Abstract
Fish skin, one type of wastes generated from Nile tilapia processing, is still a good source of collagen and gelatin. Bioactive peptides can be obtained from Nile tilapia skin gelatin by trypsin digestion. Trypsin hydrolysate was subsequently purified by gel filtration chromatography. Trypsin A fraction showed the greatest reducing power (5.138 ± 1.060 μM trolox/mg peptide) among all hydrolysate fractions, while trypsin B fraction from gel filtration column was found to exhibit the best radical scavenging and angiotensin-I-converting enzyme (ACE) inhibitory activities 8.16 ± 2.18 μg trolox/mg peptide and 59.32 ± 9.97 % inhibition, respectively. The most active fraction was subjected to MALDI-TOF/TOF MS/MS. After annotation by Mascot sequence matching software (Matrix Science) with Ludwig NR Database, two peptide sequences were identified; GPEGPAGAR (MW 810.87 Da) and GETGPAGPAGAAGPAGPR (MW 1490.61 Da). The docking analysis suggested that the shape of the shorter peptide may be slightly more proper, to fit into the binding cleft of the ACE. However, the binding affinities calculated from the docking showed no significant difference between the two peptides. In good agreement with the in silico data, results from the in vitro ACE inhibitory activity with synthetic peptides also showed no significant difference. Both peptides are thus interesting novel candidates suitable for further development as ACE inhibitory and antioxidant agents from the natural source.
Keywords: Bioactive peptide, ACE inhibition, Nile tilapia, Angiotensin-I converting enzyme, Gelatin
Introduction
Peptides that are partially and enzymatically hydrolyzed from food proteins are important in food science studies (Chalé et al. 2014). Food proteins not only provide amino acids for the growth and health maintenance of humans, they also serve as a precursor for physiologically active peptides. Many health promoting peptides have also been identified from food protein hydrolysates. These bioactive peptides are generally short peptides (2–20 amino acids) and their native proteins often have no functional activity. Upon certain proteolysis, their specific bioactive roles can be achieved and exert beneficial effects at target sites in the body after absorption (Chi et al. 2015; De Gobba et al. 2014). Many different parameters, such as the source of protein, degree of hydrolysis, peptide structure, amino acid composition, molecular weight (MW), and type of protease used, can affect the bioactivity of these peptides (Li et al. 2013; Memarpoor-Yazdi et al. 2013). The relationship between the peptide MW and their biological activities has been reported in several studies (Vandanjon et al. 2009). Low MW fractions (<3000 Da) have been found as the most interesting bioactive peptides for nutritional and pharmaceutical applications (Saidi et al. 2014).
Nile tilapia, Oreochromis niloticus, is popular in freshwater aquaculture. In the global market, the demand for tilapia in all forms is increasing rapidly (Fitzsimmons 2004). More by-products have been produced from the expansion of the tilapia processing industry. More than 60 % of these by-products, including skin, head, fins and bones, are considered as waste (Dekkers et al. 2011). However, a significant amount of protein still remains in these by-products. Fish skin, in particular, is a rich source of collagen and gelatin (Hsu 2010).
To acquire gelatin, collagen must be heated to at least 45 °C to convert it into gelatin (Gόmez-Guillén et al. 2011). Enzymatic hydrolysis of gelatin is a method of choice that produces bioactive peptides without using organic solvents or toxic chemicals (Vercruysse et al. 2005). For most peptide bioactivities, including their inhibitory effect against angiotensin-I-converting enzyme (ACE) and their antioxidant, immunomodulatory and antimicrobial activities, the amino acids composition and structure of peptide are critical factors (Raghavan and Kristinsson 2009; Zhang et al. 2008).
Hypertension or high blood pressure, which is a serious condition of cardiovascular disease, is associated with myocardial infarction, coronary heart disease, stroke, kidney failure, heart failure and vascular dementia (Sharp et al. 2011). The renin-angiotensin system (RAS) is a vital system that controls blood pressure. In this system, the main regulators are renin and ACE (Fernández-Musoles et al. 2013). Many enzymatic hydrolysates of various marine by-products have been studied for their anti-hypertensive activity, such as Hoki skin (Mendis et al. 2005), squid skin (Alemán et al. 2011a) and Pacific cod skin (Himaya et al. 2012).
This research focused on purification and identification of bioactive peptide fractions having the greatest antioxidant and anti-hypertensive activities prepared by trypsin hydrolysis of Nile tilapia skin gelatin.
Materials and methods
Materials
Lyophilized Nile tilapia gelatin was obtained according to previous research (Choonpicharn et al. 2015). Chemicals 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox; 238813), trypsin (T8003), ACE from rabbit lung (A6778), hippuryl-histidyl-leucine (HHL; 859052), Sephadex® G-50 (G50150) and SP Sephadex® (SPC25120) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Peptides were synthesized by China Peptides Co Ltd. (Shanghai, China). All other reagents used were of standard laboratory grade.
Gelatin hydrolysate production
Freeze-dried gelatin was dissolved in 0.1 M sodium phosphate buffer and hydrolyzed with trypsin (pH 8.0, 37 °C) for 4 h with continuous shaking (Alemán et al. 2011b). The ratio for enzyme:substrate was 1:100 (w/w) (One microgram of trypsin contains 12.238 N-Alpha-Benzoyl-L-Arginine ethyl ester (BAEE) unit using BAEE as a substrate, as described by the supplier). These mixtures were boiled for 10 min to deactivate the enzyme and subsequently centrifuged at 4000 × g for 15 min. This supernatant was collected to lyophilize and kept at 4 °C until used.
Purification of bioactive peptide from Nile tilapia skin gelatin hydrolysate
Hydrolyzed tilapia skin gelatin was purified by gel filtration chromatography and ion exchange chromatography with slight modification of Zhang et al. (2012). Briefly, the lyophilized gelatin hydrolysate was dissolved in DI water and loaded onto Sephadex® G-50 column (2.0 cm × 60 cm). The eluent was DI water with a flow rate of 1 mL/min. Two mL of eluate was collected per tube and monitered at 280 nm using a UV/VIS spectrophotometer (Genesys 20, Thermo Fischer Scientific, Waltham, Massachusetts, United States). The collecting tubes that had high absorbance were pooled and lyophilized. Each fraction was tested for antioxidant and ACE inhibitory activities. The highest antioxidant and ACE inhibitory activity fractions were further purified by ion exchange chromatography. The freeze-dried fraction of each hydrolysate was dissolved in 20 mM sodium acetate buffer (pH 4.0) and applied to SP sephadex® C-25 column (2.0 cm × 30 cm). The peptide was eluted with 20 mM sodium acetate buffer at 0.6 mL/min flow rate with a linear gradient of NaCl of 0–1.0 M. The fractions were collected at 2 mL per tube, measured at 280 nm and tested for their bioactivity. Fractions with highest activity were lyophilized again for further sequence analysis.
ABTS radical scavenging assay
This assay is based on the reduction of the 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid), ABTS, radical cation by antioxidants (Arts et al. 2004). The ABTS˙ solution was prepared by mixing 7 mM ABTS solution in Milli-Q water with 2.45 mM potassium persulfate in a ratio of 1:1 (v/v) and kept it in the dark for 16–18 h. Before further use, the ABTS˙ solution was diluted with Milli-Q water to give an A734 around 0.700 ± 0.200. Following this, 20 μL of sample was added to 980 μL of ABTS˙ and left at room temperature for 10 min in dark. The absorbance was measured spectrophotometrically at 734 nm. Trolox was used as standard antioxidant and the results were calculated as trolox equivalent antioxidant capacity (TEAC) value (μg trolox/mg peptide).
Ferric reducing antioxidant power (FRAP) assay
The reducing power of trypsin hydrolysate fractions was determined according to the method described by Griffin and Bhagooli (2004) with slight modification. Freshly prepared FRAP reagent, containing 10 mM 2,4,6-tripyridyl-s-triazine (TPTZ), 20 mM FeCl2 and 300 mM acetate buffer pH 3.6 in the ratio of 1:1:10, was pre-incubated at 37 °C for 30 min. Then, 900 μL of FRAP reagent was added to 100 μL of sample and kept in the dark for 10 min. The absorbance was measured at 593 nm using spectrophotometer.
Determination of ACE inhibitory activity
The ACE inhibitory activity was evaluated using a modified method of Park et al. (2003). The sample (50 μL) was mixed with 25 mU/mL ACE (50 μL) and pre-incubated at 37 °C for 10 min. Then, 100 μL of 6 mM hippuryl-histidyl-leucine (HHL) in 50 mM Tris with 300 mM NaCl was added and incubated for 30 min. The reaction was stopped by adding 200 μL of 1.0 M HCl. Hippuric acid was extracted by ethyl acetate (600 μL), followed by centrifugation at 4000 × g for 15 min. The supernatant (200 μL) was transferred to a test tube and evaporated at 95 °C to remove ethyl acetate. One mL of distilled water was added to dissolve the hippuric acid and the absorbance was spectrophotometrically determined at 228 nm. The ACE inhibition was calculated from this equation:
where Acontrol was the absorbance of blank mixture (phosphate buffer instead of sample) and Asample was the absorbance of mixture with hydrolysate sample.
Peptide sequencing
The sequence of bioactive peptide was analyzed by Proteomics International Pty Ltd. (Nedlands, Western Australia). Briefly, the most active fraction obtained by purification was applied to MALDI-TOF/TOF mass spectrometer using a 5800 Proteomics Analyzer (AB Sciex, Framingham, Massachusetts, United States). MS spectra were collected in the molecular mass range of 800–8000 Da. For MS spectra, peak detection signal to noise (S/N) ratio was set to 15. MS/MS spectra were analyzed to identify protein of interest using Mascot sequence matching software (Matrix Science) with Ludwig NR Database.
Molecular docking
Two peptides that were identified from MALDI-TOF/TOF were designated as peptide 1 and peptide 2. The molecular structures of these two peptides were built and energy minimized using SYBYL X Molecular Modeling Software version 1.2 (Tripos Inc., St. Louis, Missouri, United States). The three-dimensional crystal structure of ACE (PDB ID: 4BZR) (Kramer et al. 2014) was retrieved from the Protein Data Bank and used as the target receptor for the docking. AutoDock Vina program (Trott and Olson 2010) was selected for the docking experiment because of its versatility and flexibility, which easily allow modification and optimization of the docking parameters. Structure of peptide 1 and peptide 2 was docked onto the rigid active site of ACE, where a grid box location was centered at the original ligand (K-26) binding site. Subsequently, 40 conformations of the docked complexes were automatically ranked based on their estimated binding affinities. Each ligand pose was closely inspected and the molecular interactions between peptide 1 or peptide 2 and the ACE were analyzed to confer the most plausible binding mode of the ligands.
Statistical analysis
All experiments were performed in triplicates and the data were presented as mean ± S.D. Statistical comparison was done by the SPSS statistic program (Version 17.0). The significance level (p < 0.05) was determined by one-way ANOVA with Duncan’s test.
Results and discussions
Purification of bioactive peptides and their characterization
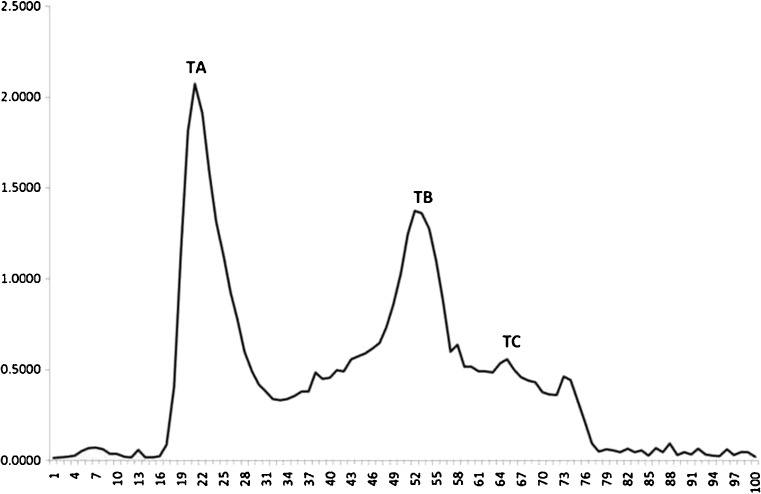
From our previous research, enzymatic incubation for 4 h resulted in the maximum degree of hydrolysis (DH) (Choonpicharn et al. 2015). The production yield of freeze-dried hydrolysate was around 90 % of freeze-dried gelatin weight basis. Trypsin hydrolysate was dissolved in DI ultra-pure water and loaded onto Sephadex® G-50 gel filtration column to fractionate the peptides according to their molecular weights. Three peaks were noticed and labelled as TA (tube 18–28), TB (tube 45–60) and TC (tube 63–68) (Fig. 1). The production yield for each fraction was 1.89 ± 0.02 %, 38.97 ± 0.04 % and 34.18 ± 0.04 % w/w, respectively. The antioxidant and ACE inhibitory activities were determined for each fraction (Table 1). No significant difference in the radical scavenging activity was observed among the three samples (p > 0.05). For FRAP assay, TA fraction, which contained peptides of the largest size among the three fractions, exhibited the highest reducing power among all fractions with 5.138 ± 1.060 μM trolox/mg peptide, while the very low reducing power potentials were observed in the TB and TC fractions (0.752 ± 0.019 and 0.599 ± 0.065 μM trolox/mg peptide, respectively). Zhang et al. (2012) reported that low MW tilapia skin gelatin hydrolysate from properase E and multifect neutral had the highest antioxidant activity among all fractions (IC50 = 110.80 μg/mL). Saidi et al. (2014) prepared a low molecular weight tuna muscle hydrolysate using alcalase and fractionated with ultrafiltration and nanofiltration membrane. The nanofiltration permeate (MW < 1 kDa) exhibited the highest 2,2′-diphenyl-1-picrylhydrazyl (DPPH), as well as the hydroxyl radical scavenging activities. The retentate (MW 1-4 kDa) also had the greatest superoxide radical and reducing power activities. Sudhakar and Nazeer (2015) reported that the peptide prepared from shotclub cuttlefish with trypsin hydrolysis exhibited highest antioxidant activity in DPPH, ABTS and superoxide anion radical scavenging assay and greatest total antioxidant capacity. Besides, the purified peptide with low MW (679.5 Da) showed significant antioxidant capacity in DPPH radical scavenging and reducing power assays. Cheung et al. (2012) hydrolyzed Pacific hake fillet by different proteases and found that those hydrolysates were good at chelating the metal ion, scavenging the ABTS and DPPH radicals, reducing the ferric ion and suppressing lipid peroxidation. Furthermore, they also noted that the peptide size (especially <1.4 kDa) was important for ABTS radical scavenging activity.
Fig. 1.
Chromatogram of trypsin gelatin hydrolysate from Nile tilapia skin fractionated by Sephadex® G-50 column that was eluted with DI water at flow rate of 1 mL/min. Groups of fractions are indicated: TA, tubes 18–28; TB, tubes 45–60; and TC, tubes 63–68
Table 1.
Bioactivities of purified peptides using a Sephadex® G-50 column
| Sample | TEAC* (μg trolox/mg peptide) | FRAP* (μM trolox/mg peptide) | %ACE* inhibition at 5 mg/mL |
|---|---|---|---|
| Crude trypsin hydrolysate | 10.291 ± 1.466a | 2.990 ± 0.503b | 56.218 ± 2.192a |
| TA** | 6.080 ± 3.887a | 5.138 ± 1.060a | 28.796 ± 9.105b |
| TB** | 8.156 ± 2.182a | 0.752 ± 0.019c | 59.325 ± 9.971a |
| TC** | 9.340 ± 1.910a | 0.599 ± 0.065c | 40.588 ± 5.889a,b |
Different letters represent significant differences (p < 0.05)
*TEAC, FRAP and ACE stand for trolox equivalent antioxidant capacity, ferric reducing antioxidant power and angiotensin I converting enzyme, respectively
**TA, TB and TC were the fractions obtained from Sephadex® G-50 gel filtration column
In the case of ACE inhibitory activity, the highest inhibition occured in TB fraction with 59.325 ± 9.971 %, followed by crude hydrolysate (56.218 ± 2.192 %), TC (40.588 ± 5.889 %) and TA (28.796 ± 9.105 %) fraction, respectively. These results suggested that a low MW fraction possesses more ACE inhibitory activity than a high MW fraction. Chalé et al. (2014) reported that the peptide fraction from the hydrolysate obtained by pepsin-pancreatin with MW lower than 1 kDa was the most active ACE inhibitory fraction (IC50 = 10.2 μg/mL). Espejo-Carpio et al. (2013) studied the ACE inhibition activity in goat milk hydrolyzed by subtilisin, trypsin and in combination of both enzymes. Their results showed that the highest activity was exhibited in the fraction having MW lower than 2.3 kDa. Similar results were also found in other researches. Mao et al. (2007) studied yak milk casein hydrolysate with alcalase and found that the highest ACE inhibition was noticed in the low MW peptide (<6 kDa). Jiang et al. (2010) suggested that the low MW peptide obtained from bovine casein hydrolysate (<3 kDa) exhibited the greatest ACE inhibiting activity. Ko et al. (2012) prepared protein hydrolysate from Chlorella ellipsoidea, a single-celled green algae, using protamex, kojizyme, neutrase, flavourzyme, alcalase, trypsin, α-chymotrypsin, pepsin and papain. His team found out that the highest ACE inhibitory activity was noticed from the alcalase hydrolysate with MW below 5 kDa. Raghavan and Kristinsson (2009) evaluated the ACE inhibitory potential of bioactive peptide from tilapia protein hydrolyzed with flavourzyme and cryotin-F. Their results showed that the maximum ACE inhibitory activity was noticed in both cryotin and flavourzyme hydrolysates with 25 % DH and, in addition, the greater inhibition was indicated in low MW peptides than high MW peptides.
However, the ACE inhibitory activity did not exhibit only in the bioactive peptide derived from the food proteins. Some medicinal plants also possessed pharmacological activities. Prathapan et al. (2011) informed that the methanolic extract of Boerhaavia diffusa, a vegetable medicinal plant widely used in South Asia, manifested the great antioxidant potency. The extract of B. diffusa could significantly scavenge the hydroxyl and superoxide radicals, inhibit lipid peroxidation, reduce ferric ion and chelate ferrous ion. Furthermore, in the experiment of H9c2 cardiac myoblast cells, the ethanolic extract of B. diffusa not only exhibited the ACE inhibitory activity but also increased various antioxidant enzyme activities and the concentration of glutathione (Prathapan et al. 2013).
Consequently, the TB fraction, which is the fraction with good radical scavenging activity and the highest ACE inhibitory activity together with highest production yield, was selected for further purification by ion exchange chromatography SP sephadex® C-25. However, the further purification with ion exchange column reduced the antioxidant and ACE inhibitory activities of bioactive peptide (data not shown). This may due to the loss of synergistic peptide to cooperate with their antioxidant and ACE inhibitory functions (Raghavan and Kristinsson 2009). Therefore, based on these results, TB fraction was chosen to identify for its sequence by MALDI-TOF/TOF MS/MS technique.
The result from MALDI-TOF/TOF technique showed that two significant peptides, 9- and 18-amino acids, are contained in the TB fraction. Their sequences were GPEGPAGAR (henceforth ‘peptide 1’, MW 810.87 Da) and GETGPAGPAGAAGPAGPR (henceforth ‘peptide 2’, MW 1490.61 Da).
Molecular docking of the peptide onto ACE
The molecular structures of peptide 1 and peptide 2 were built and energy minimized using SYBYL X Molecular Modeling Software and the structure of peptide 1 and peptide 2 was docked onto the rigid active site of ACE. In order to further evaluate the possibility of distinguishing between the binding modes of peptide 1 and peptide 2 bound onto ACE, molecular docking was performed on an X-ray complex structure of the target enzyme that has been co-crystalized with an K-26 (PDB 4BZR) inhibitor. This was reported by Kramer et al. (2014).
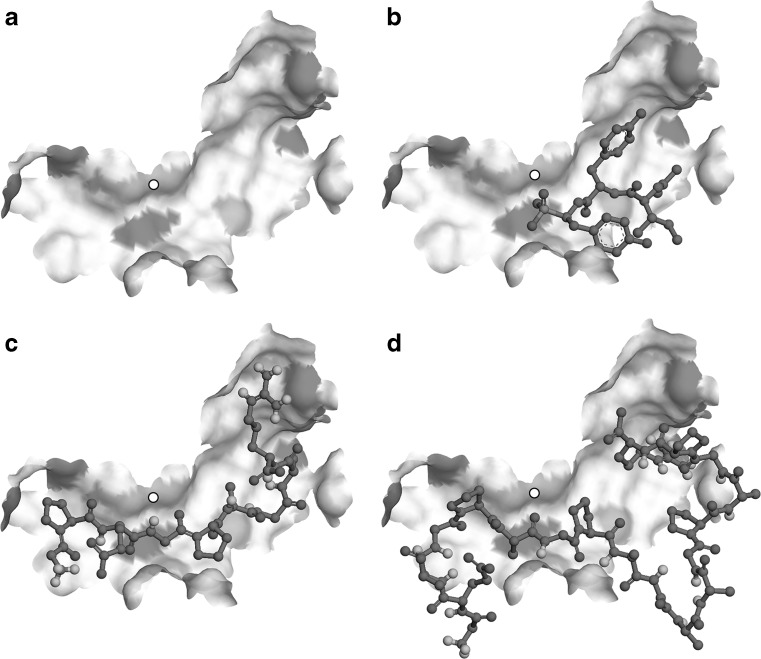
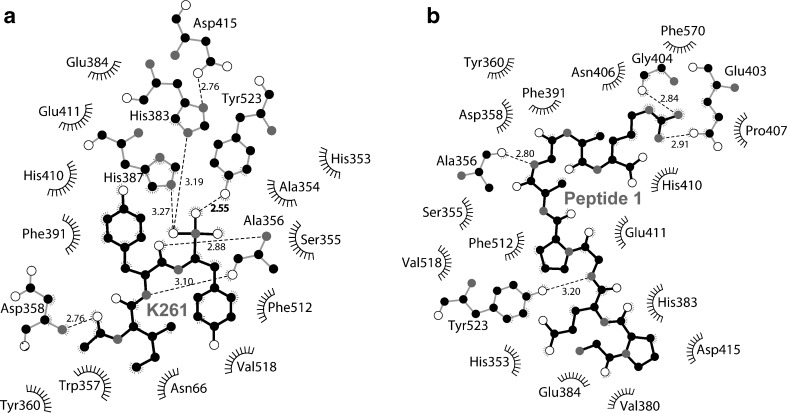
To confirm the validity of the docking, molecular interactions, including hydrogen bond, electrostatic and hydrophobic interactions found between the ligands and the active site of ACE were compared with those of inhibitor K-26. These specific interactions are summarized in Table 2. Molecular docking of K-26 inhibitor onto the active site of the ACE resulted in a pose (Fig. 2b) that, among the 40 conformations, has the lowest binding affinity of −8.2 kcal/mol (Table 2). This conformation revealed particular intermolecular interactions between the K-26 ligand and the ACE pocket (Fig. 3a). Peptide 1 docked onto the active site of the ACE revealed the best conformation (Fig. 2c) and a binding affinity (−9.5 kcal/mol), even better than of K-26. This may be due to a larger surface area of the binding interactions and, hence, a higher number of hydrogen bonds occurred at the carbonyl, amino, and hydroxyl groups of peptide 1 (Fig. 3b). For peptide 2, the best pose that docked onto the active site of the ACE (Fig. 2d) yielded a similar level of binding affinity (−9.3 kcal/mol), even though it has a longer chain and more surface area. However, some of the residues of peptide 2 appeared as sharp turns, which could generally render the overall complex less stable than that of peptide 1. Furthermore, when compared to similar interactions among the ligands in the common enzyme residues (Table 2), peptide 1 also has more interactions similar to the K-26 binding, even though it is considerably shorter than peptide 2. The binding conformation of peptide 1 in the active site cleft also appeared more expanded than that found in the peptide 2 complex. This usually indicates that a more stabilized conformation with potentially fewer stearic clashes occurred while the peptide threaded through the ACE. These characteristics suggested that peptide 1 is slightly more superior to peptide 2 in terms of fitting into the binding pocket of ACE, though having a similar level of binding affinity. Taken together, the docking study implies that the ACE-peptide 1 and ACE-peptide 2 complexes are more stable than the ACE-K-26 complex. Peptide 1 may be a better ligand for the ACE active site cleft than peptide 2, though not significantly.
Table 2.
Intermolecular interactions of the docked complexes between peptide 1, peptide 2, and the inhibitor K-26, and the active site residues of ACE. Ligand atoms are highlighted in bold letters. Similar interactions among the ligands of the common ACE residues are highlighted in the same style. Binding affinity of each complex calculated by the docking protocol is also indicated. OC = carbonyl, NH = amine/amide, OPO = phosphate, N+ = nitrogen positive charge, O- = oxygen negative charge
| Compounds | Binding affinity (kcal/mol) |
Protein-ligand interactions | ||
|---|---|---|---|---|
| Hydrogen bond | Electrostatic | Hydrophobic | ||
| K-26 | −8.2 |
ALA356:
OC
ALA356: NH ASP358 : OC GLU384:OPO T Y R 5 2 3 : O P O |
Zn 2+ : OPO | TRP357:pi-sigma HIS387 : pi-pi HIS410:pi-pi VAL518:pi-sigma |
| Peptide1 | - 9.5 | GLN281:OC(Glu3)
GLU411:OH(Gly4) T Y R 5 2 3 : O H (Gly4) ALA356: NH (Gly7) TYR360:OC(Gly7) GLU403:NH(Arg9) GLY404:NH(Arg9) |
GLU376:N
+
(Gly1)
ASP453:N + (Pro2) GLU411:N + (Pro5) GLU403:N + (Arg9) ARG522:O − (Arg9) Zn 2+ : OH (Gly4) |
HIS383:pi-sigma(Pro2)
HIS387 : pi-sigma (Pro5) PHE570:pi-sigma(Arg9) |
| Peptide2 | −9.3 | GLU162:NH(Gly1)
ASP377:NH(Gly1) CYS370:NH(Gly1) ALA354:OC(Gly1) LYS511:OC(Glu2) THR282:OH(Thr3) HIS353:NH(Gly7) ASN66:NH(Ala11) ARG124:OC(Ala12) TYR360:NH(Ala15) TYR394:NH(Arg18) ASP358 : NH (Arg18) TYR360:NH(Arg18) |
GLU162:N
+
(Gly1)
ASP377:N + (Gly1) LYS511:O − (Glu2) ASP358:N + (Arg18) Zn 2+ : OC (Ala6) |
HIS383:pi-sigma(Pro5)
HIS410:pi-cation (Arg18) |
Fig. 2.
Docking poses of ligands onto the active site of ACE. a Apo-enzyme generated by a manual removal of original (K-26) ligand from the co-crystal complex structure is shown in a surface model where ionic residues are highlighted in gray patches. A small white circle indicates the location of Zn2+ co-factor of the enzyme. b K-26 molecule re-docked onto the active site of the enzyme is shown in a ball-and-stick model. Peptide 1 (c) or peptide 2 (d) were also docked into the active site of the same apo-enzyme structure. Light gray atoms are oxygen
Fig. 3.
Two-dimensional schematics summarizing the key interactions between the ligands (black sticks) and the active site of ACE (gray sticks) analyzed by Ligplot (Wallace et al. 1995); (a) K-26 inhibitor, and (b) peptide 1. Dashed lines indicate hydrogen bonding and ‘eyelash’ curves show unbonded hydrophobic interactions. Key amino acid residues of the enzyme for the binding are indicated. White atoms are oxygen and gray atoms are nitrogen. Distances of key interactions are indicated
To confirm the ACE inhibitory activity of these two peptides, synthesized peptide 1 and 2 were used as an inhibitor sample at 1.0 mg/mL concentration. The results showed that peptide 2 could inhibit the ACE function with 40.521 ± 0.005 %, similar to peptide 1 inhibition (37.357 ± 0.003 %). Thus, no significant difference in ACE inhibitory activity between these two peptides was observed in vitro.
Conclusions
The results suggest that trypsin B fraction, being the low molecular weight peptide fraction, possesses high radical scavenging (TEAC value of 8.156 ± 2.182 μg trolox/mg peptide) and ACE inhibitory (59.325 ± 9.971 % inhibition) activities along with high productivity yield (38.97 ± 0.04 % w/w). The MALDI-TOF/TOF results show that the trypsin B (TB) fraction contained 2 main peptide sequences. These are GPEGPAGAR (peptide 1, MW 810.87 Da) and GETGPAGPAGAAGPAGPR (peptide 2, MW 1490.61 Da). Docking analysis shows that the characteristic of peptide 1 is slightly more suitable to fit into the binding pocket of ACE than peptide 2, though there is no significant difference in their binding energies. The experimental validation results of both synthetic peptides on the ACE inhibitory activity also indicated that they both have the same level of inhibitory activities. Thus, it can be concluded that both peptide 1 and 2 are new candidates towards the development of the ACE inhibitory and antioxidant agent isolated from the natural sources.
Acknowledgments
The authors thank the National Research Council of Thailand, the Graduate School of Chiang Mai University, and the Faculty of Science Publication Boosting Grant for the financial support.
Footnotes
Research highlights
• The research makes use of the fish skin waste with biotechnological knowledge.
• Peptides from Trypsin-digested gelatin are purified and sequenced.
• In silico ACE binding ability of the peptides was in agreement with in vitro ACE inhibitory assay.
• Two peptides were potential candidates for natural antihypertensive and antioxidant agents.
References
- Alemán A, Giménez B, Montero P, Gόmez-Guillén MC. Antioxidant activity of several marine skin gelatins. LWT Food Sci Technol. 2011;44:407–413. doi: 10.1016/j.lwt.2010.09.003. [DOI] [Google Scholar]
- Alemán A, Pérez-Santín E, Bordenave-Juchereau S, Arnaudin I, Gómez-Guillén MC, Montero P. Squid gelatin hydrolysates with antihypertensive, anticancer and antioxidant activity. Food Res Int. 2011;44:1044–1051. doi: 10.1016/j.foodres.2011.03.010. [DOI] [Google Scholar]
- Arts MJTJ, Haenen GRMM, Voss HP, Bast A. Antioxidant capacity of reaction products limits the applicability of the trolox equivalent antioxidant capacity (TEAC) assay. Food Chem Toxicol. 2004;42:45–49. doi: 10.1016/j.fct.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Chalé FGH, Ruiz JCR, Fernández JJA, Ancona DAB, Campos MRS. ACE inhibitory, hypotensive and antioxidant peptide fractions from mucuna pruriens proteins. Process Biochem. 2014;49:1691–1698. doi: 10.1016/j.procbio.2014.06.021. [DOI] [Google Scholar]
- Cheung IWY, Cheung LKY, Tan NY, Li-Chan ECY. The role of molecular size in antioxidant activity of peptide fractions from Pacific hake (merluccius productus) hydrolysates. Food Chem. 2012;134:1297–1306. doi: 10.1016/j.foodchem.2012.02.215. [DOI] [PubMed] [Google Scholar]
- Chi CF, Wang B, Wang YM, Zhang B, Deng SG. Isolation and characterization of three antioxidant peptides from protein hydrolysate of Bluefin leatherjacket (navodon septentrionalis) heads. J Funct Foods. 2015;12:1–10. doi: 10.1016/j.jff.2014.10.027. [DOI] [Google Scholar]
- Choonpicharn S, Jaturasitha S, Rakariyatham N, Suree N, Niamsup H. Antioxidant and antihypertensive activity of gelatin hydrolysate from Nile tilapia skin. J Food Sci Technol. 2015;52:3134–3139. doi: 10.1007/s13197-014-1581-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobba C, Tompa G, Otte J. Bioactive peptides from caseins released by cold active proteolytic enzymes from arsukibacterium ikkense. Food Chem. 2014;165:205–215. doi: 10.1016/j.foodchem.2014.05.082. [DOI] [PubMed] [Google Scholar]
- Dekkers E, Raghavan S, Kristinsson HG, Marshall MR. Oxidative stability of Mahi mahi red muscle dipped in tilapia protein hydrolysates. Food Chem. 2011;124:640–645. doi: 10.1016/j.foodchem.2010.06.088. [DOI] [Google Scholar]
- Espejo-Carpio FJ, De Gobba C, Guadix A, Guadix EM, Otte J. Angiotensin I-converting enzyme inhibitory activity of enzymatic hydrolysates of goat milk protein fractions. Int Dairy J. 2013;32:175–183. doi: 10.1016/j.idairyj.2013.04.002. [DOI] [Google Scholar]
- Fernández-Musoles R, Salom JB, Martínez-Maqueda D, Lόpez-Díez JJ, Recio I, Manzanares P. Antihypertensive effects of lactoferrin hydrolyzates: inhibition of angiotensin and endothelin-converting enzyme. Food Chem. 2013;139:994–1000. doi: 10.1016/j.foodchem.2012.12.049. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons KM (2004) Development of new products and markets for the global tilapia trade. Proceedings of ISTA 6. Manila, Philippine, pp 624–633
- Griffin SP, Bhagooli R. Measuring antioxidant potential in corals using the FRAP assay. J Exp Mar Biol Ecol. 2004;302:201–211. doi: 10.1016/j.jembe.2003.10.008. [DOI] [Google Scholar]
- Gόmez-Guillén MC, Giménez B, López-Caballero ME, Montero MP. Functional and bioactive properties of collagen and gelatin from alternative sources: a review. Food Hydrocoll. 2011;25:1813–1827. doi: 10.1016/j.foodhyd.2011.02.007. [DOI] [Google Scholar]
- Himaya SWA, Ngo DH, Ryu B, Kim SK. An active peptide purified from gastrointestinal enzyme hydrolysate of Pacific cod skin gelatin attenuates angiotensin-1 converting enzyme (ACE) activity and cellular oxidative stress. Food Chem. 2012;132:1872–1882. doi: 10.1016/j.foodchem.2011.12.020. [DOI] [Google Scholar]
- Hsu K. Purification of antioxidative peptides prepared from enzymatic hydrolysates of tuna dark muscle by-product. Food Chem. 2010;122:42–48. doi: 10.1016/j.foodchem.2010.02.013. [DOI] [Google Scholar]
- Jiang Z, Tian B, Brodkorb A, Huo G. Production, analysis and in vivo evaluation of novel angiotensin-I-converting enzyme inhibitory peptides from bovine casein. Food Chem. 2010;123:779–786. doi: 10.1016/j.foodchem.2010.05.026. [DOI] [Google Scholar]
- Ko SC, Kang N, Kim EA, Kang MC, Lee SH, Kang SM, Lee JB, Jeon BT, Kim SK, Park SJ, Park PJ, Jung WK, Kim D, Jeon YJ. A novel angiotensin I-converting enzyme (ACE) inhibitory peptide from a marine chlorella ellipsoidea and its antihypertensive effects in spontaneously hypertensive rats. Process Biochem. 2012;47:2005–2011. doi: 10.1016/j.procbio.2012.07.015. [DOI] [Google Scholar]
- Kramer GJ, Mohd A, Schwager SL, Masuyer G, Acharya KR, Sturrock ED, Bachmann BO. Interkingdom pharmacology of angiotensin-I converting enzyme inhibitor phosphonates produced by actinomycetes. ACS Med Chem Lett. 2014;5:346–351. doi: 10.1021/ml4004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang B, Chi C, Gong Y, Luo H, Ding G. Influence of average molecular weight on antioxidant and functional properties of cartilage collagen hydrolysates from Sphyrna lewini, dasyatis akeji and raja porosa. Food Res Int. 2013;51:283–293. doi: 10.1016/j.foodres.2012.12.031. [DOI] [Google Scholar]
- Mao XY, Ni JR, Sun WL, Hao PP, Fan L. Value-added utilization of yak milk casein for the production of angiotensin-I-converting enzyme inhibitory peptides. Food Chem. 2007;103:1282–1287. doi: 10.1016/j.foodchem.2006.10.041. [DOI] [Google Scholar]
- Memarpoor-Yazdi M, Mahaki H, Zare-Zardini H. Antioxidant activity of protein hydrolysates and purified peptides from zizyphus jujube fruits. J Funct Foods. 2013;5:62–70. doi: 10.1016/j.jff.2012.08.004. [DOI] [Google Scholar]
- Mendis E, Rajapakse N, Kim SK. Antioxidant properties of a radical-scavenging peptide purified from enzymatically prepared fish skin gelatin hydrolysate. J Agric Food Chem. 2005;53:581–587. doi: 10.1021/jf048877v. [DOI] [PubMed] [Google Scholar]
- Park PJ, Je JY, Kim SK. Angiotensin I converting enzyme (ACE) inhibitory activity of hetero-chitooligosaccharides prepared from partially different deacetylated chitosans. J Agric Food Chem. 2003;51:4930–4934. doi: 10.1021/jf0340557. [DOI] [PubMed] [Google Scholar]
- Prathapan A, Singh MK, Anusree SS, Kumar DRS, Sundaresan A, Raghu KG. Antiperoxidative, free radical scavenging and metal chelating activities of boerhaavia diffusa L. J Food Biochem. 2011;35:1548–1554. doi: 10.1111/j.1745-4514.2010.00477.x. [DOI] [Google Scholar]
- Prathapan A, Vineetha VP, Abhilash PA, Raghu KG. Boerhaavia diffusa L. attenuates angiotensin II-induced hypertrophy in H9c2 cardiac myoblast cells via modulating oxidative stress and down-regulating NF-κβ and transforming growth factor β1. Br J Nutr. 2013;110:1201–1210. doi: 10.1017/S0007114513000561. [DOI] [PubMed] [Google Scholar]
- Raghavan S, Kristinsson HG. ACE-inhibitory activity of tilapia protein hydrolysates. Food Chem. 2009;117:582–588. doi: 10.1016/j.foodchem.2009.04.058. [DOI] [Google Scholar]
- Saidi S, Deratani A, Belleville MP, Amar RB. Antioxidant properties of peptide fractions from tuna dark muscle protein by-product hydrolysate produced by membrane fractionation process. Food Res Int. 2014;65:329–336. doi: 10.1016/j.foodres.2014.09.023. [DOI] [Google Scholar]
- Sharp SI, Aarsland D, Day S, Sonnesyn H, Ballard C, Syst A. Hypertension is a potential risk factor for vascular dementia: systematic review. Int J Geriatr Psychiatry. 2011;26:661–669. doi: 10.1002/gps.2572. [DOI] [PubMed] [Google Scholar]
- Sudhakar S, Nazeer RA. Preparation of potent antioxidant peptide from edible part of shortclub cuttlefish against radical mediated lipid and DNA damage. LWT Food Sci Technol. 2015;64:593–601. doi: 10.1016/j.lwt.2015.06.031. [DOI] [Google Scholar]
- Trott O, Olson AJ. AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanjon L, Grignon M, Courois E, Bourseau P, Jaouen P. Fractionating white fish fillet hydrolysates by ultrafiltration and nanofiltration. J Food Eng. 2009;95:36–44. doi: 10.1016/j.jfoodeng.2009.04.007. [DOI] [Google Scholar]
- Vercruysse L, Van CJ, Smagghie G. ACE inhibitory peptides derived from enzymatic hydrolysates of animal muscle protein, a review. J Agric Food Chem. 2005;53:8106–8115. doi: 10.1021/jf0508908. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Zou AH, Manchu RG, Zhou YC, Wang SF. Purification and antimicrobial activity of antimicrobial protein from brown-spotted grouper, epinephelusfario. J Zool Syst Evol Res. 2008;29:627–632. [Google Scholar]
- Zhang Y, Duan X, Zhuang Y. Purification and characterization of novel antioxidant peptides from enzymatic hydrolysates of tilapia (Oreochromis niloticus) skin gelatin. Peptides. 2012;38:13–21. doi: 10.1016/j.peptides.2012.08.014. [DOI] [PubMed] [Google Scholar]