Abstract
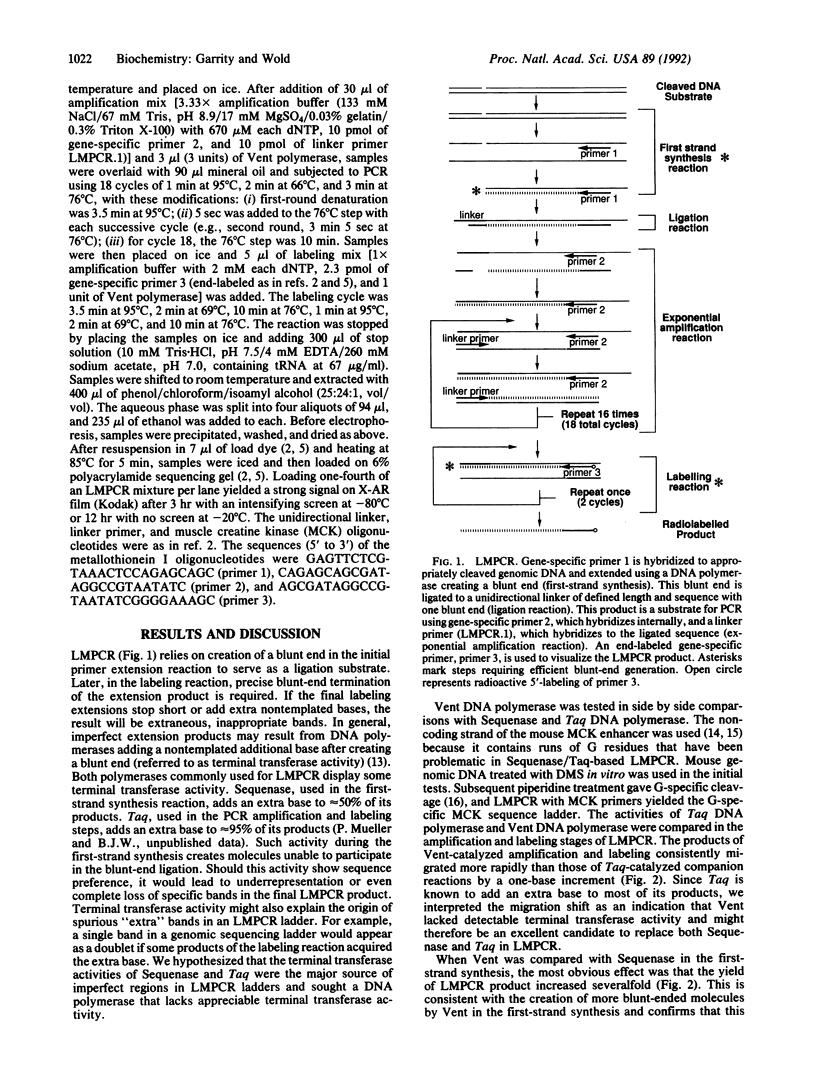
We have developed a simplified procedure for the ligation-mediated polymerase chain reaction (LMPCR) using Thermococcus litoralis DNA polymerase (Vent DNA polymerase). We show that Vent DNA polymerase produces correct, blunt-ended primer extension products with substantially higher efficiency than Thermus aquaticus (Taq) DNA polymerase or modified T7 DNA polymerase (Sequenase). This difference leads to significantly improved genomic sequencing, methylation analysis, and in vivo footprinting with LMPCR. These improvements include representation of all bands with more uniform intensity, clear visualization of previously difficult regions of sequence, and reduction in the occurrence of spurious bands. It also simplifies the use of DNase I cut DNA for LMPCR footprinting.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988 Oct 25;16(20):9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., Johnson J. E., Buskin J. N., Gartside C. L., Hauschka S. D. The muscle creatine kinase gene is regulated by multiple upstream elements, including a muscle-specific enhancer. Mol Cell Biol. 1988 Jan;8(1):62–70. doi: 10.1128/mcb.8.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara C. J., Glimcher L. H. In vivo footprinting of MHC class II genes: bare promoters in the bare lymphocyte syndrome. Science. 1991 May 3;252(5006):709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- Lyon M. F. The quest for the X-inactivation centre. Trends Genet. 1991 Mar;7(3):69–70. doi: 10.1016/0168-9525(91)90271-Q. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Darnell J. E., Jr Rapid in vivo footprinting technique identifies proteins bound to the TTR gene in the mouse liver. Genes Dev. 1991 Jan;5(1):83–93. doi: 10.1101/gad.5.1.83. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Salser S. J., Wold B. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev. 1988 Apr;2(4):412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Riggs A. D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991 Jun;5(6):1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Rideout W. M., 3rd, Coetzee G. A., Olumi A. F., Jones P. A. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990 Sep 14;249(4974):1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Solter D. Differential imprinting and expression of maternal and paternal genomes. Annu Rev Genet. 1988;22:127–146. doi: 10.1146/annurev.ge.22.120188.001015. [DOI] [PubMed] [Google Scholar]
- Sternberg E. A., Spizz G., Perry W. M., Vizard D., Weil T., Olson E. N. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988 Jul;8(7):2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguay R. L., Pfeifer G. P., Riggs A. D. PCR-aided DNaseI footprinting of single copy gene sequences in permeabilized cells. Nucleic Acids Res. 1990 Oct 11;18(19):5902–5902. doi: 10.1093/nar/18.19.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Gralla J. D. In situ nucleoprotein structure at the SV40 major late promoter: melted and wrapped DNA flank the start site. Genes Dev. 1989 Nov;3(11):1814–1822. doi: 10.1101/gad.3.11.1814. [DOI] [PubMed] [Google Scholar]







