Abstract
The growth of functional MRI has led to development of human brain atlases derived by parcellating resting-state connectivity patterns into functionally independent regions of interest (ROIs). All functional atlases to date have been derived from resting-state fMRI data. But given that functional connectivity between regions varies with task, we hypothesized that an atlas incorporating both resting-state and task-based fMRI data would produce an atlas with finer characterization of task-relevant regions than an atlas derived from resting-state alone. To test this hypothesis, we derived parcellation atlases from twenty-nine healthy adult participants enrolled in the Cognitive Connectome project, an initiative to improve functional MRI’s translation into clinical decision-making by mapping normative variance in brain-behavior relationships. Participants underwent resting-state and task-based fMRI spanning nine cognitive domains: motor, visuospatial, attention, language, memory, affective processing, decision-making, working memory, and executive function. Spatially constrained n-cut parcellation derived brain atlases using (1) all participants’ functional data (Task) or (2) a single resting-state scan (Rest). An atlas was also derived from random parcellation for comparison purposes (Random). Two methods were compared: (1) a parcellation applied to the group’s mean edge weights (mean), and (2) a two-stage approach with parcellation of individual edge weights followed by parcellation of mean binarized edges (two-stage). The resulting Task and Rest atlases had significantly greater similarity with each other (mean Jaccard indices JI= 0.72–0.85) than with the Random atlases (JI=0.59–0.63; all p<0.001 after Bonferroni correction). Task and Rest atlas similarity was greatest for the two-stage method (JI=0.85), which has been shown as more robust than the mean method; these atlases also better reproduced voxelwise seed maps of the left dorsolateral prefrontal cortex during rest and performing the n-back working memory task (r=0.75–0.80) than the Random atlases (r=0.64–0.72), further validating their utility. We expected regions governing higher-order cognition (such as frontal and anterior temporal lobes) to show greatest difference between Task and Rest atlases; contrary to expectations, these areas had greatest similarity between atlases. Our findings indicate that atlases derived from parcellation of task-based and resting-state fMRI data are highly comparable, and existing resting-state atlases are suitable for task-based analyses. We introduce an anatomically labeled fMRI-derived whole-brain human atlas for future Cognitive Connectome analyses.
1. Introduction
The recent growth of functional neuroimaging research has led to development of human brain atlases that accurately reflect the brain’s functional organization. Several such atlases have been generated by applying parcellation approaches to functional magnetic resonance imaging (fMRI) data [1–7]. These approaches identify functionally independent brain regions by first calculating the functional connectivity between all voxels (i.e. the correlation of each voxel’s activity timeseries with all other voxels), then using parcellation algorithms (such as the n-cut method, cite) that maximize within-cluster voxels’ correlations while minimizing between-cluster voxels’ correlations.
All functional atlases to date – whether encompassing the entire brain [1–4] or specific cortical regions [5–7] – have been derived from resting-state fMRI scans in which participants lie awake in the scanner while not engaged in overt tasks. But functional connectivity patterns change with cognitive task, raising the possibility that atlases derived solely from resting-state data may be suboptimal for studying task-dependent brain activity. As examples, functional connectivity between Broca’s and Wernicke’s areas dramatically increases during a reading task compared to tongue-movement or rest [8], and connectivity among motor regions increases with finger tapping compared to rest [9]. In both examples, functional connectivity seed maps show clearer boundaries during task than rest, suggesting that a parcellation approach incorporating both resting-state and task-based functional connectivity may produce an atlas with finer characterization of task-relevant regions than an atlas derived solely from resting-state data.
We addressed this potential barrier by deriving two whole-brain atlases from fMRI data acquired through the Cognitive Connectome project [10], an initiative to translate fMRI into patient care by bridging clinical neuropsychology and functional neuroimaging. We derived two atlases: one atlas incorporating data from a single resting-state scan (similar to existing atlases), and a comparison atlas incorporating data from resting-state and task-based scans encompassing motor performance, visual perception, visuospatial judgment, emotional processing, verbal memory, visual memory, working memory, language fluency, attentional conflict, reward processing, and executive function. We hypothesize that a parcellation incorporating both resting-state and task-based fMRI data (a Task atlas) will substantially differ from an atlas derived from resting-state data alone (Rest). Specifically, we predict that regions recruited during higher-order cognition (such as prefrontal and temporal regions for executive function, language, and memory) will substantially differ in size and shape between atlases, whereas regions involved in less demanding tasks (such as visual and motor regions) will be similar across atlases. We also predict that voxelwise connectivity seed maps of task-based fMRI scans will show stronger spatial correspondence to seed maps derived from the task-based atlas than the resting-state atlas. Finally, we provide the atlas as an anatomically labeled tool for future analyses of the Cognitive Connectome project data.
2. Materials and Methods
2.1. Cognitive Connectome
All Cognitive Connectome study procedures were conducted in the Brain Imaging Research Center at the University of Arkansas for Medical Sciences. Study participation was typically conducted in two sessions on separate days. Session 1 included informed consent, a structured clinical interview (SCID-IV/NP) to determine exclusionary criteria, behavioral surveys and questionnaires (such as the State-Trait Anxiety Inventory and Big Five Personality Inventory), and the first of two hour-long neuroimaging session (with neuroimaging session order counterbalanced across subjects). Session 2 included neuropsychological assessment and the second neuroimaging session. Cognitive Connectome project study procedures are described in full detail elsewhere [10].
2.2. Participants
Thirty-five participants completed both fMRI sessions. (Five additional pilot participants completed both fMRI sessions, but are excluded due to substantial task redesign based upon their feedback.) Table 1 and Supplementary Materials provide descriptions of each fMRI scan, including task design and scan duration. Scans with excessive head motion (i.e. greater than 3mm lateral movement in any direction) were excluded from analyses. Of these 35 participants, 29 were included in the final analysis: 7 with useable data from 12 scans, and 22 with useable data from all 13 scans. The excluded scans included a second resting-state scan (n=2), visual memory (n=2), motor performance (n=1), visual perception (n=1), and executive function (n=1). The twenty-nine participant sample had the following demographics: mean(sd) age = 31(9.9), range 20–50; 10 (34%) male, 19 (66%) female; 19 (66%) self-reporting as White or Caucasian, 12 (41%) as Black or African-American, 1 (3%) as Hispanic or Latino, including 1 participant who self-identified as both Caucasian and African-American; mean(sd) education = 16(2.2) years, range 10–19. All participants were recruited with approval and oversight by the UAMS Institutional Review Board (protocol #130825).
Table 1.
Descriptions of fMRI tasks
| Task | Session† | Duration | Description |
|---|---|---|---|
| Controlled Oral Word Association Task (COWAT) | A | 5m 0s | Alternating 15s blocks of Letter or Category word generation separated by 15s rest |
| Rating affective images (IAPS affect) | A | 5m 14s | 45 IAPS images (negative, neural, positive) presented in random order for 2.5s with 2–6s inter-stimulus interval |
| Recognizing affective images (IAPS recognition) | A | 10m 12s | 90 IAPS images (45 previously seen, 45 new) presented in random order for 2.5s with 2–6s interstimulus interval |
| Judgment of Line Orientation task (JLOT) | A | 4m 16s | 15 JLOT trials (self-paced, up to 15s duration) with rest at start of task and each trial completion |
| N-back | A | 8m 0s | Alternating 45s blocks of 0-back or 2-back trials separated by 15s rest |
| Resting-state | A | 7m 30s | Passive viewing of a black fixation cross upon light gray background |
| Iowa Gambling Task | B | 8m 6s – 11m 42s (me an 9m 14s) | Self-paced, participant draws 100 cards of varying reward/loss from 4 decks |
| Finger tapping | B | 3m 0s | 18s blocks of index finger tapping (left-right-right-left-right-left) separated by 10s rest |
| Multi-source Interference Task (MSIT) | B | 8m 0s | Alternating 48s blocks of Congruent or Incongruent MSIT trials with 30s rest at start, middle, and end of task |
| Verbal Paired Associates | B | 2m 0s | Ten word pairs presented as consecutive 6s blocks with 30s rest at start and end of task |
| Resting-state | B | 7m 30s | Passive viewing of a black fixation cross upon light gray background |
| Tower of London | B | 3m 26s – 7m 30s (me an 4m 20s) | Self-paced, 24 Tower of London trials (2-, 3- and 4-move) starting with 5s planning phase and ending with 6s rest |
| Flashing visual checkerboard | B | 2m 0s | Four 18s blocks of 4Hz flashing checkerboard separated by 10s rest |
Session order was counterbalanced across participants. Each session began with a resting-state scan, and IAPS affect/IAPS recognition were the second and final scans of session A; otherwise, within-session task order was also counterbalanced across participants.
2.3. Image Acquisition and Preprocessing
2.3.1. Image Acquisition
Imaging data were acquired using a Philips 3T Achieva X-series MRI scanner (Philips Healthcare, Eindhoven, The Netherlands). Anatomic images were acquired with a MPRAGE sequence (matrix = 256 × 256, 220 sagittal slices, TR/TE/FA = shortest/shortest/8°, final resolution =0.94 × 0.94 × 1 mm3 resolution). Functional images for early participants (001–050) were acquired using an 8-channel head coil with an echo planar imaging (EPI) sequence (TR/TE/FA = 2000 ms/30 ms/90°, FOV=240 × 240 mm, matrix = 80 × 80, 37 oblique slices parallel to orbitofrontal cortex to reduce sinus artifact, interleaved ascending slice acquisition, slice thickness = 4 mm, final resolution 3.0 × 3.0 × 4.0 mm3). For these subjects, one session’s resting-state scan was acquired with 3-mm slice thickness to be consistent with data acquired for other BIRC studies. Functional images for later participants (051–079) were acquired using a 32-channel head coil with the following EPI sequence parameters: TR/TE/FA = 2000 ms/30 ms/90°, FOV = 240 × 240 mm, matrix = 80 × 80, 37 oblique slices, ascending sequential slice acquisition, slice thickness = 2.5 mm with 0.5 mm gap, final resolution 3.0 × 3.0 × 3.0 mm3. Parameters for the 32-channel coil were selected to reduce orbitofrontal signal loss due to sinus artifact. To assess head coil as a potential confound, we regressed head coil against participants’ functional connectivity data (measured as voxelwise seed maps with the dorsolateral prefrontal cortex, see below) to evaluate if choice of head coil significantly influenced functional connectivity (and thus, atlas generation).
2.3.2. Image Preprocessing
All MRI data preprocessing was conducted in AFNI unless otherwise noted [11]. Anatomic data underwent skull stripping, spatial normalization to the icbm452 brain atlas, and segmentation into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) with FSL [12]. Functional data underwent despiking; slice correction; deobliquing (to 3×3×3 mm3 voxels); motion correction (using the 10th timepoint); transformation to the spatially normalized anatomic image; regression of motion parameters, mean timecourse of WM voxels, and mean timecourse of CSF voxels; spatial smoothing with a 6-mm FWHM Gaussian kernel; and scaling to percent signal change. Resting-state scans also underwent bandpass filtering (0.01–0.10 Hz) to remove physiological artifact relating to noise. fMRI scans with head motion exceeding 3mm lateral movement were excluded from subsequent scans. Participants’ binarized GM masks were averaged to generate a group-level gray matter mask; voxels with a group mean GM value ≥ 0.5 were included in the parcellation approach described below.
2.4. Analyses
2.4.1. Parcellation
MRI data parcellation utilized the normalized cut (n-cut) approach [1]. This approach used a refinement of graph cutting algorithms that attempt to partition undirected, weighted graphs by assigning the graph’s partitions according to a minimized cut cost. The cut cost, cut(A,B), represents the sum of the weights of edges that must be removed from the graph to completely partition the subset of nodes, A, from a disjoint subset of nodes, B. The n-cut algorithm modifies the cut cost by dividing the total sum of edge weights associated with subsets A and B, respectively, thus normalizing the influence between densely and sparsely connected nodes. In application to MRI, graph nodes were represented by individual voxels and graph edges include only those voxels in the 3-dimensional (face and edge adjacent) neighborhood, resulting in 26 edges per voxel. Edge weights were set equal to voxel pair-wise Pearson correlations over the voxels’ time-courses. In other words, n-cut searched for “fissures” of weak connectivity between neighboring voxels, and set these fissures as boundaries to maximize within-cluster correlations among voxels and minimize correlations of voxels belonging to neighboring clusters.
This n-cut approach also allows two algorithms for group-level parcellation. The group mean algorithm averages edge weights across the group prior to the n-cut parcellation. The group two-stage algorithm first parcellates each subject’s MRI based on their edge weights, then digitizes the first-stage parcellations to produce binary (0 or 1) edge weights which are averages across the group prior to the second n-cut parcellation. Both methods were explored here.
Three region of interest (ROI) group atlases were generated for each method. The first atlas (Rest) was constructed entirely from one resting-state scan (comparable to experiments conducted in [1]). The second atlas (Task) was constructed from combined resting-state and task scans, which are concatenated into a single image as follows. Each individual scan was first z-scored voxel-wise; then the scans were concatenated in the time dimension; then the combined scan was z-scored. The third atlas (Random) was constructed by setting the edge weights between neighboring voxels to 1 rather than the voxels’ Pearson correlation. Thus there are no “fissures” of weak connectivity to guide parcellation, causing voxels to randomly parcellate into equally sized clusters.
All n-cut parcellations were conducted using the experimental source code for the work published in [1] available at https://www.nitrc.org/projects/cluster_roi/. Normalization and concatenation calculations were conducted using Matlab. All experiments were executed on a Hewlett Packard ProLiant DL980 G7 Server (80 processors and 4TB single-addressable memory). Scripts and data are available upon request.
2.4.2. Comparing ROIs across atlases
We compared the similarity (homology) of ROIs across atlas parcellations as follows. Let set I equal all voxels comprising an ROI in Atlas A (ROIA). The voxels spatially corresponding to the voxels in set I were identified in Atlas B. The values of these voxels indicate the ROI(s) in Atlas B with partial overlap with ROIA, and the mode of these voxels’ values identify the ROI in Atlas B (ROIB) with the greatest overlap of ROIA. Letting set J equal all voxel comprising ROIB, the similarity of ROIB and ROIB was calculated using the Jaccard index, or the number of voxels shared by ROIA and ROIB divided by the total number of voxels in ROIA and ROIB (i.e. intersection I∩J/union I∪J).
2.4.3. Comparing connectivity seed-maps across atlases
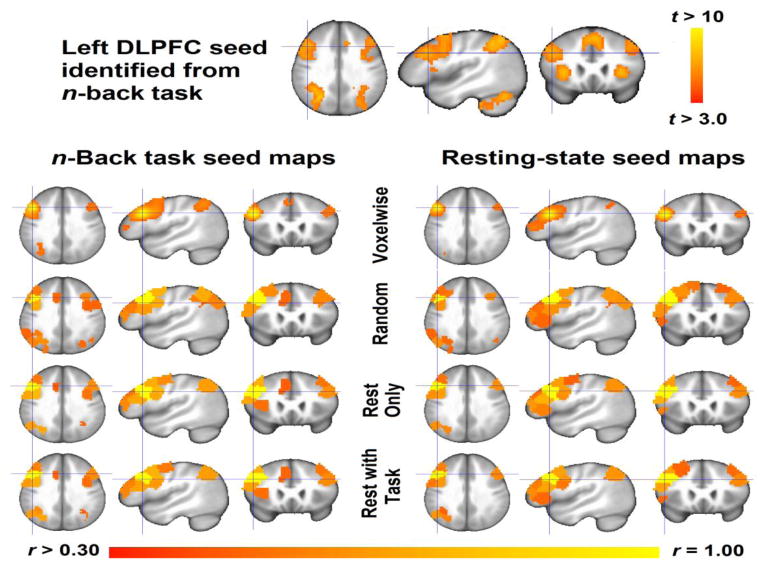
We compared the atlases’ ability to replicate connectivity seed maps for the left dorsolateral prefrontal cortex (LDLPFC) across two conditions: during wakeful rest and during performance of the n-back working memory task. The LDLPFC was identified from group-level analysis of the n-back task: brain activity was contrasted between 2-back and 0-back conditions for each participant using general linear modeling (GLM) with AFNI’s 3dDeconvolve, residual maximum likelihood (REML) analyses accounted for influence of temporal autocorrelation with 3dREMLfit, and mixed-effects meta-analysis identified group-level differences between 2-back and 0-back conditions with 3dMEMA (all scripts available upon request). A 6mm radius spherical ROI was centered upon the LDLPFC (MNI coordinates −44, 23, 31; Figure 3), and voxelwise seed maps were generated by correlating each voxels’ activation timeseries with the mean activation timeseries of voxels within the ROI. Atlas seed maps were generated by identifying the atlas seed ROI containing the most voxels from the task-defined ROI, extracting the mean timeseries of voxels comprising each ROI, correlating each ROI timeseries with the atlas seed ROI timeseries, and backprojecting these correlations to GM voxels comprising each atlas. The resulting seed maps (voxelwise, Task atlas, Rest atlas) were Fisher z-transformed so that the voxels’ correlations approximated linearity, and the three seed maps were compared via pairwise spatial correlation.
Figure 3. Validation of atlases via task-based and resting-state replications of seed maps.
(Top) The n-back task elicited greater group-level bilateral dorsolateral prefrontal (DLPFC) and parietal activity for 2-back condition than 0-back condition. Results are displayed a p<0.005 uncorrected and contiguous cluster size>2,050mm3 (76 contiguous voxels) for AlphaSim corrected q<0.01. We identified the left DLPFC seed as a 6mm radius ROI centered upon MNI coordinates (−44, 23, 21), indicated by blue crosshairs. (Bottom) Correlation seed maps were generated for the n-back and resting-state data using voxelwise data and mean timecourses of the Random, Resting, and Task-based parcellation atlases. All three atlas seed maps correlated with the voxelwise seed map, although correlations were significantly higher for the Resting and Task atlases (r=0.75–0.80) than the Random atlas (r=0.64–0.72). Note that the left DLPFC ROIs identified from Resting and Task atlases are centered upon the n-back task derived ROI, whereas the Random atlas DLPFC ROI is not, probably owing to the incorporation of fMRI data.
3. Results
3.1. Evaluating ROI sizes
ROIs encompassing fewer than 5 voxels (135 mm3) were removed from each parcellation, as these ROIs were too small to be biologically meaningful. ROIs which were three standard deviations larger than the mean were also removed from each parcellation. This includes a cluster composed of over 2,300 voxels (62,100 mm3) identified for the Task, group two-stage method which covered much of the brain’s circumference. Similar implausibly large ROIs have been identified and omitted from other brain atlases [1]. ROI sizes did not significantly differ between atlases, either before removal of artifactual clusters [F(5,1169)=0.09, p<0.99] or after removal [F(5,1161)=1.08, p<0.37]. After removal, mean ROI size ranged from 177–186 voxels across parcellations, with standard deviations ranging from 42–50 voxels.
3.2. Evaluating similarity
3.2.1. ROI similarity across all atlases
Table 1 provides mean Jaccard indices (JI) for ROI similarity between atlases. Each combination of dataset and parcellation method yielded an atlas with strong similarity to the random parcellation (mean JI 0.59–0.63). Jaccard indices were skewed toward the highest possible value of 1, prompting use of nonparametric statistics to compare ROI similarity between atlases. ROI JIs were much greater for atlases generated using the same method or same data than for the randomly generated atlases (all Wilcoxon rank sum tests values > 4, all p < 0.001 after Bonferonni correction for 8 comparisons). This was particularly true for the group two-stage method Task and Rest atlases; ROI JIs were greater for these atlases (μ=0.846) than for the random parcellation (μ=0.645; z-value>10, p<0.001). We replicated these analyses using the Dice coefficient similarity index and found almost perfect correlation (r=0.99), supporting our use of the Jaccard index.
3.2.2. Similarity between All and Rest atlas ROIs
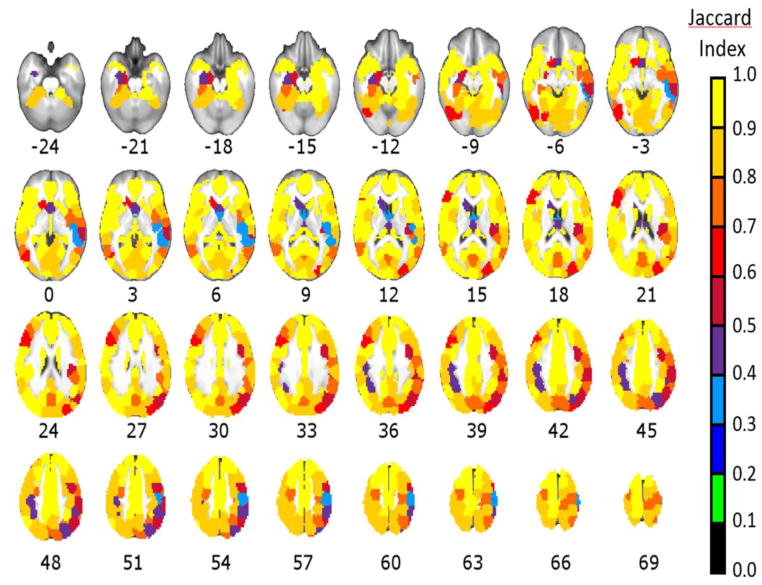
Figure 1 depicts ROIs for the Task, group two-stage atlas, and Figure 2 depicts the JI for each ROI compared to its homolog in the Rest, group two-stage atlas. 47% of these ROIs had a JI ≥0.90, and 79% had JI ≥0.80. Contrary to hypotheses, greatest similarity was observed for prefrontal cortex, cingulate gyrus, left parietal lobe, and left temporal lobe. Only 5% of ROIs had a JI <0.50. Table 2 lists all ROIs of the Task, group two-stage atlas ranked by their similarity to the Rest, group two-stage atlas. Regions with lowest similarity (JI<0.50) included right sensorimotor area (middle primary sensory cortex (S1), JI=0.34; inferior S1, JI=0.41; lateral premotor area, JI=0.37;), left sensorimotor area (left S1, JI=0.48; and adjacent left inferior parietal lobule, JI=0.49), regions bordering ventricles (left caudate, JI=0.42; septum pellucidum, JI=0.47; thalamus, JI=0.42), right middle temporal gyrus (medial region, JI=0.33; lateral region, JI=0.33), and right posterior superior parietal lobule (JI=0.40).
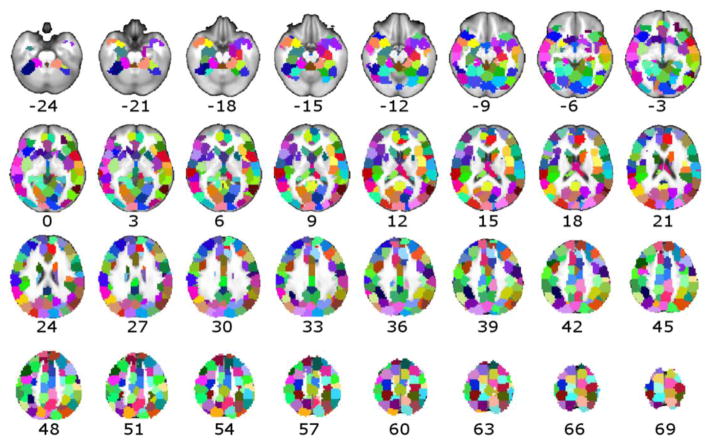
Figure 1. Regions comprising the Task, group two-stage atlas.
The parcellation derived from 29 healthy participants using the Group two-stage method and all task-based and resting-state data is depicted across axial slices (MNI coordinates z=−24 to z=69). Regions of interest (ROIs) are color-coded to maximize contrast between parcellation boundaries. ROIs demonstrate strong bilateral symmetry of ROIs between hemispheres.
Figure 2. Strong similarity between Task and Rest atlases.
The parcellation derived using the group, two-stage method and all task-based and resting-state data was compared to the parcellation derived using the group, two-stage method but a single resting-state session for each participant. The Jaccard Index (JI) reported overall strong similarity between parcellations (mean JI= 0.85). Contrary to hypotheses, regions associated with higher-order cognition (such as prefrontal and temporal regions) showed strong similarity across task-based and resting-state parcellations. Poorest similarity was observed for ROIs in right sensorimotor area, right superior temporal sulcus, and regions bordering left lateral ventricle.
Table 2.
Jaccard Similarity Indices across Atlases
| Atlas 1 | Atlas 2 | Jaccard Index
|
|
|---|---|---|---|
| mean | sd | ||
|
| |||
| Comparison to Random atlas | |||
| Rest, Random | All, Random | 0.645 | 0.230 |
| Rest, group two-stage | Rest, Random | 0.632 | 0.216 |
| Rest, mean | Rest, Random | 0.611 | 0.228 |
| Task, group two-stage | Task, Random | 0.607 | 0.211 |
| Task, mean | Task, Random | 0.588 | 0.203 |
| Comparison of Atlases by Method | |||
| Task, group two-stage | Rest, group two-stage | 0.846 | 0.144 |
| Task, mean | Rest, mean | 0.716 | 0.221 |
| Comparison of Atlases by Data | |||
| Rest, group two-stage | Resting, mean | 0.720 | 0.200 |
| Task, group two-stage | Task, mean | 0.719 | 0.203 |
3.3. Seed map comparisons
Figure 3 and Table 3 compare left DLPFC seed maps derived via parcellation-atlases or voxelwise approaches. All parcellation-derived seed maps were significantly correlated with voxewise seed maps (r= 0.64–0.80, all p<0.001). Voxelwise seed maps had stronger correlation (and thus higher replication) with Task and Rest atlas seed maps (r= 0.75–0.80) than Random atlas seed maps (r= 0.64–0.72). Additionally, Task and Rest atlas seed maps were highly correlated for resting-state (r=0.89) and n-back task (r=0.92), further emphasizing these atlases’ similarity.
Table 3.
Regions of Interest for Task, Group Two-Stage atlas ranked by similarity to Rest, Group Two-stage Atlas
| Region | Jaccard Index | MNI Coordinates | Lobe | Volume (in mm3) | Label | ||
|---|---|---|---|---|---|---|---|
| X | Y | Z | |||||
| 38 | 100 | −32.5 | 35.9 | −3 | Visual | 2106 | LaOFG_38 |
| 59 | 100 | −0.2 | −0.6 | 34.9 | Cingulate | 3618 | MidCingulate_59 |
| 132 | 100 | −17.1 | −13.3 | 25.3 | Subcortical | 729 | Lpo_Caudate_132 |
| 35 | 99 | −9.2 | −20.4 | 43.8 | Motor | 4482 | Lpo_pericingulate_35 |
| 60 | 99 | −9.5 | 59.8 | 24.9 | Frontal | 4428 | LrmPFC/LvmPFC_60 |
| 102 | 99 | −47.4 | 31.7 | 3.5 | Frontal | 4509 | LIFG_BA45_102 |
| 155 | 99 | −0.3 | 23.4 | 26.3 | Cingulate | 4158 | dACC_155 |
| 73 | 98 | −25.5 | 56.5 | 15.6 | Frontal | 3726 | LavlPFC_73 |
| 80 | 98 | −25.8 | 5.8 | −13.8 | Temporal | 4239 | LOlfC_80 |
| 91 | 98 | 18.8 | −8.4 | −14.8 | Temporal | 3807 | RHPC_CA/Amyg_91 |
| 94 | 98 | −11.2 | −27.8 | 10.5 | Subcortical | 2808 | Lthalamus_94 |
| 103 | 98 | 17.3 | −34.3 | −18.6 | Visual | 5373 | Rculmen_103 |
| 125 | 98 | −34.5 | 22.4 | 10.5 | Frontal | 4617 | Lsup_AIns_125 |
| 128 | 98 | 29.5 | −19.6 | −13.2 | Temporal | 3699 | RHPC_body_128 |
| 197 | 98 | −13.8 | 3.1 | 21.2 | Subcortical | 1809 | Lacaudate_197 |
| 49 | 97 | 45.1 | 29.3 | 3.4 | Frontal | 4293 | RIFG_BA45_49 |
| 115 | 97 | 5.7 | 33 | 35.5 | Frontal | 4752 | RAperiCing_95 |
| 131 | 97 | −40.1 | 46.2 | 12.4 | Frontal | 4212 | LvlPFC_131 |
| 143 | 97 | 35.1 | 19.9 | −6.8 | Frontal | 4320 | Rinf_AIns_143 |
| 186 | 97 | 12.2 | 19.2 | 8.6 | Subcortical | 2619 | RaCaudate_186 |
| 15 | 96 | 24.4 | 8.5 | 3.6 | Subcortical | 2646 | RAputamen_15 |
| 26 | 96 | 9.2 | −28 | 10.1 | Subcortical | 2349 | Rthalamus_26 |
| 34 | 96 | −1.2 | −40.3 | −14.6 | Cerebellum | 2997 | Vermis_34 |
| 41 | 96 | 6.2 | 42.4 | 19.8 | Cingulate | 5130 | RrostralACC_MPFC_41 |
| 47 | 96 | −7.9 | 40.5 | 26.5 | Cingulate | 5265 | LrostralACC_MPFC_47 |
| 54 | 96 | −10.5 | 34.6 | 53.5 | Frontal | 3942 | LsupFG_54 |
| 57 | 96 | 6.3 | 58.5 | 12.6 | Frontal | 3321 | RvmPFC_57 |
| 62 | 96 | −39.1 | 6.9 | 35.1 | Frontal | 4833 | LpodlPFC_62 |
| 71 | 96 | 39.1 | 46.7 | 6.3 | Frontal | 3213 | RvlPFC_71 |
| 82 | 96 | −10 | −39.5 | 45.9 | Parietal | 5454 | LSPL_5C |
| 85 | 96 | −10 | 49.4 | 42 | Frontal | 3861 | LdmPFC_85 |
| 113 | 96 | −36.9 | 20 | −5.5 | Frontal | 5265 | Linf_AIns_113 |
| 118 | 96 | 8.3 | 7.2 | 63.7 | Cingulate | 4509 | RpreSMA_118 |
| 130 | 96 | 34.4 | 19.6 | 10.1 | Temporal | 4509 | Rsup_AIns_130 |
| 136 | 96 | −7.4 | 0.2 | 48.3 | Cingulate | 4941 | LpreSMA/dACC_136 |
| 181 | 96 | −25.4 | 8.3 | 4.3 | Subcortical | 3024 | LAputamen_181 |
| 193 | 96 | 29.2 | 6.4 | −15.4 | Temporal | 3834 | ROlfC_193 |
| 185 | 92 | 46.1 | 5.2 | −14.5 | Temporal | 4104 | RTempPole_185 |
| 188 | 92 | −26.5 | −36.7 | −0.4 | Temporal | 3294 | LHPC_tail_188 |
| 190 | 92 | −39.6 | −7.9 | −0.4 | Temporal | 5913 | Linf_MidIns_190 |
| 5 | 91 | −1.1 | −51 | 13 | Cingulate | 5292 | retrosplenial_cortex_5 |
| 14 | 91 | −46.8 | −55.9 | 41.3 | Parietal | 5940 | LIPC_PGa_14 |
| 19 | 91 | 24.4 | 47.2 | 34.4 | Frontal | 5292 | RrlPFC_19 |
| 40 | 91 | −59.6 | −19.7 | 27.6 | Parietal | 5751 | LIPC_Pfop_40 |
| 53 | 91 | −53.8 | 6.4 | 27.2 | Motor | 5400 | LdlPFC_53 |
| 89 | 91 | 8.2 | −31.9 | 43.9 | Cingulate | 5373 | RSPL_5C |
| 158 | 91 | −9.5 | −54 | 35.4 | Visual | 5265 | Lprecu_158 |
| 160 | 91 | −26.3 | 6.1 | 58 | Motor | 5778 | LFEF_160 |
| 27 | 90 | −12.6 | −19.7 | 69.5 | Motor | 5049 | LlatSMA_27 |
| 56 | 90 | 23.5 | −51.2 | −10.9 | Visual | 4887 | Rfusiform_56 |
| 66 | 90 | 42.9 | −54.9 | 18.2 | Parietal | 5238 | RTPOJ_66 |
| 70 | 90 | −41.6 | −18 | 14.9 | Temporal | 4833 | LIns_OP1_70 |
| 112 | 90 | −54.6 | −2.3 | −5.2 | Temporal | 5130 | LAinfMTG_112 |
| 120 | 90 | −25 | 23.5 | 50 | Frontal | 5454 | LsupDLPFC_120 |
| 134 | 90 | −25.3 | −86.1 | 17.8 | Visual | 5049 | Locc_BA18_134 |
| 140 | 90 | 57.1 | −9.2 | 14.1 | Motor | 5913 | R_OP4_140 |
| 149 | 90 | 32.6 | −36.5 | −14.9 | Visual | 4239 | Rfusiform/PHG_149 |
| 168 | 90 | −50.8 | 13.2 | 8 | Frontal | 5832 | LIFG_BA44_168 |
| 11 | 89 | 10.5 | −43.1 | −3.1 | Visual | 4698 | RALingual_11 |
| 31 | 89 | −29.2 | −53.6 | 57.1 | Parietal | 6156 | LSPL_7A_31 |
| 42 | 89 | 22.7 | −68.9 | −9.1 | Visual | 5022 | Rpofusiform_42 |
| 43 | 89 | 38.2 | −22.5 | 43.3 | Motor | 5670 | RM1_43 |
| 52 | 89 | 10.7 | −64.2 | 58.3 | Parietal | 5022 | RSPL_52 |
| 167 | 89 | 50.4 | 8.5 | 2.6 | Frontal | 5454 | RIFG_BA44_167 |
| 174 | 89 | 41.2 | −50.5 | −15.8 | Visual | 5913 | Rfusiform/RITG_174 |
| 33 | 88 | −39.7 | 1.1 | 12.4 | Motor | 4401 | Lsup_MidIns_33 |
| 76 | 88 | −30.6 | −66.4 | 41.6 | Parietal | 6615 | LpoIPL_76 |
| 87 | 88 | −26.9 | −33.6 | 61.4 | Motor | 6102 | Lmedial_CentralSulcus_87 |
| 148 | 88 | 3.7 | −52.4 | 48.8 | Visual | 6129 | SPL_7A_148 |
| 151 | 88 | −26.5 | 50.4 | 29.6 | Frontal | 4968 | LrlPFC_151 |
| 163 | 88 | 25.3 | −36 | 60.5 | Motor | 6021 | Rmedial_CentralSulcus_163 |
| 170 | 88 | 0.6 | −60.7 | −3 | Cerebellum | 6534 | medcbell_170 |
| 22 | 87 | −17.5 | −33 | −17.1 | Cerebellum | 5076 | Lculmen_22 |
| 36 | 87 | 51 | 20.1 | 16.5 | Frontal | 5076 | RIFG_BA44_36 |
| 13 | 86 | −12.1 | −53.4 | 61.1 | Parietal | 5292 | LSPL_7A_13 |
| 16 | 86 | 23.5 | 13.2 | 56.5 | Frontal | 5778 | RFEF_16 |
| 61 | 86 | 52.9 | 5.3 | 25.3 | Motor | 5589 | RdlPFC_61 |
| 86 | 86 | −56.9 | −44.6 | 25.5 | Parietal | 6102 | LpoSTS_121 |
| 95 | 95 | −7.4 | 21.1 | 40.6 | Cingulate | 4968 | LAperiCing_95 |
| 96 | 95 | −7.1 | 48.7 | 6.8 | Frontal | 4482 | LvmPFC/LpgACC_96 |
| 105 | 95 | 40 | 40.4 | 21.4 | Frontal | 5454 | RrlPFC_105 |
| 139 | 95 | 27.2 | 37.8 | −2.8 | Frontal | 1404 | RaOFG_139 |
| 144 | 95 | 9.3 | 46.4 | 45 | Frontal | 4266 | RdmPFC_144 |
| 156 | 95 | −0.4 | −21.7 | 2.5 | N/A | 2025 | 3rdVent_156 |
| 162 | 95 | 6.2 | 13.6 | 44.1 | Cingulate | 4995 | RdorsalperiCing_162 |
| 172 | 95 | 27.3 | 53.8 | 17.7 | Frontal | 4374 | RavlPFC_172 |
| 28 | 94 | −0.3 | −22.9 | 32.2 | Cingulate | 3348 | PCC_28 |
| 88 | 94 | 14.6 | −10.4 | 24.9 | N/A | 1080 | Rpo_Caudate_88 |
| 117 | 94 | 7.2 | 45.6 | 4.2 | Frontal | 3240 | RvmPFC/RpgACC_117 |
| 119 | 94 | 19.8 | −57 | 4.2 | Visual | 4401 | Rlingual_119 |
| 129 | 94 | 23 | 30.1 | 48.4 | Frontal | 5724 | RsupDLPFC_129 |
| 138 | 94 | −12.3 | −47.3 | −3.1 | Visual | 5157 | Llingual_138 |
| 141 | 94 | −18.1 | −67.3 | 23.9 | Visual | 6561 | LlatPrecuneus/V3A_141 |
| 166 | 94 | −10.9 | 16.2 | 61.7 | Frontal | 4482 | RsupFG_166 |
| 177 | 94 | −1.3 | −24.7 | 57.6 | Cingulate | 5373 | midline_SMA_177 |
| 178 | 94 | −43.8 | 8.4 | −14.3 | Temporal | 4482 | LTempPole_178 |
| 179 | 94 | 15.6 | −62.2 | 18.8 | Visual | 4590 | RlatPrecuneus/V3A_179 |
| 198 | 94 | −0.5 | −2.6 | −5.5 | N/A | 2295 | 3rdVent_198 |
| 199 | 94 | −45.3 | −72.2 | 9.3 | Visual | 5238 | Llatocc_199 |
| 18 | 93 | −3.9 | −91.1 | 8.3 | Visual | 6507 | LpoV1_18 |
| 21 | 93 | −59.7 | −25.6 | 11.6 | Temporal | 5724 | LSTG_TE3_21 |
| 37 | 93 | −1 | 31.5 | 0.3 | Cingulate | 3429 | rostralACC_37 |
| 64 | 93 | 14.7 | 10.5 | −7.5 | Subcortical | 2970 | RNacc_64 |
| 67 | 93 | −20.4 | −59.8 | 6.2 | Visual | 5022 | Llingual_67 |
| 104 | 93 | 9.4 | 57.6 | 30.1 | Frontal | 3780 | RrmPFC_104 |
| 106 | 93 | −11.4 | −2.1 | 66.8 | Cingulate | 4590 | LpreSMA_106 |
| 133 | 93 | −57.6 | −5.8 | 13.3 | Motor | 5913 | L_OP4_133 |
| 152 | 93 | −46 | −59.2 | 21 | Parietal | 5940 | LTPOJ_152 |
| 175 | 93 | 12.2 | 7.2 | 18.9 | Subcortical | 2457 | RpoCaudate_175 |
| 183 | 93 | 7.5 | 28.1 | 56.8 | Frontal | 4779 | RsupFG_54 |
| 195 | 93 | 32.9 | −45.9 | 41.6 | Parietal | 5913 | RSMG_195 |
| 200 | 93 | 5.5 | −8.7 | 49.9 | Cingulate | 5400 | RpreSMA/dACC_200 |
| 9 | 92 | −56.5 | −20.3 | −4 | Temporal | 4617 | LmidMTG_9 |
| 50 | 92 | −39 | −74.9 | 26.9 | Visual | 6480 | Llat_midOcc_50 |
| 58 | 92 | 33.1 | −83.6 | −1.1 | Visual | 3618 | RV3/V4_58 |
| 68 | 92 | 39.3 | −1.6 | 13.3 | Temporal | 4212 | RsupMidIns_68 |
| 75 | 92 | 24.6 | −36.9 | 1.5 | Temporal | 2727 | RHPC_tail_75 |
| 142 | 92 | −11.4 | −38.3 | 67.8 | Motor | 4752 | LSPL_5L_142 |
| 165 | 92 | −43 | −32.6 | 17.9 | Motor | 5130 | LpoIns_165 |
| 93 | 86 | −54.6 | −35.9 | 0.6 | Motor | 4995 | LpoMTG_93 |
| 107 | 86 | 7 | −85.8 | −2.6 | Visual | 8829 | RpoV1_107 |
| 32 | 85 | −43.3 | −21.4 | 57.5 | Motor | 4779 | LlateralCentral_Sulcus_32 |
| 78 | 85 | 9.9 | −74.7 | 7.6 | Visual | 5967 | RV1_78 |
| 79 | 85 | 9.5 | −11 | 68.4 | Cingulate | 4698 | RlatSMA_79 |
| 81 | 85 | −44.8 | −46.3 | −14.3 | Temporal | 3726 | LITG_81 |
| 83 | 85 | 22.6 | −67.9 | 31.9 | Visual | 6048 | RlatPrecuneus_V3A_83 |
| 101 | 85 | 45.6 | 23.6 | 31.4 | Frontal | 5373 | RrostMPFC_101 |
| 154 | 85 | 58.5 | −26.6 | 13.3 | Temporal | 4779 | RSTG_TE3_154 |
| 189 | 85 | 58.2 | −22.8 | 30.1 | Visual | 5697 | RIPC_Pfop_189 |
| 10 | 84 | 39.4 | 15.8 | 47.7 | Frontal | 5616 | RsupdlPFC_10 |
| 17 | 84 | −32.1 | −43.6 | −24.4 | Cerebellum | 5913 | LlatCbell_17 |
| 29 | 84 | −26.4 | 37.1 | 40.2 | Frontal | 5427 | LsupFG_29 |
| 63 | 84 | −31.9 | −84.2 | 1 | Visual | 4320 | LV3/V4_63 |
| 110 | 84 | −1 | −40.7 | 28.9 | Cingulate | 3618 | dPCC_110 |
| 116 | 84 | −15.1 | −68.2 | 52.7 | Parietal | 5778 | LSPL_116 |
| 145 | 84 | −41.3 | −4.1 | 51.7 | Motor | 5076 | LLPM_lat_145 |
| 146 | 84 | 41.9 | −74.6 | 13.6 | Visual | 5400 | Rlatocc_146 |
| 44 | 83 | −42.5 | −39.4 | 54.4 | Motor | 4644 | LsupS1_44 |
| 69 | 83 | −8.9 | −86.4 | 26.1 | Visual | 5211 | Lcuneus_69 |
| 97 | 83 | 9.3 | −46.3 | 65.6 | Motor | 5643 | RSPL_5L_97 |
| 123 | 83 | −53.7 | −7.4 | 39.1 | Motor | 5400 | LlatM1_BA4a_123 |
| 159 | 83 | 43.7 | −67.6 | −4.2 | Visual | 5319 | Rinf_OccG_159 |
| 171 | 83 | 33 | 32.4 | 36.4 | Frontal | 5373 | RsupFG_171 |
| 191 | 83 | 6.4 | −53.9 | 32.6 | Visual | 4941 | Rprecu_191 |
| 46 | 82 | −27.3 | −50.3 | −11.1 | Visual | 5049 | Lfusiform_46 |
| 84 | 82 | −41.2 | 13.8 | 48.3 | Frontal | 4806 | LsupdlPFC_84 |
| 126 | 82 | −11.2 | −75.7 | 5.2 | Visual | 6048 | RV1_126 |
| 164 | 82 | 10.7 | −86.5 | 27.6 | Visual | 6210 | RlatCuneus_164 |
| 65 | 81 | −54.3 | −32.4 | 41.1 | Parietal | 6507 | LIPC_PF_65 |
| 121 | 81 | 58.4 | −43.7 | 21.2 | Temporal | 5562 | RpoSTS_121 |
| 150 | 80 | −20.9 | −80.8 | 38 | Visual | 6075 | LmidOccG_150 |
| 180 | 80 | 24.3 | −4.3 | 58.5 | Motor | 5994 | RLPM_med_180 |
| 7 | 79 | −26.9 | −12.9 | 59.7 | Motor | 6534 | LLPM_med_7 |
| 20 | 79 | 9.8 | −29.6 | 70 | Motor | 4320 | RsupM1_20 |
| 90 | 79 | −39.2 | 38.3 | 27.8 | Frontal | 5589 | LsupFG_90 |
| 77 | 78 | 25.8 | −19.3 | 64.1 | Motor | 5157 | RLPM_77 |
| 153 | 78 | 38.5 | −5.7 | −1.1 | Temporal | 5265 | Rinf_MidIns_153 |
| 74 | 77 | 54.8 | −7.9 | −4.7 | Temporal | 4212 | RAinfMTG_74 |
| 127 | 77 | −32.2 | −26.7 | −13.3 | Temporal | 4266 | LHPC_body/PHG_127 |
| 192 | 77 | −0.9 | −68.7 | 20.7 | Visual | 5697 | midV2_192 |
| 99 | 75 | 13.8 | −76.1 | 45.2 | Parietal | 4941 | RSPL_99 |
| 6 | 74 | 53.6 | −57.4 | 6.8 | Temporal | 5211 | RpoMTG_6 |
| 182 | 74 | −1.5 | −72.3 | 39.5 | Visual | 5589 | midV3/V3A_182 |
| 98 | 73 | −54.1 | −51.3 | 8.2 | Temporal | 5319 | LinfTPOJ_98 |
| 39 | 72 | 25.1 | −55.8 | 57.2 | Parietal | 6561 | RSPL_7P_39 |
| 111 | 72 | 52.2 | −50.5 | 35.6 | Parietal | 5265 | RIPC_Pga_111 |
| 194 | 72 | 42.7 | −33.4 | 20.6 | Temporal | 4860 | RpoIns_194 |
| 48 | 71 | 51.7 | −5.5 | 38 | Motor | 5643 | RlatM1_BA4a_48 |
| 122 | 69 | 24.7 | −85.9 | 15.1 | Visual | 4752 | Rocc_BA18_122 |
| 184 | 69 | 30 | −81.5 | 30.8 | Visual | 5454 | RmidOccG_30 |
| 187 | 68 | −45.8 | −62.9 | −5.1 | Visual | 5751 | Linf_OccG_159 |
| 3 | 67 | −49.2 | 27 | 21.4 | Frontal | 4779 | LAdlPFC_3 |
| 23 | 66 | −42.8 | 22.9 | 35.6 | Frontal | 4860 | LrostMPFC_23 |
| 196 | 62 | −11.4 | 15.7 | −2.3 | Subcortical | 2889 | LNAcc_196 |
| 100 | 56 | 41.7 | −52 | 51 | Parietal | 5103 | RIPL_PGa_100 |
| 109 | 56 | 39.3 | −19.4 | 14 | Temporal | 4887 | RIns_OP1_109 |
| 2 | 55 | 36.9 | 6.2 | 36.1 | Frontal | 5049 | RpodlPFC_2 |
| 45 | 55 | 57.2 | −22.4 | −3.6 | Temporal | 3321 | RmidMTG_45 |
| 157 | 55 | 47.5 | −67.3 | 28.3 | Parietal | 5913 | Rlat_midocc_157 |
| 51 | 54 | −25 | −10.1 | −16.1 | Temporal | 6048 | LHPC_CA/Amyg_51 |
| 147 | 53 | 53 | −34.7 | 43 | Parietal | 5454 | RIPC_PF_147 |
| 169 | 49 | −35.4 | −45.2 | 40.4 | Parietal | 5238 | LSMG_169 |
| 124 | 48 | −12.9 | 16.8 | 12 | N/A | 2943 | Lpocaudate_124 |
| 176 | 48 | −40.7 | −21.6 | 42.8 | Motor | 5319 | LmidS1_176 |
| 114 | 47 | 0.1 | 7 | 5 | N/A | 3807 | SubgenCing_114 |
| 135 | 42 | −0.5 | −13.1 | 15.3 | Subcortical | 2403 | thalamus_135 |
| 12 | 41 | 41 | −31.9 | 56.7 | Motor | 4428 | Rlateral_CentralSulcus_41 |
| 30 | 40 | 33.2 | −68.6 | 44.7 | Visual | 6291 | RpoIPL_30 |
| 24 | 38 | 57.8 | −39.8 | 0.7 | Temporal | 3996 | RpoMTG_24 |
| 161 | 37 | 39.5 | −2.7 | 52.5 | Motor | 5211 | RLPM_lat_161 |
| 92 | 34 | 47.5 | −17.5 | 50.9 | Motor | 4347 | RlatM1/S1_92 |
| 173 | 33 | 47.3 | −30.4 | 2.5 | Temporal | 4644 | RpoMTG_173 |
| 55 | 1 | −23 | −68.7 | −9 | Visual | 4617 | Lpofusiform_55 |
| 1 | 0 | 0 | 0 | 0 | N/A | 0 | Garbage_1 |
| 4 | 0 | 0 | 0 | 0 | N/A | 0 | Garbage_4 |
| 8 | 0 | 0 | 0 | 0 | N/A | 0 | Garbage_8 |
| 25 | 0 | 0 | 0 | 0 | N/A | 0 | Garbage_25 |
| 72 | 0 | 0 | 0 | 0 | N/A | 0 | Garbage_72 |
| 108 | 0 | 0 | 0 | 0 | N/A | 0 | Garbage_108 |
| 137 | 0 | 0 | 0 | 0 | N/A | 0 | Garbage_137 |
Finally, the regression of head coil against DLPFC connectivity seed maps showed no consistent pattern of coil-related differences in connectivity (Table 4). During the n-back task, the 32-channel head coil was associated with greater LDLPFC connectivity among two regions: one located at pre-SMA/dACC and another in right SMA (AlphaSim corrected q<0.05). These regions corresponded with the Task atlas’s ROIs #200 and #79 – which were highly replicable across Task and Rest atlases (Table 3, JI=0.93 and 0.85, respectively). Furthermore, no regions showed coil-related significant differences in DLPFC connectivity for the resting-state scans. The lack of replicable, systemic coil-related differences in connectivity across these two tasks suggests that head coil is not influencing functional connectivity patterns, and thus not confounding atlas generation. This finding is consistent with our past findings that head-coil does not significantly influence task-related brain activity [10]
Table 4.
Regions of increased voxelwise connectivity to left DLPFC for 32- vs 8-channel head coil (q<0.05)
| Cluster | MNI Coordinates | Volume (in mm3) | Corresponding Region in Task Atlas | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | Region | Label | Jaccard index with Rest Atlas | ||
| 1 | 8 | −16 | 42 | 8046 | 200 | Right pre-SMA/dorsal cingulate | 93 |
| 2 | 5 | −16 | 66 | 2,133 | 79 | Right SMA | 85 |
Differences reported for n-back task only; no significant coil-related differences observed during resting-state
4. Discussion
We report strong similarity between atlases derived via parcellation of resting-state data and atlases derived via parcellation of both resting-state and task-based data. Our findings are consistent with past research suggesting that brain networks are consistently organized across task and rest [13]; we expand upon those findings to suggest that the brain’s functionally independent subunits (“nodes”) are also consistently represented across task and rest. The similarity between Task and Resting atlases may partially stem from the necessary incorporation of baseline conditions in fMRI tasks to model task-related changes in brain activity or connectivity. These baseline conditions are low-level cognitive control tasks (such as the 0-back condition of the n-back task) and/or resting-state epochs, which may enforce similar connectivity structure between task-based and resting-state fMRI scans. Nonetheless, the similarity between atlases remains striking given that resting-state epochs compose less than half of the tasks’ timepoints.
We also report strong similarity of these atlases to the random parcellations (mean JI=0.59–0.63). Although initially surprising, this may be explained by our decision to constrain the parcellation approach to gray matter voxels, which causes white matter to form consistent boundaries across all parcellations. For example, regions with multiple white matter boundaries (such as the cingulate, which is bounded by white matter to the left, right, and inferior surfaces) have greater constraint in how they may be parcellated, potentially explaining the strong Jaccard Indices between atlases observed in Figure 2.
Leave-one out cross-validation has shown that the group two-stage approach generates atlases that are more representative across individuals than the group mean approach [1]. We thus limited our comparison of task-based and resting-state parcellations to atlases generated using the group two-stage approach. Contrary to hypotheses, task-based and resting-state parcellations had strong similarity (JI≥0.90) for regions involved in higher-order cognition, including prefrontal cortices, cingulate, and left temporal lobe. We interpret this as evidence that the cognitive processes occurring in the absence of overt task (such as rumination, autobiographical memory retrieval, introspection and theory of mind) are sufficiently similar across rest and diverse tasks to map these regions with high consistency [14;15].
Conversely, we report least similarity for (a) three regions in right sensorimotor cortex, (b) two regions in right superior temporal sulcus, and (c) two regions bordering the left lateral ventricle. The sensorimotor cortex encodes the neural representation of the hand, with significantly greater activity for contralateral hand movement but greater variability for ipsilateral hand movement [16], particularly in the context of changing task demand [17]. Given the variety of tasks performed (with 7 tasks requiring right hand responses and 2 requiring both left and right hand responses), greater heterogeneity (less similarity) in right sensorimotor cortex is not surprising given this predominantly (90%) right-hand dominant sample.
The role of the right STG is not as clearly established as the left STG, which is strongly associated with auditory processing [18;19]. Right STG has been implicated in diverse processes such as encoding auditory rhythms [20], multisensory integration [21], and contextual awareness [22]. These processes may be more strongly engaged during task than rest, resulting in variable recruitment of the right STG across tasks and thus dissimilar representation of right STG between task and rest. Perhaps more surprising is the dissimilarity between atlases for striatal regions bordering left lateral ventricle, which show bilateral recruitment for processes such as learning, reward processing, and motor processing [23–28]. Future work will evaluate asymmetric striatal recruitment across tasks to evaluate which cognitions could be leading to the discrepancy in striatal representation between atlases.
Finally, we demonstrate that voxelwise seed maps of left dorsolateral prefrontal cortex (LDLPFC) functional connectivity are more consistently reproduced by the All and Rest atlas than the Random atlas. Interestingly, the All and Rest parcellations produced a LDLPFC ROI that strongly corresponds to the LDLPFC region identified by the n-back task, as indicated by the blue crosshairs in Figure 3. Conversely, the Random atlas LDLPFC ROI shows poor correspondence with the task-defined ROI. This finding both reinforces the similarity of the Task and Rest atlases while demonstrating their superiority over the Random atlas in capturing underlying neural organization.
An important caveat is that our parcellations were constrained to brain regions covered across all participants and sessions. Our conservative approach led to incomplete coverage of the inferior orbitofrontal cortex, a region notoriously difficult to image with fMRI due to proximity of air in the sinus cavity which distorts magnetic signal in this region. While combined spin-echo and echo-planar sequences have been developed to optimally image this region [29], these sequences suffer a 50% reduction in temporal resolution, prompting our selection of the standard echo-planar sequence. We contend that our atlas is well-suited for analyzing Cognitive Connectome project data and other fMRI datasets, the majority of which also rely upon echo-planar sequences. However, given the strong similarity between atlases derived from task-based and resting-state data, we conclude that previously published atlases derived from combined spin-echo/echo-planar sequences are likewise suitable for task-based analyses.
5. Conclusions
We have uploaded a fully labeled atlas to the Neuroinformatics Tool and Resources Center (https://www.nitrc.org/) for public use.1 Our findings indicate that this atlas is well-suited for analysis of both resting-state and task-based data. Our findings further suggest that existing atlases derived solely from resting-state data are equally suitable for resting-state and task-based analyses.
Acknowledgments
This research was supported by the Translational Research Institute (TRI) at the University of Arkansas for Medical Sciences (UAMS) which is funded by the National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program (UL1TR000039); the CTSA KL2 Scholars Program (KL2TR000063; to GAJ). We additionally thank Mr. Jonathan Young and Mrs. Sonet Smitherman for assistance with data collection and maintaining institutional compliance. All authors contributed to the interpretation and writing of this manuscript.
Footnotes
Atlas will be uploaded to NITRC upon acceptance of manuscript and/or made available upon the Magnetic Resonance Imaging website, pending further discussion with editors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Craddock RC, James GA, Holtzheimer PE, III, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33(8):1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, et al. Non-stationarity in the “resting brain’s” modular architecture. PLoS One. 2012 Jul 5;:e39731. doi: 10.1371/journal.pone.0039731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison SJ, Woolrich MW, Robinson EC, Glasser MF, Beckmann CF, Jenkinson M, et al. Large-scale probabilistic functional modes from resting state fMRI. Neuroimage. 2015;109:217–231. doi: 10.1016/j.neuroimage.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen X, Tokoglu F, Papademetris X, Constable RT. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage. 2013;82:403–415. doi: 10.1016/j.neuroimage.2013.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goulas A, Uylings HB, Stiers P. Unravelling the intrinsic functional organization of the human lateral frontal cortex: a parcellation scheme based on resting state fMRI. J Neurosci. 2012;32(30):10238–10252. doi: 10.1523/JNEUROSCI.5852-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, et al. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67(1):156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He AG, Tan LH, Tang Y, James GA, Wright P, Eckert MA, et al. Modulation of neural connectivity during tongue movement and reading. Hum Brain Mapp. 2003 Feb 25;:222–232. doi: 10.1002/hbm.10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton AT, Morgan VL, Gore JC. Task demand modulation of steady-state functional connectivity to primary motor cortex. Hum Brain Mapp. 2007;28(7):663–672. doi: 10.1002/hbm.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gess JL, Fausett JS, Kearney-Ramos TE, Kilts CD, James GA. Task-dependent Recruitment of Resting State Networks Reflects Normative Variance in Cognition. Brain and Behavior. 2014;4:650–664. doi: 10.1002/brb3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Computers and biomedical research, an international journal. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 12.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl Neuroimage. 2011 Oct 08;:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009 Jul 22;:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008 Apr 11;:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- 15.Andrews-Hanna JR, Saxe R, Yarkoni T. Contributions of episodic retrieval and mentalizing to autobiographical thought: evidence from functional neuroimaging, resting-state connectivity, and fMRI meta-analyses. Neuroimage. 2014;91:324–335. doi: 10.1016/j.neuroimage.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dassonville P, Zhu XH, Uurbil K, Kim SG, Ashe J. Functional activation in motor cortex reflects the direction and the degree of handedness. Proc Natl Acad Sci U S A. 1997;94(25):14015–14018. doi: 10.1073/pnas.94.25.14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wexler BE, Fulbright RK, Lacadie CM, Skudlarski P, Kelz MB, Constable RT, et al. An fMRI study of the human cortical motor system response to increasing functional demands. Magn Reson Imaging. 1997;15(4):385–396. doi: 10.1016/s0730-725x(96)00232-9. [DOI] [PubMed] [Google Scholar]
- 18.Peeva MG, Guenther FH, Tourville JA, Nieto-Castanon A, Anton JL, Nazarian B, et al. Distinct representations of phonemes, syllables, and supra-syllabic sequences in the speech production network. Neuroimage. 2010;50(2):626–638. doi: 10.1016/j.neuroimage.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bitan T, Cheon J, Lu D, Burman DD, Gitelman DR, Mesulam MM, et al. Developmental changes in activation and effective connectivity in phonological processing. Neuroimage. 2007;38(3):564–575. doi: 10.1016/j.neuroimage.2007.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung SJ, Chen JL, Zatorre RJ, Penhune VB. Interacting cortical and basal ganglia networks underlying finding and tapping to the musical beat. J Cogn Neurosci. 2013;25(3):401–420. doi: 10.1162/jocn_a_00325. [DOI] [PubMed] [Google Scholar]
- 21.Beauchamp MS. See me, hear me, touch me: multisensory integration in lateral occipital-temporal cortex. Curr Opin Neurobiol. 2005;15(2):145–153. doi: 10.1016/j.conb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Zucker NL, Green S, Morris JP, Kragel P, Pelphrey KA, Bulik CM, et al. Hemodynamic signals of mixed messages during a social exchange. NeuroReport. 2011;22(9):413–418. doi: 10.1097/WNR.0b013e3283455c23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grahn JA, Parkinson JA, Owen AM. The role of the basal ganglia in learning and memory: neuropsychological studies. Behav Brain Res. 2009;199(1):53–60. doi: 10.1016/j.bbr.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 24.Knutson B, Cooper JC. Functional magnetic resonance imaging of reward prediction. Curr Opin Neurol. 2005;18(4):411–417. doi: 10.1097/01.wco.0000173463.24758.f6. [DOI] [PubMed] [Google Scholar]
- 25.Hikosaka O, Kim HF, Yasuda M, Yamamoto S. Basal ganglia circuits for reward value-guided behavior. Annu Rev Neurosci. 2014;37:289–306. doi: 10.1146/annurev-neuro-071013-013924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ena S, de Kerchove dA, Schiffmann SN. Unraveling the differential functions and regulation of striatal neuron sub-populations in motor control, reward, and motivational processes. Front Behav Neurosci. 2011;5:47. doi: 10.3389/fnbeh.2011.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Hairston J, Schrier M, Fan J. Common and distinct networks underlying reward valence and processing stages: a meta-analysis of functional neuroimaging studies. Neurosci Biobehav Rev. 2011;35(5):1219–1236. doi: 10.1016/j.neubiorev.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balleine BW, O’Doherty JP. Human and rodent homologies in action control: corticostriatal determinants of goal-directed and habitual action. Neuropsychopharmacology. 2010;35(1):48–69. doi: 10.1038/npp.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heberlein KA, Hu X. Simultaneous acquisition of gradient-echo and asymmetric spin-echo for single-shot z-shim: Z-SAGA. Magn Reson Med. 2004;51:212–216. doi: 10.1002/mrm.10680. [DOI] [PubMed] [Google Scholar]