Abstract
Background:
Tuberculosis (TB) is a chronic wasting inflammatory disease characterized by multisystem involvement, which can cause metabolic derangements in afflicted patients. Metabolic signatures have been exploited in the study of several diseases. However, the serum that is successfully used in TB diagnosis on the basis of metabolic profiling is not by much.
Methods:
Orthogonal partial least-squares discriminant analysis was capable of distinguishing TB patients from both healthy subjects and patients with conditions other than TB. Therefore, TB-specific metabolic profiling was established. Clusters of potential biomarkers for differentiating TB active from non-TB diseases were identified using Mann–Whitney U-test. Multiple logistic regression analysis of metabolites was calculated to determine the suitable biomarker group that allows the efficient differentiation of patients with TB active from the control subjects.
Results:
From among 271 participants, 12 metabolites were found to contribute to the distinction between the TB active group and the control groups. These metabolites were mainly involved in the metabolic pathways of the following three biomolecules: Fatty acids, amino acids, and lipids. The receiver operating characteristic curves of 3D, 7D, and 11D-phytanic acid, behenic acid, and threoninyl-γ-glutamate exhibited excellent efficiency with area under the curve (AUC) values of 0.904 (95% confidence interval [CI]: 0863–0.944), 0.93 (95% CI: 0.893–0.966), and 0.964 (95% CI: 00.941–0.988), respectively. The largest and smallest resulting AUCs were 0.964 and 0.720, indicating that these biomarkers may be involved in the disease mechanisms. The combination of lysophosphatidylcholine (18:0), behenic acid, threoninyl-γ-glutamate, and presqualene diphosphate was used to represent the most suitable biomarker group for the differentiation of patients with TB active from the control subjects, with an AUC value of 0.991.
Conclusion:
The metabolic analysis results identified new serum biomarkers that can distinguish TB from non-TB diseases. The metabolomics-based analysis provides specific insights into the biology of TB and may offer new avenues for TB diagnosis.
Keywords: Metabolites, Orthogonal Partial Least-squares Discriminant Analysis, Serum, Tuberculosis, Ultra-performance Liquid Chromatography-mass Spectrometry
INTRODUCTION
Metabolomics is the quantitative measurement of the dynamic multiparametric metabolic responses of living systems to pathophysiological stimuli or genetic modifications.[1] Metabolites can be viewed as a close recapitulation of disease phenotypes, and they may represent the ongoing pathogenesis in an organism to a greater extent than changes in gene expression.[2] Consequently, it has deepened our understanding of the biological mechanisms involved in several noninfectious diseases and provided a platform for the identification of new biomarkers.[3] Metabolic signatures have been exploited in the study of several diseases, such as Alzheimer's disease,[4] Parkinson's disease,[5] myocardial ischemia,[6] hypertension,[7] cancer,[8] and diabetes.[9,10,11,12] However, this powerful analytical tool has not been widely applied to infectious diseases for the development of diagnostic biomarkers.[13,14,15]
Tuberculosis (TB) is a major worldwide health problem, with a global estimate of 9.4 million incidences in 2010 (range; 8.9 × 106–9.9 × 106). Of the more than 2 billion people infected with Mycobacterium tuberculosis (Mtb) globally, more than one-tenth are likely to develop active TB during their lifetime. In many regions, notably in developing countries with high TB incidences, diagnosis is neither sensitive nor specific, and an estimated 40% of TB patients fail to be correctly diagnosed.[16] In recent years, advancement in the field of metabolomics has mainly been applied to metabolic profiling about TB drug metabolism, rather than diagnosis of TB.[17,18,19,20,21] However, the serum that is successfully used in TB diagnosis on the basis of metabolic profiling is not by much.[22,23] A previous study by Weiner et al. using gas chromatography-mass spectrometry (MS) showed differences in metabolic profiles among the uninfected individuals, individuals with latent (inactive) TB, and patients with active TB.[24] In a recent study, nuclear magnetic resonance spectroscopy was used to characterize the metabolism of the host during Mtb infection.[25] However, relatively few methods are considered to be capable of distinguishing between active TB and diseases other than TB, such as lung cancer, pneumonia, and so forth. It can be expected that several of the observed changes are the result of a general inflammatory process rather than a specific response to TB. In this study, 120 patients with active TB and 251 controls who were either healthy or had diseases other than TB (non-TB group) were enrolled. Using ultra performance liquid chromatography-MS (UPLC-MS), we investigated the feasibility of identifying small molecule biochemical profiles in serum for gaining novel biological insights into the mechanisms underlying TB. The unique feature of this study was to employ the largest number of patients included in any study of a similar nature to date (120 with active TB and 251 controls). Multivariate statistical analysis was implemented to discriminate patients with active TB from the control subjects on the basis of their metabolic profiles. We identified 12 distinct biomarkers, including clusters that could be categorized as amino acids, fatty acids, lysophosphatidylcholine (lysoPC), and terpenoid compounds. Finally, these new metabolite markers, which can distinguish TB from non-TB diseases, led to a number of hypotheses. Our results provide specific insights into the biology of TB and may offer new avenues for TB diagnosis or therapy.
METHODS
Chemicals
Acetonitrile was purchased from Merck and Co. (Merck KGaA, Germany) and formic acid was obtained from Shanghai Nanxiang People Chemical Factory (China). All other solvents were of high-performance liquid chromatography grade. Reference standards were purchased from Sigma-Aldrich (St. Louis, MO, USA) and ultrapure water was prepared by Milli-Q system (Millipore Co., USA).
Serum sample collection
Serum samples were collected at Tianjin Haihe Hospital from 105 healthy individuals who visited the hospital for medical check-up, 146 patients with lung diseases that were due to non-TB conditions, and 120 patients with clinical signs of TB [Table 1]. TB patients had the following symptoms: Cough for more than 2 weeks and at least two additional symptoms (hemoptysis [coughing up blood], breathing difficulty, fever, night sweats, weight loss, chest pain, or fatigue). All patients were diagnosed based on chest X-rays and sputum samples obtained from each TB patient were analyzed by Ziehl-Neelsen staining and mycobacterium growth indicator tube culture. Active TB was diagnosed when (1) Mtb was cultured, (2) a caseating granuloma was found in the lung tissue by transthoracic needle biopsy and showed an appropriate response to treatment; or (3) clinical findings were compatible with TB, no clinical improvement was seen following treatment with empirical antibiotics, and treatment with anti-TB medication resulted in clinical and radiological improvement. The 251 controls without comorbidities were matched to the TB group with respect to age and sex. Whole-blood samples were drawn from a peripheral vein between 7:00 am and 9:00 am. Sera from patients and healthy volunteers were acquired from ethylenediaminetetraacetic acid-preserved whole blood samples following centrifugation and were stored at −80°C until analysis. At the time of sample collection, none of the patients were receiving treatment. Before the UPLC-MS analysis, serum samples were defrosted at room temperature for <20 minutes and 200 μl aliquots were combined with 300 μl of saline (0.9% NaCl in 20% D2O/80% H2O) and then centrifuged at 12,000 ×g for 5 minutes. A volume of 500 μl aliquot of the supernatant was pipetted into a 5 mm tube, and samples were stored at −80°C until further use.
Table 1.
Characteristics of TB patients and the control groups
| Items | Active TB | Healthy control | Non-TB group |
|---|---|---|---|
| Total individuals (n) | 120 | 105 | 146 |
| Age (years)* | 48.32 ± 18.15 | 42.71 ± 15.31 | 50.35 ± 17.41 |
| Gender (female/male) | 55/65 | 49/56 | 61/85 |
There was no significant difference in demographic data between TB patients and the controls. *Data are presented as mean ± SD. SD: Standard deviation; TB: Tuberculosis.
Sample extraction
Before further metabolomics analysis, samples were defrosted at room temperature for <20 minutes. Briefly, 400 μl of acetonitrile was added to the samples in a 4:1 (v/v) ratio. These sample mixtures were then homogenized by shaking them for 30 seconds and centrifuged at 15,000 ×g for 20 minutes at −4°C. The supernatant was collected and divided into three fractions: One for analysis by UPLC-MS, one for quality control (QC) analysis, and one was stored for further analysis (if necessary). A 500 μl aliquot of the supernatant was pipetted into a 5 mm test tube and then loaded into the UPLC-MS/MS system for analysis.
Instruments and conditions
In the first part of the study, samples were measured with a nano liquid chromatography system (Thermo Fisher Scientific, Germaring, Germany) coupled on-line to a hybrid linear ion trap/Orbitrap™ mass spectrometer (LTQ-Orbitrap-XL, Thermo Fisher Scientific). Samples were loaded onto a trap column (PepMap C18, 300 μm internal diameter [ID], 5 mm length, 5 μm particle size, 100 Å pore size; Thermo Fisher Scientific), then washed and desalted for 15 minutes using 0.1% trifluoroacetic acid in water as the loading solvent. Next, the trap column was switched in-line with the analytical column (PepMap C18, 75 μm ID, 250 mm length, 3 μm particle size, 100 Å pore size; Thermo Fisher Scientific) and peptides were eluted with the following binary gradient: Starting with 100% solvent A and B, where solvent A consisted of 2% acetonitrile and 0.1% formic acid in water, and solvent B consisted of 80% acetonitrile and 0.08% formic acid in water. The column flow rate was set at 200 nl/min. For electro-spray ionization (ESI), nano ESI emitters (New Objective, Woburn, MA, USA) were used, and a spray voltage of 4.5 kV was applied. For MS detection, a data-dependent acquisition method was used: High-resolution survey scan from 50 to 1000 (m\z). Orbitrap full scan spectra and ion trap MS/MS fragmentation spectra were acquired partially simultaneously.
Quality control solution composition
To observe the stability of the machine, QC solution was formulated from the mixed supernatants of all of the samples. The mixed samples solution was divided into 31 aliquots. Ten copies of QC solution were detected continuously before analyzing, and then the rest of copies were randomly inserted between each of the 10 sample runs [Figure 1]. The order in which samples were analyzed was randomized using the Excel 2013 software (Microsoft, USA), and a posttest blank sample was run after each of the 10 samples to avoid cross-contamination.
Figure 1.
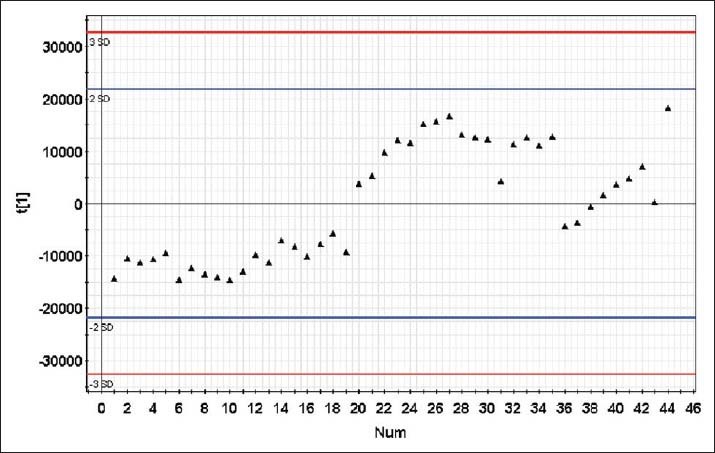
Quality control (QC) map. ▲: QC point. First ten QC points were run to and then the rest of the QC points were interleaved between the ten test samples. None of the QC points produced results outside of the control range.
Compound identification, quantification, and data curation
The UPLC-MS raw data were converted and processed by using MZmine 2.10 (http://www.biomedcentral.com/1471-2105/11/395). Briefly, chromatograms were built and peaks were recognized using the local minimum search function, and the ion intensities, matching m/z, and retention time was grouped into peak lists. Later, these peak lists were exported individually and imported into MetaboAnalyst 2.0 (http://nar.oxfordjournals.org/content/early/2012/05/01/nar.gks374.full). The peaks were aligned and normalized to the sum of all detected peaks. The processed and normalized data were imported into SIMCA-P (Umetrics, Umeε, Sweden) for multivariate statistical analysis. To distinguish TB from the controls, orthogonal partial least squares discriminant analysis (OPLS-DA) was performed. Based on the OPLS-DA model, the specific metabolites were determined by applying Mann–Whitney U-test (SPSS 15.0, IBM, NY, USA) with P value threshold of 0.05. For each biomarker, a receiver operating characteristic (ROC) curve was generated. The area under curve (AUC) value and 95% confidence interval (CI) were calculated to determine the specificity and sensitivity of TB. To increase the diagnostic accuracy of combined changes in serum metabolites levels, multiple logistic regression analysis was carried out.
RESULTS
Baseline characteristics
Table 1 lists the baseline characteristics of enrolled patients and the controls. The active TB patients were 48.32 ± 18.15 (mean ± standard deviation) years old on average and consisted of 65 males and 55 females. The healthy controls were 42.71 ± 15.31 years old and consisted of 56 males and 49 females. The non-TB controls were 50.35 ± 17.41 years old and consisted of 85 males and 61 females. Of the 146 patients with non-TB controls, 51 patients (60.1%) had lung cancer, comprising the cancer subgroup (L); 45 patients (39.9%) made up the pneumonia subgroup (P); 28 patients were in the chronic obstructive pulmonary disease (COPD) subgroup (C); and 22 patients comprised the bronchiectasis subgroup (B).
Ultra performance liquid chromatography-mass spectrometry platform performance
Representative UPLC-MS chromatograms of the serum samples of the patients with active and the subjects with the healthy and non-TB controls were shown in Figure 2. The peaks were very well resolved and were evenly dispersed across the entire retention time domain, showing the high quality of the raw data. There were many significant differences in the areas and heights of peaks among groups, as a matter of fact, it is inferred that differences in peaks resulted from metabolic derangements along with diseases.
Figure 2.
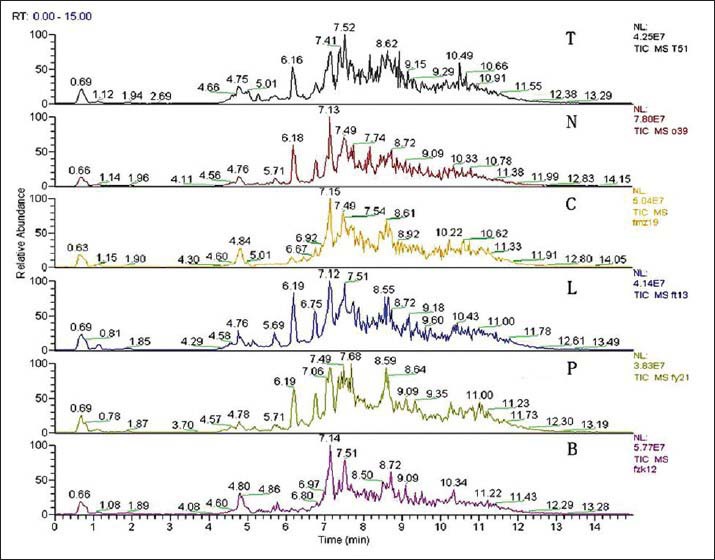
High performance liquid chromatography-mass spectrometry chromatograms of the serum samples of patients from the tuberculosis (TB) group, the healthy group, and the four subgroups of patients with non-TB diseases. Abscissa: Retention time. Ordinate: Relative abundance. The numbers on the point of the peaks indicate the retention time of each substance. T indicates TB group; N indicates healthy control; P indicates pulmonitis subgroup; L indicates lung cancer subgroup; C indicates chronic obstructive pulmonary disease subgroup; B indicates bronchiectasis subgroup.
Differentiation between tuberculosis patients and controls, and among the subgroups based on OPLS-DA
As mentioned above, the overall peak profiles of the three groups looked quite different, which suggested that these profiles could be used to discriminate TB from the controls. For the holistic treatment of these data, multivariate analysis was used to identify the metabolomic differences between the groups. For data reduction and pattern recognition (PR), a series of PR methods were applied using SIMCA-P 12.1 software (Umetrics, Sweden). Principal component analysis (PCA) was initially applied to the data to visualize inherent clustering between the three groups. PCA involves the transformation of a multidimensional set of possibly correlated variables into two linearly uncorrelated dimensions. This model explained an estimated 41.4% of the original data (R2 = 0.414 and Q2 = 0.334) [Figure 3]. However, in addition to the effects of the diseases on the metabolome, there were other factors that are known to contribute to differences of endogenous metabolites, such as age and diet.
Figure 3.
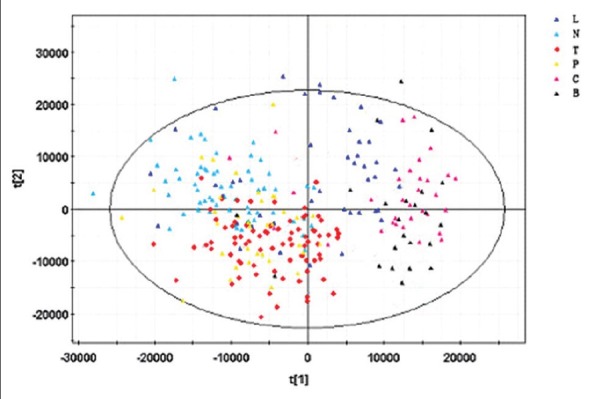
Principal component analysis (PCA) model. PCA scores plots of the SIMCA-P + 12.0.1.0 generated data, showing tuberculosis (TB) patients versus healthy controls and the non-TB group collected serum samples before the removal of ‘noise’ and interfering compounds from the dataset.  : TB group;
: TB group;  : Healthy control;
: Healthy control;  : Pulmonitis subgroup;
: Pulmonitis subgroup;  : Lung cancer subgroup;
: Lung cancer subgroup;  : COPD subgroup;
: COPD subgroup;  : Bronchiectasis subgroup.
: Bronchiectasis subgroup.
Therefore, orthogonal signal correction technology was used to filter out unrelated variable information and retain related variables. Hence, OPLS-DA model was introduced, which is used to determine the maximum separation between different kinds of samples according to the sample classification information. The OPLS-DA model was used to identify biomarkers that accounted for the differences between the three groups, and it clearly distinguished between the TB group and the two control groups (non-TB disease subgroups were combined) [Figure 4]. The results showed that 82.1% of samples were consistent with the discrimination of the model, and the predictive ability of the model was 58.2% (Q2Y = 0.582). These findings indicate that the OPLS-DA model may pave the way for the diagnosis of TB and permit differentiation between other kinds of related diseases.
Figure 4.
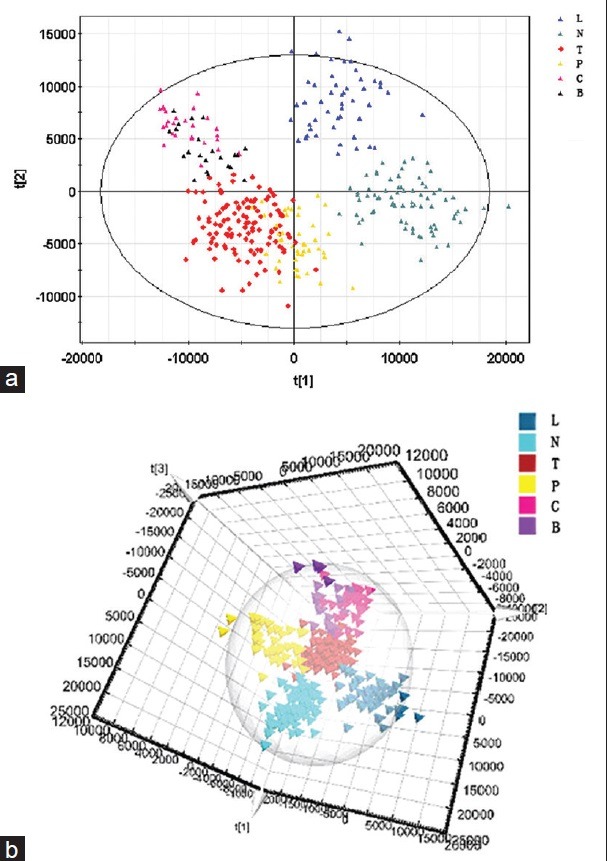
(a) OPLS-DA two-dimensional model.  : TB group;
: TB group;  : Healthy control;
: Healthy control;  : Pulmonitis subgroup;
: Pulmonitis subgroup;  : Lung cancer subgroup;
: Lung cancer subgroup;  : COPD subgroup;
: COPD subgroup;  : Bronchiectasis subgroup. (b) OPLS-DA three-dimensional model. OPLS-DA scores plot discriminating serum samples of tuberculosis (TB) patients, healthy control and non-TB group based on the metabolite profiling data.
: Bronchiectasis subgroup. (b) OPLS-DA three-dimensional model. OPLS-DA scores plot discriminating serum samples of tuberculosis (TB) patients, healthy control and non-TB group based on the metabolite profiling data.  : TB group;
: TB group;  : Healthy control;
: Healthy control;  : Pulmonitis subgroup;
: Pulmonitis subgroup;  : Lung cancer subgroup;
: Lung cancer subgroup;  : COPD subgroup;
: COPD subgroup;  : Bronchiectasis subgroup.
: Bronchiectasis subgroup.
Identification of tuberculosis-specific metabolites
The successful use of UPLC-MS metabolomics analysis and the OPLS-DA model described above to distinguish between the TB group and control groups led us to search for the specific metabolites that contributed to the metabolomic differences. Based on the OPLS-DA model, the signals that were highly correlated and had high signal-to-noise ratio values were selected. The metabolites of >400 small molecules in the sera of patients in the three groups were explored. The molecules responsible for these signals were identified and differences in the abundance of these small molecules were determined by applying Mann–Whitney U-test for each of the three possible comparisons, using a P value threshold of 0.05. Twenty-seven metabolites were detected at significantly different levels between the active TB group and the control groups. Of those metabolites identified to differ significantly between groups, 12 metabolites [Table 2 and Figure 6], were clustered in the fatty acid, phospholipids, amino acids, and terpenoid compounds metabolite sets. To confirm that the three groups differed in terms of the serum levels of these metabolites, a heat map, a graphical representation of data where the individual values are represented as colors, was drawn [Figure 5]. Heat maps are 2D displays of the measured experimental values in the data matrix. The relatively high abundance of any specific metabolite is represented by yellow-colored squares (pixels) and a low abundance is represented by orange-colored squares.
Table 2.
Details of the 12 metabolites best describing the variation between patients with active TB and the controls
| Metabolite | Preferred adducts | Molecular weight (m/z)* | P | Retention time | Molecular formula† | Trend (O/T)‡ |
|---|---|---|---|---|---|---|
| Palmitic acid | M + NH4 | 274.274 | 0.000053 | 6.17544 | C16H32O2 | ↓ |
| LysoPC (16:0) | M + H | 496.339 | 0.000765 | 7.5063 | C24H50NO7P | ↓ |
| LysoPC (18:0) | M + Na | 546.352 | 0.001066 | 8.6785 | C26H54NO7P | ↓ |
| 3D, 7D, 11D-phytanic acid | M + NH4 | 330.336 | 0.000654 | 7.3454 | C20H40O2 | ↓ |
| Behenic acid | M + NH4 | 358.367 | 0.000954 | 7.92855 | C22H44O2 | ↓ |
| Phytal | M + NH4 | 312.329 | 0.002911 | 7.35257 | C20H38O | ↓ |
| Threoninyl-γ-glutamate | M + H | 248.124 | 0.000097 | 5.66712 | C9H17N3O5 | ↓ |
| Kynurenine | M + NH4 | 341.24 | 0.003481 | 4.80868 | C10H12N2O3 | ↑ |
| Quinolinic acid | M + H | 565.283 | 0.002253 | 4.46054 | C7H5NO4 | ↑ |
| Presqualene diphosphate | M + Na | 609.31 | 0.001955 | 4.49312 | C30H52O7P2 | ↑ |
| LysoPC (P-18:1 (9Z)) | M + H | 506.36 | 0.000501 | 8.67014 | C26H52NO6P | ↓ |
| LysoPC (P-16:0) | M + H | 480.344 | 0.000851 | 7.82707 | C24H50NO6P | ↓ |
*Molecular weight (m/z) is denoted by its monoisotopic mass, †The chemical formulas were predicted based on accurate mass by using the molecular formula generator algorithm of Mass Frontier 6.0 software (Thermo Fisher Scientific), ‡The trend of marker levels in the active TB group. ↑ and ↓ indicate increased and decreased levels, respectively, compared with the healthy group. TB: Tuberculosis; LysoPC: Lysophosphatidylcholine.
Figure 6.

Relative abundances of metabolites in tuberculosis patients (T), healthy controls (O) and nontuberculosis patients (N). The metabolites changed in relative abundance between groups were: Palmitic acid, LysoPC (16:0), LysoPC (18:0), phytanic acid, behenic acid, phytal, threoninyl-γ-glutamate, kynurenine, quinolinic acid, presqualene diphosphate, LysoPC (P-18:1 (9Z)), and LysoPC (P-16:0). Black lines indicates sample means. Asterisks indicate significant differences between 2 groups (results from t-tests corrected for multiple testing; *P < 0.05; **P < 0.01).
Figure 5.
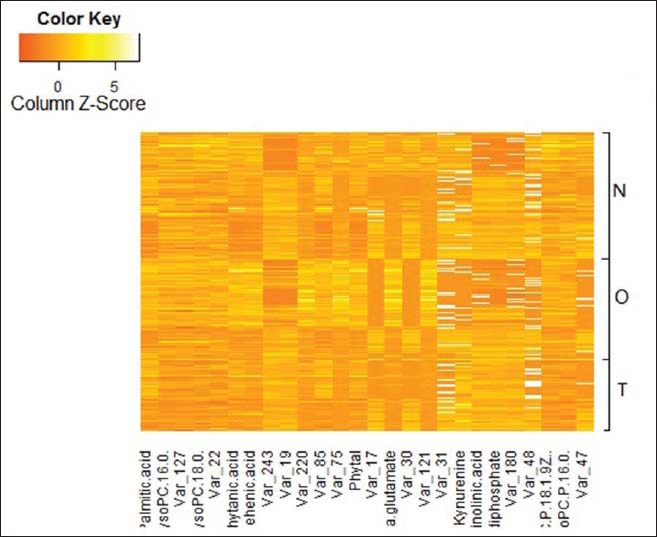
Heat map based on the differences in the abundance of small metabolic compounds among sera from the tuberculosis (TB) group, healthy group and non-TB group. Yellow color indicates higher abundance of metabolites; Orange color indicates lower abundance of metabolites.
The results showed that the three groups were significantly different in terms of the abundances of these biomarker metabolites in the patient sera.
Differences in small metabolites can be used as specific and sensitive biosignatures of tuberculosis status
To investigate whether the characteristics of the metabolites that significantly differed among the three groups could be efficiently exploited for building a sensitive biosignature of TB status, ROC curves, which have been conventionally used to evaluate diagnostic performance in clinical research, were calculated. In the specific metabolites that were decreased in active TB patients, the ROC curves of 3D, 7D, 11D-phytanic acid, behenic acid and threoninyl-γ-glutamate exhibited excellent efficiency with a AUC values of 0.904 (95% CI: 0863–0.944), 0.93 (95% CI: 0.893–0.966) and 0.964 (95% CI: 00.941–0.988), respectively [Table 3 and Figure 7a]. Kynurenine, quinolinic acid (QUIN), and presqualene diphosphate (PSDP) showed significant up-regulation in patients with active TB (P < 0.05). The AUC values for these metabolites were more than 0.8 [Table 3 and Figure 7b], showing moderate performance of diagnostic value. The largest and smallest resulting AUC values were 0.964 and 0.720 [Table 3, Figure 7a and b], which indicated that these biomarkers may potentially be involved in the disease mechanisms. Using multiple logistic regression analysis of these 12 metabolites, the combination of lysoPC (18:0), behenic acid, threoninyl-γ-glutamate, and PSDP was used to represent a suitable biomarker group that allowed efficient differentiation of active TB from the controls. The resulting ROC curve of the biomarker combination had an AUC value of 0.991 (95% CI: 0.982–1.000) [Table 3 and Figure 7c], which reflects strong significant difference between active TB and the control patient groups.
Table 3.
The ROC curves of the 12 metabolites found to represent potential biomarkers of active TB
| Test result variable (s) | Area | Standard error§ | Asymptotic significant|| | Asymptotic 95% confidence intervals | |
|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||
| Palmitic acid* | 0.772 | 0.033 | 0.000 | 0.708 | 0.837 |
| LysoPC_16_0* | 0.786 | 0.032 | 0.000 | 0.723 | 0.850 |
| LysoPC_18_0* | 0.720 | 0.036 | 0.000 | 0.650 | 0.790 |
| 3D, 7D, 11D-phytanic acid* | 0.904 | 0.021 | 0.000 | 0.863 | 0.944 |
| Behenic acid* | 0.930 | 0.019 | 0.000 | 0.893 | 0.966 |
| Phytal* | 0.820 | 0.031 | 0.000 | 0.761 | 0.880 |
| Threoninyl-γ-glutamate* | 0.964 | 0.012 | 0.000 | 0.941 | 0.988 |
| LysoPC P18* | 0.770 | 0.033 | 0.000 | 0.706 | 0.834 |
| LysoPC P16* | 0.732 | 0.036 | 0.000 | 0.662 | 0.802 |
| Kynurenine† | 0.826 | 0.032 | 0.000 | 0.764 | 0.888 |
| Quinolinic acid† | 0.854 | 0.033 | 0.000 | 0.788 | 0.919 |
| Presqualene diphosphate† | 0.856 | 0.03 | 0.000 | 0.791 | 0.920 |
| LysoPC (18:0), behenic acid, threoninyl- γ-glutamate, and presqualene diphosphate‡ | 0.991 | 0.005 | 0.000 | 0.982 | 1.000 |
*Decreased metabolites, †Increased metabolites, ‡The biomarker combination, §Under the nonparametric assumption, ||Null hypothesis: True area = 0.5. TB: Tuberculosis; LysoPC: Lysophosphatidylcholine; ROC: Receiver operating characteristic.
Figure 7.
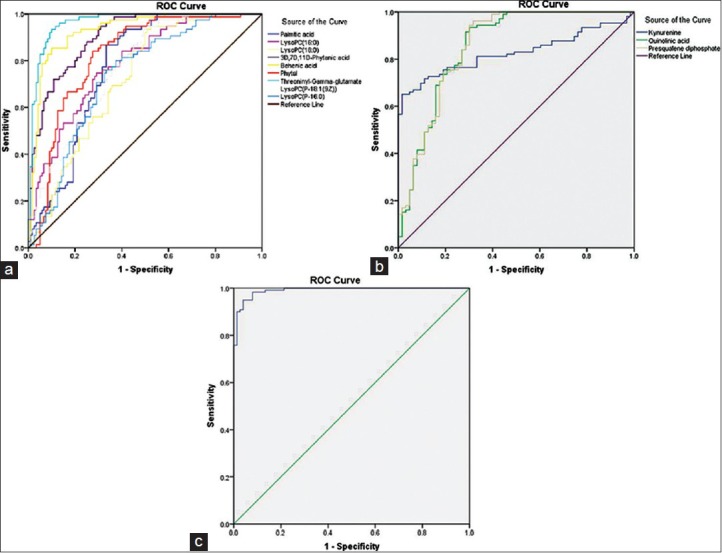
(a) The receiver operating characteristic (ROC) curve of metabolites that were decreased in the active tuberculosis (TB) group compared with controls. ROC curves of metabolites for which the serum concentrations were significantly decreased in the active TB group compared with controls. The ROC curves of each metabolite that was decreased in concentration in the TB group sera showed a moderate distinguishing efficiency. (b) The receiver operating characteristic (ROC) curve of metabolites that were increased in the active tuberculosis (TB) group compared with controls. ROC curves of metabolites for which the serum concentrations were significantly increased in the active TB group compared with controls. The ROC curves of each metabolite that was increased in concentration in the TB group sera showed a moderate distinguishing efficiency. (c) The receiver operating characteristic (ROC) curve of the biomarker combination identified as a putative serum signature of active tuberculosis (TB). The ROC curve of the combination of lysophosphatidylcholine (18:0), behenic acid, threoninyl-γ-glutamate, and presqualene diphosphate showed excellent distinguishing efficiency between patients with active TB and controls.
Comparison between patients with active tuberculosis and nontuberculosis disease subgroups based on serum metabolic profiling
To determine whether our metabolomic approach could be used to make a distinction between the active TB and non-TB groups, Mann–Whitney U-test was applied for comparison between the active TB group and the subgroups of non-TB. The findings indicated that the UPLC-MS analysis of serum may help in the diagnosis of TB and non-TB as remarked above. Finally, the specific markers between active TB and the subgroups were explored as below [Table 4]:
Table 4.
P values for comparisons between the active TB group and each subgroup of the Non-TB group
| Items | LC | COPD | Pulmonitis | Bronchiectasis |
|---|---|---|---|---|
| Palmitic acid | 0.0723 | 0.0000† | 0.0010† | 0.0230* |
| LysoPC (16:0) | 0.0000† | 0.0000† | 0.0030† | 0.0040* |
| LysoPC (18:0) | 0.0000† | 0.4820 | 0.2000 | 0.5460 |
| 3D, 7D, 11D-phytanic acid | 0.0000† | 0.0000† | 0.0380* | 0.0080* |
| Behenic acid | 0.0000† | 0.0000† | 0.3320 | 0.0150 |
| Phytal | 0.0010* | 0.0000† | 0.0790 | 0.0030* |
| Threoninyl-γ-glutamate | 0.0000† | 0.0120* | 0.0690 | 0.2890 |
| Kynurenine | 0.0020* | 0.0060† | 0.0210* | 0.0080* |
| Quinolinic acid | 0.0000† | 0.1110 | 0.2800 | 0.2420 |
| Presqualene diphosphate | 0.0000† | 0.0820 | 0.1900 | 0.2250 |
| LysoPC (P-18:1 (9Z)) | 0.0000† | 0.0320* | 0.8820 | 0.1940 |
| LysoPC (P-16:0) | 0.0000† | 0.0000* | 0.1870 | 0.0020* |
*P < 0.05, †P < 0.01. Application Mann-whitney U-test in TB group and each subgroup. TB: Tuberculosis; LysoPC: Lysophosphatidylcholine; COPD: Chronic obstructive pulmonary disease; LC: Liquid chromatography.
The serum levels of PSDP, lysoPCs, 3D, 7D, 11D-phytanic acid, behenic acid, hypoglycin B, phytal and threoninyl-γ-glutamate were significantly different between the active TB and lung cancer subgroups (Π < 0.05)
Palmitic acid, lysoPC (16:0), 3D, 7D, 11D-phytanic acid, behenic acid, phytal, lysoPC (P-16:0), and kynurenine were more effective in discriminating active TB from bronchiectasis (Π < 0.05)
Palmitic acid, lysoPC (16:0), 3D, 7D, 11D-phytanic acid, behenic acid, phytal, threoninyl-γ-glutamate, kynurenine, and lysoPC (P-16:0) were expected to become more effective in diagnosis and differentiation of active TB from COPD (Π < 0.05)
The serum levels of palmitic acid, lysoPC (16:0), 3D, 7D, 11D-phytanic acid, and kynurenine were significantly different between the active TB and pneumonia subgroups
LysoPC (16:0) was the only single metabolite to significantly distinguish active TB from the diseases other than TB (Π < 0.05).
DISCUSSION
The present study used UPLC-MS to profile active TB and build a statistical model that enabled the identification of biomarkers for disease diagnosis based on metabolomic research. Findings indicated that 12 metabolites were unambiguously altered in serum of active TB patients as compared with two other groups (healthy controls and non-TB disease patients) [Table 2]. In the following discussion, we consider the biological relevance of these prominent metabolites and their potential biosignatures for the diagnosis of active TB.
Fatty acids
It is universally acknowledged that Mtb preferentially relies on fatty acid metabolism to maintain chronic infection.[26] When there is persistent infection in the lung tissue, the fatty acids, which are metabolized in two ways (in β-oxidation decomposition and the glyoxylate cycle) may be a source of carbon and energy of Mtb.[27] In this study, palmitic acid, phytanic acid and behenic acid, which decreased significantly in the sera of TB patients compared with the healthy group, bronchiectasis, and COPD subgroups, may be among the products of the fatty acid consumption. In addition, palmitic acid, as the most abundant free fatty acid in the human body, can induce inhibition of the electron transport chain of macrophage, which, in turn, reduces adenosine triphosphate production in the mitochondria and stimulates the activity of the mitochondrial apoptotic pathway. Another study that further supports our findings was that the palmitic acid, as one of the active compounds from the plant extraction, has some toxicity toward mycobacteria, but it is not highly toxic.[28]
Phytanic acid, a 20-carbon branched chain fatty acid, can be used as the substrate of CYP124A1,[29] which is a heme-containing enzyme that belongs to the cytochrome p450 superfamily. When CYP124A1 was combined with the substrate of phytanic acid, the molecular conformation of CYP124A1 changed in favor of lipid oxidation, to play a better role in the important biological functions of Mtb.[30] Meanwhile, phytanic acid, as a substrate of CYP124A1, was consumed.
In our study, behenic acid was detected in a lower concentration in the TB group than the healthy and non-TB groups, except for the pneumonia subgroup. Behenic acid has been identified as a fatty acid that increases cholesterol in humans.[31] Cholesterol has a significant role in the development of TB, because it is necessary for the good functioning of macrophages and lymphocytes.[32] In the macrophage cell membrane, cholesterol directly participates in the function of phagocytizing Mtb, while, in the lymphocyte membrane, it is involved in the differentiation and proliferation of cytotoxic cells. Apart from this, cholesterol also has the functions of activating lymphocytes, CD4+ T cells, CD8+ T cells, and γ-T cells, and promoting the release of interferon and tumor necrosis factor-α, all of which effects are effective in killing Mtb. A previous study showed that TB patients universally had a lowered concentration of total serum cholesterol.[33] Due to this, the decreased concentration of behenic acid, as a cholesterol-raising fatty acid, in sera of TB patients may further explain the hypocholesteremia status in TB.[34]
Lysophosphatidylcholines
Lysophosphatidylcholine (P-18:1(9Z)), lysoPC (P-16:0), lysoPC (16:0), and lysoPC (18:0) formed one cluster of metabolites that were present at lower levels in the active TB group and significantly differed between the healthy and non-TB groups. LysoPCs impair nitric oxide (NO) production and endothelium-dependent vasorelaxation. Moreover, lysoPC levels have been shown to be negatively correlated with NO levels.[36] Other research has pointed out that the generation of NO by activated macrophages could control mycobacterial infection in the murine system.[35] A low lysoPC level may accelerate Mtb infection by reducing the generation of NO. Additionally, another finding that TB can induce macrophage apoptosis by inhibition of phospholipase A2[37,38,39] could also explain the low concentrations of lysoPCs in patients with active TB, a finding that is consistent with the previous report by Weiner et al.[24]
Amino acids
Our data showed an increase in the levels of two amino acids (kynurenine and QUIN), and a decrease in threoninyl-γ-glutamate in sera from TB patients relative to those of healthy controls, suggesting alterations in protein metabolism during active TB. Amino acid metabolism is complicated, involving a large number of metabolites. Gluconeogenesis, proteolysis, and oxidative catabolism contribute to amino acid balance. Tryptophan (TRP) is an essential amino acid that has various important biological functions. Kynurenine, as the downstream metabolite of TRP, can be accumulated by enhancing the activity of indoleamine 2, 3-dioxygenase-1 (IDO-1), which is widely known as the rate-limiting enzyme in TRP metabolism.[40] In addition, kynurenine can regulate human T cells, which are well known as the main anti-TB immunological cell type.
Quinolinic acid, a neuroactive metabolite of the kynurenine pathway, can induce the expression of several proinflammatory cytokines and chemokines. These findings support the concept that the TRP-kynurenine pathway can influence immune functions through the effects of TRP depletion and the accumulation of kynurenine and QUIN, suggesting that the TRP degradation pathway controlled by IDO-1 is involved in the pathogenesis of pulmonary TB.[41] QUIN, a neuroactive metabolite of the kynurenine pathway, can induce the expression of several proinflammatory cytokines and chemokines.[42] In addition, imbalances in TRP metabolism have been linked to cancer-related immune escape and implicated in lung cancer.[43]
Threoninyl-γ-glutamate exhibited the most excellent efficiency with an AUC value of 0.964 (95% CI: 00.941–0.988) [Table 3 and Figure 7a], is a dipeptide composed of threonine and γ-glutamate. Some dipeptides are known to have physiological or cell-signaling effects, although most are simply short-lived intermediates on their way to specific amino acid degradation pathways following further proteolysis. This dipeptide has not yet been identified in human tissues or biofluids; hence, it is classified as an “expected” metabolite.
Terpenoid compounds
One of the most prominent clusters of metabolites in our study represented terpenoid compounds, including PSDP and phytal. The latter compound, which to date has never been identified or associated with TB, belongs to the family of diterpenes. However, it is reported that elisapterosin B, one kind of diterpenes extracted from plants, could inhibit the growth of Mtb.[44] Thus, the relationship between phytal and disease as well as its involvement in disease mechanisms is open to further study.
Presqualene diphosphate, an intermediate in the biosynthesis of terpenoids, was found at higher levels in the active TB group and significantly differed between the active TB and cancer subgroups by an order of magnitude. PSDP exerts an intercellular signal for the down-regulation of superoxide in neutrophils,[45] which is widely known to play a central role in host defense, inflammation, and tissue damage.[46] PSDP, an endogenous PI3K inhibitor, directly inhibited recombinant human p110-pI3K activity, which can activate polymorphonuclear neutrophils (PMN). PMN is the primary initial immune effectors of acute inflammation, and potentially PMN products released into surrounding tissues contribute to lung and respiratory injury. A negative correlation between PSDP and PMN was observed. The observed increase in the serum concentration of PSDP may be an indicator of TB inflammatory progression. PSDP, after the initial acute inflammatory state, may act to limit tissue injury by inhibiting phospholipase D, phosphoinositol-3 kinase, and superoxide anion generation, thus providing protection for the host tissues. In addition, PSDP, a well-recognized intermediate of cholesterol biosynthesis, exists in immune effector cells and is a potential regulator of the cellular response in host defense. The increased PSDP in TB patients may be a response to Mtb in host defense, which could also explain the result of this study.
CONCLUSIONS
Metabolic profiling approaches based on UPLC-MS were successfully used to distinguish TB patients from the controls and establish a TB-specific metabolite profiling. Twelve metabolites were identified to be significantly different among patient groups, with moderate performance of diagnostic value indicating that these biomarkers may potentially be involved in disease mechanisms. Most of the identified metabolites were mainly involved in fatty acid, amino acid, and lipid metabolism pathways, leading to a number of new hypotheses to better clarify the reasons for their differential abundance in active TB, including:
The decrease of palmitic acid, 3D, 7D, 11D-phytanic acid and behenic acid supported that the fatty acid was preferentially utilized by Mtb, and these metabolites may be one of the results of fatty acid consumption
Terpenoid compounds may have an important role in the processes of infection and host resistance to TB infection
Mtb infection may produce enhanced activity of the TRP -kynurenine pathway, and kynurenine and QUIN, as the TRP downstream metabolites, may affect the immune function of patients with TB
The lower levels of lysoPCs observed in the active TB group may accelerate Mtb infection by reducing the generation of NO, which is produced by activated macrophages
Multiple logistic regression analysis was performed in the 12 metabolites identified as a signature of active TB, and the combination of lysoPC (18:0), behenic acid, threoninyl-γ-glutamate, and PDSP was calculated to represent the best diagnostic value with the AUC value of 0.991 (95% CI: 0.982–1.000). Thus, this combination of metabolites may prove to be a metabolic profile that can be used for the diagnosis of TB.
ACKNOWLEDGMENTS
The authors would like to thank She-ye Liu, Ph.D., from the Third Center Hospital of Tianjin for his helpful discussions and comments.
Footnotes
Edited by: Li-shao Guo
Source of Support: This work was supported by a grant from Tianjin Municipal Health Bureau of China (No. 2013KZ042).
Conflict of Interest: None declared.
REFERENCES
- 1.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Illig T, Gieger C, Zhai G, Römisch-Margl W, Wang-Sattler R, Prehn C, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–41. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: A global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 4.Han X, M Holtzman D, McKeel DW, Jr, Kelley J, Morris JC. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer's disease: Potential role in disease pathogenesis. J Neurochem. 2002;82:809–18. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanov M, Matson WR, Wang L, Matson T, Saunders-Pullman R, Bressman SS, et al. Metabolomic profiling to develop blood biomarkers for Parkinson's disease. Brain. 2008;131:389–96. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- 6.Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, Wiegand R, et al. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112:3868–75. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- 7.Brindle JT, Nicholson JK, Schofield PM, Grainger DJ, Holmes E. Application of chemometrics to 1H NMR spectroscopic data to investigate a relationship between human serum metabolic profiles and hypertension. Analyst. 2003;128:32–6. doi: 10.1039/b209155k. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Xu G, Zheng Y, Kong H, Pang T, Lv S, et al. Diagnosis of liver cancer using HPLC-based metabonomics avoiding false-positive result from hepatitis and hepatocirrhosis diseases. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;813:59–65. doi: 10.1016/j.jchromb.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Kong H, Guan Y, Yang J, Gu J, Yang S, et al. Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis. Anal Chem. 2005;77:4108–16. doi: 10.1021/ac0481001. [DOI] [PubMed] [Google Scholar]
- 10.Yuan K, Kong H, Guan Y, Yang J, Xu G. A GC-based metabonomics investigation of type 2 diabetes by organic acids metabolic profile. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;850:236–40. doi: 10.1016/j.jchromb.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 11.Salek RM, Maguire ML, Bentley E, Rubtsov DV, Hough T, Cheeseman M, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics. 2007;29:99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- 12.Suhre K, Wallaschofski H, Raffler J, Friedrich N, Haring R, Michael K, et al. A genome-wide association study of metabolic traits in human urine. Nat Genet. 2011;43:565–9. doi: 10.1038/ng.837. [DOI] [PubMed] [Google Scholar]
- 13.Laiakis EC, Morris GA, Fornace AJ, Howie SR. Metabolomic analysis in severe childhood pneumonia in the Gambia, West Africa: Findings from a pilot study. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Utzinger J, Saric J, Li JV, Burckhardt J, Dirnhofer S, et al. Global metabolic responses of mice to Trypanosoma brucei brucei infection. Proc Natl Acad Sci U S A. 2008;105:6127–32. doi: 10.1073/pnas.0801777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slupsky CM, Rankin KN, Fu H, Chang D, Rowe BH, Charles PG, et al. Pneumococcal pneumonia: Potential for diagnosis through a urinary metabolic profile. J Proteome Res. 2009;8:5550–8. doi: 10.1021/pr9006427. [DOI] [PubMed] [Google Scholar]
- 16.Lönnroth K, Castro KG, Chakaya JM, Chauhan LS, Floyd K, Glaziou P, et al. Tuberculosis control and elimination 2010-50: Cure, care, and social development. Lancet. 2010;375:1814–29. doi: 10.1016/S0140-6736(10)60483-7. [DOI] [PubMed] [Google Scholar]
- 17.de Carvalho LP, Darby CM, Rhee KY, Nathan C. Nitazoxanide disrupts membrane potential and intrabacterial pH homeostasis of Mycobacterium tuberculosis. ACS Med Chem Lett. 2011;2:849–54. doi: 10.1021/ml200157f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng J, Krausz KW, Li F, Ma X, Gonzalez FJ. CYP2E1-dependent elevation of serum cholesterol, triglycerides, and hepatic bile acids by isoniazid. Toxicol Appl Pharmacol. 2013;266:245–53. doi: 10.1016/j.taap.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halouska S, Fenton RJ, Barletta RG, Powers R. Predicting the in vivo mechanism of action for drug leads using NMR metabolomics. ACS Chem Biol. 2012;20(7):166–71. doi: 10.1021/cb200348m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisson GP, Mehaffy C, Broeckling C, Prenni J, Rifat D, Lun DS, et al. Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis. J Bacteriol. 2012;194:6441–52. doi: 10.1128/JB.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahapatra S, Hess AM, Johnson JL, Eisenach KD, DeGroote MA, Gitta P, et al. A metabolic biosignature of early response to anti-tuberculosis treatment. BMC Infect Dis. 2014;14:53. doi: 10.1186/1471-2334-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin JH, Yang JY, Jeon BY, Yoon YJ, Cho SN, Kang YH, et al. (1) H NMR-based metabolomic profiling in mice infected with Mycobacterium tuberculosis. J Proteome Res. 2011;10:2238–47. doi: 10.1021/pr101054m. [DOI] [PubMed] [Google Scholar]
- 23.Somashekar BS, Amin AG, Rithner CD, Troudt J, Basaraba R, Izzo A, et al. Metabolic profiling of lung granuloma in Mycobacterium tuberculosis infected guinea pigs: ex vivo 1H magic angle spinning NMR studies. J Proteome Res. 2011;10:4186–95. doi: 10.1021/pr2003352. [DOI] [PubMed] [Google Scholar]
- 24.Weiner J, 3rd, Parida SK, Maertzdorf J, Black GF, Repsilber D, Telaar A, et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS One. 2012;7:e40221. doi: 10.1371/journal.pone.0040221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou A, Ni J, Xu Z, Wang Y, Lu S, Sha W, et al. Application of (1) h NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J Proteome Res. 2013;4(12):4642–9. doi: 10.1021/pr4007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai W, Liu Z. p38 mitogen-activated protein kinase mediates palmitate-induced apoptosis but not inhibitor of nuclear factor-kappaB degradation in human coronary artery endothelial cells. Endocrinology. 2007;148:1622–8. doi: 10.1210/en.2006-1068. [DOI] [PubMed] [Google Scholar]
- 27.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–8. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 28.Sandoval-Montemayor NE, García A, Elizondo-Treviño E, Garza-González E, Alvarez L, del Rayo Camacho-Corona M. Chemical composition of hexane extract of Citrus aurantifolia and anti-Mycobacterium tuberculosis activity of some of its constituents. Molecules. 2012;17:11173–84. doi: 10.3390/molecules170911173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston JB, Singh AA, Clary AA, Chen CK, Hayes PY, Chow S, et al. Substrate analog studies of the ω-regiospecificity of Mycobacterium tuberculosis cholesterol metabolizing cytochrome P450 enzymes CYP124A1, CYP125A1 and CYP142A1. Bioorg Med Chem. 2012;20:4064–81. doi: 10.1016/j.bmc.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubnau E, Chan J, Mohan VP, Smith I. Responses of Mycobacterium tuberculosis to growth in the mouse lung. Infect Immun. 2005;73:3754–7. doi: 10.1128/IAI.73.6.3754-3757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cater NB, Denke MA. Behenic acid is a cholesterol-raising saturated fatty acid in humans. Am J Clin Nutr. 2001;73:41–4. doi: 10.1093/ajcn/73.1.41. [DOI] [PubMed] [Google Scholar]
- 32.Malyankar UM. Tumor-associated antigens and biomarkers in cancer and immune therapy. Int Rev Immunol. 2007;26:223–47. doi: 10.1080/08830180701402496. [DOI] [PubMed] [Google Scholar]
- 33.Taylor GO, Bamgboye AE. Serum cholesterol and diseases in Nigerians. Am J Clin Nutr. 1979;32:2540–5. doi: 10.1093/ajcn/32.12.2540. [DOI] [PubMed] [Google Scholar]
- 34.Peteroy-Kelly M, Venketaraman V, Connell ND. Effects of Mycobacterium bovis BCG infection on regulation of L-arginine uptake and synthesis of reactive nitrogen intermediates in J774.1 murine macrophages. Infect Immun. 2001;69:5823–31. doi: 10.1128/IAI.69.9.5823-5831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy RA, Bureyko TF, Mourtzakis M, Chu QS, Clandinin MT, Reiman T, et al. Aberrations in plasma phospholipid fatty acids in lung cancer patients. Lipids. 2012;47:363–9. doi: 10.1007/s11745-011-3641-2. [DOI] [PubMed] [Google Scholar]
- 36.Oestvang J, Anthonsen MW, Johansen B. LysoPC and PAF trigger arachidonic acid release by divergent signaling mechanisms in monocytes. J Lipids 2011. 2011 doi: 10.1155/2011/532145. 532145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan L, Gan H, Arm J, Remold HG. Cytosolic phospholipase A2 participates with TNF-alpha in the induction of apoptosis of human macrophages infected with Mycobacterium tuberculosis H37Ra. J Immunol. 2001;166:7469–76. doi: 10.4049/jimmunol.166.12.7469. [DOI] [PubMed] [Google Scholar]
- 38.Mehta D, Gupta S, Gaur SN, Gangal SV, Agrawal KP. Increased leukocyte phospholipase A2 activity and plasma lysophosphatidylcholine levels in asthma and rhinitis and their relationship to airway sensitivity to histamine. Am Rev Respir Dis. 1990;142:157–61. doi: 10.1164/ajrccm/142.1.157. [DOI] [PubMed] [Google Scholar]
- 39.Fallah A, Pierre R, Abed E, Moreau R. Lysophosphatidylcholine-induced cytotoxicity in osteoblast-like MG-63 cells: Involvement of transient receptor potential vanilloid 2 (TRPV2) channels. Mol Membr Biol. 2013;30:315–26. doi: 10.3109/09687688.2013.828855. [DOI] [PubMed] [Google Scholar]
- 40.Blumenthal A, Nagalingam G, Huch JH, Walker L, Guillemin GJ, Smythe GA, et al. M. tuberculosis induces potent activation of IDO-1, but this is not essential for the immunological control of infection. PLoS One. 2012;7:e37314. doi: 10.1371/journal.pone.0037314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song H, Park H, Kim YS, Kim KD, Lee HK, Cho DH, et al. L-kynurenine-induced apoptosis in human NK cells is mediated by reactive oxygen species. Int Immunopharmacol. 2011;11:932–8. doi: 10.1016/j.intimp.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Apalset EM, Gjesdal CG, Ueland PM, Midttun Ø, Ulvik A, Eide GE, et al. Interferon (IFN)-γ-mediated inflammation and the kynurenine pathway in relation to bone mineral density: The Hordaland Health Study. Clin Exp Immunol. 2014;176:452–60. doi: 10.1111/cei.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chuang SC, Fanidi A, Ueland PM, Relton C, Midttun O, Vollset SE, et al. Circulating biomarkers of tryptophan and the kynurenine pathway and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2014;23:461–8. doi: 10.1158/1055-9965.EPI-13-0770. [DOI] [PubMed] [Google Scholar]
- 44.Molina-Salinas GM, Bórquez J, Said-Fernández S, Loyola LA, Yam-Puc A, Becerril-Montes P, et al. Antituberculosis activity of alkylated mulinane diterpenoids. Fitoterapia. 2010;81:219–22. doi: 10.1016/j.fitote.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Koohang A, Bailey JL, Coates RM, Erickson HK, Owen D, Poulter CD. Enantioselective inhibition of squalene synthase by aziridine analogues of presqualene diphosphate. J Org Chem. 2010;16(75):4769–77. doi: 10.1021/jo100718z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonnans C, Fukunaga K, Keledjian R, Petasis NA, Levy BD. Regulation of phosphatidylinositol 3-kinase by polyisoprenyl phosphates in neutrophil-mediated tissue injury. J Exp Med. 2006;203:857–63. doi: 10.1084/jem.20052143. [DOI] [PMC free article] [PubMed] [Google Scholar]


