Abstract
Background:
Polycystic ovary syndrome (PCOS) is the commonest endocrinopathy in women of reproductive age. The patients often develop insulin resistance (IR) or hyperinsulinemia despite manifesting anovulation and signs of hyperandrogenism. The cause and effect relationship of hyperinsulinemia and hyperandrogenemia (HA) is still debated. Micro-ribonucleic acids (miRNAs) have recently been shown to play a role in regulation of ovarian function. Our current study focused on the altered expression of miRNAs with PCOS.
Methods:
Ovarian theca interna tissues were obtained from 10 PCOS patients and 8 controls that were non-PCOS and had normal insulin sensitivity undergoing laparoscopy and/or ovarian wedge resection. Total RNA of all samples was extracted. We studied the repertoire of miRNAs in both PCOS and non-PCOS women by microarray hybridization. Bioinformatic analysis was performed for predicting targets of the differentially expressed miRNAs. Furthermore, selected miRNAs were validated by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
Results:
A total of 27 miRNAs were differentially expressed in PCOS patients with respect to the controls in our discovery evaluationand two (miR-92a and miR-92b) of them were significantly downregulated in PCOS women in followed validation (P < 0.05). Targets prediction revealed that miR-92a targeted both GATA family of zinc finger transcription factor GATA-binding factor 6 (GATA6) and insulin receptor substrate proteins 2 (IRS-2).
Conclusions:
MiRNAs are differentially expressed between PCOS patients and controls. We identified and validated two miRNAs-miR-92a and miR-92b. They are significantly downregulated and may be involved in the pathogenesis of PCOS.
Keywords: Hyperandrogenism, Insulin Resistance, Microrna, Polycystic Ovary Syndrome
INTRODUCTION
Polycystic ovary syndrome (PCOS) is the commonest endocrine disease of women in reproductive age, affecting 5%-10% in this population.[1] The patients often suffer from metabolic disorders or even develop metabolic syndrome (MS) despite manifesting anovulation and signs of hyperandrogenism, such as hirsutism and acne. Disturbance of carbohydrate metabolism, which is manifested by insulin resistance (IR) and/or compensatory hyperinsulinemia, is now known to be intrinsic to PCOS, and contributes in a major way to its pathogenesis. It was reported that 50%–70% of PCOS patients have various degree of IR.[2] There is considerable evidence suggesting a causal link between hyperinsulinemia and increased ovarian androgen production.[3] Ovarian theca cells, which express key enzymes of androgen production, such as 17-hydroxylase/C17–20 lyase cytochrome P50 (CYP17), and their transcription factors, for example, GATA family of zinc finger transcription factor GATA-binding factor 6 (GATA6), are the major place of androgen producing in ovary. Theca cells from PCOS ovaries which have been correlated to increased expression and/or activity of CYP17 synthesize increased levels of androgens compared with normal theca cells. GATA6 gene transcription and the stability of messenger RNA (mRNA) were reported to be increased in PCOS theca cells.[4]
Studies have been conducted regarding the mechanisms of insulin action in women with PCOS using fibroblasts,[5] adipocytes[6,7] and skeletal muscle cells.[8] Activation of the insulin receptor increases phosphorylation of intracellular substrates, principally insulin receptor substrate (IRS) proteins. Four IRS proteins; IRS-1, IRS-2, IRS-3, and IRS-4; have been identified. Phosphorylated IRS links activated insulin receptors with mediators of downstream signaling, including phosphatidylinositol-3-kinase (PI3K), Fyn, Grb-2, and Crk. And IRS-1 and IRS-2 are major mediators of insulin action. There are significant differences in insulin signaling among various insulin-responsive tissues.[9] Although there is evidence for alterations in the proximal insulin signaling events in PCOS, the effects of IR in PCOS on theca cell androgen production remain exclusive.
MiRNAs are endogenous 22 nucleotide-long, highly conserved, noncoding RNAs which, in general, negatively regulate gene expression. Functional analysis of miRNAs has revealed their significant regulatory influence on the expression of target genes involved in both physiologic and pathologic conditions. miRNAs are also known to contribute to female reproductive function.[10] However, recent studies focus mainly on the relationship of miRNAs expression and the reproductive disorders of PCOS patients.[11,12] And the miRNA profile of PCOS ovaries, especially theca tissue has not been reported.
Herein, we hypothesized that the miRNA profile of PCOS ovaries were different from those of non-PCOS ones. We screened miRNAs in theca tissue by miRNA microarray and then performed quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assays to validate them. The specific targets of those differentially expressed miRNAs were identified. Results of the present study revealed that PCOS is associated with differential expression of regulatory noncoding miRNAs in human ovarian theca tissue.
METHODS
Patient selection
All procedures for collection of tissue were approved by the ethics board of Memorial Hospital of Sun Yat-Sen, Sun Yat-Sen University. Ovarian theca tissue was obtained with written informed consent from premenopausal women (aged 20–35 years) undergoing laparoscopy and/or ovarian wedge resection. All the subjects were screened with an emphasis on menstrual regularity, gynecologic and endocrinologic history, hirsutism, acne, use of birth control pills, or hormone medications (continuous progestin, glucocorticoids, or an insulin sensitizer) in 3 months before surgery. Physical exams were also performed at the time of hospital admission.
The diagnosis of PCOS was made according to the established guidelines from revised 2003 consensus on diagnostic criteria.[13] In our study, PCOS subjects should meet at least two of the following criteria: Oligo- and/or anovulation (e.g., ≤ eight menstrual periods in a year or menstrual cycles more than 35 days in length); clinical hyperandrogenism (e.g., acne or modified Ferriman–Gallwey (mFG) scores ≥ 6); and/or biochemical hyperandrogenism (i.e., serum total testosterone (TT) ≥ 2.6 nmol/L, free testosterone (FT) ≥ 6.0 pg/ml, and TT and FT normal values were determined by the clinical laboratory of the gynecology department at the Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University); and polycystic ovaries (e.g., the presence of ≥12 follicles in each ovary measuring 2–9 mm in diameter) after exclusion of other etiologies (e.g., congenital adrenal hyperplasia, androgen-secreting tumors, and Cushing's syndrome).
Homeostasis model assessment (HOMA) was applied to estimate the degree of IR. The equation used to obtain this value is as follows: HOMA-IR = (fasting plasma glucose (mmol/L) × insulin (mU/ml))/22.5. HOMA-IR > 2.14 were considered insulin resistant according to our previous study.[14]
Tissue collection
Ovarian theca tissue samples were obtained from subjects included in this study based on the criteria mentioned above. All biopsies were washed with cold phosphate buffered saline (PBS). Theca interna was stripped from the follicle wall, and the granulosa cells were removed. Samples were immediately placed in RNALater (Invitrogen, Carlsbad, CA) and stored at −20°C.
RNA extraction
Total RNA are harvested using TRIzol (Invitrogen) and RNeasy mini kit (QIAGEN) according to manufacturer's instructions. RNA concentration was assessed using Nanodrop ND-1000, accepting a ratio of 2.0 for sample absorbance at 260/280.
Microarray analysis
Total RNA samples were analyzed by KangChen (KangChen Bio-tech) for miRNA microarray experiments. Procedures were performed as described in detail on the website of KangChen (http://www.kangchen.com.cn). The samples are labeled using the miRCURY™ Hy3™/Hy5™ Power labeling kit and hybridized on the miRCURY™ LNA Array (v. 16.0). This most recent version of the array (v. 16.0 - hsa, mmu, and rno array) contains more than 1,700 capture probes, covering all miRNAs annotated in miRBase 16.0, as well as all viral miRNAs, related to these species. Scanning is performed with the Axon GenePix 4000B microarray scanner. GenePix pro V6.0 is used to read the raw intensity of the image. Signal intensities were normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm. Median normalization method are used to obtain “normalized data”, normalized data = (foreground-background)/median, the median is 50% quantile of miRNA intensity, which is larger than 50 in all samples after background correction.
Array data processing and analysis
The analysis of miRNA predicted targets was determined using the algorithms of TargetScan 6.2, PicTar, miRBase, and miRanda.
Key genes interacting miRNAs prediction
MiRNAs which may interact with CYP17, GATA6, IRS-2, and glycogen synthase kinase-3β (GSK3β) were searched using TargetScan 6.2.
qRT-PCR analysis of miRNA expression
qRT-PCR analysis was performed to validate the miRNA microarray results in this study. The specific stem-looped RT-PCR primers to representatives of differentially expressed miRNAs (miR-200a, miR-141, miR-200c, miR-502-3p, miR-32, miR-92a, miR-92b, miR-19a, miR-1, and let-7g) were designed according to a previous study,[15] and are summarized in Table 1. The RT-PCR reaction was performed according to a previous report.[16] PCR primers for amplification of the human mature miRNAs are summarized in Table 2.
Table 1.
miRNA-Specific RT primers
| miRNA name | RT primers 5’→3’ |
|---|---|
| hsa-miR-200a | TGCGTGTCGTGGAGTC |
| hsa-miR-141 | TGCGTGTCGTGGAGTC |
| hsa-miR-200c | CAGTGCGTGTCGTGGAGT |
| hsa-miR-502-3p | CAGTGCGTGTCGTGGAGT |
| hsa-miR-32 | CAGTGCGTGTCGTGGAG |
| hsa-miR-92a | TGCGTGTCGTGGAGTC |
| hsa-miR-92b | TGCGTGTCGTGGAGTC |
| hsa-miR-19a | CAGTGCGTGTCGTGGAG |
| hsa-miR-1 | TGCGTGTCGTGGAGTC |
| hsa-let-7g | TGCGTGTCGTGGAGTC |
miRNA: Micro-ribonucleic acid; RT: Reverse transcriptase.
Table 2.
miRNA-Specific antisense primers
| miRNA name | Antisense primers 5’→3’ |
|---|---|
| hsa-miR-200a | GGGGTAACACTGTCTGGTAG |
| hsa-miR-141 | GGGGTAACACTGTCTGGTAA |
| hsa-miR-200c | GGTAATACTGCCGGGTAAT |
| hsa-miR-502-3p | GGAATGCACCTGGGCA |
| hsa-miR-32 | GGGGCTATTGCACATTACTA3 |
| hsa-miR-92a | CACCTATATTGCACTTGTCC |
| hsa-miR-92b | CGGTATTGCACTCGTCC3 |
| hsa-miR-19a | GGCAGTGTGCAAATCTATG |
| hsa-miR-1 | GGGGTGGAATGTAAAGAAG |
| hsa-let-7g | GGGGTGAGGTAGTAGTTTGT |
miRNA: Micro-ribonucleic acid.
Real-time PCR analysis was performed on Gene Amp PCR System 970 (Applied Biosystem) with 8 μl volume reaction containing 2 μl reverse transcription product, 0.8 μl PCR specific primers (10 μmol/L), and 25 mmol/L MgCl2, and H2O. The reactions were incubated in 96-well plates at 95°C for 3 minutes, following by 40 cycles (95°C for 15 seconds, 60°C for 20 seconds, 72°C for 20 seconds, and 78°C for 20 seconds), then ramped from 72°C to 99°C to obtain the melting curve. U6 snRNA was measured by the same method and used for normalization. The relative quantity of each miRNA in PCOS ovarian theca tissues, normalized to U6 RNA and relative to the expression in non-PCOS theca tissues, was calculated using the equation RQ = 2–ΔΔCT, where ΔΔCT = (CTmiRNA–CTU6 RNA) PCOS–(CTmiRNA–CTU6 RNA) Meannon-PCOS and CT is the threshold cycle to detect fluorescence.
Statistical analysis
For microarray data, the threshold value we used to screen up and down regulated miRNAs is fold change ≥1.50 and fold change ≤ 0.67. For RT-PCR data, statistical analysis was performed in Statistical Package for Social Sciences (SPSS) 16.0 for Windows (SPSS Inc). Data are presented as fold change relative to the control group. Two-tailed t-test was used for statistical evaluation of miRNA expression differences with significance accepted at P ≤ 0.05.
RESULTS
Human subject characteristics
Ovarian theca tissue samples were obtained from 10 PCOS patients with IR and eight age- and body mass index (BMI)-matched non-PCOS infertile women who had normal insulin sensitivity and normal serum androgen level. The characteristics of patients were listed below [Table 3].
Table 3.
Characteristics of PCOS patients and controls
| PCOS (n = 10) | Control (n = 8) | P | |
|---|---|---|---|
| Age | 28.80 ± 3.97 | 32.00 ± 2.16 | 0.086 |
| BMI, kg/m2 | 24.42 ± 4.84 | 20.51 ± 2.06 | 0.051 |
| mFG score | 4.3 ± 4.74 | 0.43 ± 0.787 | 0.004 |
| TT, nmol/L | 2.47 ± 0.64 | 1.27 ± 0.60 | 0.005 |
| FT, pg/mL l | 3.99 ± 2.75 | 1.85 ± 0.62 | 0.025 |
| FIN, mU/L | 13.45 ± 7.25 | 4.52 ± 1.65 | 0.001 |
| FBG, mmol/L | 4.71 ± 0.44 | 4.24 ± 0.69 | 0.201 |
| HOMA-IR | 2.85 ± 1.70 | 0.86 ± 0.39 | 0.001 |
Values are expressed as mean ± standard deviation. P < 0.05 is considered statistically significant. PCOS: Polycystic ovary syndrome; BMI: Body mass index; mFG: Modified Ferriman–Gallwey; TT: Total testosterone; FT: Free testosterone; FIN: Fasting insulin; FBG: Fasting blood glucose; HOMA-IR: Homeostasis model assessment-insulin resistance.
miRNA microarray and targets prediction
Using miRNA microarray analysis, we evaluated miRNA expression profiles of ovarian theca tissues of PCOS with IR patients and non-PCOS, non-IR controls. A total of 27 miRNAs were differentially expressed with a fold change of ≥1.5 or ≤0.67. Given that biological significance of miRNA deregulation relies on the effect upon their cognate protein-coding gene targets, we analyzed the predicted targets of the most significantly up- and downregulated miRNAs: miR-200a, miR-141, miR-200c, miR-502-3p, miR-32, miR-92a, miR-92b, miR-19b, miR-1, and let-7g. The analysis was done using four algorithms, TargetScan 6.2, PicTar, miRBase, and miRanda, which are commonly used to predict human miRNA gene targets. Prediction results [Figure 1 and Table 4] demonstrated that the putative target genes of the above miRNAs include CYP17, GATA6, and IRS-2.
Figure 1.
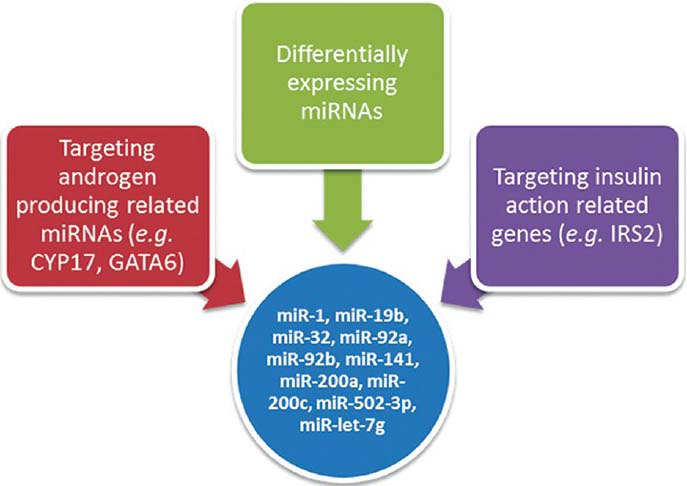
Strategies of selecting miRNAs. miRNA = Micro-ribonucleic acid; IRS-2 = insulin receptor substrate 2; GATA6 = GATA-binding factor 6.
Table 4.
Predicted targets of differentially expressed miRNAs
| miRNA | ID | Up-/downregulated | Predicted target† |
|---|---|---|---|
| hsa-miR-1 | 10916 | ↓ | IGF1, IGFBP5 |
| hsa-miR-19b | 10998 | ↓ | MAPK6 |
| hsa-miR-32 | 11053 | ↓ | IRS-2, INSIG1 |
| hsa-miR-92a | 145693 | ↓ | IRS-2 |
| hsa-miR-92b | 693235 | ↓ | IRS-2 |
| hsa-miR-141 | 10946 | ↑ | IGF1R, IGF2 |
| hsa-miR-200a | 11000 | ↑ | GATA6, IRS-2 |
| hsa-miR-200c | 17427 | ↑ | GATA2 |
| hsa-miR-502-3p | 46654 | ↑ | CYP17 |
| hsa-let-7g | 46438 | ↑ | STARD13 |
†Just part of the targeted genes of the micro-ribonucleic acids (miRNAs) are shown here. IGF1: Insulin-like growth factor 1; IGFBP5: Insulin-like growth factor binding protein 5; IGFBP3: Insulin-like growth factor binding protein 3; IGF2R: Insulin-like growth factor 2 receptor; INSIG1: Insulin-induced gene 1; INSIG2: Insulin-induced gene 2; IRS-2: Insulin receptor substrate 2; IGF1R: Insulin-like growth factor 1 receptor; GATA6: GATA-binding factor 6; GATA2: GATA-binding factor 2; STARD13: StAR-related lipid transfer (START) domain containing 13.
miRNA expression validation
To validate our array expression findings, 10 differentially expressed miRNAs (miR-1, miR-19b, miR-32, miR-92a, miR-92b, miR-141, miR-200a, miR-200c, miR-502-3p, and let-7g) were chosen for qRT-PCR analysis. The miRNAs were selected on the basis of their target genes. The trends for downregulation of miRNA expression were consistent in five of the 10 (miR-19b, miR-92a, miR-92b, miR-141, and 200a) qRT–PCR measurements [Figure 2 and Table 5].
Figure 2.
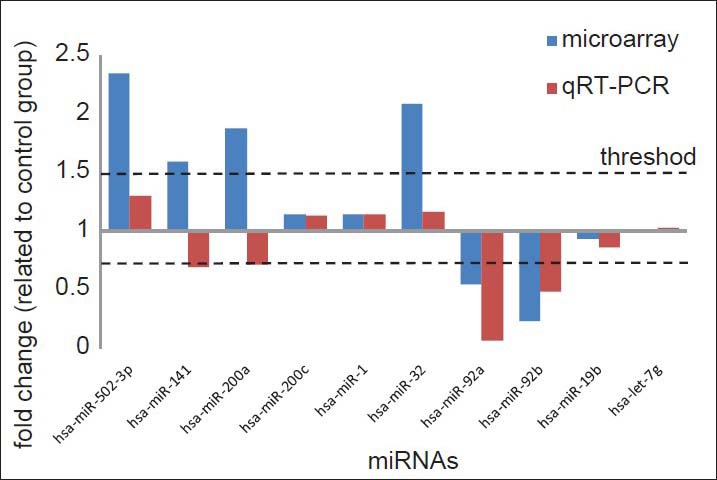
Relative expression of miR-200a, miR-141, miR-200c, miR-502-3p, miR-32, miR-92a, miR-92b, miR-19b, miR-1, and let-7g in microarray (blue) and qRT-PCR (red). Up regulation miRNAs were represented as fold change ≤1 and down regulation ones as fold change ≤1 at the same time. The dotted lines represented the thresholds (≥1.5 and ≤0.67) of fold change we have set in our study for screening differentially expressed miRNAs. miRNA = Micro-ribonucleic acid; qRT-PCR = Quantitative reverse transcriptase polymerase chain reaction.
Table 5.
Results of miRNA validation
| miRNA | Fold change (related to control group) | P | |
|---|---|---|---|
| Microarray | qRT-PCR | ||
| hsa-miR-502-3p | 2.348932585 | 1.298364 | 0.472438 |
| hsa-miR-141 | 1.593759475 | 0.694171 | 0.05319 |
| hsa-miR-200a | 1.88407 | 0.713856 | 0.113249 |
| hsa-miR-200c | 1.150974967 | 1.138108 | 0.688817 |
| hsa-miR-1 | 1.141086713 | 1.147553 | 0.736046 |
| hsa-miR-32 | 2.084112536 | 1.168739 | 0.471442 |
| hsa-miR-92a | 0.54447 | 0.0656608 | 0.005534 |
| hsa-miR-92b | 0.23541 | 0.486416 | 0.017138 |
| hsa-miR-19b | 0.940351 | 0.859191 | 0.515809 |
| hsa-let-7g | 1.02172 | 1.032705 | 0.900602 |
The column of fold change (related to control group) shows the result of qRT-PCR. The distribution of the data which was calculated by RQ = 2−ΔΔCT, whereΔΔCT = (CTmiRNA− CTU6 RNA) PCOS− (CTmiRNA− CTU6 RNA). miRNA: Micro-ribonucleic acid; qRT-PCR: Quantitative reverse transcriptase polymerase chain reaction.
DISCUSSION
In present study, we have for the first time tried to identify the differential expression of miRNAs in the ovary of women with and without PCOS, HA, and IR; and integrated the findings with the expression patterns of certain specific genes from the same cohort of specimens. In addition, qRT-PCR validation confirmed downregulation of miR-141, miR-200a, miR-92a, miR-92b, and miR-19b. Among these miRNAs, miR-92a, and miR-92b were significantly downregulated.
Both miR-92a and miR-92b belong to the miR-17-92 miRNA cluster located at 13q31.3. MiR-92a has been associated with cancer pathogenesis and has been reported being significantly downregulated in patients with acute myeloid leukemia (AML) and acute lymphocytic leukemia (ALL), hepatocellular carcinoma (HCC),[17] ovarian cancer,[18] chronic lymphatic leukemia,[19] and during myeloid differentiation.[20] Recently, miR-92a was reported to play a role in non-tumor diseases, such as ischemia.[21] And collaborating with several other miRNAs, for example, let-7a, miR-19a, miR-19b, miR-24, and miR-93; miR-92 has been reported to be significantly downregulated in the blastocysts derived from patients with polycystic ovaries.[22] In addition, dysregulated expression of miR-92 was also correlated to the development of leiomyomas.[23,24] However, the expression pattern of miR-92a in PCOS ovarian tissue has not yet been illustrated.
PCOS is a complex and heterogeneous endocrine condition characterized by HA, hyperinsulinemia, and/or IR. IR accompanied by mild compensatory hyperinsulinemia is a common feature of women with PCOS.[25] Our data showed significantly higher fast insulin concentration in PCOS patients (P = 0.001), who thus had higher HOMA-IR value [Table 1]. At the same time, two significantly downregulated miRNAs, miR-92a and miR-92b, had been predicted to target IRS-2, a key factor in insulin signaling pathway. It indicated increasing insulin signaling in ovarian tissue of PCOS patients. This result was consistent with the relative hyperinsulinemia and IR characteristics of PCOS patients in our study.
Hyperinsulinemia and HA are two principal features of PCOS and their cause and effect relationship is still being debated. Two important androgen producing-related genes, CYP17, GATA6, and one insulin receptor gene, IRS-2 were shown to be expressed significantly higher in PCOS theca tissue compared with non-PCOS ones in our previous study. It was consistent with the results of earlier reports.[4,26,27] Being worthy of note, miR-92a targeted both GATA6 and IRS-2, which might indicate that there were cross-talk between androgenic and insulin signaling pathways.
The other five miRNAs which we had validated by qRT-PCR were also reported to be very important in gynecologic physiology and pathophysiology. The let-7 family, which let-7g belonged to, was reported to be the most commonly abundant miRNA populations in the ovary.[28] And bioinformatics prediction, screening, and gene ontology analysis of its targeting genes in the mammalian ovary has identified several biological processes and pathways or molecular networks underlying ovarian functions.[29] Furthermore, a variant SNP in the LCS6 let-7 miRNA binding site of the KRAS 3’ untranslated region was found in 31% of women with severe treatment-resistant endometriosis,[30] showing the relationship of let-7 expressing and the development of endometriosis. MiR-200c and miR-141 have been reported to be differently expressed in the ovary tissues.[31] And the aberrant expression of the latter was correlated to the development of leimyoma.[23,24] MiR-200a expression results in decreased expression of dual-specificity phosphatase-2 that subsequently results in prolonged extracellular-signal-regulated kinases activation through hypoxia-inducible factor, contributing to inflammation in the pathogenesis of endometriosis.[32] According to our analysis, the five miRNAs mentioned above might target androgen or insulin-like growth factor signaling pathway factors [Table 4]. However, the expression level of them remained barely unchanged in PCOS ovarian tissue. This was likely due to the small sample size of our study and the significantly different serum level of androgen (TT and FT) among the PCOS patients.
In conclusion, our study showed that miRNAs have played an important role in insulin action in local ovarian theca interna. And the downregulation of miR-92a and miR-92b might lead to augmentation of signal transduction in both androgen pathway and insulin pathway. Significantly downregulation of insulin signaling factors targeted miRNAs in the ovarian theca tissue of PCOS patients with HA indicated that hyperinsulinemia and/or IR was perhaps the cause of HA.
In addition, although miRNAs are known to regulate multiple gene targets in different system,[33,34] they can also be regulated by other paracrine/autocrine factors.[35,36] The precise target(s) for the reported dysregulated miRNA and their immediate downstream processes need to be defined. We are going to focus on function analysis of certain miRNAs, for example, miR-92a, in the future.
Although with advantage, we recognize that relatively low number of tissues (n = 18) collected as a limitation of our study. However, the careful selection of patients and well-matched control subjects from a larger cohort of well-characterized individuals reduced the ‘noise’ in our expression profiling experiments. As such, further study is necessary to investigate the mechanisms of androgen-insulin signaling cross-talk.
Footnotes
Edited by: De Wang
Source of Support: This work was supported by grants from the National Natural Science Foundation of China (No. 81370680), Key-projects of Clinical Research of Ministry of Health (No. WGCH[2010]439), The Specialized Research Fund for the Doctoral Program of Chinese Ministry of Education (No. 20130171130009), and Science and Technology Project of Guangdong Province (No. 2013020012660).
Conflict of Interest: None declared.
REFERENCES
- 1.Diamanti-Kandarakis E. Polycystic ovarian syndrome: Pathophysiology, molecular aspects and clinical implications. Expert Rev Mol Med. 2008;10:e3. doi: 10.1017/S1462399408000598. [DOI] [PubMed] [Google Scholar]
- 2.Diamanti-Kandarakis E. Insulin resistance in PCOS. Endocrine. 2006;30:13–7. doi: 10.1385/ENDO:30:1:13. [DOI] [PubMed] [Google Scholar]
- 3.Dunaif A. Insulin resistance and the polycystic ovary syndrome: Mechanism and implications for pathogenesis. Endocr Rev. 1997;18:774–800. doi: 10.1210/edrv.18.6.0318. [DOI] [PubMed] [Google Scholar]
- 4.Ho CK, Wood JR, Stewart DR, Ewens K, Ankener W, Wickenheisser J, et al. Increased transcription and increased messenger ribonucleic acid (mRNA) stability contribute to increased GATA6 mRNA abundance in polycystic ovary syndrome theca cells. J Clin Endocrinol Metab. 2005;90:6596–602. doi: 10.1210/jc.2005-0890. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Youngren JF, Dunaif A, Goldfine ID, Maddux BA, Zhang BB, et al. Decreased insulin receptor (IR) autophosphorylation in fibroblasts from patients with PCOS: Effects of serine kinase inhibitors and IR activators. J Clin Endocrinol Metab. 2002;87:4088–93. doi: 10.1210/jc.2002-020363. [DOI] [PubMed] [Google Scholar]
- 6.Corbould A, Dunaif A. The adipose cell lineage is not intrinsically insulin resistant in polycystic ovary syndrome. Metabolism. 2007;56:716–22. doi: 10.1016/j.metabol.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mannerås-Holm L, Leonhardt H, Kullberg J, Jennische E, Odén A, Holm G, et al. Adipose tissue has aberrant morphology and function in PCOS: Enlarged adipocytes and low serum adiponectin, but not circulating sex steroids, are strongly associated with insulin resistance. J Clin Endocrinol Metab. 2011;96:E304–11. doi: 10.1210/jc.2010-1290. [DOI] [PubMed] [Google Scholar]
- 8.Peppa M, Koliaki C, Nikolopoulos P, Raptis SA. Skeletal muscle insulin resistance in endocrine disease. J Biomed Biotechnol 2010. 2010 doi: 10.1155/2010/527850. 527850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunaif A, Thomas A. Current concepts in the polycystic ovary syndrome. Annu Rev Med. 2001;52:401–19. doi: 10.1146/annurev.med.52.1.401. [DOI] [PubMed] [Google Scholar]
- 10.Carletti MZ, Christenson LK. MicroRNA in the ovary and female reproductive tract. J Anim Sci. 2009;87:E29–38. doi: 10.2527/jas.2008-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sang Q, Yao Z, Wang H, Feng R, Zhao X, Xing Q, et al. Identification of microRNAs in human follicular fluid: Characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;98:3068–79. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- 12.Roth LW, McCallie B, Alvero R, Schoolcraft WB, Minjarez D, Katz-Jaffe MG. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. J Assist Reprod Genet. 2014;31:355–62. doi: 10.1007/s10815-013-0161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Yang D, Li L, Feng S, Wang L. Abnormal glucose tolerance in Chinese women with polycystic ovary syndrome. Hum Reprod. 2006;21:2027–32. doi: 10.1093/humrep/del142. [DOI] [PubMed] [Google Scholar]
- 15.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60:351–7. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 18.Ohyagi-Hara C, Sawada K, Kamiura S, Tomita Y, Isobe A, Hashimoto K, et al. miR-92a inhibits peritoneal dissemination of ovarian cancer cells by inhibiting integrin α5 expression. Am J Pathol. 2013;182:1876–89. doi: 10.1016/j.ajpath.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 19.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chroniclymphocytic leukemia. Blood. 2007;109:4944–51. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 20.Chen A, Luo M, Yuan G, Yu J, Deng T, Zhang L, et al. Complementary analysis of microRNA and mRNA expression during phorbol 12-myristate 13-acetate (TPA)-induced differentiation of HL-60 cells. Biotechnol Lett. 2008;30:2045–52. doi: 10.1007/s10529-008-9800-8. [DOI] [PubMed] [Google Scholar]
- 21.Daniel JM, Penzkofer D, Teske R, Dutzmann J, Koch A, Bielenberg W, et al. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovasc Res. 2014;103:564–72. doi: 10.1093/cvr/cvu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCallie B, Schoolcraft WB, Katz-Jaffe MG. Aberration of blastocyst microRNA expression is associated with human infertility. Fertil Steril. 2010;93:2374–82. doi: 10.1016/j.fertnstert.2009.01.069. [DOI] [PubMed] [Google Scholar]
- 23.Chuang TD, Panda H, Luo X, Chegini N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr Relat Cancer. 2012;19:541–56. doi: 10.1530/ERC-12-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–7. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee S, Maitra A. Molecular and genetic factors contributing to insulin resistance in polycystic ovary syndrome. Indian J Med Res. 2010;131:743–60. [PubMed] [Google Scholar]
- 26.Wu X, Sallinen K, Anttila L, Makinen M, Luo C, Pollanen P, et al. Expression of insulin-receptor substrate-1 and -2 in ovaries from women with insulin resistance and from controls. Fertil Steril. 2000;74:564–72. doi: 10.1016/s0015-0282(00)00688-9. [DOI] [PubMed] [Google Scholar]
- 27.Yen HW, Jakimiuk AJ, Munir I, Magoffin DA. Selective alterations in insulin receptor substrates-1, -2 and -4 in theca but not granulosa cells from polycystic ovaries. Mol Hum Reprod. 2004;10:473–9. doi: 10.1093/molehr/gah066. [DOI] [PubMed] [Google Scholar]
- 28.Hossain MM, Sohel MM, Schellander K, Tesfaye D. Characterization and importance of microRNAs in mammalian gonadal functions. Cell Tissue Res. 2012;349:679–90. doi: 10.1007/s00441-012-1469-6. [DOI] [PubMed] [Google Scholar]
- 29.Imbar T, Eisenberg I. Regulatory role of microRNAs in ovarian function. Fertil Steril. 2014;101:1524–30. doi: 10.1016/j.fertnstert.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Grechukhina O, Petracco R, Popkhadze S, Massasa E, Paranjape T, Chan E, et al. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol Med. 2012;4:206–17. doi: 10.1002/emmm.201100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baley J, Li J. MicroRNAs and ovarian function. J Ovarian Res. 2012;5:8. doi: 10.1186/1757-2215-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin SC, Wang CC, Wu MH, Yang SH, Li YH, Tsai SJ. Hypoxia-induced microRNA-20a expression increases ERK phosphorylation and angiogenic gene expression in endometriotic stromal cells. J Clin Endocrinol Metab. 2012;97:E1515–23. doi: 10.1210/jc.2012-1450. [DOI] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 34.Toloubeydokhti T, Bukulmez O, Chegini N. Potential regulatory functions of microRNAs in the ovary. Semin Reprod Med. 2008;26:469–78. doi: 10.1055/s-0028-1096127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–63. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–9. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]


