verdeil@cirad.fr.
Fabienne Lapeyre-Montes, Email: fabienne.lapeyre-montes@cirad.fr.
Marc Lartaud, Email: marc.lartaud@cirad.fr.
Thierry Leroy, Email: thierry.leroy@cirad.frl.
Fabien De Bellis, Email: fabien.de_bellis@cirad.fr.
David Pot, Email: david.pot@cirad.fr.
Gustavo Costa Rodrigues, Email: gustavo.rodrigues@embrapa.br.
Marcelo Falsarella Carazzolle, Email: mcarazzo@lge.ibi.unicamp.br.
Gonçalo Amarante Guimarães Pereira, Email: goncalo@unicamp.br.
Alan Carvalho Andrade, Email: alan.andrade@embrapa.br.
Pierre Marraccini, Email: marraccini@cirad.fr.
References
- 1.Pay E. The market for organic and fair-trade coffee. FAO Rome. 2009.
- 2.ICO (International Coffee Organization), Statistics of production 2015: http://www.ico.org/prices/po-production.pdf.
- 3.DaMatta FM, Ramalho JC. Impact of drought and temperature stress on coffee physiology and production: a review. Braz J Plant Physiol. 2006;18:55–81. doi: 10.1590/S1677-04202006000100006. [DOI] [Google Scholar]
- 4.Silva EA, Mazzafera P, Brunini O, Sakai E, Arruda FB, Mattoso LHC, et al. The influence of water management and environmental conditions on the chemical composition and beverage quality of coffee beans. Braz J Plant Physiol. 2005;17:229–38. doi: 10.1590/S1677-04202005000200006. [DOI] [Google Scholar]
- 5.Bunn C, Läderach P, Pérez Jimenez JG, Montagnon C, Schilling T. Multiclass classification of agro-ecological zones for Arabica coffee: an improved understanding of the impacts of climate change. PLoS One. 2015;10 doi: 10.1371/journal.pone.0140490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montagnon C, Leroy T. Réaction à la sécheresse de jeunes caféiers Coffea canephora de Côte-d’Ivoire appartenant à différents groupes génétiques. Café Cacao Thé. 1993;37:179–90. [Google Scholar]
- 7.Pinheiro HA, DaMatta FM, Chaves ARM, Loureiro ME, Ducatti C. Drought tolerance is associated with rooting depth and stomatal control of water use in clones of Coffea canephora. Ann Bot. 2005;96:101–8. doi: 10.1093/aob/mci154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beining A, Burkhardt J, Fetene M. Water relations of Ethiopian wild coffee populations: genetic fixation and phenotypic plasticity. In Proceedings of the 22nd International Scientific Colloquium on Coffee, Campinas, International Scientific Association on Coffee, Paris, CDROM-A105, 2008.
- 9.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–7. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 10.Marraccini P, Vinecky F, Alves GSC, Ramos HJO, Elbelt S, Vieira NG, et al. Differentially expressed genes and proteins upon drought acclimation in tolerant and sensitive genotypes of Coffea canephora. J Exp Bot. 2012;63:4191–212. doi: 10.1093/jxb/ers103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vieira NG, Carneiro FA, Sujii PS, Alekcevetch JC, Freire LP, Vinecky F, et al. Different molecular mechanisms account for drought tolerance in Coffea canephora var. Conilon. Trop Plant Biol. 2013;6:181–90. doi: 10.1007/s12042-013-9126-0. [DOI] [Google Scholar]
- 12.Lin C, Mueller LA, McCarthy J, Crouzillat D, Pétiard V, Tanksley SD. Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts. Theor Applied Genet. 2005;112:114–30. doi: 10.1007/s00122-005-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poncet V, Rondeau M, Tranchand C, Cayrel A, Hamon S, de Kochko A, et al. SSR mining in coffee tree EST databases: potential use of EST-SSRs as marker across Coffea genus. Mol Gen Genet. 2006;276:436–49. doi: 10.1007/s00438-006-0153-5. [DOI] [PubMed] [Google Scholar]
- 14.Vieira LGE, Andrade AC, Colombo CA, Moraes AAH, Metha A, Oliveira AC, et al. Brazilian coffee genome project: an EST-based genomic resource. Braz J Plant Physiol. 2006;18:95–108. doi: 10.1590/S1677-04202006000100008. [DOI] [Google Scholar]
- 15.Guzzo SD, Harakava R, Tsai SM. Identification of coffee genes expressed during systemic acquired resistance and incompatible interaction with Hemileia vastatrix. J Phytopathol. 2009;157:625–38. doi: 10.1111/j.1439-0434.2008.01538.x. [DOI] [Google Scholar]
- 16.Fernandez D, Santos P, Agostini C, Bon MC, Petitot AS, Silva CM, et al. Coffee (Coffea arabica L.) genes early expressed during infection by the rust fungus (Hemileia vastatrix) Mol Plant Pathol. 2004;5:527–36. doi: 10.1111/j.1364-3703.2004.00250.x. [DOI] [PubMed] [Google Scholar]
- 17.Traas J, Vernoux T. The shoot apical meristem: the dynamics of a stable structure. Philos T Roy Soc B. 2002;357:737–47. doi: 10.1098/rstb.2002.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres TT, Metta M, Ottenwalder B, Schlotterer C. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–7. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho CHS, Fazuoli LC, Carvalho GR, Guerreiro-Filho O, Pereira AA, de Almeida SR, et al. Cultivares de café arábica de porte baixo. In: Carvalho CHS, et al., editors. Cultivares de Café: origem, características e recomendações. Brasilia: Embrapa Café; 2008. pp. 157–226. [Google Scholar]
- 20.Ratter JA, Ribeiro JF, Bridgewater S. The Brazilian cerrado vegetation and threats to its biodiversity. Ann Bot. 1997;80:223–30. doi: 10.1006/anbo.1997.0469. [DOI] [Google Scholar]
- 21.Lécolier A, Noirot M, Escoute J, Chrestin H, Verdeil JL. Early effects of the mutation laurina on the functioning and size of the shoot apex in coffee tree and analysis of the plastochron phases: relationships with the dwarfism of leaves. Trees. 2009;23:1043–51. doi: 10.1007/s00468-009-0346-8. [DOI] [Google Scholar]
- 22.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–9. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mondego JMC, Vidal RO, Carazzolle MF, Tokuda EK, Parizzi LP, Costa GGL, et al. An EST-based analysis identifies new genes and reveals distinctive gene expression features of Coffea arabica and Coffea canephora. BMC Plant Biol. 2011;11:30. doi: 10.1186/1471-2229-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudet C, Dias Z. New EST trimming strategy in: Brazilian Symposium on Bioinformatics, 2005. Lect Notes Bioinf. 2005;3594:206–9. [Google Scholar]
- 26.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–59. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 28.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira NG. Identificação e caracterização de genes órfãos (“NO HITS”) de café (Coffea spp.). Master Thesis, University of Lavras (UFLA), Lavras-MG, Brazil. 2013. 130p. http://repositorio.ufla.br/handle/1/4928.
- 31.Vinecky F, da Silva FR, Andrade AC. Análise in silico das bibliotecas de cDNA SH2 e SH3 para a identificação de genes responsivos à seca em cafeeiro. Coffee Sci. 2012;7:1–19. [Google Scholar]
- 32.Cotta MG, Barros LMG, de Almeida JD, de Lamotte F, Barbosa EA, Vieira NG, et al. Lipid transfer proteins in coffee: isolation of Coffea orthologs, Coffea arabica homeologs, expression during coffee fruit development and promoter analysis in transgenic tobacco plants. Plant Mol Biol. 2014;85:11–31. doi: 10.1007/s11103-013-0166-5. [DOI] [PubMed] [Google Scholar]
- 33.Marraccini P, Freire LP, Alves GSC, Vieira NG, Vinecky F, Elbelt S, et al. RBCS1 expression in coffee: Coffea orthologs, Coffea arabica homeologs, and expression variability between genotypes and under drought stress. BMC Plant Biol. 2011;11:85. doi: 10.1186/1471-2229-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–6. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 35.Schwendiman J, Pannetier C, Michaux-Ferriere N. Histology of somatic embryogenesis from leaf explants of the oil palm Elaeis guineensis. Ann Bot. 1988;62:43–52. [Google Scholar]
- 36.Buffard-Morel J, Verdeil JL, Pannetier C. Embryogenèse somatique du cocotier (Cocos nucifera L.) à partir d’explant foliaire: étude histologique. Can J Bot. 1992;70:735–41. doi: 10.1139/b92-094. [DOI] [Google Scholar]
- 37.Fisher DB. Protein staining of ribonned epon section for light microscopy. Histochemie. 1968;16:92–6. doi: 10.1007/BF00306214. [DOI] [PubMed] [Google Scholar]
- 38.Sokal RR, Rohlf JF. Biometry: the principles and practice of statistics in biological research. 3. New York: WH Freeman and Company; 1995. [Google Scholar]
- 39.Lashermes P, Andrade AC, Etienne H. Genomics of coffee, one of the world’s largest traded commodities. In: Moore H, Ming R, editors. Genomics of tropical crop plants. Berlin: Springer; 2008. pp. 203–26. [Google Scholar]
- 40.Bardil A, de Almeida JD, Combes MC, Lashermes P, Bertrand B. Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytologist. 2011;192:760–74. doi: 10.1111/j.1469-8137.2011.03833.x. [DOI] [PubMed] [Google Scholar]
- 41.Mishra NS, Tuteja R, Tuteja N. Signaling through MAP kinase networks in plants. Arch Biochem Biophys. 2006;452:55–68. doi: 10.1016/j.abb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Burnett EC, Desikan R, Moser RC, Neill SJ. ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J Exp Bot. 2000;51:197–205. doi: 10.1093/jexbot/51.343.197. [DOI] [PubMed] [Google Scholar]
- 43.Shen H, Liu C, Zhang Y, Meng X, Zhou X, Chu C, et al. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol Biol. 2012;80:241–53. doi: 10.1007/s11103-012-9941-y. [DOI] [PubMed] [Google Scholar]
- 44.Krupková E, Immerzeel P, Pauly M, Schmülling T. The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J. 2007;50:735–50. doi: 10.1111/j.1365-313X.2007.03123.x. [DOI] [PubMed] [Google Scholar]
- 45.von Groll U, Berger D, Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell. 2002;14:1527–39. doi: 10.1105/tpc.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pompelli MF, Martins SCV, Celin EF, Ventrella MC, DaMatta FM. What is the influence of ordinary epidermal cells and stomata on the leaf plasticity of coffee plants grown under full-sun and shady conditions? Braz J Biol. 2010;70:1083–8. doi: 10.1590/S1519-69842010000500025. [DOI] [PubMed] [Google Scholar]
- 47.Kufa T, Burkhardt J. Stomatal characteristics in Arabica coffee germplasm accessions under contrasting environments at Jimma, southwestern Ethiopia. Intern J Bot. 2011;7:63–72. doi: 10.3923/ijb.2011.63.72. [DOI] [Google Scholar]
- 48.Rodrigues GC, Rojas JSD, Roupsard O, Leroy T, Pot D, Moreira MZ, et al. Preliminary results on phenotypic plasticity of coffee (Coffea arabica cv. Rubi and Iapar59) plants in response to water constraint under field conditions. In Proceedings of the 23rd International Scientific Colloquium on Coffee, Bali, International Scientific Association on Coffee, Paris, CDROM-PB731, 2010.
- 49.Schwartz SH, Léon-Kloosterziel KM, Koornneef M, Zeevaart JAD. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;114:161–6. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santino A, Taurino M, De Domenico S, Bonsegna S, Poltronieri P, Pastor V, et al. Jasmonate signaling in plant development and defense response to multiple (a) biotic stresses. Plant Cell Rep. 2013;32:1085–98. doi: 10.1007/s00299-013-1441-2. [DOI] [PubMed] [Google Scholar]
- 51.Kim EH, Kim YS, Park SH, Koo YJ, Choi YD, Chung YY, et al. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009;149:1751–60. doi: 10.1104/pp.108.134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alcázar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, et al. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta. 2010;231:1237–49. doi: 10.1007/s00425-010-1130-0. [DOI] [PubMed] [Google Scholar]
- 53.Bae H, Kim SH, Kim MS, Sicher RC, Lary D, Strem MD, et al. The drought response of Theobroma cacao (cacao) and the regulation of genes involved in polyamine biosynthesis by drought and other stresses. Plant Physiol Biochem. 2008;46:174–88. doi: 10.1016/j.plaphy.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Peremarti A, Bassie L, Christou P, Capell T. Spermine facilitates recovery from drought but does not confer drought tolerance in transgenic rice plants expressing Datura stramonium S-adenosylmethionine decarboxylase. Plant Mol Biol. 2009;70:253–64. doi: 10.1007/s11103-009-9470-5. [DOI] [PubMed] [Google Scholar]
- 55.Duque AS, Almeida AM, Silva AB, Silva JM, Farinha AP, Santos D, et al. Abiotic stress responses in plants: unraveling the complexity of genes and networks to survive. In: Vahdati K, Leslie C, et al., editors. Abiotic Stress - Plant responses and applications in agriculture. Rijeka (Croatia): InTech Publisher; 2013. pp. 49–101. [Google Scholar]
- 56.Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M. Early light induced proteins protect Arabidopsis from photoxidative stress. Proc Natl Acad Sci U S A. 2003;100:4921–6. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Araújo SS, Duque AS, Silva JM, Santos D, Silva AB, Fevereiro P. Water deficit and recovery response of Medicago truncatula plants expressing the ELIP-like DSP22. Biol Plantarum. 2013;57:159–63. doi: 10.1007/s10535-012-0235-7. [DOI] [Google Scholar]
- 58.Alvarenga SM, Caixeta ET, Hufnagel B, Thiebaut F, Zambolim EM, Zambolim L, et al. In silico identification of coffee genome expressed sequences potentially associated with resistance to diseases. Gen Mol Biol. 2010;33:795–806. doi: 10.1590/S1415-47572010000400031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol J. 2007;5:339–53. doi: 10.1111/j.1467-7652.2007.00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345:1181–4. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- 61.Salmona J, Dussert S, Descroix F, de Kochko A, Bertrand B, Joët T. Deciphering transcriptional networks that govern Coffea arabica seed development using combined cDNA array and real-time RT-PCR approaches. Plant Mol Biol. 2008;66:105–24. doi: 10.1007/s11103-007-9256-6. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez D, Tisserant E, Talhinhas P, Azinheira H, Vieira A, Petitot AS, et al. 454-pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix reveals in planta-expressed pathogen-secreted proteins and plant functions in a late compatible plant-rust interaction. Mol Plant Pathol. 2012;13:17–37. doi: 10.1111/j.1364-3703.2011.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu LX, Djebrouni M, Chamberland H, Lafontaine JG, Tabaeizadeh Z. Chitinase: differential induction of gene expression and enzyme activity by drought stress in the wild (Lycopersicon chilense Dun.) and cultivated (L. esculentum Mill.) tomatoes. J Plant Physiol. 1998;153:745–53. doi: 10.1016/S0176-1617(98)80230-7. [DOI] [Google Scholar]
- 64.Dana MM, Pintor-Toro JA, Cubero B. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 2006;142:722–30. doi: 10.1104/pp.106.086140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hincha DK, Oliver AE, Crowe JH. Lipid composition determines the effects of arbutin on the stability of membranes. Biophys J. 1999;77:2024–34. doi: 10.1016/S0006-3495(99)77043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iwai K, Kishimoto N, Kakino Y, Mochida Y, Fujita T. In vitro antioxidative effects and tyrosinase inhibitory activities of seven hydroxycinnamoyl derivatives in green coffee beans. J Agric Food Chem. 2004;52:4893–8. doi: 10.1021/jf040048m. [DOI] [PubMed] [Google Scholar]
- 67.Will T, van Bel AJE. Physical and chemical interactions between aphids and plants. J Exp Bot. 2006;57:729–37. doi: 10.1093/jxb/erj089. [DOI] [PubMed] [Google Scholar]
- 68.Tjallingii WF. Salivary secretions by aphids interacting with proteins of phloem wound responses. J Exp Bot. 2006;57:739–45. doi: 10.1093/jxb/erj088. [DOI] [PubMed] [Google Scholar]
- 69.Kehr J. Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J Exp Bot. 2006;57:767–74. doi: 10.1093/jxb/erj087. [DOI] [PubMed] [Google Scholar]
- 70.Zhang C, Shi H, Chen L, Wang X, Lü B, Zhang S, et al. Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol. 2011;11:11. doi: 10.1186/1471-2229-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang C, Bao Z, Liang Y, Yang X, Wu X, Hong X, et al. Abscisic acid mediates Arabidopsis drought tolerance induced by HrpNEa in the absence of ethylene signaling. Plant Mol Biol Rep. 2007;25:98–114. doi: 10.1007/s11105-007-0012-5. [DOI] [Google Scholar]
- 72.Walz C, Juenger M, Schad M, Kehr J. Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J. 2002;31:189–97. doi: 10.1046/j.1365-313X.2002.01348.x. [DOI] [PubMed] [Google Scholar]
- 73.Nishizawa A, Yabuta Y, Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147:1251–63. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.dos Santos TB, Budzinski IGF, Marur CJ, Petkowicz CLO, Pereira LFP, Vieira LGE. Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiol Biochem. 2011;49:441–8. doi: 10.1016/j.plaphy.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 75.Freire LP, Marraccini P, Rodrigues GC, Andrade AC. Análise da expressão do gene da manose 6 fosfato redutase em cafeeiros submetidos ao déficit hídrico no campo. Coffee Sci. 2013;8:17–23. [Google Scholar]
- 76.de Carvalho K, Petkowicz CLO, Nagashima GT, Bespalhok Filho JC, Vieira LGE, Pereira LFP, et al. Homeologous genes involved in mannitol synthesis reveal unequal contributions in response to abiotic stress in Coffea arabica. Mol Genet Genomics. 2014;289:951–63. doi: 10.1007/s00438-014-0864-y. [DOI] [PubMed] [Google Scholar]
- 77.DaMatta FM, Chaves ARM, Pinheiro HA, Ducatti C, Loureiro ME. Drought tolerance of two field-grown clones of Coffea canephora. Plant Sci. 2003;164:111–7. doi: 10.1016/S0168-9452(02)00342-4. [DOI] [Google Scholar]
- 78.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 79.Treviño MB, O’Connell MA. Three drought-responsive members of the non-specific lipid-transfer protein gene family in Lycopersicon pennellii show different patterns of expression. Plant Physiol. 1998;116:1461–8. doi: 10.1104/pp.116.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jung HW, Kim W, Hwang BK. Three pathogen-inducible genes encoding lipid transfer protein from pepper are differentially activated by pathogens, abiotic, and environmental stresses. Plant Cell Environ. 2003;26:915–28. doi: 10.1046/j.1365-3040.2003.01024.x. [DOI] [PubMed] [Google Scholar]
- 81.Jang CS, Lee HJ, Chang SJ, Seo YW. Expression and promoter analysis of the TaLTP1 gene induced by drought and salt stress in wheat (Triticum aestivum L.) Plant Sci. 2004;167:995–1001. doi: 10.1016/j.plantsci.2004.05.019. [DOI] [Google Scholar]
- 82.Cameron KD, Teece MA, Smart LB. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006;140:176–83. doi: 10.1104/pp.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schweizer P, Felix G, Buchala A, Muller C, Metraux JP. Perception of free cutin monomers by plant cells. Plant J. 1996;10:331–41. doi: 10.1046/j.1365-313X.1996.10020331.x. [DOI] [Google Scholar]
- 84.Cameron KD, Moskal WA, Smart LB. A second member of the Nicotiana glauca lipid transfer protein gene family, NgLTP2, encodes a divergent and differentially expressed protein. Funct Plant Biol. 2006;33:141–52. doi: 10.1071/FP05170. [DOI] [PubMed] [Google Scholar]
- 85.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–42. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 86.Vidal RO, Mondego JMC, Pot D, Ambrósio AB, Andrade AC, Pereira LFP, et al. A high-throughput data mining of single nucleotide polymorphisms in Coffea species expressed sequence tags suggests differential homeologous gene expression in the allotetraploid Coffea arabica. Plant Physiol. 2010;154:1053–66. doi: 10.1104/pp.110.162438. [DOI] [PMC free article] [PubMed] [Google Scholar]


