Abstract
Background and Objective
We live our lives by the calendar and the clock, but time is also an abstraction, even an illusion. The sense of time can be both domain-specific and complex, and is often left implicit, requiring significant domain knowledge to accurately recognize and harness. In the clinical domain, the momentum gained from recent advances in infrastructure and governance practices has enabled the collection of tremendous amount of data at each moment in time. Electronic Health Records (EHRs) have paved the way to making these data available for practitioners and researchers. However, temporal data representation, normalization, extraction and reasoning are very important in order to mine such massive data and therefore for constructing the clinical timeline. The objective of this work is to provide an overview of the problem of constructing a timeline at the clinical point of care and to summarize the state-of-the-art in processing temporal information of clinical narratives.
Methods
This review surveys the methods used in three important area: modeling and representing of time, Medical NLP methods for extracting time, and methods of time reasoning and processing. The review emphasis on the current existing gap between present methods and the semantic web technologies and catch up with the possible combinations.
Results
the main findings of this review is revealing the importance of time processing not only in constructing timelines and clinical decision support systems but also as a vital component of EHR data models and operations.
Conclusions
Extracting temporal information in clinical narratives is a challenging task. The inclusion of ontologies and semantic web will lead to better assessment of the annotation task and, together with medical NLP techniques, will help resolving granularity and co-reference resolution problems.
Keywords: Clinical Temporal Information, Temporal Representation, Temporal Extraction, Ontologies of time, Medical NLP
1. Introduction
Time is a universal phenomenon that has interested many disciplines of science for many years. It provides basic elements for understanding the world in its dynamics: (a) in mining actions and changes to recognize pattern evolution, and (b) describing time-oriented relations for intelligent decision-making. Similarly, time plays a major role in the clinical domain by helping understanding chronological development of clinical procedures such as diagnosis (e.g. the order in which symptoms develop), treatment (e.g. time of taking medications), and prevention (e.g., signals for pre-disease). Remarkably, researchers have avidly studied time concepts and their representations. Mathematicians formulate time theories in order to abstract elements of time and temporal entities; philosophers contest changes and dynamics since ancient time; physicists, in both Newton’s and Einstein’s physics, debate the notions of special time and the dimension of time.
In the clinical domain, patients’ data have been collected over time and recorded in Electronic Health Record (EHR) systems; the ever growing complexity of such patterns of data reveals challenges in handling its high dimensionality taking into consideration complex parameters such as amplitude modifications, time warping and noise. In addition, clinical data differs from other time series by the fact that observations are made at irregular time intervals, and some of them may be missed or disrupted [1]. Therefore, temporal mining will need to provide solutions and innovations in both, theoretical view, such as time parameterization and abstraction, and methodological view, such as temporal relations extraction and event calculation. Moreover, as we are heading towards the “Big Clinical Data” era, we are faced with a torrent of data generated and captured in digital form as a result of the advancement of sciences, engineering and information technology. Consequently, there is a great potential of new waves of innovation to be aroused on detailed trend analyses by taking advantage of this large-scale and high-resolution data sets. Nevertheless, the heterogeneity and the complex nature of big data make it challenging to be leveraged directly by any algorithms without intensive and manual analysis.
Additionally, approximately 80% of EHR is unstructured [2] [3]. Correspondingly, temporal reasoning and interpretation will confront additional challenges as reasoning about time requires so-called “common knowledge” which can be notoriously difficult to establish (e.g. the yesterday in “yesterday she experienced some pain” could be implemented rigidly as a microsecond after midnight). Furthermore, idiosyncratically structured and disparate health information suitable for large-scale analyses as well as inference, needs to be dynamically transformed into standards. Many grounded researches have been established for better extraction of temporal information and providing guidelines for the standardization of temporal statements. In this connection, there is yet a gap between semantic inference technologies research and clinical and medical NLP approaches. We posit that the fundamental challenges that hinder the secondary use of EHR temporal data include: 1) temporal information exists in different formats (structured, semi-structured and non-structured) [4]; 2) mechanisms to harvest time for specific purposes are not formalized or readily available; and 3) Medical Natural Language Processing (Medical NLP) tools are of variable quality and completeness with respect to a given extraction purpose.
In this survey, we provide the background approaches of representation and reasoning about time-oriented aspects. It is devised in a way that even a non-specialized reader will be able to grasp. The current state of this research is inspired by state-of-the-art work in temporal annotation and extraction and at the same time reveals relevant issues of research in the near future. The paper is organized as follows. The following section overviews the modeling of time in the clinical domain; it presents some ontology-based representations and discusses temporal reasoning and some applications of time modeling. Section 3 describes the handling of time by standardized clinical models of structured data. Section 4 focuses on the extraction of temporal information in clinical narratives: it overviews temporal annotation schemas and the methods of temporal extraction. Section 5 presents recent work on time processing techniques such as time normalization and temporal abstraction and problems such as time granularity and temporal co-reference. The last section discusses the proposition of our approach for combining semantic web standards and medical NLP tools and it concludes this work.
2. Temporal modeling in the clinical domain
The conception of time relies on how we perceive it. People easily distinguish between past, present and future, but also, the time is tenseless when defined through a mathematical structure. For example, time may be discrete, dense, or continuous according to the arithmetical definitions of discreteness, denseness, and continuity respectively. The EHR data are rich with statements filled with assertions about time such as patient visits, laboratory tests, and disease symptoms and also statements about procedures such as diagnosis, prognosis, and medical therapy. Thus, the temporal modeling will provide the element to capture all these clinical variables and minimize the risk of information loss [5]. In this section we present an overview of methods of representing time and how these methods impact the reasoning and other interfaces in the clinical applications.
2.1 Multifaceted aspects of temporal concepts
The requirements of time representation in the clinical domain are many and diverse because time is recognized in different ways. Zhou et al [6] realized that time falls into manifold categories determined by structure/construct and reference/position and can be represented under different aspects such as periodicity and granularity. Table 1 illustrates principal time categories and their specifications.
Table 1.
Principal categories and sub categories of temporal expressions
| Category | Types | Definition | Features |
|---|---|---|---|
| Primitive of time | Point | Ideal for specifying accurate positions in time | Scheduling, planning, temporal constraints and temporal relations are barely supported [7] [8] |
| Interval | Ideal for representing coarse and incomplete temporal knowledge [9] | Used by Allen’s temporal logic [10]. | |
| Linear or branching | Linear time | Time flows from past to future in a timeline order | Used by time-lining events |
| Branching time | Time is linear from the past till present, after it divides into several futures | Used for hypothesizing. Suitable for diagnosis and prognosis | |
| Circular time | Time turns around a circle | Used to describe recurrent events, such as “administration of regular insulin every morning” | |
| Reference of time | absolute/Anchored date and time | Accurate position in the time/day clock | Has limited temporal reasoning tasks |
| Relative / unanchored date and time | More expressive, comprises more information such as temporal relation to other expressions | Entails prevalent time-based knowledge [11] and requires linguistic analysis tools | |
| Duration of time | Quantitative duration | Fixed quantity in time | Not flexible in reasoning |
| Qualitative duration | Ideal for the specification of temporal constraints | Flexible for qualitative reasoning over events and temporal expressions |
2.2 Temporal representation in the semantic web
The Semantic Web provides a suitable environment for representing the multifaceted structures of temporal clinical data. It has gathered the consensuses in time representation in many domains for its formality and rich expressiveness [12]. W3C most recommended metadata models and languages have been vividly involved. Web Ontology Language (OWL) [13] has been proven to easily incorporate time entities into existing ontologies by representing temporal knowledge and time-based information. Temporal Description Logic (TDL) [14] has extended standard description logic to capture the evolving behavior of dynamic domains. Semantic Web Rule Language (SWRL) [15] is an inferencing rule language that has been used for ordering and other forms of reasoning. Temporal RDF extended standard RDF by adding an additional dimension of temporal annotation and allows for reasoning over incomplete temporal information by combining fuzzy logic [16] and undefined intervals techniques [17]. Reification techniques [18] [19] [20] have extended OWL’s restriction of unary and binary specification in time relationships by expressing nary relations. Temporal querying languages such as t-SPARQL [21] and other extensions of SPARQL allow the questioning of RDF triple stores based on temporal descriptions. Many ontologies have picked up from these tools for temporal representations. The table 2 illustrates some of these ontologies.
Table 2.
Principal ontologies for modeling time
| Ontology | Description |
|---|---|
| OWL-Time ontology [22] | Provides vocabularies for instants and intervals, durations, datetime, and Allen’s Interval Temporal Logic [10]. |
| SWRL temporal ontology [23] | Contains SWRL built-ins to reason about defined temporal information |
| DAML ontology of time [24] | Integrates First-order predicate calculus for topological temporal relations such as intervals, events, dates and times. |
| CHRONOS [25] | Stand-alone ontology and Allen’s Interval Temporal Logic framework that enable to infer implied relations, detect inconsistencies and ensure path consistency |
| PSI-Time Ontology [26] | Represents the concepts of relativist and absolutist durations, periodic time intervals, interval phases, open time, linear time, discrete time, anisotropic time, and the relations, point-to-point, point-to-interval and interval-to-interval. |
| RSCDF Ontology [27] | Temporal and contextual extension of RDF |
| Reusable Time ontology [28] | Represents time granularity by reusing a Physical-Quantities ontology that belongs to Ontolingua library [29]. |
By and large, these ontologies taken together do not satisfactorily cover some important features, such as phase, granular time, and modality. In addition they are meant for the general domain and does not fulfill the need of clinical and healthcare applications. For these reasons, Clinical Narrative Temporal Relation Ontology (CNTRO) [30] was developed towards filling the semantic gap of temporal modeling in the clinical domain. CNTRO is meant to annotate temporal expressions and relations in clinical narratives. CNTRO provides vocabularies such as Event, Time, Duration, Granularity, Precision, and TemporalRelationStatement. The Event is defined to sort occurrences, states, perceptions, procedures, symptoms and situations. The Time concept is inherited by some constructs of time, like points of time, intervals of time, and periodical time and associated with properties such as hasGranularity, hasOrigTime and hasNormalizedTime. TemporalRelationStatement is used to describe temporal relations between two events or between an event and a time instance. The concepts TimePhase and TimePeriod specify Event concept for representing intermittent events. CNTRO has been used for time-oriented question answering in Clinical Narratives [31]. The framework provides a query API for users to query represented knowledge.
2.3 Temporal reasoning and mining
In recent years, temporal reasoning has gained momentum in clinical applications, partly due to the increasing demand for time-related clinical decision support. Many efforts have picked up from the latest works on reasoning about temporal relations. Allen’s interval algebra, first event ordering systems, has been chosen by many ontology-based reasoning systems [31]; however; this theory doesn’t provide accurate precision in ordering intervals (e.g. the overlaps relations suffices for the intersection on 1 point in time). The Quality Data Model (QDM) [32] has proposed 25 different temporal ordering relations between intervals. The constraint propagation algorithm for temporal reasoning [33] and temporal constraint networks [34] have extended network-based methods of constraint satisfaction by permitting the processing of metric information over continuous variables, and assessing the time difference between events. For statistical reasoning, Long [35] used a pseudo-Bayesian probabilistic reasoning method to eliminate diagnostic errors such as findings with longer chronic diseases. To manage temporal uncertainty, Palma et al [36] presented a temporal model-based diagnosis approach using the fuzzy temporal constraints network (FTCN) [127][128]. The proposed model defines temporal patterns that capture temporal and causal relations between elements describing the evolution of a disease. Therefore, the diagnostic solutions are presented and evaluated in the form of a causal network and possibility theory. One emerging solution from that would be designing a medical model that effectively combines causal and temporal knowledge, and enable the dynamic derivation of different forms of interactions. Table 3 summarizes some Clinical Decision Support systems (CDSS), which include temporal reasoning in their core decision systems.
Table 3.
CDSS and temporal reasoning. This table describes the most the best well-known decision support systems in real use in the clinical domain that support temporal based decision-making
| CDSS | Owner/ Architecture | Applications | Knowledge base |
|---|---|---|---|
| IndiGO, individualized guidelines and outcomes | Archimedes, Inc. | At-risk patient populations and patient-specific care plans | EHRs, and disease registries |
| Autonomy Health | Cambridge University HP Healthcare Analytics (Subsidiary) | Diagnostic | Clinical big data stores. |
| DiagnosisOne | Microsoft, Oracle and, RedHatt | Real-time patient and population assessment | Disease packages Cypress patient test inputs |
| DxPlain | Massachusettes General Hospital | Diagnostic | Contains 2400 Diseases, 5000 clinical findings, and 230 data points. |
| Elsevier Clinical Decision Support | CDS developers in Elsevier | Analytics and reporting; Predictive analytics | Drug database |
| Isabel Healthcare | Jason and Charlotte Maude | Diagnosis for uncommon or rare disease | 100,000 documents and “knowledge kernels” |
| Problem-Knowledge Coupling PKC | Dr. Lawrence Weed | Diagnoses and care plans | EHR and the subjective, objective, analytical, and planning (SOAP) approach |
| Micromedex V2.0 | Thomson Reuters | Medication safety, health and disease management, patient education, and toxicology | More than 3,500 hospitals in 83 countries |
| ProVation | Wolters Kluwer Health | Evidence-based clinical content | Up-to-date knowledge base |
| Zynx Health | Cedars-Sinai Health System and Zynx Health | Evidence-based clinical content | 500 clinical decision support rules and 1,100 templates |
Temporal Data Mining (TDM) [43][44] aimed at extending data mining techniques and methods to explicitly handle temporal reasoning which will help find better decisions plans. Temporal data mining refers to the extraction of implicit, non-trivial, and potentially useful abstract information from large collections of temporal data. Many applications have been developed. KarmaLego[41] is a method that exploits the transitivity inherent in temporal relations. The usefulness of KarmaLego was proven by finding meaningful temporal patterns within a set of records of diabetic patients that were used for classifying multivariate time series. Sequential pattern mining [42] and sequential rule mining algorithms [43] have been developed to discover sequential rules common to several sequences. The Electronic Medical Records (EMR) mining system EMRView[44] enables exploration of the relationships precedence between temporal events to identify partial order information of patients. We argue that one of the important roles of data mining is to help discovering hidden periodic patterns in temporal data.
3. Time representation by standardized clinical models
Many organizations indeed, such as Health Level Seven (HL7), Centre for European Normalization (CEN), and Good European Health Record (GEHR) have emphasized on structuration and standardization of EHR data in the healthcare arena [45] [46][47]. In this section, we highlight the methods of handling time in some standardized clinical data models, and we compare their approaches in handling the multifaceted characteristics of medical and clinical data.
3.1 Standard clinical models
Rector et al [48] has proposed the method of two-level model, which consists of separating clinical data records into two levels: direct observations and the meta-statements [49][50][51]. This method has been considered by OpenEHR archetypes [52], Clinical Element Models (CEM) [53], and Detailed Clinical Model (DCM) [53]. For example, DCM separates out the data generated by functions of record systems from the clinical details, which are specific to each record. OpenEHR separates the data level, which is meant to generate artifacts from concrete expressions and a processing level, which models the higher-level concepts of instructions and actions. In the following we overview the specifications of the most important clinical models and we compare their methods of handling time.
Health Level Seven [54] [55] [56] is an international organization developing standards to provide a comprehensive frameworks for the exchange, integration, sharing, and retrieval of electronic health information. HL7 standards aim to facilitate transfer of clinical and administrative data between hospital information systems[54]. The HL7 Clinical Document Architecture (CDA) document is a defined and complete information object that can include many types of contents and be conveyed in a HL7 message. The Cancer Data Standards Repository caDSR (caDSR CDE) [57] [58] is an open-source license distribution that support creating, editing, controlling, deploying, monitoring, and finding reusable medical and clinical metadata. It provides a semantic bridge between the data elements in registered data objects and standard vocabularies and ontologies. Initially developed to enable quality measurement in EHRs, the Quality Data Model (QDM) [32] is intended to enable automation of structured data captured during routine care in electronic health records. It provides a structure for describing clinical concepts contained within quality measures in a standardized format, allowing individuals who monitor clinical performance and outcomes (e.g., providers, researchers, or measure developers) to communicate information concisely and consistently. Clinical Element Model (CEM) [59] provides an abstract instance model, which defines a structure to represent instances of medical data and an abstract constraint model, which defines constraints about the abstract instance model. The model uses qualifies to represent time and the Coupling Strength as the semantic linking of constraints. OpenEHR archetypes [60] allow to specify complex data in an understandable format. Archetypes separate informatics concerns and clinical content discussion, enabling therefore clinicians to focus on the clinical content instead of technical details. Also they efficiently manage the specification of information to share between health care systems [61]
3.2 Comparison of standard models in handling time
The clinical models represent time information in different ways: combining different abstraction levels, and using various concepts and relations. The HL7 V 3 provides a comprehensive model in terms of covering time and temporal entity concepts. The syntax of time is based on the ISO 8601 standard [9] and it has five defined concepts: Point in time, Interval, Duration, Periodic time, and Periodic time as sets. Point in time defines a point on the axis of natural time; it can be specified as a day and time in the specific calendar or as a physical quantity using an epoch and a counter. The definition of points in time is very uniform and concise and its granularity is unbounded (i.e. given a precise measuring method one can specify the time exact to the millisecond, nanosecond, picosecond, and more). The conceptualization of timestamps is independent from any special calendar, and thus can be used with many different calendars. In addition, the translations between epoch-granularity-counter systems (clocks) are simple linear translations between coordinate systems. The intervals in HL7 are the generic data type to express a range of values of consecutive points in time. An interval is thus a continuous subset of its base data type. Interval is defined by at least two of the three properties low boundary, high boundary, and width. Duration is a physical quantity that represents a measurement in the dimension of time. Periodic events are perfectly represented using periodic continuous functions analysis, which are the counterpart of congruence in number theory. Periodic time is represented by periods and phases as modulus and remainders respectively, with consideration of an initial time since a calendar divides the even flow of time into cycles and counts full cycles in integer numbers. The more complex periodic times are expressed based on the simple period/phase model (e.g. business hours of a service). This stems from the fact that periodic points in time and periodic intervals of time are special kinds of sets that might be infinite, as the periodic time will be defined along the entire time axis from prehistoric past to distant future. HL 7 represents those temporal entities by combining sets using the operations for union (∪) and intersection (∩) to form each complex specification.
On the other hand, time specification in OpenEHR is about potentiality rather than actuality (certainty). The time-related datatypes used comprise date, time, date-time, and duration. The first three concepts fall into the absolute category while the relative category contains only the duration used for expressing durations of clinical phenomena and differences between absolute times. One feature taken into account here is that partial or uncertain dates/times are maintained. Thus, timestamp concept tolerates missing information (day, month, or year) or being represented in the format of date-intervals to allow indicating uncertainty. This can be useful in many situations, for example when the uncertainty about her date of birth; imprecise onsets: 10 ARE +/− 15 min; or when the periodicity of time is not meant to be in the strict sense (e.g. “three times a day” not meant to be literally each 8 hours). This works with the logic of many applications in the clinical domain when precision in time is not a concern or when representing probable occurrences of future events. The table 4 illustrates a comparison of the standard models mentioned above in handling time. The comparison is made according to five entities: point in time, interval, duration, periodic time, and periodic time as sets.
Table 4.
Description of temporal elements specification by five clinical information models. The description includes the elements of representation and the operators.
| Temporal element / aspect | Point in time | Duration | Intervals | Periodic time | Cumulative periodic time |
|---|---|---|---|---|---|
| Model | |||||
| HL7 V 3 | Compatible with ISO 8601 standard. Canonical format: YYYY-MM-DDThh:mm:ss. Time zones: “+hh:mm”, “− hh:mm” relative to UTC. Operators: = =,+,−. | Is a Physical quantity of time, measured by seconds, minutes, hours, days, months and years. | Provides choices between: open interval, closed interval, high open, high closed, low open, low closed. | Use Period (derived from mathematical modulus) and phase (derived from mathematical remainder) to represent periodic points and intervals in time. | Represented using Union (∪) and or Intersection (∩) of periodic times |
| QDM | Derived from ISO 8601:2004. Format: Date/Time stamp from calendar: my/mm/did/hh:mm:so | Use DateTimeDiff function, which returns the quantity of time between two points. | Covers 25 different temporal operators between intervals. | N/A | N/A |
| CEM | Use the ISO 8601 format with the syntax: YYYYMMDDHHMMSS. UUUU [+|−ZZzz] Operator: >, <, >=, <= | It’s a Physical Quantity (PQ), which has a value and unit. | Represents closed interval of physical quantities by using properties, Low and High. | Represented by Ratio Physical Quantity with a numerator and denominator | N/A |
| OpenEHR | Represents absolute time as Conform to ISO 8601. Allow missing information to represent approximate time stamps. | Represents period of time in customary format, i.e. days, hours, minutes etc. | Derived from a generic interval defined for physical quantity package | Loosely expressed by common specifications. | N/A |
| caDSR CDE | Time and Date from calendar. | It’s a length of time between specified events. | Represented by Time or Time/Date. | Limited to specific information about frequency such as daily, weekly. | N/A |
To sum up, the standardized clinical models have lot of common similarities as they have some differences in the representation of temporal clinical data. These discrepancies can be explained by the divergence of their objectives. We realize that both HL7 V3 and QDM present some alternatives for representing non-anchored time expressions, only HL 7 V3 provides representation of cumulative periodic times, and QDM provides an elaborated list (25 operators) of temporal interval relations. These models define data types that can be used to specify the complex timing of events and processes such as those that occur in clinical and medical applications. However, their time models are meant for structured data with limited choices of time representation, which remain untapped with the unstructured and semi-structured data.
4. Extraction of temporal information
80% of actual clinical data is narrative in nature [2] [3]. One reason is that text is the most preferable to humans to keep track of their records. To extract time information existing in text and render it machine-readable, computer scientists and linguistics use Medical NLP techniques. However, temporal information can be vaguely and implicitly conveyed in clinical narratives and discharge summaries, which makes the automation of temporal annotation a complex process.
4.1 Temporal clinical guidelines
Many researchers in clinical guidelines inception have picked up from temporal representation and modeling advances. From the theoretical view, applications have been using clinical data models with standards such specification languages and temporal logic to achieve their goals. Time specification languages provide pattern expressions for specified and underspecified temporal expressions. They have been used to annotate events, time expressions and temporal relations. The markup language TimeML [62] is the most commonly used guideline for temporal annotation. TimeML consists of three types of entities EVENTS, TIMEX3s and Signals and three types of relations TLINKS (Temporal Links), ALINKS (Aspectual Links) and SLINKS (Subordinate Links). The markup is primarily designed to stamp events in absolute timestamp and reason with contextually underspecified temporal expressions (e.g. last week, previous visit) in order to sequence the events in a chronological timeline and validate their persistence [63]. The TimeML specification has been illustrated and proven for the first time using TimeBank [64], an English corpus of 186 news articles. Further, the markup has been used in three temporal analysis evaluation tasks in the SemEval competitions, namely TempEval-1[65], TempEval-2[66], and TempEval-3 [67]. In addition, TimeML has been standardized to ISO-TimeML [68] to avoid confusion caused by the difference between national notations and to increase the portability of the markup language. The ISO-TimeML complies with the international standard ISO 8601which specifies numeric representations of date and time. Therefore TimeML has been the base for many other annotation schemas. The THYME-TimeML [69] is an annotation guideline that is developed to create robust gold standards for semantic information in clinical notes. A simplified version of this guideline formed the basis for the 2012 Informatics for Integrating Biology and the Bedside (i2b2) medical-domain temporal relation challenge. The TRIOS system [70] added the missing TimeBank events and temporal expressions after annotation with TimeML. The addition consisted of some semantic links (SLINKs) and some relations between events (RLINK). OntoTimeFL annotator [71] is a formalism for reasoning about complex events, it categorizes events as narrative, intentional, and causal [72]. TARSQI Toolkit (TTK) [73] is a modular system for automatic temporal and event annotation of natural language texts built on top of TimeML. TTK includes a module that combines potentially conflicting temporal relations into a consistent temporal graph of a document, which can be succinctly displayed using the TBox representation [74]. MED-TTK[75] is an extension of TTK for medical narratives. The TTK’s time tagger was modified to comply with temporal references in medical notes, also the notion of narrative containers (i.e. for event reasoning and ordering) in medical applications has been introduced [76].
In the other hand, clinical guidelines entail significant amounts of temporal-logic statements that need to be checked for semantics errors and possible extensions. In this connection, many formal languages have been used to describe the time logic namely Linear Temporal Logic (LTL)[77] which allows to reason about time on single paths, Computation Tree Logic (CTL) [78] which quantifies time on sets of paths and Action Computation Tree Logic (ACTL) [79] which suitably describes the occurrence of transitions. Perez et al [80] design a Model Driven Development based framework to enable authoring and verification of clinical guidelines. The framework lies on a model checker to verify guidelines against semantic errors and inconsistencies and enables automatically processing manually created guideline specifications. The framework considers the LTL to specify temporal properties and temporal-logic statements in order to be used for the checking and verification process. The same temporal formalism has been used by Bottrighi et al [81] who have adopted an approach based on the integration of a computerized guideline management system with a model-checker. For this purpose, they have used GLARE (GuideLine Acquisition, Representation and Execution) [82] to represent temporal constraints in clinical guidelines. The GLARE allows to represent temporal constraints and to check their consistency during the guideline acquisition phase, and to check the consistency between action execution-times and the constraints in the guidelines during the execution phase [83]. Kamsu-Foguem et al [84] propose a formal modeling of temporal knowledge using Computational Tree Logic (CTL) in order to introduce the semantic interpretation of the temporal logic expressions in models of conceptual graphs. The method was used in formal medical guideline specification and background knowledge representation. Groot et al. [85] propose a method for critiquing using model checking. Given patient data and a treatment plan as input (temporal specifications), the critiquing system uses a model checker to verify consistency with respect to a guideline model and generates a critique. As a cast to a critiquing system, the temporal logic is used to formally describe the actions taken by a medical doctor in the management of the disease of a patient. Also the properties of guidelines are specified using both CTL and LTL. In case of non-compliance with a guideline, the critic is then generated using the model checker. Also an example of using LTL, Hommersom et al [86] have set up a general framework for the verification of medical guidelines consisting of three components : medical guideline, medical background knowledge, and quality requirements. Schmitt et al [87] have developed KIV (Karlsruhe Interactive Verifier) a model for background knowledge support and task verification based on linear temporal logic. The application can handle large-scale formal models by efficient proof techniques, multi-user support, and an ergonomic user interface. Mor [88] presented an interesting work covering the reviews of methods and papers presenting Computer Interpretable Guidelines (CIG). The author has found that among 8 different topics in the lifecycle of the CIGs development, the higher number of publications focus on two topics: CIGs modeling languages and knowledge acquisition and specification tools.
Overall, even though the state of the art in temporal guideline is very developed, the temporal annotation schemas often cover specific types of clinical narratives and are not suitable for others. In this connection, some efforts have been made to merge multiple temporal annotations considering both the annotator performance and domain particularities [89]. We realize that the combination of the annotation standards with a formalism of time domain knowledge such as ontologies will lead to high-quality semantically annotated corpora. In addition, this hybrid representation will provide the mean to support temporal-based reasoning into the temporal annotation process.
4.2 Extraction of temporal expressions, evens, and temporal relations
The annotation of temporal events and expressions needs to take into consideration the peculiarities of the clinical domain. Indeed, the clinical text can be of different structures and formats, for example report-style documents such as patient summary exhibits few tense and aspect variations and contains limited absolute time markers whereas narrative-style documents such as clinical note is rich of temporal information. Furthermore, while the human brain is capable of processing temporal information very efficiently, identifying temporal relations between events remains a difficult task due to the diversity of linguistic mechanisms for expressing temporal information and the complex interplay of explicit and implicit inference required to understand such information [90]. We distinguish three temporal entities that need to be annotated in a clinical text: temporal expression, event, and temporal relation.
Temporal expression extractors are various. The Heidelberg University’s tool of HeidelTime extracts temporal expressions from documents and normalizes them according to the TIMEX3 annotation standard. MedTime [91] is a temporal information extraction system that uses rule-based and machine-learning pattern recognition procedures. TIPSem/TIPSem-B [92], and TRIOS [93] have been used in temporal expressions tagging of clinical narratives as well. For event extraction, the Mayo’s cTAKES (clinical Text Analysis and Knowledge Extraction System) pipeline [94] uses well trained components such as sentence boundary detector, tokenizer, part-of-speech tagger, shallow parser, named entity recognizer and context discovery modules to extract events in the clinical narratives. The National Center for Biomedical Ontologies (NCBO) [95] uses transitive closure, semantic distance and mapping concepts from UMLS and NCBO ontologies to annotate temporal events in clinical narratives. Song et al [96] proposed Semantator, a protégé-based semi-automatic tool to connect ontology representation to existing clinical NLP tools for events annotation. The 2010 i2b2/VA competition task about event named entity recognition has resulted in many approaches for extracting medical entities (i.e. 22 systems for concept extraction evaluated on held out test data) [97]. Gurulingappa et al [98] and Jonnalagadda and Gonzalez [99] applied a semi-supervised Conditional Random Fields CRF that used ‘distributional semantics’ features to implement a semi-automatic tools for events extraction. Min et al’s system [100] employed a machine learning approach using standard features to classify main events in consecutive sentences. Puşcaşy’s system [101] inferred temporal relations from temporal reasoning applied on a temporally tagged parse tree formed from heuristic inferences based on semantic properties and syntactic types.
As for annotation of temporal relations, the i2b2 2012 Challenge for clinical records has focused on temporal relation classification as one of the most important temporal information extraction (IE) tasks. This challenge has classified temporal relation into three sets, namely: (a) temporal relations between EVENTs and TIMEX3s within the same sentence; (b) temporal relations between the main EVENTs in adjacent sentences; and (c) temporal relations between two EVENTs where one dominates the other [102]. Many works in classifying temporal relations evaluated their results on the i2b2 clinical temporal relations challenge corpus [103]. Wang et al. [104] demonstrate the feasibility of the tasks defined by the i2b2 organizer and develop an end-to-end temporal relation system that includes three subsystems: an event extraction system, a temporal extraction system and a temporal relation system. Koyla et al [105] studied the identification of relations between events and their document creation times and developed two systems, one based on machine learning using Conditional Random Field (CRF) and the other based on constructed handcrafted rules. The Evaluation results showed that the rule-based system performs better compared to the machine learning. Mirroshandel et al [106] used Support Vector Machine (SVM) to improve the accuracy of classification of temporal relation using the automatically generated syntactic features. The study demonstrated that adding syntactic features results in a considerable improvement over the state-of-the-art methods of temporal relations classification. D’Souza et al [107] classified TLINKs using PropBank-style predicate-argument relations, and discourse relations. TLINK represents the temporal relation that holds between events, times or between an event and a time with different subsets of values (simultaneous, before, after, etc.) [108]. The authors used semantic and discourse relations and a combination between machine learning and rule based systems. In another work, D’Souza et al [109], have identified and classified temporal relations to 12 relation types rather than focusing on ‘three’ temporal relations as in the shared task, the experiments on the i2b2 corpus showed the effectiveness of the approach over the state-of-the-art 3-class classification results reported in the 2012 i2b2 challenge. The temporal relation extraction is also the process of identifying the chronological order of entities. For ordering events based on temporal relations, Allen [10] proposed thirteen relations are exhaustive and any unspecified qualitative relation can be designed by a combination of them [10]. Chang et al [110] stated that many rules were left out or not stated formally in many extracting systems in i2b2 and proposed TEMPTING (TEMPoral relaTion extractING) that integrates the results of both rule-based and supervised learning systems. Nikfarjam [111] proposed a system extracting the temporal relations from clinical notes. The system designed a separate extraction components for different types of temporal relations and utilized machine-learning and graph-based inference to extract the links between events and temporal expressions in the clinical notes. Santos et al [112] proposed a framework to reason about uncertainty over temporal constraints using Temporal Bayesian Knowledge Base (TBKB). TBKB permits to manage incompleteness and cycles in temporal knowledge and represent highly dynamic events.
4.3 Temporal corpora
Annotated corpora are an important asset in the clinical domain, standing as readily available resources for training clinical language processing algorithms and their evaluation. Several datasets manually annotated with temporal relations were produced in the past decade, including the TimeBank corpus [64], works on the 2012 i2b2 shared task on temporal relation extraction [102], in addition to TempEval-1 [65] and TempEval- 2 [66]. The TempEval-2 task involved identifying temporal relations between events and temporal expressions in the clinical text. The i2b2 project developed a temporally annotated corpus of clinical narratives for temporal relation extraction to promote research. The corpus is 310 de-identified discharge summaries annotated with clinical events, temporal expressions and temporal relations. Besides their nature and their purpose (extraction, evaluation, research, etc.) clinical corpora differ in their ways of handling the temporal annotation (temporal tagger) and their methodology of annotating temporal relations (i.e. intra-sentential, inter-sentential). CLEF (Clinical E-Science Framework) [113] has used the TimeML annotation schema to annotate 566K of documents using GUTime [114][115][116] as the temporal tagger. The corpus of annotated History of Present Illness (HPI) [117] uses i2b2 annotation to annotate 44 sections with 410 sentences and 7704 tokens using the HeidelTime temporal tagger with emphasis on intra-sentential annotation. Sun et el [118] have developed an annotated corpus of 40 discharge summaries with intra-sentential and intra documents support. THYME corpus [119], has examined 1,254 de-identified notes from a large healthcare practice (the Mayo Clinic), which has been made publicly available, and proposed for use in a SemEval 2015 task.
4.4 Temporal annotation challenges
A most prominent work in corpus annotation comes through shared tasks and temporal extraction challenges. The TempEval challenges have been motivated by the importance of temporal annotation for Medical NLP tasks and to advance research on temporal information processing, which could eventually help applications like question answering, textual entailment, and summarization. The first challenge task TempEval-1 [65], called also TempEval-Task 15, was organized at the SemEval workshop 2007 and was deliberately focused on subtasks of the larger problem of automatic temporal annotation. The TempEval-Task 15 corpus used the same documents as TimeBank 1.2 corpus [120] but used a simplified set of temporal relations, grouped into three separate tasks. Task A involved classification of temporal relation between an event and temporal expression in the same sentence; Task B involved classification of temporal relation between an event and the document creation time (DCT); and Task C involved classification between main events in consecutive sentences. TempEval-2 [66] is a follow-up on TempEval-1 and is a multilingual task that comprises additional evaluation tasks related to temporal classification which has included automatic classification of sub-ordinated event relations within the same sentence (i.e., relations between two events where one event syntactically dominates the other). TempEval-3 picks up from the two past TempEval events and incorporates a three-part task structure covering event, temporal expression and temporal relation extraction. TempEval-3 has included relevant features such as the use of the complete set of TimeML temporal relations instead of a simplified version as used in previous editions; a 10-times larger dataset; and single overall performance scores which allow the ranking of the participating systems in each task and also in general.
The 2012 Informatics for Integrating Biology and the Bedside (i2b2) challenge [121] marked a shift in the wide community in the sense that it refocused the research initiative towards temporal relation extraction from newswire data to data from the clinical domain [107]. Twenty teams representing 23 organizations and nine countries have participated in the medication challenge. The teams produced rule based, machine learning, and hybrid systems targeted to the task. The challenge focused specifically on the identification of clinically relevant events in the patient record, and the relative ordering of the events with respect to each other and with respect to time expressions included in the records. Tang, Wu, Jiang, et al [122] proposed a cascaded classifier for event extraction and their attributes. The system uses CRFs and SVM to assign polarity and modularity respectively for each event. For the extraction of TIMEXs, the system uses separate modules: normalized TIMEX3, type, value, and modifier. The extraction of TLINKs is done on two phases: TLink candidates generation and TLink candidates classification. The system was ranked first for both End to End TLink track and TLink only track and fourth in Temporal Expression Extraction. [123] Grouin, Grabar, Hamon, et al used a machine learning library to predict modality and polarity of events. The system presented a suggestion to adapt HeidelTime to clinical domain and used a post processing normalization of temporal expressions to fit i2b2 requirements. Sun, Rumshisky, Uzuner, et al [102] pointed out that event detecting is more challenging in 2012 i2b2 due to the addition of three new EVENT types finding out that the most hardest to detect are acronyms and anaphoric expressions and suggest that better co-reference and acronyms handling may improve the result. The authors also pointed out that relative time normalization remains challenging problem indicating that context-aware temporal expression understanding requires further research. Sunghwan, Kavishwar, Dingcheng et al [124] described MayoTime, a TIMEX3 system, as a comprehensive temporal information extraction and classification system for i2b2 2012 NLP challenge. Roberts, Rink, Harabagiu, et al [125] have proposed a method for recognition of medical events and expressions, normalizing temporal expressions, and detecting temporal relations. The methods are based on supervised and unsupervised learning and they performed well in the 2012 i2b2 shared task.
5. Temporal processing in the clinical domain
Temporal information processing in medicine is a task that draws from many fields, including philosophy, artificial intelligence, database management, computational linguistics, and biomedical informatics. The significant impact of computer technologies in this area is to normalize and abstract temporal information and resolve problems like granularity and co-reference. In this section of the survey we shed some light in some advances researches on these aspects.
5.1 Temporal information normalization
With the variety of representations of time and time-related concepts comes the necessity for a system of normalization of time. Different types of information are required to determine the normalized meaning of an expression, for example, if it’s a relative time (e.g., this morning 7 a.m.) we need the reference of time, if it has a positional offset (e.g., next month) we need the quantity of time, if it’s durative (3 days 4 hours) we need the quantity of time or if simply it has uncommon temporal expression formats (e.g., “April 28=12”), we will need to look for similar pattern resources. This disparity of formats makes it difficult to process time expressions for normalization tasks. The norm ISO8601 [126] requires date/time TIMEX3s to be normalized to [YYYY-MM-DD]T[HH:MM] format and duration/frequency TIMEX3s to be normalized to R[#1 times]P[#2][Units] (repeat for #1 times during #2 units of time). For example, ‘twice every three weeks’ is normalized as R2P3W. Also, similarly to TimeML TIMEX3s, the i2b2 TIMEX3s have a modifier attribute (MOD) that represents a subset of TimeML TIMEX3 modifier values: MORE, LESS, APPROX, START, END, MIDDLE and the default NA[119]. There are a number of temporal taggers that include a temporal normalization component such as TempEx, GUTime, CHRONOS, Terseo, TimexTag, TEA, Dante, HeidelTime, TRIPS, TRIOS, TIPSem, and TIPSemB. For example, HeidelTime extracts temporal expressions from documents and normalizes them according to the TIMEX3 annotation standard. CHRONOS sets the values of all TIMEX2 attributes based on the context information collected during the detection phase. However, these systems include their own custom rule sets (e.g. Heideltime has its own rule resources, pattern resources and normalization resources) and also much of the normalization effort is not inherently language independent (e.g. newswire). Because of that, separating the logic of dealing with a specific domain from language-specific requirements will enable effective normalization across languages [127]. In an attempt to use the ontological reasoning, Tao et al [30] applied a normalizer in their CNTRO ontology that converts commonly used time notations to the xsd DateTime Data Type format by defining two data properties, hasOrigTime and hasNormalizedTime. The normalization is meant to keep track of the time instant in both its original form and in the normalized form. With regard to complex datasets and automation of query processing, the TEXer [128] combined heuristic rule and pattern learning methods for temporal expression identification that use TimeML and its XML-based format of TimeX3 to normalize temporal expressions. TIMEN [127] is an open extensible and state-of-the-art temporal normalization library for building and sharing knowledge and rules for TimeML temporal expression normalization subtask, it can be easily integrated as a module in temporal information processing system.
For the temporal event annotation it is recommended to consider words or phrases that have a meaningful and contextually relevant match in the Unified Medical Language System UMLS, which can be accessible to annotators via UMLS Terminology Services. In this connection, events can be split into two subsets, top-down category, which designate the common health items, such as diseases, disorders, treatments, procedures and drugs prescribed to the patient as well as normal health situations like pregnancy that may affect the patient’s health and the bottom-up category which gathers detailed events in a defined sequence of care such as patient arriving and leaving, patient declared symptoms, pre-measurement of temperature and so on. This last category is focused on local observations and actions and depends on particular medical applications.
5.2 Temporal Abstraction
Temporal abstraction (TA) is a well known data analysis technique that is frequently applied in clinical domains to analyze complex multivariate clinical histories [129] [130], i.e. the series of relevant clinical events occurring to a patient (e.g. hospitalizations, visits, drug intakes, sudden variations of arterial blood pressure and glycemic control). Temporal patterns detection can be particularly useful for a variety of medical analyses, such as data exploration and summarization, temporal reasoning, evaluation of the response to specific treatments, anomaly detection, and prediction of clinical outcomes. Temporal patterns can be extracted in different ways and several methodologies have been proposed in the literature to achieve this goal [131]. Sacchi et al [131] proposed JTSA (Java Time Series Abstractor), a Java-based framework providing a library of algorithms for Temporal Abstraction detection, which can be easily extended and integrated into other applications. JTSA can be used both as a standalone tool for data summarization and as a module to be embedded into a complex architecture to select specific phenotypes based on TAs in a large dataset. Alvarez et al [132] proposed an algorithm for discovering frequent temporal patterns from a set of time-stamped event sequences called ASTPminer. This algorithm represents temporal patterns as metric temporal constraint networks for a set of events, where precise or imprecise information could be induced between each pair of events represented in the network. The ASTPminer allows to search for frequent temporal patterns by considering a pattern to be a temporal arrangement between a set of event types that satisfies some similarity criteria through different occurrences. Despite the relevance of TA as a methodology for temporal reasoning, only few efforts have been made to create a framework that is general, easy to use and simple to integrate into other applications [131]. Conceptual graphs (CGs) are used to represent clinical guidelines because they support visual reasoning with a logical background, making them a potentially valuable representation for guidelines [133]. Kamsu-Foguem et al [134] address problem of improving the integration of the visual and analytical methods applied to medical monitoring systems. They have described a methodology for the provision of a user-centered visual analysis to medical decision support systems that builds on an existing methodology and an existing Intensive Care Units monitoring system. In another effort, Kamsu-Foguem et al [135] proposed a conceptual graph formalism to facilitate sharing and reusing of medical practices and provided a visual reasoning mechanism for selecting best procedures for treating diseases. The nested conceptual graphs are used to visually express the semantic meaning of computation tree logic (CTL) constructs that are used for formal specification of temporal properties of the domain of knowledge. This approach mitigates knowledge loss with conceptual development assistance to enable the automated verification of compliances requirements models of the domain knowledge and to improve the quality of care and patient safety. Juarez et al [136] proposed a visualization model based on multiple temporal axes (MTA) model. MTA was evaluated using a controlled experiment approach, and demonstrated in a tool called 8VISU used in a real-world ambient assisted living system for elderly people living alone. The results of the experiments show the advantages of the MTA model over other models (timeline, Gantt, spiral) in different scenarios.
5.3 Time granularity
The granularity of temporal information is the level of abstraction at which the information is exposed. Medical and clinical events may be annotated using time points and time spans with different granularities. The granular value of time is an important factor that cannot be underestimated. It doesn’t only add valuable detail to the temporal expression but also handles some semantic information intrinsic to the expression. For instance, an event that is counted by minutes carry a semantic precision higher than a one that can last for months. Moreover, the projection from finer to coarser granularity or the reverse involves complex semantic issues [137]. That is, it sounds mathematically correct to convert all entities of date time to the smallest unit of time, but this will also erase the semantics of expressions, so in the end it will be uncomfortable to say that a treatment has lasted for seconds or minutes. Table 5 shows that switching from finer to coarser (left to right) or coarser to finer (right to left) granularities cause semantic problems.
Table 5.
Granular time examples. This table illustrates time granularity in the clinical text. The sentences in the left denote some durations and intervals of time, which, when converted to finer granularities (right column), have caused the semantic to change.
| 1. The patient has tested blood sugar every week. | a. The patient has tested blood sugar every 7 days. |
| 2. The x-rays are valid within 19 June 2015. | b. The x-rays are valid within 19 June 2015 at midnight. |
| 3. The treatment has to be completed within 1 day. | c. The treatment has to be completed within 24 hours. |
Thus, the resolution of granularity depends not only on the time expressions themselves but on the contextual information as well. Iqbal et al [138] concluded that in order to solve temporal granularity issues in discharge summaries, one should be able to (a) represent and store time instants with different and mixed granularities, (b) handle granularity mismatches in operations between temporal primitives with different granularities, (c) convert a temporal primitive from one granularity to another, and (d) consider different interpretations for time labels [139].
5.4 Co-reference resolution problem
The coreference in clinical text refers to the problem of relating together all medical mentions that refer to the same medical entity. This latter can be an event, state or even a marked temporal entity associated with the patient’s medical condition and healthcare. Resolving this task has proven essential for many types of problems such as temporal annotations [140], temporal questions/ answering [31], and redundant information within and across clinical narratives [141]. One question that may arise thus far is the possible existence of multiple syntactic derivations of the same entity (e.g. cauterize, cauterization). One proposed answer[141] is to refer to the UMLS which includes a large Metathesaurus of concepts and terms from many biomedical vocabularies and a lexicon which contains syntactic, morphological, and orthographic information for biomedical and common words in the English language. In fact, UMLS has 2,404,937 concept unique identifiers and 15,333,246 links between them as seen in the full UMLS graph structure. A medical concept in UMLS represents a single meaning and contains all atoms in the UMLS that express that meaning in any way, whether formal or casual, verbose or abbreviated. In the i2b2/VA challenge on co-reference resolution [142], a large amount of records have been fully de-identified and manually annotated for co-reference. The challenge provided the community with two annotated ground truth corpora and evaluated systems on co-reference resolution in two ways: first, it evaluated systems for their ability to identify mentions of concepts and to link together those mentions. Second, it evaluated the ability of the systems to link together ground truth mentions that refer to the same entity [142].
Resolving the co-reference problem has been done quite satisfactorily manually [143], however this task not only seems to be costly in human effort but also lacks some abilities such as annotation of distributed temporal concepts at topic level or document level [144]. For that purpose, extracting semantic and temporal features helps identify conditionally independent views of the medical event which is an important step in order to co-train classifiers and make the co-reference decision. Additionally, the semantic and temporal feature sets are naturally independent (i.e. semantic features help identify synonymous medical concepts and the temporal feature helps identify time of occurrence). For the calculation of the semantic feature, k-Neighborhood decentralization method [145] seems to outperform breadth-first and depth-first searches between concepts in the UMLS graph structure[145]. KNDM can be used to index and transitively traverse associated relations between concept unique identifiers in the UMLS graph and reveal reachability, distance, and summary of paths, between two concepts in the UMLS graph structure. On the other hand, for calculating the temporal feature, it is well known that medical concept instances, cited throughout a document, that have occurred at the same time (stamped with the same time expression) tend to be similar. However, in clinical narratives, many temporal concepts are left with implicit reference. In this case, a successful path to determining temporal features uses the headed information of patient admission and discharge date to learn about assigning medical concepts to time periods referred to as time-bins. For this purpose, nine of the top 10 systems used conditional random fields (CRFs), which is a statistical modeling method for sequential data labeling [146]. This method consists of using sections as logical, and at times, temporal grouping of information in the narrative (e.g. way before admission, before admission, on admission, after admission, after discharge). Thus sequencing the observation of medical concepts in the order in which they appear in a clinical narrative help leveraging the temporal feature.
Although co-reference resolution is a well-studied problem in computational linguistics [141], there have been very few efforts at tackling this problem in longitudinal clinical narratives[143]. Ragjavan et al [141] have used the supervised binary classification task for medical concepts coreference resolution with MaxEnt classifiers through semi-supervised methods that co-train MaxEnt classifiers and MaxEnt models using posterior regularization. The approach is based on using two independent views of the data - semantic view and temporal view. The first tested method consists of co-training two MaxEnt classifiers, one on the semantic features and the other for temporal features of the data, to classify pairs of medical concepts as co-refer or no-co-refer in a semi-supervised fashion[147]. The algorithms used allow for labeling unlabeled data for both classifiers (semantic and temporal features set) to augment the training data. The problem here is the need to set a threshold for an unlabeled sample to be added to the labeled pool. This issue is overcome by proposing to repeatedly update the size of the pool of the medical pairs that co-refer and that don’t co-refer. The second method consists of using a learning method applied to MaxEnt with posterior regularization using expectation constraints [148]. Semi-supervised posterior regularization is used to derive a multiview-learning algorithm (semantic view and temporal view) while specifying constraints that the models should agree on (i.e. desired level of precision and recall).
6. Observations and conclusion
The objective of this review was to find out how theories, models, and ontologies of time, available to date, cover the representation of time and time related concepts in the clinical domain, particularly in clinical narratives. It investigated on how the Medical NLP approaches catch up with the available reasoning and inferencing systems. We found that both point and interval constructs are used in time specification and, the periodicity and cumulative periodicity are commonly used and need to be implemented in accurate and unambiguous way. The Allen’s theory of interval relations has been widely applied for temporal ordering and partitioning. On the other hand, uncertainty and granularity of time, and co-reference of events are relevant features that necessitate adequate interpretation and representation. The information models of structured data have been described and compared on their methods of handling time. Based on the comparison of five clinical data models, namely, HL7 V3, QDM, CEM, caDSR CDE and OpenEHR, it is found that these models fulfill different objectives and they use different tools (object-oriented, archetypes, etc.) Also, these models cover only basic aspects of time and cannot represent unstructured data, which represent 80 % of clinical data. The uncertainty and granularity in time instants and intervals are barely supported by temporal models. It has been proved that ontologies of time are potential candidate to fulfill these facets. It is found that, except for CNTRO ontology that has represented clinical intrinsic features such as normalization, granularity, and periodicity, most of developed ontologies remain for only restricted and general use. On the other hand, the co-reference remains poorly covered. Some contributions in co-reference resolution and the narrative containers are valuable and need to be implemented integrated with existing ontologies of time. For that we posit that a cross-disciplinary effort is required.
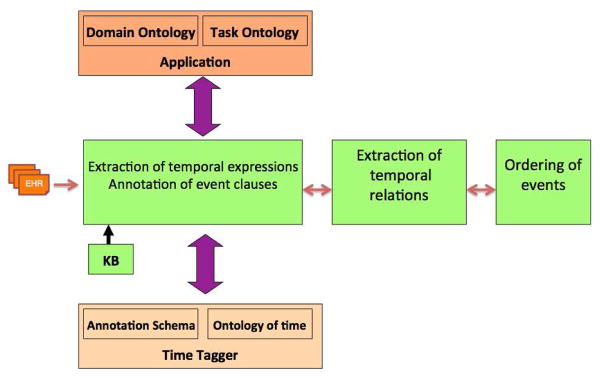
The automatic temporal extraction is a promising and active research field in the clinical domain. In the last few years, temporal taggers have matured and acquired reuse capabilities. However, there are competing goals that hinder the obtaining of high-quality annotated corpora, namely to fully comply with the domain specification and the avoidance of unnecessary annotations. In order to resolve this issue, we need a dynamic adaptation of extraction rules and use of machine learning for annotator training and automation. In addition, precise and concise extraction of temporal events in clinical narratives has been a long-standing interest in the clinical domain. There are three types of approaches namely, data-driven approaches, knowledge-driven approaches, and hybrid approaches. The data-driven approach develops models that approximate linguistic phenomena from large text corpora using quantitative techniques. Some methods of the approach are support vector machine, multi-class classifier, and hierarchical clustering techniques. However, this approach has been criticized for the large amount of data needed to generate statistically significant results. The knowledge driven approach, by contrast, exploits syntactic and semantic patterns encoded in the form of rules to extract desired information. Even though this approach has proven its consistency, it remains inaccurate when some knowledge of linguistics or domain expertise is missing. In hybrid approach, methods have been encoding lexical knowledge as features for statistical learning to extract events. Overall, although these approaches have showed good results in many extraction contests and challenges, there is still a large semantic gap between Medical NLP techniques and rules-based systems. As a proposition to solve this problem, we suggest, in figure 1, a new temporal extraction pipeline for the clinical domain. This architecture will allow for a tight combination of Medical NLP based methods such as Hidden Markov models, Maximum Entropy, SVM, and conditional random field (CRF) and semantic web technologies such as OWL, RDF, and techniques of querying.
Figure 1.
Temporal-extraction pipeline. The ontologies of both application and time tagger gather their knowledge base (KB) for annotating the temporal expressions and events; the temporal relations and ordering of events can be then inferred by using rules or/and machine learning techniques.
In conclusion, combining Medical NLP and semantic web techniques to construct a timeline from medical records is promising. In essence, the Medical NLP provides the necessary tools and methods to extract, normalize, classify and summarize temporal data in clinical narratives whereas, ontologies and semantic web techniques are proven tools for knowledge driven approaches that allow for the use and management of information and domain knowledge. One challenge here is how to combine the information and knowledge of both temporal extraction process and application domain into one knowledge base. One way to do so is by considering mapping between concepts. Furthermore, in order to establish a counterbalance of using some specific techniques or methods along with keeping the flexibility required in such situations, the Markov Logic Network (MLN) [149] is a first-order knowledge base used to combining first-order logic and probabilistic graphical models in a single representation. Using such an approach will enrich the interaction, for example, of rules and machine learning system for the temporal relations extraction.
Highlights.
Multifaceted aspects in time and time-oriented concepts
Comparison of clinical data models in handling time
Ontologies of representation and reasoning about time in the clinical domain
Constructing the timelines for the medical histories of patients
Temporal concept coreference resolution problem
Acknowledgments
This research is partially supported by the National Institutes of Health under Award Numbers R01LM011829 and R01GM103859.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mohcine Madkour, Email: mohcine.madkour@uth.tmc.edu.
Driss Benhaddou, Email: dbenhadd@Central.uh.edu.
Cui Tao, Email: cui.tao@uth.tmc.edu.
References
- 1.Zitao Liu MH, Wu Lei. Modeling Clinical Time Series Using Gaussian Process Sequences. SIAM Int Conf data mining. 2013 [Google Scholar]
- 2.Joe P. Natural language processing in electronic health records. 2011 [Google Scholar]
- 3.ENRICH Cl. The Application of CNLP (Clinical Natural Language Processing) for Improved Analytics. White Pap. 2014 [Google Scholar]
- 4.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Informatics Assoc. 2010;17(2):131–135. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman S. Impact of electronic health record systems on information integrity: quality and safety implications. Perspect Health Inf Manag. 2013 Jan;10(1c) [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou L, Melton GB, Parsons S, Hripcsak G. A temporal constraint structure for extracting temporal information from clinical narrative. J Biomed Inform. 2006 Aug;39(4):424–39. doi: 10.1016/j.jbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Vilain M, Kautz H, van Beek P. Constraint propagation algorithms for temporal reasoning: a revised report. 1989 Dec;:373–381. [Google Scholar]
- 8.James JAK, Allen F. Planning using a temporal world model. IJCAI. 1983;8 [Google Scholar]
- 9.Freksa C. Temporal reasoning based on semi-intervals. Artif Intell. 1992;54(1–2):199–227. [Google Scholar]
- 10.Allen JF. Maintaining knowledge about temporal intervals. Commun ACM. 1983;26(11):832–843. [Google Scholar]
- 11.Shahar Y, Combi C. Timing is everything. Time-oriented clinical information systems. West J Med. 1998 Feb;168(2):105–13. [PMC free article] [PubMed] [Google Scholar]
- 12.Ermolayev V, Batsakis S, Keberle N, Tatarintseva O, Antoniou G. Ontologies of Time: Review and Trends. Int J Comput Sci Appl. 2014 Dec;11(3):57–115. [Google Scholar]
- 13.OWL Web Ontology Language Reference. [Online]. Available: http://www.w3.org/TR/owl-ref/
- 14.Lutz C, Wolter F, Zakharyaschev M. Temporal Description Logics: A Survey. 2008 15th International Symposium on Temporal Representation and Reasoning; 2008. pp. 3–14. [Google Scholar]
- 15.“SWRL(A Semantic Web Rule Language Combining OWL and RuleML),.”
- 16.Straccia U, Lopes N, Lukacsy G, Polleres A. A General Framework for Representing and Reasoning with Annotated Semantic Web Data.,” in. AAAI. 2010 [Google Scholar]
- 17.Hurtado C, Vaisman A. Reasoning with temporal constraints in RDF. Springer; 2006. [Google Scholar]
- 18.Champin PA, Passant A. SIOC in action representing the dynamics of online communities. Proceedings of the 6th International Conference on Semantic Systems - I-SEMANTICS ’10; 2010; p. 1. [Google Scholar]
- 19.Shaw R, Troncy R, Hardman L. The Semantic Web. Vol. 5926. Berlin, Heidelberg: Springer Berlin Heidelberg; 2009. [Google Scholar]
- 20.Wang Y, Zhu M, Qu L, Spaniol M, Weikum G. Timely YAGO. Proceedings of the 13th International Conference on Extending Database Technology - EDBT ’10; 2010; p. 697. [Google Scholar]
- 21.Grandi F. T-SPARQL: A TSQL2-like Temporal Query Language for RDF.,” in. ADBIS (Local Proceedings) 2010:21–30. [Google Scholar]
- 22.Time Ontology in OWL Available. [Online]. Available: http://www.w3.org/TR/owl-time/
- 23.SWRL Temporal Ontology. [Online]. Available: http://protege.cim3.net/cgibin/
- 24.Hobbs MD, Ferguson J, Allen G, Fikes J, Hayes RP. A DAML Ontology of time. 2002 [Online]. Available: http://www.cs.rochester.edu/~ferguson/daml/
- 25.Anagnostopoulos E, Batsakis S, Petrakis EGM. CHRONOS: A Reasoning Engine for Qualitative Temporal Information in OWL. Procedia Comput Sci. 2013;22:70–77. [Google Scholar]
- 26.Ermolayev V, Keberle N, Matzke W-E. An Ontology of Environments, Events, and Happenings. 2008 32nd Annual IEEE International Computer Software and Applications Conference; 2008; pp. 539–546. [Google Scholar]
- 27.Kaykova O, Khriyenko O, Naumenko A, Terziyan V, Zharko A. RSCDF: A Dynamic and Context Sensitive Metadata Description Framework for Industrial Resources. 2005 Jan; [Google Scholar]
- 28.Qing Zhou QZRF. A Reusable Time Ontology [Google Scholar]
- 29.Standford. Ontololigua. [Online]. Available: http://www.ksl.stanford.edu/software/ontolingua/
- 30.Tao C, Wei W-Q, Solbrig HR, Savova G, Chute CG. CNTRO: a semantic web ontology for temporal relation inferencing in clinical narratives,” in. AMIA Annual Symposium Proceedings; 2010. p. 787. [PMC free article] [PubMed] [Google Scholar]
- 31.Tao C, Solbrig HR, Sharma DK, Wei W-Q, Savova GK, Chute CG. The Semantic Web--ISWC 2010. Springer; 2010. Time-oriented question answering from clinical narratives using semantic-web techniques,” in; pp. 241–256. [Google Scholar]
- 32.N. Q. Forum. Quality measures (NQF QDM) [Online]. Available: http://www.qualityforum.org/qualitydatamodel.aspx.
- 33.Vilain MB, Kautz HA. Constraint Propagation Algorithms for Temporal Reasoning.,” in. Aaai. 1986;86:377–382. [Google Scholar]
- 34.Dechter R. Temporal constraint networks. Artif Intell. 1991 May;49(1–3):61–95. [Google Scholar]
- 35.Long W. Temporal reasoning for diagnosis in a causal probabilistic knowledge base. Artif Intell Med. 1996;8(3):193–215. doi: 10.1016/0933-3657(95)00033-x. [DOI] [PubMed] [Google Scholar]
- 36.Palma J, Juarez JM, Campos M, Marin R. Fuzzy theory approach for temporal model-based diagnosis: An application to medical domains. Artif Intell Med. 2006;38(2):197–218. doi: 10.1016/j.artmed.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Barro S, Marín R, Mira J, Patón AR. A model and a language for the fuzzy representation and handling of time. Fuzzy Sets Syst. 1994;61(2):153–175. [Google Scholar]
- 38.Marín R, Cárdenas MA, Balsa M, Sanchez JL. Obtaining solutions in fuzzy constraint networks. Int J Approx Reason. 1997;16(3):261–288. [Google Scholar]
- 39.Post AR, Harrison JH. Temporal data mining. Clin Lab Med. 2008;28(1):83–100. doi: 10.1016/j.cll.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Roddick JF, Spiliopoulou M. A survey of temporal knowledge discovery paradigms and methods. Knowl Data Eng IEEE Trans. 2002;14(4):750–767. [Google Scholar]
- 41.Moskovitch R, Shahar Y. Medical temporal-knowledge discovery via temporal abstraction. AMIA annual symposium proceedings; 2009. p. 452. [PMC free article] [PubMed] [Google Scholar]
- 42.Agrawal R, Srikant R. Mining sequential patterns. Data Engineering, 1995. Proceedings of the Eleventh International Conference on; 1995; pp. 3–14. [Google Scholar]
- 43.Das G, Lin K-I, Mannila H, Renganathan G, Smyth P. Rule Discovery from Time Series.,” in. KDD. 1998;98:16–22. [Google Scholar]
- 44.Debprakash Patnaik LPPBBJKNRDAH. Experiences with Mining Temporal Event Sequences from Electronic Medical Records: Initial Successes and Some Challenges [Google Scholar]
- 45.Ingram D. The good european health record. In: Laires MF, Ladeira MF, Christ JP, editors. Heal new Commun age. IOS; 1995. pp. 66–74. [Google Scholar]
- 46.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012 Jun;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 47.Wiki H. HL7 Detailed Clinical Models. 2015 [Online]. Available: http://wiki.hl7.org/index.php?title=Detailed_Clinical_Models]
- 48.Al Rector WNSKCGTH. A Framework for Modelling the Electronic Medical Record. [PubMed] [Google Scholar]
- 49.Rector AL, Nowlan WA, Kay S, Goble CA, Howkins TJ. A framework for modelling the electronic medical record. Methods Inf Med. 1993 Apr;32(2):109–19. [PubMed] [Google Scholar]
- 50.Rector AL, Nowlan WA, Kay S. Foundations for an electronic medical record. Methods Inf Med. 1991 Aug;30(3):179–86. [PubMed] [Google Scholar]
- 51.Goossen DWTF. Detailed Clinical Models : Kennis en semantieK weergeven met uml en Xml. Elements. 2011;17(1):11–16. [Google Scholar]
- 52.Beale T. Archetypes and the EHR. Stud Health Technol Inform. 2003 Jan;96:238–44. [PubMed] [Google Scholar]
- 53.Huff SM, Rocha RA, Coyle JF, Narus SP. Integrating detailed clinical models into application development tools. Stud Health Technol Inform. 2004 Jan;107(Pt 2):1058–62. [PubMed] [Google Scholar]
- 54.Health Level seven. 2015 [Online]. Available: http://www.hl7.org/
- 55.N. edition of the H. S. Health Level 7. Ann Arbor (MI): Health Level Seven International; 2010. [Online]. Available: http://www.hl7.org. [Google Scholar]
- 56.Dolin RH, Alschuler L, Beebe C, Biron PV, Boyer SL, Essin D, Kimber E, Lincoln T, Mattison JE. The HL7 clinical document architecture. J Am Med Informatics Assoc. 2001;8(6):552–569. doi: 10.1136/jamia.2001.0080552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiley A. CTEP Common Data Elements. 2013 [Online]. Available: https://wiki.nci.nih.gov/display/caDSR/CTEP+Common+Data+Elements.
- 58.caDSR. 2015 [Online]. Available: https://cbiit.nci.nih.gov/ncip/biomedical-informatics-resources/interoperability-and-semantics/metadata-and-models.
- 59.Coyle J, Heras Y, Oniki T, Huff S. Clinical element model. Univ. Utah; 2008. [Google Scholar]
- 60.OpenEHR. Ocean Informatics. New South Wales: Ocean Informatics; c2007–2010. 2010 [Google Scholar]
- 61.OpenEHR. OpemEHR. 2015 [Online]. Available: http://www.openehr.org/programs/clinicalmodels/
- 62.Sauri R, Littman J, Knippen B, et al. TimeML annotation guidelines, V.1.2.1. 2005. [Google Scholar]
- 63.Ligozat G. Extracting, Annotating and Reasoning about Time and Space in Texts and Discourse. [Google Scholar]
- 64.Pustejovsky J, Hanks P, Saurí R, See A, Gaizauskas R, Setzer A, Radev D, Sundheim B, Day D, Ferro L, Lazo M. The timebank corpus. [Google Scholar]
- 65.Verhagen M, Gaizauskas R, Schilder F, Hepple M, Katz G, Pustejovsky J. SemEval-2007 task 15: TempEval temporal relation identification. Jun, 2007. pp. 75–80. [Google Scholar]
- 66.Verhagen M, Saurí R, Caselli T, Pustejovsky J. SemEval-2010 task 13: TempEval-2. 2010 Jul;:57–62. [Google Scholar]
- 67.UzZaman N, Llorens H, Allen J, Derczynski L, Verhagen M, Pustejovsky J. Tempeval-3: Evaluating events, time expressions, and temporal relations. 2012 arXiv Prepr. arXiv1206.5333. [Google Scholar]
- 68.James P, Lee K, Bunt H, Romary L. ISO-TimeML: An International Standard for Semantic Annotation. 2010 Jan; [Google Scholar]
- 69.Styler WF, IV, Bethard S, Finan S, Palmer M, Pradhan S, de Groen PC, Erickson B, Miller T, Lin C, Savova G, et al. Temporal annotation in the clinical domain. Trans Assoc Comput Linguist. 2014;2:143–154. [PMC free article] [PubMed] [Google Scholar]
- 70.UzZaman N, Allen JF. TRIOS-TimeBank Corpus: Extended TimeBank Corpus with Help of Deep Understanding of Text.,” in. LREC. 2010 [Google Scholar]
- 71.Mele F, Sorgente A. New Challenges in Distributed Information Filtering and Retrieval. Springer; 2013. OntoTimeFL--A Formalism for Temporal Annotation and Reasoning for Natural Language Text; pp. 151–170. [Google Scholar]
- 72.Lai C, Semeraro G, Vargiu E. New Challenges in Distributed Information Filtering and Retrieval. Vol. 439. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. [Google Scholar]
- 73.Verhagen M, Pustejovsky J. Temporal processing with the TARSQI toolkit. Aug, 2008. pp. 189–192. [Google Scholar]
- 74.Verhagen M. Drawing TimeML Relations with TBox. Springer; 2007. [Google Scholar]
- 75.Reeves RM, Ong FR, Matheny ME, Denny JC, Aronsky D, Gobbel GT, Montella D, Speroff T, Brown SH. Detecting temporal expressions in medical narratives. Int J Med Inform. 2013 Feb;82(2):118–27. doi: 10.1016/j.ijmedinf.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 76.MV, Pustejovsky J. The TARSQI Toolkit. Proceedings of the Eight International Conference on Language Resources and Evaluation (LREC’12); 2012; pp. 23–25. [Google Scholar]
- 77.Pnueli A. The temporal logic of programs. 18th Annual Symposium on Foundations of Computer Science (sfcs 1977); 1977; pp. 46–57. [Google Scholar]
- 78.Clarke EM, Emerson EA. Design and synthesis of synchronization skeletons using branching time temporal logic. Springer; 1982. [Google Scholar]
- 79.Meolic R, Kapus T, Brezocnik Z. An action computation tree logic with unless operator. Proceedings of the 1st South-East European workshop on formal methods SEEFM; 2003; 2003. pp. 100–114. [Google Scholar]
- 80.Pérez B, Porres I. Authoring and verification of clinical guidelines: A model driven approach. J Biomed Inform. 2010;43(4):520–536. doi: 10.1016/j.jbi.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Bottrighi A, Giordano L, Molino G, Montani S, Terenziani P, Torchio M. Adopting model checking techniques for clinical guidelines verification. Artif Intell Med. 2010 Jan;48(1):1–19. doi: 10.1016/j.artmed.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 82.Terenziani P. Toward a Unifying Ontology Dealing with Both User--Defined Periodicity and Temporal Constraints About Repeated Events. Comput Intell. 2002;18(3):336–385. [Google Scholar]
- 83.Anselma L, Terenziani P, Montani S, Bottrighi A. Towards a comprehensive treatment of repetitions, periodicity and temporal constraints in clinical guidelines. Artif Intell Med. 2006;38(2):171–195. doi: 10.1016/j.artmed.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 84.Kamsu-Foguem B, Tchuenté-Foguem G, Foguem C. Verifying a medical protocol with temporal graphs: the case of a nosocomial disease. J Crit Care. 2014 Aug;29(4):690.e1–9. doi: 10.1016/j.jcrc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 85.Groot P, Hommersom A, Lucas PJF, Merk R-J, ten Teije A, van Harmelen F, Serban R. Using model checking for critiquing based on clinical guidelines. Artif Intell Med. 2009;46(1):19–36. doi: 10.1016/j.artmed.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 86.Hommersom A, Groot P, Lucas P, Balser M, Schmitt J. Combining task execution and background knowledge for the verification of medical guidelines. Knowledge-Based Syst. 2007;20(2):113–119. [Google Scholar]
- 87.Schmitt J, Balser M, Reif W. Verification of medical guidelines in KIV. Stud Health Technol Inform. 2008 Jan;139:253–62. [PubMed] [Google Scholar]
- 88.Peleg M. Computer-interpretable clinical guidelines: a methodological review. J Biomed Inform. 2013 Aug;46(4):744–63. doi: 10.1016/j.jbi.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 89.Llorens H, Uzzaman N, Allen JF. Merging Temporal Annotations. 2012 19th International Symposium on Temporal Representation and Reasoning; 2012; pp. 107–113. [Google Scholar]
- 90.Sun W, Rumshisky A, Uzuner O. Temporal reasoning over clinical text: the state of the art. J Am Med Inform Assoc. Jan;20(5):814–9. doi: 10.1136/amiajnl-2013-001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin Y-K, Chen H, Brown RA. MedTime: a temporal information extraction system for clinical narratives. J Biomed Inform. 2013 Dec;46(Suppl):S20–8. doi: 10.1016/j.jbi.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 92.Llorens H, Saquete E, Navarro B. Tipsem (english and spanish): Evaluating crfs and semantic roles in tempeval-2. Proceedings of the 5th International Workshop on Semantic Evaluation; 2010; pp. 284–291. [Google Scholar]
- 93.UzZaman N, Allen JF. TRIPS and TRIOS system for TempEval-2: Extracting temporal information from text. Proceedings of the 5th International Workshop on Semantic Evaluation; 2010; pp. 276–283. [Google Scholar]
- 94.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. Jan;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Musen MA. Encyclopedia of Systems Biology. Springer; 2013. National Center for Biomedical Ontology; p. 1492. [Google Scholar]
- 96.Song D, Chute CG, Tao C. Semantator: annotating clinical narratives with semantic web ontologies. AMIA Jt Summits Transl Sci Proc AMIA Summit Transl Sci. 2012 Jan;2012:20–9. [PMC free article] [PubMed] [Google Scholar]
- 97.Uzuner Ö, South BR, Shen S, DuVall SL. i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2010 Jan;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gurulingappa FJ, Hofmann-Apitius HM. Concept identification and assertion classification in patient health records. 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data; 2010. [Google Scholar]
- 99.Jonnalagadda S, Gonzalez G. Can distributional statistics aid clinical concept extraction. Proceedings of the 2010 i2b2/VA workshop on challenges in natural language processing for clinical data; Boston, MA, USA. 2010.p. i2b2. [Google Scholar]
- 100.Min FA, Srikanth CM. LCC-TE: a hybrid approach to temporal relation identification in news text. Proceedings of the fourth international workshop on semantic evaluations (SemEval-2007); 2007; pp. 219–22. [Google Scholar]
- 101.Puşcaşu G. WVALI: temporal relation identification by syntactico-semantic analysis. Jun, 2007. pp. 484–487. [Google Scholar]
- 102.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc. Jan;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.I2b2. Informatics for Integrating Biology and the Bedside. 2012. [Google Scholar]
- 104.Xu Y, Wang Y, Liu T, Tsujii J, Chang EI-C. An end-to-end system to identify temporal relation in discharge summaries: 2012 i2b2 challenge. J Am Med Inform Assoc. 2013 Jan;20(5):849–58. doi: 10.1136/amiajnl-2012-001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kolya AK, Ekbal A, Bandyopadhyay S. Event-time relation identification using machine learning and rules. 2010 Sep;:117–124. [Google Scholar]
- 106.Mirroshandel SA, Ghassem-Sani G, Khayyamian M. Using syntactic-based kernels for classifying temporal relations. J Comput Sci Technol. 2011;26(1):68–80. [Google Scholar]
- 107.D’Souza J, Ng V. Classifying temporal relations in clinical data: a hybrid, knowledge-rich approach. J Biomed Inform. 2013 Dec;46(Suppl):S29–39. doi: 10.1016/j.jbi.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Savova G, Bethard S, Styler W, Martin J, Palmer M, Masanz J, Ward W. Towards temporal relation discovery from the clinical narrative. AMIA Annu Symp Proc. 2009 Jan;2009:568–72. [PMC free article] [PubMed] [Google Scholar]
- 109.D’Souza J, Ng V. Knowledge-rich temporal relation identification and classification in clinical notes. Database (Oxford) 2014 Jan;2014:bau109. doi: 10.1093/database/bau109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chang Y-C, Dai H-J, Wu JC-Y, Chen J-M, Tsai RT-H, Hsu W-L. TEMPTING system: a hybrid method of rule and machine learning for temporal relation extraction in patient discharge summaries. J Biomed Inform. 2013 Dec;46(Suppl):S54–62. doi: 10.1016/j.jbi.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 111.Nikfarjam A, Emadzadeh E, Gonzalez G. Towards generating a patient’s timeline: extracting temporal relationships from clinical notes. J Biomed Inform. 2013 Dec;46(Suppl):S40–7. doi: 10.1016/j.jbi.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Santos E, Li D, Santos EE, Korah J. Temporal Bayesian Knowledge Bases – Reasoning about uncertainty with temporal constraints. Expert Syst Appl. 2012 Dec;39(17):12905–12917. [Google Scholar]
- 113.Roberts A, Gaizauskas R, Hepple M, Davis N, Demetriou G, Guo Y, Kola J, Roberts I, Setzer A, Tapuria A, Wheeldin B. The CLEF corpus: semantic annotation of clinical text. AMIA Annu Symp Proc. 2007 Jan;:625–9. [PMC free article] [PubMed] [Google Scholar]
- 114.Mani I, Wilson G. Robust temporal processing of news. Proceedings of the 38th Annual Meeting on Association for Computational Linguistics - ACL ’00; 2000; pp. 69–76. [Google Scholar]
- 115.Verhagen M, Mani I, Sauri R, Knippen R, Jang SB, Littman J, Rumshisky A, Phillips J, Pustejovsky J. Automating temporal annotation with TARSQI. Proceedings of the ACL 2005 on Interactive poster and demonstration sessions - ACL ’05; 2005; pp. 81–84. [Google Scholar]
- 116.“GUTime.”. [Online]. Available: http://timeml.org/site/tarsqi/modules/gutime/
- 117.Galescu L, Blaylock N. A corpus of clinical narratives annotated with temporal information. Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium; 2012; pp. 715–720. [Google Scholar]
- 118.Sun W, Rumshisky A, Uzuner O. Annotating temporal information in clinical narratives. J Biomed Inform. 2013 Dec;46(Suppl):S5–12. doi: 10.1016/j.jbi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.SIV WF, Bethard S, Finan S, Palmer M, Pradhan S, de Groen PC, Erickson B, Miller T, Lin C, Savova G, Pustejovsky J. Temporal Annotation in the Clinical Domain. Transactions of the Association for Computational Linguistics. 2014 Apr 30;2:143–154. [PMC free article] [PubMed] [Google Scholar]
- 120.“TimeBank,”. 2007 [Online]. Available: http://www.timeml.org/site/timebank/timebank.html.
- 121.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc. 2013 Jan;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang B, Wu Y, Jiang M, Chen Y, Denny JC, Xu H. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc. Jan;20(5):828–35. doi: 10.1136/amiajnl-2013-001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grouin C, Grabar N, Hamon T, Rosset S, Tannier X, Zweigenbaum P. Eventual situations for timeline extraction from clinical reports. J Am Med Inform Assoc. Jan;20(5):820–7. doi: 10.1136/amiajnl-2013-001627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sohn S, Wagholikar KB, Li D, Jonnalagadda SR, Tao C, Komandur Elayavilli R, Liu H. Comprehensive temporal information detection from clinical text: medical events, time, and TLINK identification. J Am Med Inform Assoc. Jan;20(5):836–42. doi: 10.1136/amiajnl-2013-001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roberts K, Rink B, Harabagiu SM. A flexible framework for recognizing events, temporal expressions, and temporal relations in clinical text. J Am Med Inform Assoc. 2013 Jan;20(5):867–75. doi: 10.1136/amiajnl-2013-001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolf M, Wicksteed C. W3C NOTE NOTE-datetime-19980827. Aug, 1998. Date and time formats. [Google Scholar]
- 127.Llorens H, Derczynski L, Gaizauskas RJ, Saquete E. TIMEN: An Open Temporal Expression Normalisation Resource. LREC. 2012:3044–3051. [Google Scholar]
- 128.Hao T, Rusanov A, Weng C. Smart Health. Springer; 2013. Extracting and normalizing temporal expressions in clinical data requests from researchers; pp. 41–51. [Google Scholar]
- 129.Orphanou K, Stassopoulou A, Keravnou E. Temporal abstraction and temporal Bayesian networks in clinical domains: a survey. Artif Intell Med. 2014 Mar;60(3):133–49. doi: 10.1016/j.artmed.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 130.Stacey M, McGregor C. Temporal abstraction in intelligent clinical data analysis: a survey. Artif Intell Med. 2007 Jan;39(1):1–24. doi: 10.1016/j.artmed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 131.Sacchi L, Capozzi D, Bellazzi R, Larizza C. JTSA: an open source framework for time series abstractions. Comput Methods Programs Biomed. 2015 Oct;121(3):175–88. doi: 10.1016/j.cmpb.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 132.Álvarez MR, Félix P, Cariñena P. Discovering metric temporal constraint networks on temporal databases. Artif Intell Med. 2013 Jul;58(3):139–154. doi: 10.1016/j.artmed.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 133.Kamsu-Foguem B, Tchuenté-Foguem G, Foguem C. Conceptual graph operations for formal visual reasoning in the medical domain. IRBM. 2014 Oct;35(5):262–270. [Google Scholar]
- 134.Kamsu-Foguem B, Tchuenté-Foguem G, Allart L, Zennir Y, Vilhelm C, Mehdaoui H, Zitouni D, Hubert H, Lemdani M, Ravaux P. User-centered visual analysis using a hybrid reasoning architecture for intensive care units. Decis Support Syst. 2012 Dec;54(1):496–509. [Google Scholar]
- 135.Kamsu-Foguem B, Diallo G, Foguem C. Conceptual graph-based knowledge representation for supporting reasoning in African traditional medicine. Eng Appl Artif Intell. 2013 Apr;26(4):1348–1365. [Google Scholar]
- 136.Juarez JM, Ochotorena JM, Campos M, Combi C. Spatiotemporal data visualisation for homecare monitoring of elderly people. Artif Intell Med. 2015 Oct;65(2):97–111. doi: 10.1016/j.artmed.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 137.Zhou L, Hripcsak G. Temporal reasoning with medical data--a review with emphasis on medical natural language processing. J Biomed Inform. 2007 Apr;40(2):183–202. doi: 10.1016/j.jbi.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iqbal D, Goralwalla A, Leontiev Yuri, Tamer Özsu M. Temporal Granularity: Completing the Puzzle. J Intell Inf Syst [Google Scholar]
- 139.Zhou L, Melton GB, Parsons S, Hripcsak G. A temporal constraint structure for extracting temporal information from clinical narrative. J Biomed Inform. 2006;39(4):424–439. doi: 10.1016/j.jbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 140.Sun W, Rumshisky A, Uzuner O. Annotating temporal information in clinical narratives. J Biomed Inform. 2013 Dec;46(Suppl):S5–12. doi: 10.1016/j.jbi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Raghavan P, Fosler-Lussier E, Lai AM. Exploring semi-supervised coreference resolution of medical concepts using semantic and temporal features. 2012 Jun;:731–741. [Google Scholar]
- 142.Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc. Jan;19(5):786–91. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Raghavan P, Fosler-Lussier E, Lai AM. Inter-annotator reliability of medical events, coreferences and temporal relations in clinical narratives by annotators with varying levels of clinical expertise. AMIA Annu Symp Proc. 2012 Jan;2012:1366–74. [PMC free article] [PubMed] [Google Scholar]
- 144.Bejan CA, Harabagiu S. Unsupervised event coreference resolution with rich linguistic features. 2010 Jul;:1412–1422. [Google Scholar]
- 145.Xiang Y, Lu K, James SL, Borlawsky TB, Huang K, Payne PRO. k-Neighborhood decentralization: a comprehensive solution to index the UMLS for large scale knowledge discovery. J Biomed Inform. 2012 Apr;45(2):323–36. doi: 10.1016/j.jbi.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lafferty J, McCallum A, Pereira FCN. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. 2001 [Google Scholar]
- 147.Blum A, Mitchell T. Combining labeled and unlabeled data with co-training. Proceedings of the eleventh annual conference on Computational learning theory; 1998; pp. 92–100. [Google Scholar]
- 148.Ganchev K, Graça J, Gillenwater J, Taskar B. Posterior regularization for structured latent variable models. J Mach Learn Res. 2010;11:2001–2049. [Google Scholar]
- 149.Richardson M, Domingos P. Markov logic networks. Mach Learn. 2006;62(1–2):107–136. [Google Scholar]