SUMMARY
The pathway to generate T cells from hematopoietic stem cells guides progenitors through a succession of fate choices while balancing differentiation progression against proliferation, stage to stage. Many elements of the regulatory system that controls this process are known, but the requirement for multiple, functionally distinct transcription factors needs clarification in terms of gene network architecture. Here we compare the features of the T-cell specification system with the rule sets underlying two other influential types of gene network models: first, the combinatorial, hierarchical regulatory systems that generate the orderly, synchronized increases in complexity in most invertebrate embryos; second, the dueling “master regulator” systems that are commonly used to explain bistability in microbial systems and in many fate choices in terminal differentiation. The T-cell specification process shares certain features with each of these prevalent models but differs from both of them in central respects. The T-cell system is highly combinatorial but also highly dose-sensitive in its use of crucial regulatory factors. The roles of these factors are not always T-lineage specific, but they balance and modulate each other’s activities long before any mutually exclusive silencing occurs. T-cell specification may provide a new hybrid model for gene networks in vertebrate developmental systems.
Keywords: Gene regulatory networks, Transcription factors, Commitment, Lineage hierarchy
Introduction: T Cell Development as a Developmental Model System
From the vantage point of immunology, the program of T cell development seems unexceptional, even particularly well-ordered. The hierarchy of intermediate states in early T cell development is well defined by multiple molecular criteria and the succession of these states has been shown to progress faithfully in adult life, in fetal life, and in very useful in vitro culture systems. Compared to other hematopoietic programs, T-cell development is a distinctive showcase for interesting cell fate determination mechanisms. T cell functional maturation is very distinct from the classic terminal differentiation programs of erythrocytes, megakaryocytes, and granulocytes, for example, where maturity is tightly coupled with permanent growth arrest and commitment to die. The whole range of T-cell development also includes many more known branches than erythroid development or even B-cell development. This means that the questions of how choices are made, and whether choices are irreversible, once made, pervade any consideration of T-cell function. However, the basic principles that enable stem cells to generate T cells throughout fetal and postnatal life are normal and exemplary for the hematopoietic system.
Seen in a different light, however, T cell development has arrestingly strange properties. It has indeterminate timing; it comes in different variants depending on adult vs. fetal context; it includes a prolonged period of indeterminacy of cell fate, based on the delay in ability to repress key transcription factors of alternative lineages; and it shares different modular parts of its program with different alternative cell types (innate lymphoid cells for its functional components, B cells for its receptor-generating components), blurring the hierarchy of its relationships to other fates. Most glaringly, this program generates a huge excess of immature T cells that will be ruthlessly culled by a selection mechanism before maturation to discard all but a lucky few percent. These properties typify the difference between this developmental system based on multipotent stem cells and one based on rapid, efficient, and precise fate-determination mechanisms operating in early embryos of many organisms. Here, T cell development is less an exception than a prototype of the opportunities provided by a stem-cell based system as opposed to the rigorously scheduled program of cell type definitions created by gene network operation in the early embryos.
This comparison is timely in light of the recent death of our colleague, Eric H. Davidson, whose lively exposition of gene network rules governing early embryo development provided a framework for many far-reaching discussions. Which elements in developmental system design are essential and universal? What aspects of gene networks make parts of the developmental program more evolutionarily malleable? The answers obtained from early embryos of multiple types of invertebrates are very strong, but they are in some respects different from the ones that would emerge from an analysis based on vertebrate hematopoietic systems, as exemplified by T cell development. Thus, the question arises, how do particular rules of gene network design serve different biological needs?
This article considers how the set of gene network features that we must account for in early T cell development connect to and distinguish it from other established gene network models for development. These network features force some revisions of common paradigms, and they have broader consequences for understanding of the mechanism of T-cell specification and of hematopoietic specification more generally.
Paradigms for Development
Two kinds of gene network models have been highly influential in explaining development. One is to account for the highly ordered creation of complexity from a single totipotent cell in embryonic development, which has been investigated in key invertebrate systems. The other is for the choice between terminal differentiation fates in bipotent precursor cells, formulated as a duel between two mutually antagonistic transcription factors.
Development at high efficiency: principles from invertebrate embryos
The most universal features of multicellular, highly organized metazoan animal life can be identified as those that are found in all three main branches of animal phylogeny: ecdysozoans (e.g. insects, crustacea, nematodes), lophotrochozoans (e.g. annelid worms, and mollusks ranging from clams to snails to octopuses), and deuterostomes (e.g. echinoderms, ascidians, fish, humans). All three branches have three germ layers and bilateral symmetry in their embryos (metazoans) and have numerous points of molecular similarity, despite often-alien looking body plans. All three branches share the use of a mode of embryonic development that has been called “Type I” embryogenesis (1–3). This set of developmental rules enables embryos to become capable of feeding independently in a short time. Such embryos start subdividing into future tissue types in an organized way during the early cleavage divisions soon after fertilization. Every cell counts. Future body axes of the bilaterally symmetric embryo are established from the time the egg is fertilized, due in part to localized deposits of signaling proteins or RNA in a particular region of the egg that are laid down during oogenesis, and often combined with polarized cytoskeletal movements triggered by the entry of the sperm. A cascade of signaling begins to create spatially differential gene expression as cleavage gets under way, and the fates of the cells become highly predictable due to the canonical nature of the cleavage pattern. Cells inheriting the cytoplasm with localized maternal determinants signal to their neighbors, which in turn activate genes encoding new signaling molecules to affect the next tier of neighbors. Cell types begin to turn on fate-specific distinctive gene expression patterns many cell cycles before they differentiate morphologically, and their fates are predictable and usually invariant from this early stage. Thus, despite its burgeoning variety of cell types and their morphogenetic changes in position, the embryo develops canonically and synchronously.
The crucial feature of this kind of development is its predictability and lack of ambiguity. Gene networks in these cases are used not only to generate complexity but also to make the process fail-safe: thus cells are used parsimoniously but regulatory genes are not, and gene network circuits are typically “over-engineered” (4). These features of gene networks have been demonstrated explicitly in a non-vertebrate deuterostome, the purple sea urchin Strongylocentrotus purpuratus (e.g. (5, 6)), and most of them are mirrored by evidence from another deuterostome, the sea squirt Ciona intestinalis (7–9), and two key embryonic model systems from the ecdysozoan branch of evolution, the nematode worm Caenorhabditis elegans (10, 11) and the fruit fly Drosophila melanogaster (12–15). Although the mode of embryonic development of Drosophila is divergent from the basic type I embryo (1), many features of type I embryonic networks still pertain to it. All these gene networks use the initially localized signals to trigger a hierarchy of progressive subdivisions to specify future tissue types. To enable different cells to express different genes at the end, the key is to set up diverse, stable patterns of transcription factor expression; to make an embryo, these expression domains must be strictly spatially organized.
A transcription factor coding gene as a rule is expressed not in the same pattern as any one of its own positive regulators, but rather, in a defined subset of the region in which each of its “upstream” regulators is active. This is because each regulatory gene is activated only by a particular combination of positive regulators in the absence of a certain set of negative regulators (16). These conditions are “computed” by transcription factor binding to cis-regulatory elements of the gene, so that new transcription factors are turned on only in cells where the domains of two old ones overlap, or in the region excluded by their overlap (e.g. (17–21)). Several tiers of factors are activated as development gets under way, in a sequential Boolean hierarchy (22). The increase in complexity of the developing embryo arises because each intermediate transcription factor coding gene in the network responds to a distinct, unique combination of inputs, thus propagating different state information to its own targets. Importantly, the set of factors activated in an early embryonic cell also usually includes at least one negative regulatory factor that excludes the expression of genes that might otherwise be expressed, e.g. in a sister cell. Repressors in the best-studied embryonic systems do not simply compete with activators at an enhancer, as a rule, but rather exert a veto over the ability of that enhancer to drive the target gene’s expression, and this helps to sharpen boundaries between future tissues (16). Once a fully specific set of transcription factors is active in a cell, then these collaborate to turn on the correct battery of cell type-defining differentiation genes, and the cell’s fate is set. Because of the canonical positioning of the “upstream” factors in the embryo, though, each cell’s fate is predictable from one embryo to another even before this molecular lockdown has occurred.
This type I mode of embryonic development is different in a number of respects from the kind of development that dominates in vertebrates, which has been labeled “type II”. Vertebrate embryos during cleavage tend to establish large populations of cells whose fates are not defined until after they migrate across other domains of the embryo and experience different signals from the environments through which they move. Lineage is much less clearly specified and thus the regulatory state preconditions for particular pathways of development are less clear until much later in the developmental process. An exception that may prove the rule is the relatively early specification of trophectoderm in mammals: but this is the only part of the mammalian embryo that functionally performs like a type I embryo – an early-specified part that enables feeding. Because of the indeterminacy of the cell fates and their relationships to initial gene expression patterns in most vertebrate embryos, gene network models have not been able to capture the early events of these embryos to date. In contrast, strong and highly detailed gene network models have been developed and extensively tested for both the early sea urchin (22) and the early Drosophila embryos (14). These models have revealed the logical power of cross-regulation among transcription factor coding genes, evident even from the topological layout of the network interactions (23, 24), to specify novel organismal structures and cell type identities
The early theories about the role of diffusible signals called “morphogens” in patterning embryos led to predictions that transcription factors should cause different effects at different doses. The biophysics of transcription factor binding make it reasonable to imagine that the regulatory sites that a factor might need to bind in two different cell type-specific genes might have evolved to require different concentrations of the factor per nucleus in order to be engaged effectively. However, surprisingly, the evidence from the well-studied invertebrate embryos is ambivalent about dosage sensitivity (25, 26). Although the boundaries of expression of individual genes can be sensitive to different levels of a transcription factor, embryogenesis as a process seems to compensate, making it robust to substantial changes in absolute transcription factor activity levels so long as the right relationships are maintained between levels at different parts of the embryo. What establishes the boundaries between cell types is not the absolute level of a transcription factor, then, but the gene network relationships between one part of the embryo and another, i.e. the ability of a repressor for genes of a particular cell type to be activated in one domain while the right combination of activators is turned on elsewhere.
Thus, the spatial subdivisions that are important for such embryogenesis programs use combinatoriality and gene network architecture to define a sequence of Boolean rules to generate an organized, diversified embryo. These rules are robust to considerable noise in absolute gene expression levels. This gives embryonic systems the ability to activate a massively diverse set of genes accurately in different parts of the embryo with precisely coordinated timing, despite the dependence of different cell types on different factors. This mechanism ensures that when a migrating mesenchyme cell is eventually brought into contact with a different cell type, it will be ready to receive and the other cell type will be ready to deliver the signals needed to trigger the next step of differentiation.
Duels between “Master Regulators”
A very different view of cell type specification has come from a “bottom up” approach to characterizing lineage identity, and from the precedents set by binary fate choices in microbial systems (27, 28). In this “master regulator” model, there is an option for much more cell-to-cell diversity in fate choice and in the timing of fate decisions even within an initially equivalent precursor population.
When terminally differentiated cells execute their specific gene expression programs, there are often a small number of transcription factors that work on many of the cells’ characteristic set of genes in parallel. Whereas the actual cis-regulatory systems of these differentiation genes can receive inputs from multiple transcription factors, some of them fairly broadly distributed, attention is usually focused on the handful of factors that drive cell-type specific genes and are themselves most narrowly expressed in the cell types of interest. Then, any gene expression that depends on those factors is inferred to be the result of the factors’ “Master Regulator”1 functions. The simplification possible when seeing the world through a lens of a few “master regulators” can be enticing. Even more attractively, some “master regulators” appear to be capable of positive autoregulation, while the “master regulators” of some differentiation programs appear to be able to antagonize the action of transcription factors that are viewed as “master regulators” of alternative differentiation programs; the Thpok (Zbtb7b)--Runx3 antagonism governing CD4/CD8 T cell maturation lineage choice provides a good example (rev. by (29–31)). Thus with a very small number of key molecular actors, far fewer than in a comprehensive gene network, there is the prospect of explaining both expression of one differentiated fate and exclusion of an alternative differentiated fate.
The examples of such potent regulatory factors with simple circuits of interactions between them have prompted creation of models in which developmental choice is based on a proposed duel between the contending “master regulators” (32–34). This kind of model is very easy to convert into systems of differential equations, which makes such a model appealing across disciplines. The solutions to these systems of equations predict a state space defined by the levels of these two regulators with dynamically stable and unstable nodes resulting from their cross-regulation. Such state space diagrams have a canonical form, and predict that cells must ultimately be driven to a state in which one or the other “master regulator” dominates; the main difference among various versions is how long a cell is likely to persist in the region outside of these extremes. Note that these aspects of the model can be accurate representations of “lockdown” machinery that maintains lineage fidelity in the committed, differentiated states, no matter how the cells arrived at them. Note also that such models incorporate a stochastic element that allows substantial asynchrony between different cells in a population in their transitions to irreversible fates.
However, with such a model in hand, it is often gradually suspected that there is a stage of real development that corresponds to the unstable equilibrium between the two “master regulators”. The model predicts that such a cell type would be perfectly bipotent, and that the real developmental choice is based on the contest between the two factors in the cell to drive it to one of the stable end states. Interestingly, the idea of a perfectly bistable switch imposes on this single-cell decision the binary choice bias that is also common in embryonic systems, where for completely understandable reasons cell fates frequently segregate at mitosis (e.g. because one daughter cell remains near a signal source while the other is separated from it). The idea that metastable precursors exist that express low levels of the regulators of more than one fate has been supported by early evidence of multi-lineage priming in hematopoietic precursors. However, note that the (widespread) assumption that there should be two alternative fates in contention is not precisely required by the biological evidence. From hematopoietic multipotent progenitors to their differentiated descendants, there is no evidence for any particular mitosis at which the two daughter cells are required to adopt different fates, in contrast to the embryonic cases. Furthermore, since mitosis does not play the role here that it does in canonical embryonic cleavages, the only reason the alternatives can be assumed to be only two is because of a choice to include only two “master regulator” transcription factor genes in the model.
Key differences between gene networks for type I embryonic and dueling “master regulator” models
The cardinal features of these two kinds of models have implications for the relationship between gene network architectures and the biological phenomena that the network models seek to explain. Table 1 summarizes points of comparison between them. The features optimized by the type I embryo system are to increase complexity with precise timing and predictable cell-fate assignments. The source of the steadily increasing complexity comes from the combinatorial logic that is the rule, not the exception, for gene expression. That is, these early embryos require each newly expressed regulatory gene to be activated by a new combination of several transcription factors that were already activated in intersecting but different regions of the embryo. The precision of timing comes from a well-defined time zero point (fertilization of a mature oocyte), and from the observation that there are few if any initial silencing mechanisms to be overcome. This means that target genes rapidly respond once the correct combination of trans-acting positive regulators is available in a cell. The precision of fate assignments comes from the potency of negative regulators where they are present. In these embryonic systems, repression generally trumps activation in a noncompetitive way. Spatial patterning-associated repressors abort target gene expression in their domains, even if activators are present at high level (18).
TABLE 1.
Comparison and contrast of different classes of models for developmental fate specification
| Type I models | Dueling regulator models | Hybrid models | |
|---|---|---|---|
| Paradigmatic Examples |
Embryonic development
|
Bacteriophage
|
Hematopoiesis Vertebrate development? |
| Emergent properties | Inexorable state transitions, predictable cell fates | Choice between two stable states, not predictable | Biased outcome but individual cell fates unpredictable |
| Increasing complexity | Resolution of metastable states | Increasing complexity | |
| Transcription factor control | Weak dose-dependence; effectively boolean | Strong dosage dependence in metastable states | Strong dosage dependence |
| Combinatorial action | Two “master regulators” | Combinatorial action | |
| Deterministic | Noise sensitive | Noise sensitive |
This table compares features of two well-studied classes of models for development and contrasts them with the form of model that may be required not only for hematopoietic lineage choice in adult mammals but also possible for vertebrate embryos as well.
Fruit fly (Drosophila melanogaster) does not develop by a classical type I mechanism because of its special syncytial early cleavage stages. However, many features of its rapid and deterministically acting gene network are type I-like
By contrast, the dueling repressor models apply best to more terminal differentiation decisions when two alternative end states are already defined by the reciprocal actions of two alternative transcription factors, and where it does not matter precisely which fate is adopted by which cell. Combinatoriality is explicitly left out of these models in which the two “master” factors are used to account for the whole response. Indeed, the only genes of interest that must respond to more than one regulator in these models are the genes encoding the factors themselves. Unlike the embryonic models, the entire dynamic response of the system is highly dose-dependent as well as noise-sensitive, because of the simplicity of the regulatory inputs. At high levels of the factors, the one with higher effective activity wins all. At low levels of both factors, the system can remain balanced in a bipotent state, but unstable regulator expression accelerates the transition to differentiation in either direction. The ability of the alternative transcription factors to repress their antagonists’ positive targets may be included but need not be absolute, because the two factors will repress each other. Thus, quickly or slowly, the winning factor will starve the alternative factor’s targets of their positive inputs. An often-overlooked corollary of this kind of model is that it is strongly influenced by relative doses of the two factors but becomes dosage-insensitive as each factor approaches dominance. Because winning also allows a factor to increase its own expression, each individual factor must coherently keep driving the developmental system to the same endpoint as its own levels increase.
Key elements of both kinds of models can be found in early T-cell development, but there are sharp divergences from the assumptions of both model types as well.
T cell emergence seen from perspectives of established developmental models
Pathways, stages, and major factors: overview
Hematopoietic differentiation from stem cells in postnatal mammals is a multi-outcome, branched developmental system. Most of the developmental options are implemented in the bone marrow, where there are likely to be different local signaling environments that foster different outcomes, but these are still anatomically somewhat obscure. The thymus is an outstanding exception, since cells that are to become T cells almost always need to immigrate to the thymus from the bone marrow through the blood. The thymus provides a specialized architecture to guide T cell development of the precursors through migration through a sequence of distinct anatomical domains. Because there are very few cells taking non-T cell developmental fates in the thymus, the analysis of thymocytes over the years has provided a very fine-grained picture of T-cell developmental stages, characterized by different surface markers, gene expression patterns, cell cycle behaviors, T-cell receptor (TCR) gene rearrangement statuses, and intrathymic locations (35–37). Because the cells are viable and functional upon transfer to host mice, thymic organ cultures, or stromal co-cultures that mimic the thymic microenvironment, it has also been possible to characterize the different stages of thymocyte development in depth in terms of developmental potential at each stage and developmental kinetics of transition to the next stage.
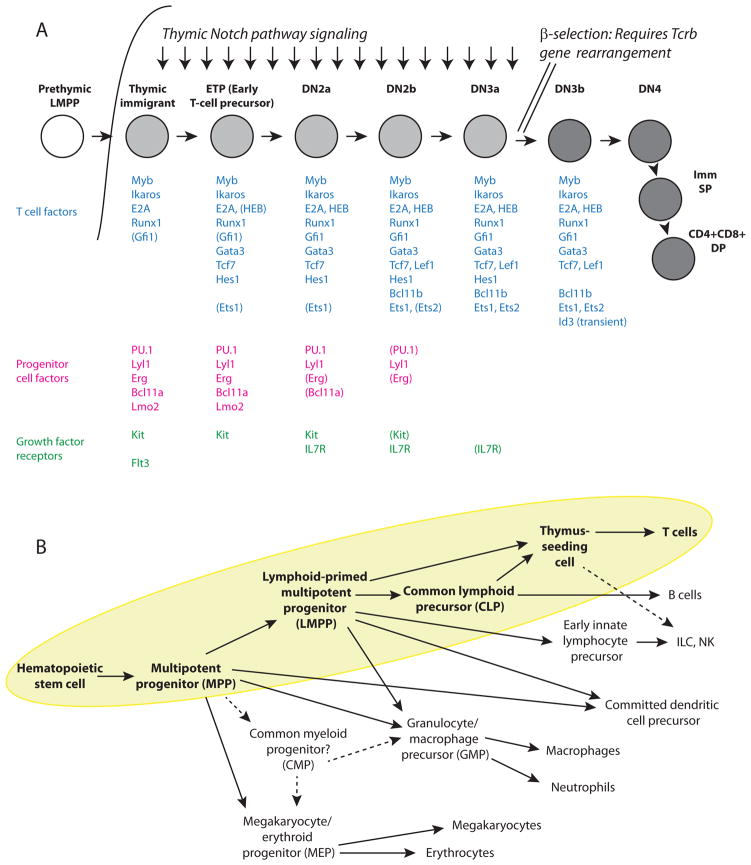
These stages and their relationships are shown in Fig. 1A. Here are defined [i] the early intrathymic precursor “ETP” stage, [ii] the immediately precommitment DN2a stage (DN=double negative for mature T-cell coreceptors CD4 and CD8), [iii] the newly committed DN2b stage, [iv] the DN3a stage that is the first major period of TCR gene rearrangement, [v] the DN3b, DN4, and Immature Single Positive stages when cells proliferate vigorously following the first successful TCR locus gene rearrangement, and finally [vi] the DP (CD4, CD8 double positive) stage when the second TCR locus is rearranged and successful cells for the first time express their final TCR recognition specificity. The DP stage is the point of departure for differentiation all the later effector subsets of T cells using the αβ form of the TCR, and considerable work is dedicated to understanding the mechanisms involved in those branching pathways. That later set of lineage specializations is outside the scope of this review. However, the identity of the cells as T cells of one kind or another is set by the end of DN3 stage (Fig. 1A).
Figure 1.
Overview of mouse T cell development. (A) Stages of T cell development and regulatory genes active at different stages. Upper schematic: progression of stages from thymic entry to completion of TCR rearrangement in DP stage. For cell-surface markers used to define stages, see recent reviews (35, 36). The β-selection transition, in which DN3a cells pass quickly through the DN3b, DN4, and immature single positive (Imm SP) stages before arriving at the DP stage, is only triggered in cells that have successfully rearranged their Tcrb gene loci to express an in-frame TCRβ protein. To focus on steps leading to confirmation of T-cell identity, the figure omits alternative branches of T-cell development leading the TCRγδ cells and the positive and negative selection events that lead to functional sub-specialization after successful rearrangement of the Tcra locus to express TCRαβ dimers in the DP stage. Transcription factors and growth factor receptors listed are chosen as those known to play functional roles in aspects of normal or leukemic T cell development (see (35, 36)). Gene names in parentheses denote expression at at lower levels or lower activity states than in stages where they are listed without parentheses. (B) Relationship of T-cell precursors to other hematopoietic fates. The figure presents a compromise view of the main inputs to the T-cell pathway relative to MPPs, LMPPs, and myeloid or erythroid-committed cell types. The yellow background and boldface type highlight stages that are in the pathway to generate T cells. For simplicity, pathways leading to eosinophils, basophils, and mast cells and the complexities of precursor relationships in the Innate Lymphoid Cell (ILC)—Natural Killer (NK) and Dendritic cell (DC) pathways are omitted. As described in the text, the roles of certain restricted progenitors in the erythromyeloid lineages are still controversial (dashed arrows). The main pathways indicated here are supported by recent single-cell analyses (55, 56).
In postnatal mice, the whole T-cell pathway emanates from a multipotent precursor that still has the ability to give rise to natural killer cells, at least one or two additional types of innate lymphoid cells, B cells, dendritic cells, granulocytes, and macrophages, and to some extent also mast cells. The relationships of cells with this combination of potentials to other hematopoietic cells are shown in Fig. 1B. The thymus-settling precursors are closest to the Lymphoid-biased Multipotent Precursors and the Common Lymphoid Precursors, i.e. uncommitted intermediates with a strong preference for lymphoid fates in vivo but a latent capacity to adopt myeloid fates in vitro. These adult progenitor cells are thought to have lost erythroid and megakaryocytic potential at an earlier stage. As noted above, it is still controversial whether other precursors may exist that maintain erythroid and granulocyte/macrophage potential but lack lymphoid potential; cells with these features are most evident in early fetal liver before the emergence of true hematopoietic stem cells. But the connection of lymphoid and myeloid fates is the one that needs to be resolved for T cell development.
Key regulatory factors that are known to be differentially important in these pathways are introduced in Fig. 1A and discussed further below. Importantly, the thymus provides a clear extrinsic signal to trigger T cell development, activating the Notch pathway by interaction of the Notch ligand Delta-like 4 on the thymic stroma with Notch1 molecules on the surfaces of the immigrant precursors and their immediate descendants. Although Notch pathway activation is not unique to the thymus, the thymus provides a particularly strong Notch signal in the context of cytokines that support lymphocyte proliferation and developmental progression. Notch signaling creates a specification mechanism that converts the cells from multipotency to committed pro-T cells, and then the committed state becomes Notch-independent.
Explanation of T cell specification by a “Type I embryo” model: combinatorial definition of identity
To think about a stem-cell differentiation process using terms developed for embryonic differentiation, it is convenient to consider lineage choice to be the equivalent of spatial demarcation of distinct embryonic territories. Thus, the identity of the cells like the identity of the embryonic territory depends on the overlap between positive regulatory inputs to the cell type gene network, and the exclusion of negative regulatory inputs. T cell differentiation from ETP stage to DP stage depends on a collection of disparate transcription factors, each with its own expression profile. Myb, Runx1 with its partner CBFβ, and Ikaros combine with GATA-3, TCF-1 (encoded by Tcf7), Gfi1, Bcl11b, some representation of Ets1-subfamily ETS factors (“E26 transformation-specific”, a large family with many members expressed), and strong, unencumbered activity of E protein dimers, either E2A-E2A (encoded by Tcf3) or E2A-HEB (encoded by Tcf12). The factors that need to be excluded are EBF1 and Pax5, which would otherwise divert the cells to B-cell lineage; antagonists of E protein dimers like Id2, which would promote an innate lymphoid type fate; and C/EBP factors and PU.1, which would otherwise promote neutrophilic granulocyte or macrophage fates (C/EBP factors and PU.1) and dendritic-cell fates (PU.1). Other factors that probably need to be excluded are Irf8, a key participant in macrophage, B-cell, and dendritic-cell development; factors involved in erythroid and megakaryocytic development; and factors that would otherwise promote self-renewal in a multipotent state. Thus, the prediction would be that if the right combination of factors could be established in a hematopoietic precursor, it would become a T cell.
In reality, the triggering and execution of the T-cell developmental program depend on Notch pathway signaling. This is needed directly to activate Tcf7 at least, and probably also to help activate Gata3 and Bcl11b, to optimize the levels of Runx1 expression, and to induce repressors like Hes1 that silence Cebpa. However, this signal is not a stable component of the regulatory state mix. Notch responsiveness cuts off before the DP stage, and once induced, most of the regulatory genes that contribute positively to the T-cell program sustain maximal levels of expression even when Notch signaling is withdrawn. This can be viewed as a classic case of the kind of embryonic circuitry that “transforms transient spatial regulatory inputs into stable regulatory state” (2, 38). The conversion from Notch-dependence to Notch-independence in this kind of model would arise from the positive regulation that the Notch-induced regulatory genes provide for each other which collectively renders the Notch input dispensable.
There are two large questions in using this kind of formalism to explain the T-cell developmental process. First is to explain how the thymic microenvironment can cause the right combination of factors to be expressed in the hematopoietic progenitor cells with other factors excluded. Lacking organized spatial axes along which different signaling cues can be provided in complex anatomical crossing patterns, the thymic stroma mainly seems to provide the single, repeated Notch signal. Thus, the developing T-cell progenitor needs to respond to Notch with its own intracellular cascade of successive, new regulatory states to enable this simple input to affect the levels of so many regulatory genes, considering that these genes otherwise have disparate expression patterns. The second question is whether the predicted combination of transcription factors really is necessary, or really is sufficient, to create a T-cell identity. Is it the simple presence of the right combination of factors, or the cell’s achieving the right relative levels of this combination of factors, or the order in which the right combination of factors is induced to work, that generates T-cell identity? The identifications of a collection of required factors and a collection of factors to be excluded still leave some distance to the answers to these questions.
Explanation of T cell specification by a “dueling master regulators” model: defining the sequence of binary choices
In a dueling regulator model each choice is seen as binary, and hematopoietic stem cells generate far more than simply two cell types, as described in more detail below. Therefore, by this kind of model the T-cell pathway must emerge from a succession of binary choices, each governed by a different regulatory antagonist pair. As discussed in more detail in the next section, this view fits well with the classic hematopoietic lineage diagrams that dominated the literature until about 2005 (39), where the hematopoietic stem cell would first commit to a lymphoid vs. erythromyeloid fate (choice 1), followed by a subdivision of B from T and NK and/or a subdivision of NK from B and T (choice 2), and then followed by the separation of T cell fate from either an NK or a B cell alternative (choice 3). Crucial antagonist pairs in this scheme could include Ikaros as the pro-lymphoid factor with E proteins vs. Id2 in the (B or T) vs. NK choice, and GATA-3 vs. EBF1 in the T vs. B choice. A modification of the model would still work with the revised lineage diagrams more widely accepted from 2005 onward that first separate erythroid/megakaryocyte precursors (GATA-1+) away from a lymphomyeloid precursor (PU.1+)(40–44). Only then would lymphoid progeny be separated from myeloid progeny of the lymphomyeloid precursor, by antagonism of “myeloid” C/EBPα by “lymphoid” EBF1 in the B cell case, and by GATA-3 and Runx-mediated silencing of PU.1 in the T cell case.
By a dueling regulator model, the correct analysis of the circuitry of T-lineage choice depends on the agonist-antagonist regulator pair at the heart of the mechanism. Thus, to explain T cell specification by such a model, the central issue is to define the most relevant alternative to T-cell differentiation in the cell immediately before it commits to the T cell fate. The “master regulator” duel itself can then be validated by showing that the agonist and antagonist transcription factors actually coexist in the cells before they choose between these options. Proving whether the system really is bistable at that point then depends on showing that the factors engaged in the struggle actually do regulate themselves positively, do antagonize each other’s expression, and do each promote one of the outcomes in the balance in a monotonically increasing way.
A closer look at the details of T cell specification in the context of hematopoietic differentiation suggests that both kinds of models incorporate useful elements, but that the real program uses a distinctive hybrid kind of mechanism.
Hematopoiesis: a distinctive terrain for gene networks
A multidimensional landscape of hematopoietic transcription factors
One way to reduce a complex developmental landscape to a form in which dueling master regulator models can work is to posit that all fates are generated by a hierarchical series of binary decisions. Indeed, this has been a classic theme in hematopoiesis research. As already noted, hematopoiesis was long assumed to begin with a split between erythromyeloid and lymphoid fates, followed by splits between erythroid and myeloid cells and splits between B and T lymphocytes, then finally myeloid cells were supposed to split between granulocyte and macrophage fates (39). Dueling master regulator models scored key successes in this binary choice hierarchy, as the divergence between erythroid and megakaryocytic fates on the one hand and macrophage and granulocytic fates on the other hand (here termed “myeloid fates”) could be explained in terms of GATA-1 and PU.1 (32, 45–48). Then, also, granulocyte vs. macrophage divergence could be explained in terms of relative expression of C/EBPα and PU.1 (49), using an additional continuous-to-discrete conversion subcircuit of Gfi1 in opposition to Egr1, Egr2 and Nab2 (33).
The founding model of this kind was the PU.1-GATA-1 relationship, which is thought to play a key role in the separation between myeloid and erythro-megakaryocytic lineages. Both PU.1 and GATA-1 are indeed potent positive regulators of these respective fates, and they both have mechanisms for positive autoregulation and for mutual repression at the transcriptional level plus protein-protein antagonism. However, through more than 15 years of intense investigation, it remained controversial whether there is any true common (erythro-)myeloid precursor in which these factors compete on a level playing field for a winner-take-all outcome (40, 43, 50–54). Evidence for biased, GATA-1-dominant or PU.1-dominant precursors is ample, and in the past year single-cell transcriptome analyses coupled with single-cell fate determination methods have showed that in the great majority of these supposed bipotent precursors the contest has already been won by one or the other (55–57). Then in the PU.1-expressing multipotent cells, at least, there is a variety of possible lymphoid and myeloid developmental options that depend on more than simply PU.1 itself (58, 59). Evidence from zebrafish embryos shows that the mutual antagonism of PU.1 and GATA-1 is not hard-wired but actually depends on a transcriptional modulating factor, Trim33 (Tif-1γ)(60). In mammalian hematopoiesis also, it has been suggested that the mutual antagonism is not a simple function of PU.1 and GATA-1 themselves, but occurs primarily when C/EBPα and FOG-1 (Zfpm1) provide “henchman” duties (61). In agreement with this, multipotent hematopoietic precursors also generate mast cells, the highly robust and sustainable regulatory state of which excludes FOG-1 and C/EBP factors but requires stable expression of PU.1 together with GATA-1 and GATA-2 (62–65). Thus, factor-factor reciprocal antagonism within network circuits is a valuable and potent mechanism for driving cells to different end states, but in reality more than two factors may be involved, and thus the decisions may involve more than two potential cell fates.
The ability to use these simple circuits to explain hematopoietic development has been further challenged as increasing evidence showed most lymphoid cells to branch off from myeloid cell precursors some time after erythroid potential had been lost or minimized (40, 41, 52). This meant that the actual GATA-1-PU.1 choice was between erythro-megakaryocytic fates and a mixture of lymphoid and myeloid potential. Thus a new binary choice needed to be postulated to explain lymphoid vs. myeloid divergence. One switch-determining factor appeared to be C/EBPα, which would be retained in myeloid lineages but silenced in B and T cell lineages, and in an alternative model Ikaros, E proteins, and Gfi1 were all postulated to work against PU.1 to confer lymphoid identity (66). However, neither Ikaros, E proteins, nor Gfi1 are unique to lymphocytes, none of them directly repress C/EBPα (which activates Gfi1 in the macrophage-granulocyte model), and the complexity of the circuitry required opened new questions. In fact, although C/EBPα is repressed in both B and T cell precursors, it is not repressed by a common pan-lymphoid mechanism: EBF1 and/or Pax5 silence it in B cells (67, 68), whereas Notch signaling via Hes1 silences it in T cells (69). Therefore, even the lymphoid-myeloid split needs to be modeled as more than a binary choice (discussed further below).
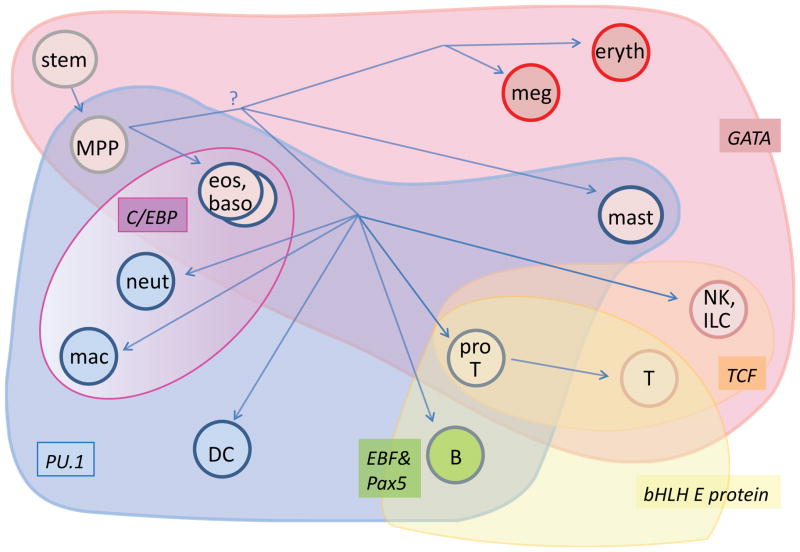
Most importantly, expression patterns of candidate “master regulators” themselves do not fall into the neat hierarchical system of binary exclusions that would be required by a system-wide use of a dueling regulator model (Fig. 2). A multidimensional picture gives a clearer representation of the domains of different transcription factors’ actions in hematopoiesis. In Fig. 2, this is approximated by overlapping colored contours that are superimposed on a putative lineage tree leading to developmental end states.
Figure 2.
Broadly combinatorial use of key transcription factors in specifying distinct hematopoietic cell fates. A schematic view is presented of mouse hematopoiesis, superimposed by the overlapping domains of activity of PU.1 (blue), GATA family factors (GATA-1, GATA-2, or GATA-3, red), C/EBP family factors (violet, graded to indicate dosage relative to PU.1), E proteins (E2A and/or HEB, yellow), TCF family factors (TCF-1/LEF-1, orange), and EBF1+Pax5 (B lineage specific, green). Whereas C/EBP family factors do not appear to overlap with the domains of E proteins and TCF-1/LEF-1, and the EBF1+Pax5 combination is restricted to a single cell type, there is extensive variation in the combinations of GATA, PU.1, C/EBP, E proteins, and TCF-1/LEF-1 that are allowed to distinguish the other cell fates shown. Eryth=erythrocyte. Meg=megakaryocyte. Eos=eosinophil. Baso=basophil. Neut=neutrophil. Mac=macrophage. DC=dendritic cell. ILC=innate lymphoid cell. NK=natural killer. ProT= T-cell precursor up to commitment (during period of PU.1 expression).
One glaring exception to the GATA-1-PU.1 antagonism, just noted, is the existence of mast cells which depend upon sustained activity of both factors, albeit with lower PU.1 levels than in mature macrophages. Dueling regulator models predict that cells can exist with low levels of both antagonists, but they predict that such cells should be developmentally unstable. By contrast, mast cells are not only stable but also capable of maintaining their phenotype through many generations of proliferative expansion. Also relevant are the classes of granulocytes that use GATA-1 and/or GATA2 along with PU.1 and high levels of C/EBP factors, such as eosinophils and basophils (64, 70, 71)(Fig. 2). If the separation between erythroid cells and myeloid cells involves a decisive choice between silencing GATA-1 and silencing PU.1 and/or C/EBPα, then the origin of these cells is obviously problematic – do they emerge before or after the split, or do they have some special mechanism to reactivate the silenced master regulators in the presence of the others? The potential compatibility between GATA factors and myeloid factors is strikingly underlined by results of manipulating developmentally arrested mutant pro-B cells. Here, despite the lymphoid-biased context, GATA factor introduction not only fails to protect the cells from direction to myeloid fates, but actually promotes conversion to myeloid fates, acting similarly to C/EBPα (72).
The domains of action of these transcription factors obviously intersect each other in different combinations across the diagram. It is also evident that the biological system exploits the freedom of this combinatoriality, often violating hierarchy, to specify multiple distinct cell types with a minimal number of different factors in play. This requires a network with greater complexity in its drivers than a simple succession of “master-regulator” duels.
Lineage-specific ordering of developmental fate exclusions
Cell fate choices may depend on cross-regulation between different potent transcription factors, but again the fate decisions do not fall into a strict hierarchy. Indeed, it appears that cells expressing C/EBP factors have access to fates that depend on this factor, such as neutrophil, macrophage, basophil or eosinophil fates, while those that express GATA-1 have access to erythroid, megakaryocytic, eosinophil, basophil, or mast cell fates. Lymphocyte precursors express none of these. However, the order in which lymphocyte fates subdivide as compared to the order in which they separate from macrophage, granulocyte, and dendritic cell fates is surprisingly dependent on the fate chosen.
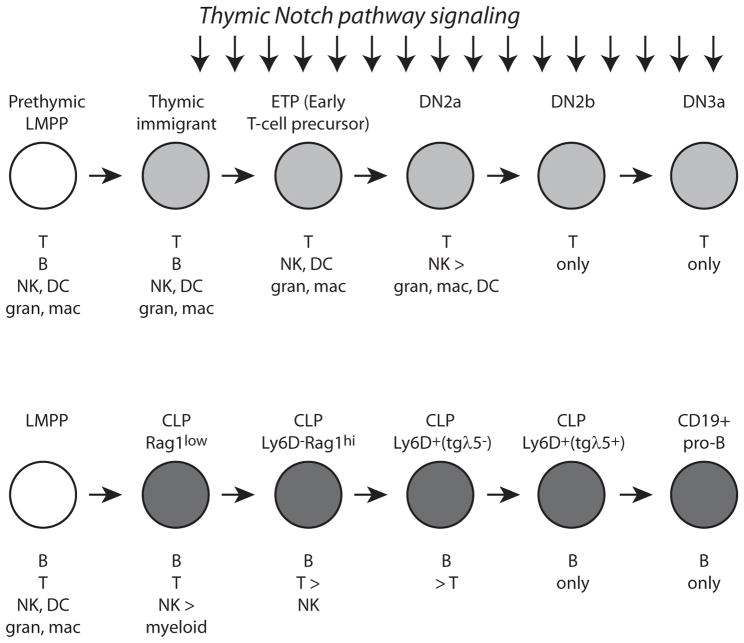
Precursors of both B and T cells have been examined in detail by flow cytometric separation followed by single-cell assays of developmental potential in broadly permissive conditions in vitro, as well as by transfer to adoptive hosts in vivo (Fig. 3). In the B cell pathway, myeloid potential is excluded quite early, whereas for another stage or two B cell precursors still appear to be able to give rise to T cells if exposed to inducing Notch ligands (73–75). Thus, from the B lineage vantage point, myeloid potential is apparently lost before the bifurcation of the B and T-cell branches. However, in the T-cell lineage, this order is reversed. Access to at least some myeloid fates continues for multiple cell divisions after entry into the thymus, and through the ETP to DN2 transition: DN2a pro-T cells can be trans-differentiated into macrophages, granulocytes, or dendritic cells simply by changing the culture conditions (44, 76–83). However, T-cell precursors lose B cell potential much earlier, within a very short time after arriving in the thymus, and despite the cells’ still retaining obvious myeloid potential (79, 80, 84–87). From the T-cell precursor perspective, then, lymphoid lineage bifurcation between B and T cells becomes cell-intrinsic even before the split between lymphoid and myeloid fates (Fig. 3).
Figure 3.
Contrast between the timings of myeloid exclusion relative to alternative lymphoid fate exclusion in the lineages leading to T cells and B cells. Top: developmental potentials demonstrable in T-cell precursors at different stages. Bottom: developmental potentials demonstrable in B-cell precursors at different stages. Figure focuses on data from defined in vitro systems where the clone sizes and cloning frequencies can be measured, rather than in vivo systems where migration bias and differential proliferation may also contribute to outcomes. In each case, to demonstrate non-T (top) or non-B (bottom) potential, cells are transferred to conditions that selectively favor manifestation of these alternative options. Data for T cell developmental alternatives are from refs. (76, 78–81, 94, 98). Data for B cell developmental alternatives are from refs. (73, 75, 178, 179). Not shown are potentials for mast cell and innate lymphoid cell type 2 development, which have also been seen in DN2 T-cell precursors (132, 180).
The discrepancy is not a contradiction; it simply shows that exclusion of a given fate alternative (a repressive mechanism) is carried out by a different mechanism than initiation of progress toward another fate alternative (an activating mechanism). B cells block the T-cell pathway only after they begin to express their own distinctive regulators, EBF1 and Pax5, which inhibit Gata3, Tcf7, and even the expression of the Notch1 signal receptor that would be required to activate either one. To exclude the myeloid fates, different mechanisms are used in most T cell precursors, which can simply turn off PU.1, than in B cell precursors, which must exclude myeloid development despite continuing to express PU.1. (It is interesting to speculate whether any of the rare pro-B cells that can be converted to T-cell precursors in vitro would transiently regain access to dendritic-cell or myeloid pathways in the process, but this would be difficult to test.) Most cells have to start down either a Notch-influenced T-cell pathway or a Notch-averse B cell pathway before it is clear which tool they can use to jettison their heritage of myeloid potential.
In general, a cell can remain in a single lineage path due to a combination of intrinsic and extrinsic factors, so that the dominant role of Notch signaling in T-cell development and the differential expression of myeloid-potentiating factor PU.1 gives B and T cells different options (Fig. 4). B and myeloid cells share the strong regulator PU.1 throughout B cell maturation, but require different dosages of PU.1 for their differentiation (88, 89). B cells sharply reduce PU.1 expression levels at the beginning of their development from multipotent precursors (58), possibly even preceding their expression of EBF1 and Pax5. Cells becoming pro-B cells from fetal liver appear to escape the mechanism that allows PU.1 to regulate itself positively (90–92), and developing B cell precursors can also dilute out higher levels of PU.1 from their precursors by maintaining a rapid rate of proliferation throughout specification: this is important because the PU.1 protein itself is very stable(58). It is known that different PU.1 regulatory sites are open in a pre B cell line than in a macrophage cell line or mature macrophages (91). It has been argued that a key negative regulator of PU.1 is Ikaros (66), although this is not consistent with all progenitor-cell gene expression data (93). Indeed, in B lineage cells Ikaros acts to repress PU.1 although in myeloid cells it can cooperate in upregulating PU.1 (90). PU.1-driven myeloid development might still pose a threat to B cell lineage fidelity until C/EBPα is fully silenced by EBF1 and Pax5, but the downregulation of PU.1 is intrinsically controlled. This could help to reduce detectable myeloid potential in cell transfer assays from an early stage.
Figure 4.
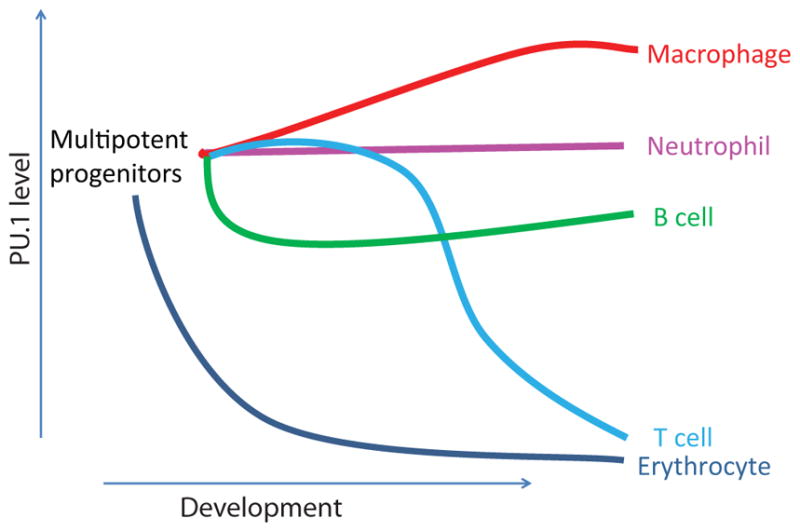
Dynamic control of PU.1 activity levels in different hematopoietic lineages. Schematic of relative protein or RNA levels at different stages of development in five different pathways of hematopoietic development. For B, neutrophil and macrophage data, see (58). Although both T cells and erythroid cells silence PU.1 expression, PU.1 is specifically required for the T-cell developmental pathway to initiate, whereas it is not required for erythroid cell generation (134).
By contrast, T cells cannot be specified without Notch signaling, so that in the microenvironment where B cells normally develop, an intrinsic blockade of T-cell potential may be optional until much later. The chromatin status of the earliest B-cell and T-cell signature regulatory genes is also notably asymmetric in the early precursors of the opposite lineages. In E2A−/− or EBF1−/− pre-pro B cells, the Gata3 and Tcf7 loci are in an accessible chromatin state similar to their states in ETP pro-T cells, whereas in early T-cell precursors, both the Ebf1 and the Pax5 genes are maintained in chromatin states repressed by Polycomb Repressive Complex 2 (H3K27me3).
Developing T cells initially express a considerable level of PU.1, but silence it completely during lineage commitment (78, 94, 95), approximately 10–14 cell divisions after entering the thymus (96, 97). Thus these cells have only a short time in which myeloid potential poses a threat to lineage fidelity. The thymic microenvironment is nonpermissive for expression of this myeloid potential. First, there is no myeloid-supporting growth factor present, and second, the extrinsic Notch signals in the thymus not only push forward the T-cell program but also restrain PU.1 from implementing the myeloid or dendritic cell program effectively in vivo (98). One aspect of the mechanism is that the Notch-activated factor Hes1 represses C/EBPα expression (69), but Notch signaling also affects the spectrum of genes regulated by PU.1 in the absence of C/EBPα (99). The persistent myeloid potential is only visible if the cells are removed from contact with Notch ligand in an experimental commitment test during these sensitive stages (99–101).
Whereas myeloid alternatives are only a transient threat for developing T cells, T cell and B cell precursors share their use of Ikaros, Myb, Gfi1, Runx factors, and especially their use of E proteins and IL-7/IL-7 receptors as major regulators and proliferation signals throughout their development. The thymus can be a very favorable site for B cell development except where cells can receive Notch signals by direct contact with Notch ligands: Notch1−/− precursors generate B cells abundantly in the thymus (102). Thus, when B-cell potential is blocked early in mammalian T-cell precursors (86, 103, 104), it is tempting to speculate that this is not only to maintain lineage fidelity but also to make sure T-cell precursors do not need to compete for the local supply of IL-7 (105–107). Imposing this block intrinsically is seen to be one of the earliest events in T-cell development, and it is mediated at least in part by the Notch-dependent induction of GATA-3 which then maintains B lineage exclusion even when Notch signals are withdrawn (84, 85). The reason B and T cells need to exclude different fate options in this piecemeal way is precisely because their precursors do not first pass through a binary choice at which all nonlymphoid fates are excluded.
Dose dependence and diverse mechanisms of dosage control
The definition of cell type by various mixtures of overlapping transcription factors is more consistent with the multiple-input combinatorial mechanisms of territorial patterning in the “type I embryo” models than with the binary fate subdivisions that would emerge from “dueling master regulator” models. However, one aspect of the hematopoietic system stands out in contrast to the models used to explain boundary formation in embryos. Transcription factors have highly dose-sensitive effects in hematopoietic fate choice; they do not work in a simple digital way. The preceding section referred to the need for B cells to keep PU.1 expression at much lower levels than those needed for this same factor in myeloid development. Lineage-specific and stage-specific dose restrictions for a given factor in different contexts are not exceptional but common in mammalian postnatal hematopoiesis. In marked contrast to the model systems in which embryonic development has been dissected genetically, in mammalian hematopoiesis transcription factor activities are frequently haplo-insufficient, with altered development resulting from even a small twofold change in expression. Stem cell establishment and self-renewal are sensitive to heterozygosity in Runx1 (108, 109) and Gata2 (110), all dendritic cell development and choices between macrophage vs. granulocyte development are sensitive to PU.1 heterozygosity(49, 111), generation and stability of B cells are sensitive to heterozygosity in EBF1 particularly when combined with heterozygosity in E2A, Runx1, or Pax5 (112–114), and early T cell development is sensitive to twofold dosage reductions in GATA-3 (84, 115, 116).
This dose dependence implies, first, that positive regulation of cell lineage features (and survival) depends on transcription factor binding to non-optimal sites at key loci, where binding is not saturated easily; and second, that the ability of one transcription factor to repress the major regulator of a competing program is likewise graded and incomplete. A clear example is the effect of modest reduction in GATA-3 expression in the early T cell precursors that coexpress GATA-3 with PU.1: reduced GATA-3 expression leads quickly to upregulation of PU.1 levels, implying that GATA-3 is already damping down PU.1 expression but in a weak, concentration-sensitive way. These weak cross-regulatory effects and the dose-dependence that pervades the system are features that are not common in deterministic embryonic systems, although they are accommodated in the dueling master regulator models within the range of expression levels where the ultimate winner is not yet decided.
Dosage differences can result from many mechanisms besides heterozygosity for a functional gene, or changes in the activity of an upstream regulator. A recent addition to the list of mechanisms for graded dosage control came from analysis of the dynamics of PU.1 expression in multipotent progenitors (58), a population which in principle contains the precursors of macrophages, granulocytes, B cells and T cells, all of which will go on to regulate PU.1 in clearly divergent patterns (Fig. 4). These cells initially express high levels of PU.1, but as they differentiate, two different regulatory mechanisms combine to drive PU.1 levels higher or lower in different progeny types. One is the maintenance or termination of PU.1 autoregulation. The myeloid descendants of the progenitors maintain positive autoregulation of PU.1 (Spi1 gene) expression mediated through at least three different site clusters in the Spi1 cis-regulatory system (91, 117), and their absolute rates of Spi1 transcription are constant at a high level. The B lymphoid descendants, on the other hand, lose sensitivity to autoregulation and reduce their own absolute rates of Spi1 transcription by nearly an order of magnitude. (Introduction of exogenous PU.1 does not appear to upregulate transcription from the endogenous locus in these cells, although this is hard to prove because many developing B cells are killed by excess PU.1.) Another mechanism is needed, however, to increase PU.1 activity from the level in the precursors to the level needed for macrophage maturation, and the surprise is that this mechanism is not transcriptional. Instead, it depends on the fact that PU.1 protein in these cells is extremely stable, to the extent that a major contributor to its turnover function in a cell is not actual decay but rather the dilution caused by cell division. Because of this stability, PU.1 levels can be increased in cells that maintain a constant rate of PU.1 synthesis simply by slowing the cell cycle (58). In the cells becoming macrophages, PU.1 itself promotes a cell cycle lengthening, in part by reducing the expression of Myc and Myb. However, this mechanism can be manipulated experimentally, with the same cells adopting macrophage or non-macrophage fates depending upon whether they are allowed or encouraged to slow their cell cycles or not (58).
Making cell cycle control a regulatory input as well as a simple effect of other factors is an important change of perspective. Interestingly, cell cycle control as an input to developmental decisions is reminiscent of the highly regulated cell cycle timings and geometries of embryos, but in the hematopoietic case it is being used in a cell autonomous, geometrically random, stochastic way very different from the rules in embryos. An important corollary of this mechanism is that the ratio of long-lived factors like PU.1 to short-lived factors such as C/EBPα in the cell nucleus should be extremely sensitive to cell cycle length changes, without change in their transcription rates, because the relative contributions of mitotic dilution to their protein steady state accumulations should be so different for the two proteins. Thus any third factor that affected cell division rates could dominantly alter the balance between two other factors, affecting cell fate in a dose-sensitive system like hematopoiesis, even when their synthesis rates remain the same. Furthermore, as single-cell RNA-seq analysis has been used to dissect stem and progenitor cell populations, it has become increasingly clear that differential expression of cell cycle genes is a major criterion of functional distinction (118). The role of cell cycle length control in developmental choice may help to distinguish different kinds of stem cells too.
Cytokine dependence and cell fate: cell cycle arrest and death for commitment and boundary formation
Cell cycle control in hematopoiesis is clearly dependent on microenvironmental conditions, especially the availability of appropriate cytokines. Thus the ability to read out any of the gene network control circuitry that is influenced by cell cycle length and continuation will perforce be microenvironment-dependent. A fundamental aspect of differentiation along each hematopoietic pathway is the activation of distinctive lineage-specific cytokine receptors that will become essential for progenitor proliferation and survival along that pathway. It is well understood that the potentials of different progenitors will not be read out accurately if some of the key growth factors needed by certain classes of descendants are not included in the assay conditions, because those descendants will not expand or survive. However, the ability to proliferate also enables cells to shift from one regulatory state to another by diluting away prior transcription factors and prior chromatin marks. In a key transition of T-cell development, β-selection, the clearance of immature characteristics only occurs normally if the cells are able to undergo a full extent of proliferation (usually >5 cell cycles)(119, 120). By the same token, when cell division stops, it enables terminal states to be stabilized by reducing chromatin state changes and allowing transcriptional regulators like PU.1 to accumulate.
Cytokine dependence does more than control the extent of clonal expansion of precursors. It is also used in hematopoiesis as a mechanism for enforcing lineage fidelity of a cell’s gene expression pattern. One aspect of this is the positive feedback circuitry connecting cytokine receptors to the transcription factors important to promote the receptors’ expression. An elegant example is the ability of PU.1 to drive the expression of two growth factor receptors, GM-CSF receptor (Csf2ra/Csf2rb) and M-CSF receptor (Csf1r), which upon ligand binding transduce signals that feed back to stimulate transcription of Spi1 (PU.1 coding gene)(121, 122). Another example, from B cell development, is the ability of IL-7R dependent signals to enhance expression of EBF1, which then reciprocally helps to sustain IL-7R expression (123–127)(although also see (128)).
But cytokine receptor dependence does not simply enhance growth: it can also set the boundary between regulatory states of permitted cell types, by creating a no-man’s land between them. Interestingly, when a cell is shifting from one developmental state to another, it not only gains responsiveness to a new cytokine, but also can lose responsiveness to the cytokine that had been supporting it till then. Typical cases are the losses of sensitivity to Kit/Kit ligand (Stem Cell Factor) signaling during lineage commitment in both early T cells and early erythroid cells (78, 129, 130). The stringency of this regulation is shown in experiments where a developing cell is perturbed by the forced expression of a transcription factor used in another lineage. If the growth factor environment is unchanged, then very commonly there is severe mortality in the transduced cells (100, 131, 132). This can occur within a day or two, even before the cells lose expression of their previously supportive growth factor receptors from the cell surface, and the death can substantially mask any transdifferentiation effects of the introduced factor. However, many of the transdifferentiating cells can be rescued simply by shifting the cells quickly into a different growth factor environment matching the one in which the introduced factor usually works (101), or more neutrally, by adding a Bcl2 transgene without changing the growth factor environment (95, 99, 100, 132). Then transdifferentiating cells with inter-lineage phenotypes at the single-cell level become readily detectable.
The implication is that in vivo, the integration of cytokine receptor functional pathways into particular cell lineage programs act as a quality control to make any improperly specified cells nonviable. Generation of cells with abnormal cytokine signaling behavior is a sensitive indicator of an abnormal regulatory state, and may immediately condemn the cell to die, unless its microenvironment dramatically changes. Note that this process can be seen as the equivalent of an auxiliary cell-type boundary formation mechanism. If cells with defective fate specification can simply be killed by their loss of response to survival promoting cytokines, there is no need to make the specification process itself perfect.
Thus, cytokine dependence as an integral aspect of fate determination enables the system to dispense with the need for the sharp, dominant repression machinery of embryonic systems. It opens the ability to use damping repression primarily, which tunes regulator levels quantitatively. This makes possible transcription factor dosage control to specify an increasing diversity of cell types, while allowing the system as a whole still to generate strikingly efficient and coherent differentiation decisions.
T-cell specification: something old, something new, something borrowed
An inheritance of multipotent precursor transcription factors in early T cells
The migration of T-cell precursors to the thymus before commitment means that the most immature thymocytes at any given time provide a clear early view of cells that are fated to become T cells eventually, long before they express many T-cell genes yet. This opportunity is rare as compared to precursors of other hematopoietic cell lineages, mixed together in the bone marrow or fetal liver, which often cannot be recognized as such until after they begin to express a distinctive lineage-specific gene expression program. In T cell precursors, perhaps uniquely, it is possible to distinguish the initiation of the developmental program from its culmination.
Molecular and functional characterization of T-cell precursors has shown that early T-cell precursors at ETP and DN2a stages maintain expression of a variety of stem and progenitor-associated regulatory genes. Some of the factors encoded by these genes remain essential parts of the T-cell gene network and are either sustained or upregulated, e.g. Myb, Runx1, Ikaros, and E2A. Others of these factors are expressed, apparently through as many as ten cell cycles, and then silenced as the cells commit intrinsically to a T-cell fate (36, 96). This early-expressed set of factors has been labeled “phase 1 regulators” and is enriched for transcription factors that would cause acute lymphoblastic leukemia if not repressed efficiently, such as SCL (Tal1), Lyl1, Lmo2, Mef2c, Bcl11a, Hhex, Erg, and Mycn, and the growth factor receptors Kit and Flt3. The myeloid-promoting factor PU.1 and the erythroid-linked factor Gfi1b are also expressed like Phase 1 genes, with robust expression in early T cell precursors, although less directly associated with leukemia. PU.1, like Bcl11a and several of the other phase 1 factors, is a clear holdover from the lymphoid-biased multipotent precursors found in the bone marrow (43, 133, 134).
This load of prior regulatory apparatus might be assumed to promote self-renewal with continuation of the multipotent state. However, it is flexible enough to permit induction of a new program, the T-cell development program, under the influence of Notch signaling and IL-7/IL-7R signaling in the thymic microenvironment.
Understanding the role of the phase 1 factors is still incomplete, but several examples are now in hand. The factors implicated in T-cell leukemia are thought to be involved normally in some of the initial proliferative expansion that the ETP and early DN2a cells undergo before lineage commitment, even though the exact target genes are not identified. The proto-oncogene Lyl1 also plays a needed positive role in activating expression of at least one other important T-cell gene, the zinc finger factor Gfi1 (135). Our own evidence focuses on PU.l, which is not only a myeloid “master regulator” but also a factor important to maintain survival, lineage identity, and growth factor receptor expression in multipotent lymphomyeloid precursors, macrophage precursors, and dendritic cells.
In early T cells, PU.1 is important to arm the cells for optimal proliferation, and PU.1 protein binds to a large fraction of active regulatory sequences in the cells, but its roles are complex (136, 137). First, in a muted version of the binary competition model, PU.1 dampens the intensity of Notch pathway signals, while at the same time the presence of continuing Notch pathway stimulation restrains PU.1 from activating a myeloid program (99). The “soft” mutual antagonism between PU.1 and Notch signaling is still being dissected in terms of mechanism, but it has the consequence that when PU.1 is finally silenced during commitment, one of the systemic impacts is an increase in most Notch target gene expression (136). Second, PU.1 drives expression of a fascinating set of cell biology genes likely to be involved in chemokine response, G protein coupled receptor signaling, and cytoskeletal dynamics, which may be important specifically in the early intrathymic migrations of the precursor cells. Third, at the same time, PU.1 cross-regulates other phase 1 transcription factor genes, sustaining some (Bcl11a, Lmo2, Mef2c) and damping down expression of others (Erg, Mycn, Gfi1b, Kit) (99, 136). Finally, it is possible that PU.1 in the precommitment stages of T-cell development acts as a pioneer factor to open certain sites for later-dominant factors that will contribute positively to T-cell differentiation, like Ets1 (138).
The split impact of PU.1 on other phase 1 genes is an important feature of the T-cell system. It shows that the progenitor-associated regulatory genes are themselves responding to multiple positive and negative regulatory inputs in a balanced progenitor state. The persistence of this pattern through multiple cell cycles seems inconsistent with a dueling regulator model. The limits of PU.1’s “master regulator” activity in the T-cell pathway are also evident from the fact that several of the genes most responsive to PU.1, including regulatory genes Mef2c and Lmo2, are downregulated at least one stage before PU.1 levels themselves drop (99, 136, 137). This implies that other factors must be modulating PU.1’s activity. At the same time, however, the reproducible, sustained co-expression of PU.1 together with some of its negative regulation targets show again that PU.1 negative regulatory effects work to reduce the amplitude of target gene expression without silencing. This kind of damping repression is typical of the relationships among many regulators in the T-cell program, but is substantially different from the silencing repression that is central to boundary formation in type I embryos.
Activation of the T-cell program
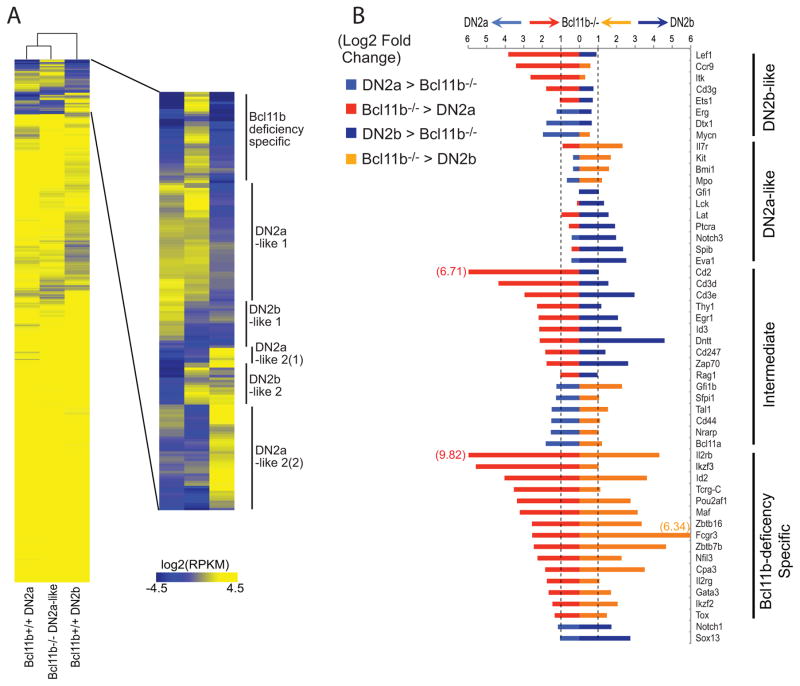
Phase 1 gene expression and regulatory activity overlaps with the activation of the T cell program itself at the single cell level. The T-cell program can be recognized by the induction of three regulatory genes: Gata3, Tcf7 (encoding TCF-1), and the Notch-triggered target gene Hes1. T cells complete their commitment multiple cell cycles later, when they progress to DN2A stage and then finally turn on expression of the Bcl11b gene. This is the most nearly T-cell specific of all the regulatory genes in the program and one of the two regulatory genes in the whole genome with the greatest increase in expression from the ETP stage to the newly-committed DN2b stage.
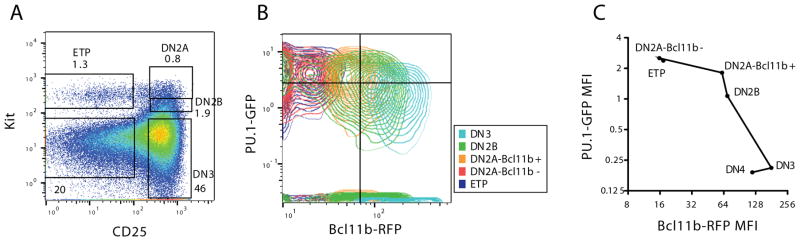
Multiple other regulatory changes take place between the initiation of the program and the success of lineage commitment; several phase 1 genes are already silent by the time that Bcl11b is turned on. However, while they are expressed, these phase 1 genes are an integral part of the T-cell program and foreshadow its activation. Single-cell analysis using fluorescent reporters for the Bcl11b locus together with reporters for phase 1 genes Bcl11a (133) or Spi1 (PU.1; Fig. 5) confirm that the cells first activating Bcl11b expression still have detectable expression of these phase 1 gene reporters, though they become downregulated afterwards. DN2b cells, by now functionally committed, still clearly retain PU.1 protein as shown by single-cell fluorescent staining (78). As discussed below, they also sustain a clear pattern of PU.1 binding at many regulatory sites throughout the genome, as shown by ChIP-seq of these purified populations (chromatin immune precipitation analyzed by deep sequencing) (137). These results make it highly unlikely that phase 1 gene expression signals are coming from non-T contaminants; instead, the T-cell program is built on a multilineage hematopoietic precursor foundation.
Figure 5.
T cell precursors in the thymus begin by expressing PU.1. Data from experiments using thymocytes from progeny of a PU.1-GFP reporter mouse (59) crossed with a Bcl11b-mCherry reporter mouse (K. K. H. Ng, H. Y. Kueh, and M. A. Yui, unpublished), in which activation of the T-cell specific Bc11b gene in DN2 stage identifies cells definitively as T-cell precursors. (A) Gating of immature T-cell precursors to separate ETP, DN2a, DN2b, DN3(a), and DN4 cells. (B) Expression of PU.1-GFP relative to Bcl11b-mCherry in the indicated populations of cells from panel A. Note that the level of GFP from this PU.1 reporter is always low in early T cells, but the pattern of expression perfectly fits the measured PU.1 RNA and protein expression patterns determined by realtime PCR, RNA-seq, and intracellular staining (78, 137). The upregulation of the Bcl11b-mCherry reporter is further used to distinguish the earlier DN2a cells (Bcl11b-mCherry-negative) from the later ones beginning to express Bcl11b (Bcl11b-mCherry positive). (C) Quantitation of PU.1-GFP levels in T-cell precursors as they activate the T-lineage specific reporter Bcl11b-mCherry. Points graphed show Mean Fluorescent Intensities for both markers at the indicated stages, from results in (B). Note that when Bcl11b is first turned on, the cells are still expressing PU.1 comparably to ETP cells. Results are representative of three independent experiments.
Molecular features of the emergent T-cell program are once again unexpected from the dichotomous perspectives of either the dueling regulator models or the boundary formation functions in type I embryos. The T-lineage specific genes to be activated are presumably being driven in their expression by the positive regulators that are newly activated during T-cell development, and their activation supposedly distinguishes these cells from their stem/progenitor antecedents. However, although only limited ChIP-seq data are available for phase 1 regulators in early T cells, the results seen to date open the possibility of an intermediate phase of active collaboration between the prior state regulators and the new regulators. Following PU.1 as a factor obviously alien to the eventual T-cell program, it is remarkable that PU.1 binds in ETP cells to a pattern of sites with numerous points of difference from its occupancy patterns in EBF1−/− pre-pro B cells, B cells, or macrophages (137). Many of the differences concern sites uniquely occupied in the ETP and DN2a cells, where PU.1 is recruited to the vicinity of genes specifically activated in the T-cell lineage (137). The functional significance of these binding events is not certain, but it is noteworthy that these genes do not require PU.1 binding to be silent in non-T lineage cells. Thus it is quite possible that PU.1 is being drawn to participate in gene regulatory complexes that activate T-cell genes or poise them for future activation. Such an interpretation is further suggested by the fact that the earliest sites where T-lineage factor GATA-3 is seen binding to the DNA, soon after its own induction in ETP cells, are often closely associated with sites of PU.1 binding as well; at later stages when the PU.1 protein levels fall, GATA-3 redistributes to other sites (137). The sustained presence of phase 1 regulators could thus promote a gradual regulatory handover to the T-cell regulatory state, rather than a boundary erected immediately, or a winner-take-all duel.
Complex regulatory interactions within the T-cell program
GATA-3: essential but troublesome regulator
There is indeed a need for cross-regulatory constraint in T-cell development, but some of it must be exerted within the T-cell program itself, and not only to exclude non-T cell program elements. Ironically, a particularly urgent example is provided by GATA-3, the T-lineage relative of GATA-1 that has such a paradigmatic role in the PU.1-GATA-1 dueling regulator model. GATA-3 is essential for T-cell development from the earliest stage throughout multiple later developmental checkpoints, and it is restricted in its hematopoietic expression to T cells and T-cell-like Innate Lymphoid Cells (116, 139–141). GATA-3 can indeed antagonize alternative developmental program regulators, through its ability to repress PU.1 and its ability to squelch access to the B-cell program (84, 85, 132, 142). But in other respects it is very unlike a “master regulator”.
GATA-3 not only has dose-dependent effects on different aspects of the commitment process (84), but also has the capacity to destroy T-lineage specification if it is overexpressed (132, 143). In this way, its role in early mouse T-cell development differs from its roles in mature Th2 effector differentiation or its role in Innate Lymphoid Cell Type 2 development, where overexpression promotes stronger differentiation along the same pathway where endogenous expression is essential (144–146). In early T cells, GATA-3 inadequacy leads to poor viability from the ETP stage, poor ETP to DN2 progression (84, 85), derangement of the DN2 stage so that DN2 and DN3-like features are intermixed (84), and failure to undergo TCRβ rearrangement (147). However, overlapping the time windows of these effects, GATA-3 overexpression also leads to gross losses of viability from ETP and CLP stages, steep losses of expression of PU.1 (transiently) and IL-7Rα, abnormalities at the ETP and DN2 stages if the viability loss is compensated, and a preference to redirect development to a mast-cell like fate (84, 132, 142, 143). More moderate degrees of overexpression transform developing thymocytes to lymphoma (148). Thus, strict dosage control is vital to keep GATA-3 levels appropriate for the developmental stage even within the T-cell program.
One aspect of the delicacy of GATA-3 function is that its pattern of DNA binding is quite variable. Other factors, including PU.1, exhibit lineage-specific differences in their binding sites across the genome, which probably reflects different chromatin accessibility at these sites in different contexts combined with different opportunities for crowd-binding with the other transcription factors available in those contexts (137, 149–154). GATA-3 takes this flexibility to an extreme, however, as its genomic occupancies redistribute considerably even between different stages of DN thymocyte development and between later branches of effector T-cell development (137, 155). Some of the sites with dynamic occupancy are associated with important regulatory genes of lympho-hematopoietic development, and although they are not among its most sensitive target genes, there is evidence that GATA-3 contributes positive regulation at least to Ets1 and Bcl11b and negative regulation to Spi1 (PU.1) and Bcl11a (84). It is likely that binding to many of these sites is intrinsically weak enough so that it depends either on cooperation with other transcription factors, or on dosage. If the system is tuned to require GATA-3 action to be targeted by the availability of other partners, then an increase in GATA-3 dosage could override this specificity, to bad effect.
Fortunately for homeostasis, in early T cells GATA-3 is not positively autoregulating. Exogenous GATA-3 cannot enhance endogenous Gata3 expression (132), and Gata3 knockout early T cells from which the exons encoding the DNA-binding domain have been deleted continue to express RNA from the Gata3 promoter at stable or even increased levels (84). Thus GATA-3 levels appear to be dependent primarily on other factors. Later in T-cell development, GATA-3 expression levels come under control of the signaling pathways responding to TCR engagement and Stat6, but these do not account for the expression dynamics in the early stages. Instead, Gata3 control after initial Notch-dependent induction is probably maintained by Myb and TCF-1 as likely positive regulators (99, 156, 157), and PU.1 and E2A as conditional or mild negative regulators (99, 136, 143). In fact, Gata3 expression at RNA and protein levels in developing mouse pro-T cells appears to be kept within a ~3x range from late ETP through DN3a stage (84), and recent evidence suggests that for much of this time only a single Gata3 allele is active, suggesting a constraint on chromatin opening as well (115). GATA-3 is powerful and important, but it cannot be the controlling protagonist in a regulatory duel for the T-cell fate.
Roles of Bcl11b: imposing commitment over shadow priming for innate lymphoid programs
In the past six years, a discrete new T-lineage commitment function has been identified that is not also needed for ETP survival, and this has been helpful to dissect components of the fate determination process. This new function is the role of Bcl11b. The combinatorial action of GATA-3, TCF-1, Notch signaling, and the inherited progenitor factor Runx1/CBFβ converge to cause the Bcl11b gene to be turned on in late DN2a stage (H. Y. Kueh, M. A. Yui, K. K. H. Ng, et al., submitted)(85, 157–160). This event is functionally linked to T-cell lineage commitment (159, 161, 162), and in vivo it is accompanied by a wave of repression of phase 1 regulatory genes (Kueh, Yui, Ng, et al., submitted)(rev in (36)). If Bcl11b is deleted before the cells undergo commitment, but strong Notch and IL-7 signals from the environment are maintained, the cells proliferate in an uncommitted, DN2a-like state. However, the cells become less sensitive to Notch signals than controls when Bcl11b fails to be expressed, and even modest reductions in Notch ligand availability then enable Bcl11b-deleted pro-T cells to shift quickly to an NK-like regulatory state (159). In the thymus in vivo, interestingly, Bcl11b-knockout pro-T cells do not have adequate access to these proliferation requirements to sustain self-renewal (163). However, the robust growth of Bcl11b knockout cells in Notch-stimulating conditions in vitro (162) offer a valuable look at the intrinsic sustainability of an uncommitted pro-T cell gene network state over many days or weeks.
Genome-wide analysis of RNA expression in Bcl11b knockout pro-T cells situates them between DN2a and DN2b stages (Fig. 6A)(164)(J. A. Zhang, L. Li, and E.V.R., unpublished data). This is a satisfying result insofar as Bcl11b itself is not turned on until late in DN2a stage, and thus should only be able to affect gene expression after that. Focusing on the genes that normally change expression from DN2a to DN2b, Bcl11b-knockout DN2 cells resemble DN2a cells in maintaining expression of genes that are normally turned off in DN2b, while failing to turn on many genes that are normally turned on in DN2b (Fig. 6A, DN2a-like groups 1 & 2). However, with respect to a smaller number of genes, Bcl11b-knockout DN2 cells appear more DN2b-like; a certain number of DN2b-specific genes are activated and a certain number of DN2a-specific genes are repressed in Bcl11b-knockout cells as though they were normal DN2b cells (DN2b-like groups 1 & 2). Interestingly, important developmental regulators are represented in both DN2a-like and DN2b-like groups (Fig. 6B). The incomplete arrest phenotype shows that the effects of Bcl11b loss are gene specific. Bcl11b is not a “master regulator” any more than GATA-3, since direct Bcl11b targets or gene network subcircuits dependent on Bcl11b are a select subcomponent of the gene expression changes that normally accompany commitment, not a switch governing the whole process.
Figure 6.
Gene expression effects of Bcl11b deletion in the context of DN2a to DN2b progression. (A) Heat map of RNA-seq phenotype of Bcl11b−/− DN2a-like pro-T cells, compared with true DN2a cells and true DN2b cells. Bcl11b−/− cells were prepared as in ref. (162), and compared by RNA-seq with DN2a and DN2b cells as in ref. (137). Bcl11b-deficient cells mimic DN2a cells with respect to up- and down-regulation of different groups of genes (DN2a-like 1, DN2a-like 2) more than they mimic DN2b cells (DN2b-like 1, DN2b-like 2), since the DN2a-like groups contain more genes than the DN2b-like groups; but the phenotype is clearly split. Also note the top group of genes (“Bcl11b-deficiency specific”) which are strongly expressed in the mutant cells but not expressed in either normal control. (B) Quantitation of effects of Bcl11b deletion on expression of specific genes, from RNA-seq analyses. Each gene is compared in expression levels between DN2a cells and Bcl11b-deficient cells (left bars), and between DN2b cells and Bcl11b-deficient cells (right bars), with the colors showing whether the expression is higher in the wildtype cells (light, dark blue) or the mutant cells (red, orange). Differential expression is shown as fold difference on a log2 scale from −6 to +6. Where differences exceed |26| fold, for Cd2, Il2rb and Fcgr3, the value (log2) is provided numerically. Similar results for these genes have been obtained in >4 independent experiments (J. A. Zhang, W. Zeng, L. Li, A. Mortazavi, E. V. Rothenberg, unpublished).
Strikingly, in addition there is also a third class of gene expression changes in the Bcl11b knockout cells that does not fit into the pattern of either DN2a or DN2b. Affected genes in this class include Notch 1, which is under-expressed in Bcl11b-knockout cells as compared to either DN2a or DN2b normal controls. However, the most common feature of the mutant-specific gene expression phenotype is the overexpression of a large and highly coherent group of genes that would not normally be expressed in early T cells at all, neither before nor after the onset of Bcl11b expression (Fig. 6B, Bcl11b-deficiency-specific) (164). These upregulated genes include many that are implicated in innate lymphoid cell function, NK cell function, and innate-like T cell specification: Id2, Il2rb, Fcgr3, Zbtb16, and Nfil3. Also precociously activated are Maf, Ikzf2, Zbtb7b, and Pou2af1, factors expressed in other mature T-cell subsets (165). At a lower level, but markedly above background, Bcl11b-knockout cells activate the highly NK-specific transcription factor Zfp105 as well (159, 162). These gene expression responses give Bcl11b-knockout cells an NK cell (or NKT-like) functional bias that distinguishes them from mainstream normal T cells. However, they also represent a surprise.
The emergence of a new program of gene expression by deletion of one transcription factor implies that that factor would normally be acting as a repressor for the program. In the case of Bcl11b, the absence of expression of these genes in normal cells would imply that Bcl11b is a very efficient repressor of these genes. However, the comparison should not just be between fully committed (DN2b) normal cells and their Bcl11b-deficient counterparts; the developmentally informative comparison is also with precursor normal cells that have not had any opportunity to express Bcl11b yet (DN2a cells). It would make sense for Bcl11b-deficient cells to continue to express phase 1 genes that are normally expressed until Bcl11b is activated, if Bcl11b were needed to terminate their expression. But the results here show that many genes that are initially silent in the cells before Bcl11b is ever expressed change to become expressed by default just around the time that Bcl11b is turned on to restrain them. In other words, the phenotype of Bcl11b-knockout cells implies that developing T cells are not only programmed to acquire T-cell characteristics, but also programmed to acquire innate-like characteristics – yet to hold those characteristics in check, using Bcl11b to prevent activation of this parallel gene expression program.
Why does this make sense? Importantly, there are few genes yet known to be truly unique to innate lymphocytes; much of the distinction between innate lymphocytes and T cells at the single-cell level has to do with the types of triggering receptors they express and the thresholds for their effector responses, which are higher for T cells. Thus, the arming of normal mature T cells for effector function is very close to the establishment of an innate lymphocyte effector gene expression program, except that in the T cell there is also a stronger restraint mechanism to keep the cells quiescent until unleashed. Thus it seems plausible that as T-cell specification proceeds toward its climax, the cells could also gain latent properties that we classify as “innate” programs.
In accord with this view, we know that at least three important T-cell regulators are shared with the innate program: GATA-3, TCF-1, and Runx1; Notch is used for ILC2 and ILC3 specification as well (rev. by (35, 166)). In fact, Bcl11b itself is also an essential enabling factor for one particular innate cell subset (ILC2) to emerge as opposed to the others (NK, ILC1, ILC3)(167–169). There are also hints that cytokine receptor stimulation, such as the stimulation that fosters early pro-T cell expansion, also stimulates expression of regulatory infrastructure for T/innate lymphocyte effector function. The mechanism that appears to control all these effector programs, from data in hand thus far, is the balance between E protein dominance and Id2 dominance. A role for Bcl11b on the E protein side of this balance is strongly hinted by the massive upsurge in Id2 expression when Bcl11b is deleted (Fig. 7)(159). Here again, the type of repression that Bcl11b is likely to be providing is not permanent silencing repression. The genes repressed by Bcl11b will permanently remain accessible in mature T cells for conditional activation in response to the appropriate combination of TCR and cytokine signals, conditions which cause Bcl11b to undergo modest downregulation (170). From the point of view of the NK, ILC1, CD8 effector, and NKT cell programs, Bcl11b activation would thus be the key event in an incoherent feedforward loop, activated by the same regulators that may contribute to the innate-cell programs, but then acting as an antagonist to them.
Figure 7.
Different types of combinatoriality for transcription factor action in T-cell specification: two theoretical possibilities. Different gene expression patterns of cells at different stages are shown as a trajectory from an initial gene expression pattern at one point in a three-principal-component space to a final gene expression pattern at another point (thick dark blue arrows). Two models are shown in which the effects of different transcription factors on gene expression are shown as different colored vectors propelling the cells’ gene expression profiles along this path, where the individual roles of the factors differ. (A) A model in which all factors promote the T-cell program faithfully (vectors all parallel to the main trajectory), but in which participating factors are required to work in AND logic to drive developmental progression from start to finish. Left, parallel but stage-dependent effects of factors push cells along the unique trajectory of gene expression change. Right, if factor represented by magenta vector is removed, development halts but is still on the canonical track. Green and yellow vectors, and part of the light blue vector, are depicted as thin arrows in this case to indicate the constraint that these factors cannot work when the magenta factor is absent. (B) An alternative model in which individual vectors do not simply work to promote the T cell fate (not individually parallel to main path), but all have a component that can contribute to the main path and/or counteract another factor’s tendency to deviate from the main path. Although the panel shows the arrows operating sequentially to show their individual directions more clearly, note that this model also works well if they often act in combinations in overlapping time frames like the factors in A. Left, under normal conditions the resultant of adding the disparate transcription factor activities together pushes the cells along the main path. Right, if factor represented by magenta vector is removed, vector addition still advances the cell to change its gene expression pattern, but now it no longer follows the T-cell path.
Transcription factor vector addition defines cell type
The evidence is strong that T-cell specification depends on multiple transcription factor combinatorial action. However, there are different ways to understand this requirement for combinatoriality in executing the T-cell developmental program. Most often the model is that of a pathway, and indications of the stages at which different factors work are based on an implicit model that there is a kind of regulatory relay. Thus, in this model each factor is working to push the cells inexorably toward the T-cell goal, even as “and” logic requires more and more of them to be active for the last genes in the pathway to achieve correct regulation. We can visualize what this could mean for the different factors if we imagine development as a series of gene expression changes that can be plotted to form a linear trajectory in principal component space (Fig. 7A). According to this additive pathway-relay model, all the gene expression changes driven by any of the individual factors would be vectors parallel to the main trajectory (Fig. 7A; different colored arrows represent effects of different factors). If this is the correct way to explain the roles of these factors, then any loss of T-cell factor expression would arrest T-cell development, but leave the regulatory state at some point on the same pathway (Fig. 7A, right).
The phenotypes of Bcl11b knockout cells and GATA-3 overexpressing cells suggest that the combinatoriality may depend on something else, however. In these cases, the cells not only become arrested but also acquire new, non-T identities, similar to what happens when a factor working mainly outside the T-cell program, like PU.1, is overexpressed. The gain of function effects of PU.1 and GATA-3 are not a simple reversal of the defects seen when these factors are deleted in early T cells. As already mentioned, the asymmetry is partly due to dose dependence, but it is manifested through the gene network effects that different regulators have on each other, partly antagonizing each other’s expression (GATA-3 vs. PU.1, even Bcl11b vs. GATA-3) and selectively blunting some of each other’s functions (Notch signals vs. PU.1). Thus, a better way to represent the action of successive factors in our gene expression principal component space may be as a vector addition among non-parallel vectors (Fig. 7B, left). In this model, the correctly coordinated action of the different factors drives the cell from the initial state to the end state, but with actions that partially synergize and partially counterbalance each other. Then in such a model, when one factor is lost or excessively activated, the cell naturally falls off the T-cell trajectory to a different end state (Fig. 7B, right). In this case, combinatoriality is not only required to complete the pathway, but to define the direction of the pathway itself.
Stabilizing fate in the T-cell system
In classic models of differentiation, the mature cell that emerges is postmitotic. This acts as a natural source of stability for the regulatory state of the cells that controls gene expression. For T-cell precursors, however, cell cycle arrest may be a checkpoint or a pause, but it is not the end of proliferation or differentiation choices, which continue long after maturation is complete. Even more interestingly, the relative expression levels of different factors among the T-cell specification core may keep changing. Some relationships can be maintained in certain T-cell subsets: for example, the positive regulatory cooperation of GATA-3 and TCF-1 appears to continue through the CD4/CD8 lineage choice into the Th2 effector T-cell differentiation program (171, 172). However, Notch signaling stops after β-selection and may only resume in the periphery as part of the antigen stimulation program. GATA-3 levels are variable among helper cell lineages and between helper and killer cells, Myb and Gfi1 are turned off after the cells leave the thymus, and the correlation between Bcl11b and TCF-1 expression levels is dramatically shattered in the Treg lineage (173, 174). E protein activity levels are particularly dynamic, as the E protein HEB (Tcf12) rises to a sharp peak in the DP stage, then declines, while all encounters with TCR stimuli throughout T cell development and function activate one or another of the E protein antagonists, Id2 or Id3 (175, 176). Rather than using T-cell transcription factors to set up a stabilizing mutual activation feedback circuitry, the T cells treat them as free variables for additional combinatorial diversification of regulatory states. Yet despite this transcription factor instability, T cells retain stable expression of their key signature genes through weeks, months, or years of function, migration, and proliferation in different environments. Thus even if the developmental process triggered by Notch signaling uses “fuzzy logic” to establish T-cell identity, this identity is surprisingly durable.
One of the consequences of the first TCR-dependent selection event, during the transition from DN3 to DP, is a general increase in H3K27me3 repressive marking of quiescent loci across the genome (177), even when these genes had been transcriptionally silenced one or two stages previously (137). The deep silencing of progenitor-associated transcription factor genes is indeed likely to contribute to stability, but it remains mysterious what really is needed to sustain positive regulation of the T-cell identity genes themselves. In at least one case, we have preliminary evidence that the regulatory requirements for activation of an important gene are considerably more complex than the requirements for keeping it on, starting almost immediately after its activation (Kueh, Yui, Ng, et al., submitted). This kind of phenomenon suggests that a major threshold could lie at the level of a process like chromatin opening.
Concluding remarks
There are many unknowns yet to be revealed in this culmination aspect of the T-cell development process. Does T-cell development require such a large number of regulatory inputs only in order to open genes, so that only a few factors are needed to sustain them later? Does the extended proliferation in the early stages of T-cell development make its own regulatory contribution, either by dilution of previously expressed transcription factors or by helping to remodel chromatin structure? How small, in the end, might be the core “maintenance” T cell gene regulatory network? Could only a few stably expressed regulatory factors in fact be enough, and are those that are needed truly T lineage-unique at all? Finally, how important is the order in which T-cell regulators are serially deployed in order to guide the process to the right endpoints? Can the process be substantially accelerated and simplified?
In this review we have drawn a picture of the T-cell development process as one guided from a multipotent stem cell state by a gradual process that depends on extensive transcription factor combinatoriality but permits fluid combinations of factors to occur and then reform with other partners through multiple stages. One enabling feature of the process is its hallmark use of damping repression rather than silencing in most transcription factor cross-repressive interactions, which both permits coexistence and restrains overexpression. The dosage restraint makes it possible for this program to use factors which at other levels of expression, or without the constraint of Notch signaling, push cells to completely different end states. This mechanism is important to understand in depth through modeling, but the right framework has to be chosen. This system has obvious differences in its functional rules of operation both from the Boolean behavior of well-characterized embryonic cell networks and from the simple bistability of the dueling regulator models.
But in the context of emergence from stem cells, the process through which T cells emerge makes sense. Recall that vertebrate embryos in general arise from a different process than type I embryos; vertebrate embryonic cells as a rule proliferate extensively and migrate to new parts of the embryo, defying fate maps, before their future identities are determined. It is possible that the rule set needed to solve the T-cell specification network will also prove to be a fine model for vertebrate development in general.
Acknowledgments
We gratefully acknowledge the late Eric Davidson for stimulating discussions of developmental gene network properties in diverse systems. We thank past and present members of the Rothenberg and Michael Elowitz groups for valuable advice and sharing of unpublished results, Maria Lerica Gutierrez Quiloan for mouse colony supervision, and Rochelle Diamond, Keith Beadle, and Diana Perez for flow cytometry. The ideas presented here were developed with support by grants from NIH: RC2CA148278, R01AI083514, R01AI095943, R01CA90233, R01HD076915 and R01HL119102 to E.V.R., R01AI064590 to M.A.Y., and K99HL119638A to H.Y.K.; also by the L. A. Garfinkle Memorial Laboratory Fund, and by the Albert Billings Ruddock Professorship to E.V.R.
Footnotes
“Master regulator” is in quotes throughout because in molecular terms, none of these transcription factors work in isolation or override the context in which they are expressed. Authors refer to them as “master regulators” in some cases to emphasize their functional contributions to the processes in which they participate, and in other cases to propose a deliberately simplified model of those processes.
Conflict of Interest: None of the authors has a conflict of interest.
References
- 1.Davidson EH. Spatial mechanisms of gene regulation in metazoan embryos. Development. 1991;113:1–26. doi: 10.1242/dev.113.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic Press; 2006. [Google Scholar]
- 3.Peter IS, Davidson EH. Genomic Control Process: Development and Evolution. San Diego, CA: Academic Press, Elsevier; 2015. [Google Scholar]
- 4.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 5.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2009;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson B, Christiaen L. Linking chordate gene networks to cellular behavior in ascidians. Cell. 2006;124:247–250. doi: 10.1016/j.cell.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Kubo A, Imai KS, Satou Y. Gene-regulatory networks in the Ciona embryos. Brief Funct Genomic Proteomic. 2009;8:250–255. doi: 10.1093/bfgp/elp018. [DOI] [PubMed] [Google Scholar]
- 9.Lemaire P, Smith WC, Nishida H. Ascidians and the plasticity of the chordate developmental program. Curr Biol. 2008;18:R620–631. doi: 10.1016/j.cub.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maduro MF. Developmental robustness in the Caenorhabditis elegans embryo. Mol Reprod Dev. 2015 doi: 10.1002/mrd.22582. [DOI] [PubMed] [Google Scholar]
- 11.Hobert O. Development of left/right asymmetry in the Caenorhabditis elegans nervous system: from zygote to postmitotic neuron. Genesis. 2014;52:528–543. doi: 10.1002/dvg.22747. [DOI] [PubMed] [Google Scholar]
- 12.Stathopoulos A, Levine M. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev Biol. 2005;280:482–493. doi: 10.1016/j.ydbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Jaeger J. The gap gene network. Cell Mol Life Sci. 2011;68:243–274. doi: 10.1007/s00018-010-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Xu Z, Mei C, Yu D, Small S. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell. 2012;149:618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 17.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 18.Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, et al. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 20.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 21.Flores GV, et al. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 22.Peter IS, Faure E, Davidson EH. Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci U S A. 2012;109:16434–16442. doi: 10.1073/pnas.1207852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta. 2009;1789:363–374. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longabaugh WJR, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 25.Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci USA. 2009;106:3823–3828. doi: 10.1073/pnas.0807878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. λ Repressor and cro--components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 28.Ptashne M, et al. How the λ repressor and cro work. Cell. 1980;19:1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- 29.Naito T, Taniuchi I. The network of transcription factors that underlie the CD4 versus CD8 lineage decision. Int Immunol. 2010;22:791–796. doi: 10.1093/intimm/dxq436. [DOI] [PubMed] [Google Scholar]
- 30.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Bosselut R. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. J Immunol. 2009;183:2903–2910. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, Guo YP, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 33.Laslo P, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 34.Roeder I, Glauche I. Towards an understanding of lineage specification in hematopoietic stem cells: a mathematical model for the interaction of transcription factors GATA-1 and PU. 1. J Theor Biol. 2006;241:852–865. doi: 10.1016/j.jtbi.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 35.De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- 36.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014;14:529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akashi K, Reya T, Dalma-Weiszhausz D, Weissman IL. Lymphoid precursors. Curr Opin Immunol. 2000;12:144–150. doi: 10.1016/s0952-7915(99)00064-3. [DOI] [PubMed] [Google Scholar]
- 40.Månsson R, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 41.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Akashi K, Traver D, Zon LI. The complex cartography of stem cell commitment. Cell. 2005;121:160–162. doi: 10.1016/j.cell.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Arinobu Y, et al. Reciprocal activation of GATA-1 and PU. 1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 45.Graf T, Zon LI. Hematopoiesis: a Developmental Approach. New York: Oxford University Press; 2001. Transcription factors that induce commitment of multipotent hematopoietic progenitors: lessons from the MEP system; pp. 355–362. [Google Scholar]
- 46.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU. 1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- 47.Rekhtman N, Radparvar F, Evans T, Skoultchi A. Direct interaction of hematopoietic transcription factors PU. 1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU. 1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahl R, et al. Regulation of macrophage and neutrophil cell fates by the PU. 1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 50.Guo G, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13:492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moignard V, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu M, Kawamoto H, Katsube Y, Ikawa T, Katsura Y. The common myelolymphoid progenitor: a key intermediate stage in hemopoiesis generating T and B cells. J Immunol. 2002;169:3519–3525. doi: 10.4049/jimmunol.169.7.3519. [DOI] [PubMed] [Google Scholar]
- 53.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 54.Katsura Y, Kawamoto H. Stepwise lineage restriction of progenitors in lympho-myelopoiesis. Int Rev Immunol. 2001;20:1–20. doi: 10.3109/08830180109056720. [DOI] [PubMed] [Google Scholar]
- 55.Paul F, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 56.Perié L, Duffy KR, Kok L, de Boer RJ, Schumacher TN. The branching point in erythro-myeloid differentiation. Cell. 2015;163:1655–1662. doi: 10.1016/j.cell.2015.11.059. [DOI] [PubMed] [Google Scholar]
- 57.Notta F, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351:aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU. 1 and the cell cycle controls myeloid differentiation. Science. 2013;341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU. 1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monteiro R, Pouget C, Patient R. The gata1/pu1 lineage fate paradigm varies between blood populations and is modulated by tif1γ. EMBO J. 2011;30:1093–1103. doi: 10.1038/emboj.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chickarmane V, Enver T, Peterson C. Computational modeling of the hematopoietic erythroid-myeloid switch reveals insights into cooperativity, priming, and irreversibility. PLoS Comput Biol. 2009;5:e1000268. doi: 10.1371/journal.pcbi.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chlon TM, Dore LC, Crispino JD. Cofactor-mediated restriction of GATA-1 chromatin occupancy coordinates lineage-specific gene expression. Mol Cell. 2012;47:608–621. doi: 10.1016/j.molcel.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cantor AB, et al. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med. 2008;205:611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iwasaki H, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walsh JC, et al. Cooperative and antagonistic interplay between PU. 1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 66.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU. 1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsu CL, King-Fleischman AG, Lai AY, Matsumoto Y, Weissman IL, Kondo M. Antagonistic effect of CCAAT enhancer-binding protein-α and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors. Proc Natl Acad Sci USA. 2006;103:672–677. doi: 10.1073/pnas.0510304103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 69.De Obaldia ME, et al. T cell development requires constraint of the myeloid regulator C/EBP-α by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14:1277–1284. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McNagny KM, Sieweke MH, Doderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 1998;17:3669–3680. doi: 10.1093/emboj/17.13.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Du J, et al. Novel combinatorial interactions of GATA-1, PU. 1, and C/EBPî isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem. 2002;277:43481–43494. doi: 10.1074/jbc.M204777200. [DOI] [PubMed] [Google Scholar]
- 72.Heavey B, Charalambous C, Cobaleda C, Busslinger M. Myeloid lineage switch of Pax5 mutant but not wild-type B cell progenitors by C/EBPα and GATA factors. EMBO J. 2003;22:3887–3897. doi: 10.1093/emboj/cdg380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zandi S, Åhsberg J, Tsapogas P, Stjernberg J, Qian H, Sigvardsson M. Single-cell analysis of early B-lymphocyte development suggests independent regulation of lineage specification and commitment in vivo. Proc Natl Acad Sci U S A. 2012;109:15871–15876. doi: 10.1073/pnas.1210144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Welinder E, Åhsberg J, Sigvardsson M. B-lymphocyte commitment: identifying the point of no return. Semin Immunol. 2011;23:335–340. doi: 10.1016/j.smim.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 75.Mansson R, et al. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115:2601–2609. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- 76.De Obaldia ME, Bell JJ, Bhandoola A. Early T-cell progenitors are the major granulocyte precursors in the adult mouse thymus. Blood. 2013;121:64–71. doi: 10.1182/blood-2012-08-451773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luc S, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 80.Wada H, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 81.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 82.Perry SS, Wang H, Pierce LJ, Yang AM, Tsai S, Spangrude GJ. L-Selectin defines a bone marrow analogue to the thymic Early T-lineage Progenitor. Blood. 2004;103:2990–2996. doi: 10.1182/blood-2003-09-3030. [DOI] [PubMed] [Google Scholar]
- 83.Wu L, Antica M, Johnson GR, Scollay R, Shortman K. Developmental potential of the earliest precursor cells from the adult mouse thymus. J Exp Med. 1991;174:1617–1627. doi: 10.1084/jem.174.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scripture-Adams DD, et al. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol. 2014;193:3470–3491. doi: 10.4049/jimmunol.1301663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.García-Ojeda ME, et al. GATA-3 promotes T cell specification by repressing B cell potential in pro-T cells. Blood. 2013;121:1749–1759. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- 86.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol. 2007;178:858–868. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- 87.Allman D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 88.Kamath MB, et al. Dose-dependent repression of T-cell and natural killer cell genes by PU. 1 enforces myeloid and B-cell identity. Leukemia. 2008;22:1214–1225. doi: 10.1038/leu.2008.67. [DOI] [PubMed] [Google Scholar]
- 89.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU. 1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 90.Zarnegar MA, Rothenberg EV. Ikaros represses and activates PU. 1 cell-type-specifically through the multifunctional Sfpi1 URE and a myeloid specific enhancer. Oncogene. 2012;31:4647–4654. doi: 10.1038/onc.2011.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leddin M, et al. Two distinct auto-regulatory loops operate at the PU. 1 locus in B cells and myeloid cells. Blood. 2011;117:2827–2838. doi: 10.1182/blood-2010-08-302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen H, et al. PU. 1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 1995;11:1549–1560. [PubMed] [Google Scholar]
- 93.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- 95.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU. 1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 96.Manesso E, Chickarmane V, Kueh HY, Rothenberg EV, Peterson C. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface. 2013;10:20120774. doi: 10.1098/rsif.2012.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu M, et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 98.Schlenner SM, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 99.Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU. 1, Notch and Gata3. Development. 2013;140:1207–1219. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Franco CB, et al. Notch/Delta signaling constrains reengineering of pro-T cells by PU. 1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU. 1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 102.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sambandam A, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 104.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 105.El-Kassar N, et al. High levels of IL-7 cause dysregulation of thymocyte development. Int Immunol. 2012;24:661–671. doi: 10.1093/intimm/dxs067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prockop SE, Petrie HT. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J Immunol. 2004;173:1604–1611. doi: 10.4049/jimmunol.173.3.1604. [DOI] [PubMed] [Google Scholar]
- 107.Munitic I, et al. Dynamic regulation of IL-7 receptor expression is required for normal thymopoiesis. Blood. 2004;104:4165–4172. doi: 10.1182/blood-2004-06-2484. [DOI] [PubMed] [Google Scholar]
- 108.Cai Z, et al. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity. 2000;13:423–431. doi: 10.1016/s1074-7613(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 109.Lacaud G, Kouskoff V, Trumble A, Schwantz S, Keller G. Haploinsufficiency of Runx1 results in the acceleration of mesodermal development and hemangioblast specification upon in vitro differentiation of ES cells. Blood. 2004;103:886–889. doi: 10.1182/blood-2003-06-2149. [DOI] [PubMed] [Google Scholar]
- 110.Rodrigues NP, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 111.Carotta S, et al. The transcription factor PU. 1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 112.O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 113.Lukin K, et al. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc Natl Acad Sci USA. 2010;107:7869–7874. doi: 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ungerbäck J, Ahsberg J, Strid T, Somasundaram R, Sigvardsson M. Combined heterozygous loss of Ebf1 and Pax5 allows for T-lineage conversion of B cell progenitors. J Exp Med. 2015;212:1109–1123. doi: 10.1084/jem.20132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ku CJ, Lim KC, Kalantry S, Maillard I, Engel JD, Hosoya T. A monoallelic-to-biallelic T-cell transcriptional switch regulates GATA3 abundance. Genes Dev. 2015;29:1930–1941. doi: 10.1101/gad.265025.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hosoya T, et al. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zarnegar MA, Chen J, Rothenberg EV. Cell type-specific activation and repression of PU. 1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol. 2010;30:4922–4939. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wilson NK, et al. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kreslavsky T, et al. β-Selection-induced proliferation is required for αβ T cell differentiation. Immunity. 2012;37:840–853. doi: 10.1016/j.immuni.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Falk I, Biro J, Kohler H, Eichmann K. Proliferation kinetics associated with T cell receptor-β chain selection of fetal murine thymocytes. J Exp Med. 1996;184:2327–2339. doi: 10.1084/jem.184.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mossadegh-Keller N, et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013;497:239–243. doi: 10.1038/nature12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shibata Y, Berclaz PY, Chroneos ZC, Yoshida M, Whitsett JA, Trapnell BC. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU. 1. Immunity. 2001;15:557–567. doi: 10.1016/s1074-7613(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 123.Tsapogas P, et al. IL-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors. Blood. 2011;118:1283–1290. doi: 10.1182/blood-2011-01-332189. [DOI] [PubMed] [Google Scholar]
- 124.Kikuchi K, Kasai H, Watanabe A, Lai AY, Kondo M. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-proB transition stage by maintaining early B cell factor expression. J Immunol. 2008;181:383–392. doi: 10.4049/jimmunol.181.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dias S, Silva H, Jr, Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J Exp Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boller S, Grosschedl R. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol Rev. 2014;261:102–115. doi: 10.1111/imr.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Malin S, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Massa S, Balciunaite G, Ceredig R, Rolink AG. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur J Immunol. 2006;36:526–532. doi: 10.1002/eji.200535760. [DOI] [PubMed] [Google Scholar]
- 130.Seshasayee D, Gaines P, Wojchowski DM. GATA-1 dominantly activates a program of erythroid gene expression in factor-dependent myeloid FDCW2 cells. Mol Cell Biol. 1998;18:3278–3288. doi: 10.1128/mcb.18.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dionne CJ, et al. Subversion of T lineage commitment by PU. 1 in a clonal cell line system. Dev Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 132.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu Y, et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012;209:2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU. 1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zohren F, et al. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol. 2012;13:761–769. doi: 10.1038/ni.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Champhekar A, Damle SS, Freedman G, Carotta S, Nutt SL, Rothenberg EV. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU. 1. Genes Dev. 2015;29:832–848. doi: 10.1101/gad.259879.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cauchy P, et al. Dynamic recruitment of Ets1 to both nucleosome-occupied and -depleted enhancer regions mediates a transcriptional program switch during early T-cell differentiation. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 140.Hosoya T, Maillard I, Engel JD. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev. 2010;238:110–125. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias AM, Chen D, Rothenberg EV. Definition of regulatory network elements for T cell development by perturbation analysis with PU. 1 and GATA-3. Dev Biol. 2002;246:103–121. doi: 10.1006/dbio.2002.0674. [DOI] [PubMed] [Google Scholar]
- 143.Xu W, Carr T, Ramirez K, McGregor S, Sigvardsson M, Kee BL. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood. 2013;121:1534–1542. doi: 10.1182/blood-2012-08-449447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee HJ, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ouyang W, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 146.Klein Wolterink RGJ, et al. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc Natl Acad Sci U S A. 2013;110:10240–10245. doi: 10.1073/pnas.1217158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 148.Nawijn MC, et al. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol. 2001;167:715–723. doi: 10.4049/jimmunol.167.2.715. [DOI] [PubMed] [Google Scholar]
- 149.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 150.Pham T-H, et al. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU. 1. Nucleic Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wontakal SN, et al. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc Natl Acad Sci U S A. 2012;109:3832–3837. doi: 10.1073/pnas.1121019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pham TH, et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012;119:e161–171. doi: 10.1182/blood-2012-01-402453. [DOI] [PubMed] [Google Scholar]
- 154.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wei G, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gimferrer I, et al. Regulation of GATA-3 expression during CD4 lineage differentiation. J Immunol. 2011;186:3892–3898. doi: 10.4049/jimmunol.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li L, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development, and cooperate with Notch signaling during T cell specification. Blood. 2008;112:480–492. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 162.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wakabayashi Y, et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 164.Zhang JA. PhD Thesis. California Institute of Technology; 2012. Global Analysis of Dynamic Epigenetic Marking and Transcriptional Regulation Underlying T-cell Lineage Commitment. [Google Scholar]
- 165.Mingueneau M, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 167.Walker JA, et al. Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med. 2015;212:875–882. doi: 10.1084/jem.20142224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yu Y, et al. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med. 2015;212:865–874. doi: 10.1084/jem.20142318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Califano D, et al. Transcription factor Bcl11b controls identity and function of mature Type 2 innate lymphoid cells. Immunity. 2015;43:354–368. doi: 10.1016/j.immuni.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cismasiu VB, et al. BCL11B participates in the activation of interleukin-2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yu Q, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-γ. Nat Immunol. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Steinke FC, et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4+ T cell fate and interact with Runx3 to silence Cd4 in CD8+ T cells. Nat Immunol. 2014;15:646–656. doi: 10.1038/ni.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vanvalkenburgh J, et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med. 2011;208:2069–2081. doi: 10.1084/jem.20102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Barra MM, et al. Transcription Factor 7 Limits Regulatory T Cell Generation in the Thymus. J Immunol. 2015;195:3058–3070. doi: 10.4049/jimmunol.1500821. [DOI] [PubMed] [Google Scholar]
- 175.De Pooter RF, Kee BL. E proteins and the regulation of early lymphocyte development. Immunol Rev. 2010;238:93–109. doi: 10.1111/j.1600-065X.2010.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vigano MA, et al. An epigenetic profile of early T-cell development from multipotent progenitors to committed T-cell descendants. Eur J Immunol. 2014;44:1181–1193. doi: 10.1002/eji.201344022. [DOI] [PubMed] [Google Scholar]
- 178.Richie Ehrlich LI, Serwold T, Weissman IL. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2011;117:2618–2624. doi: 10.1182/blood-2010-05-287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wong SH, et al. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]