Abstract
Background. An estimated 1 million children die each year before their fifth birthday from diarrhea. Previous population-based surveys of pediatric diarrheal diseases have identified the protozoan parasite Entamoeba histolytica, the etiological agent of amebiasis, as one of the causes of moderate-to-severe diarrhea in sub-Saharan Africa and South Asia.
Methods. We prospectively studied the natural history of E. histolytica colonization and diarrhea among infants in an urban slum of Dhaka, Bangladesh.
Results. Approximately 80% of children were infected with E. histolytica by the age of 2 years. Fecal anti-galactose/N-acetylgalactosamine lectin immunoglobulin A was associated with protection from reinfection, while a high parasite burden and expansion of the Prevotella copri level was associated with diarrhea.
Conclusions. E. histolytica infection was prevalent in this population, with most infections asymptomatic and diarrhea associated with both the amount of parasite and the composition of the microbiota.
Keywords: amebiasis, Entamoeba histolytica, Prevotella copri, IgA, environmental enteropathy
Children in developing countries are continually exposed to enteric pathogens [1–4]. This results in high rates of morbidity and mortality, especially in infants [5]. The outcome of an Entamoeba histolytica exposure can be diarrhea, dysentery, or amebic liver abscess, with the latter occurring mainly in men [6]. Disease, however, occurs in a minority of E. histolytica infections (approximately 20%) [6]. While both host and parasite factors contribute to outcome, they do not entirely explain susceptibility [7–9].
A prospective longitudinal study of enteric pathogens in a slum community in Dhaka, Bangladesh, has provided a rich data set to identify parameters important for protection and disease. It has been previously shown that children in this study cohort who are malnourished at birth are more susceptible to amebiasis and in general experience more-severe diarrhea [2]. There is evidence in support of both passive and active acquired immunity. Infants with mothers with high levels of breast milk anti-galactose/N-acetylgalactosamine (Gal/GalNAc) lectin immunoglobulin A (IgA) had fewer E. histolytica infections, and children with fecal IgA anti-Gal/GalNAc lectin IgA also had a lower incidence of new infection [10, 11].
Weaning and the introduction of supplementary food into the diet is known to trigger changes in the bacterial composition of the microbiome that include increases in Prevotellaceae and Bacteroides species [12, 13]. The first 2 years of life in general is a period of rapid immune and microbiome maturation, which, in this population, may be disrupted by malnutrition and environmental insult [13–15].
Here we describe the natural history of amebiasis in the first 2 years of life in infants from an urban slum in Dhaka. The cumulative incidence of amebic infection and diarrhea, the association of protection with anti-Gal/GalNAc lectin IgA, the impact of parasite burden on symptoms, and potential role of Prevotella copri in E. histolytica diarrhea are described.
MATERIALS AND METHODS
Study Area and Population
Details of the study methods have been described in depth elsewhere [3, 10, 16]. In brief, 392 children born into an urban slum of Dhaka were enrolled in the first week after birth into a community-based prospective cohort study of enteric infections. Socioeconomic information about the study households was collected upon enrollment, using a structured questionnaire (Table 1). In this article, we report the data on the children followed through the second year of life, in the study period ending 5 May 2012.
Table 1.
Characteristics of the Study Population, by Entamoeba histolytica Infection Status During the First 2 Years of Life
| Characteristic | Infected (n = 307) | Uninfected (n = 85) | P Value |
|---|---|---|---|
| Male sex, no. (%) | 169 (55.0) | 48 (56.5) | .82 |
| Family income, BDT | 6860.5 ± 3089.8 | 6534.2 ± 2900.4 | .38 |
| Maternal education duration, ya | 3.7 ± 3.6 | 3.9 ± 3.5 | .80 |
| Family size, no. | 5.4 ± 2.2 | 5.4 ± 2.3 | .96 |
| Maternal BMI | 21.4 ± 3.3 | 21.1 ± 3.1 | .37 |
| Exclusive breast feeding, d | 118.4 ± 67.4 | 129.6 ± 65.2 | .17 |
| LAZ, by age | |||
| At birth | −0.97 ± 1.17 | −0.92 ± 1.10 | .73 |
| 1 y | −1.80 ± 1.44 | −1.95 ± 1.51 | .39 |
| 2 y | −2.43 ± 1.71 | −2.53 ± 1.65 | .63 |
Data are mean ± SD, unless otherwise indicated.
Abbreviations: BDT, Bangladeshi taka (currency); BMI, body mass index; LAZ, length-for-age z score.
a A duration of >9 years indicates formal certification.
Child development was followed by length-for-age z (LAZ) score measurements every quarter. Field workers visited the child's household twice weekly to record any diarrheal event; 1 stool sample was collected at the time of the diarrheal event, and 1 was collected during monthly surveillance.
Clinical Definitions
Diarrhea was defined as having ≥3 unformed or abnormal stools (as defined by the mother) in a 24-hour period [2]. A diarrheal episode was defined as being separated from another episode by at least 3 diarrhea-free days. However, if E. histolytica was detected in samples <60 days apart, they were counted as part of a single E. histolytica infection [17]. Because infection with multiple enteric pathogens was common in our study population, an episode of diarrhea was only counted as being caused by E. histolytica if infection was coincident with symptoms and if the parasite had not been detected in the previous monthly surveillance stool sample [3].
Sampling and Specimen Testing
The diarrheal and monthly surveillance stool specimens were tested for E. histolytica by use of the stools-specific Tri-Combo enzyme-linked immunosorbent assay (ELISA; TechLab, Blacksburg, Virginia) and an in-house ELISA for anti-E. histolytica IgA anti-Gal/GalNAc lectin (TechLab) [17, 18]. Positive stool samples were retested with the E. histolytica species-specific E. histolytica II enzyme linked immunosorbent assay (ELISA; TechLab) [19]. Samples were also tested by a species-specific quantitative polymerase chain reaction (qPCR) assay on DNA extracted from feces (Supplementary Table A1) [20–22]. As previously reported, qPCR assay results with a quantification cycle (Cq) of >35 were negative for antigen in fecal specimens [3]. Anti-E. histolytica IgA against the CRD region of the Gal/GalNAc lectin (Eh-IgA) was measured in both diarrheal and surveillance stool specimens by use of an in-house ELISA assay as previously described, children were regarded as having fecal specimens positive for anti-E. histolytica IgA against the CRD region of the Gal/GalNAc lectin when the A450 value was ≥0.2 [23].
P. copri and Bacteroides thetaiotaomicron were detected using qPCR primers described by Scher et al, SensiFAST qPCR mix (Bioline, Taunton, Massachusetts), and the CFX96 PCR machine (Bio-Rad, Hercules, California; Supplementary Table A1) [24]. A qPCR assay based on the primers described by Barman et al was used to detect 16S genes of Enterobacteriaceae, and a standard curve, using DNA extracted from known amounts of Escherichia coli (ATCC 25922), was used to determine the equivalent concentration of bacteria (Supplementary Table A1) [15]. Fecal DNA was used to confirm that the amplification efficiency of Enterobacteriaceae, P. copri, and B. thetaiotaomicron qPCR assays were comparable. The common reaction conditions consisted of a 3-minute hot start at 95°C, followed by DNA amplification for 40 cycles (a denaturation step at 95°C for 15 seconds followed by an annealing/extension step at 60°C for 15 seconds and plate reading). Melting curve analysis was then used to check that the designated target was amplified. To confirm P. copri assay specificity, the amplicon from selected clinical samples was sequenced and compared to that from cultured P. copri (CB7, DSMZ; a kind gift from D. Littman; Supplementary Figure A1).
Analysis of Samples Positive for Entamoeba Species by qPCR
Samples positive by microscopy for Entamoeba species but negative by qPCR assays for E. histolytica, Entamoeba dispar, and Entamoeba moshkovskii were analyzed using a modification of the broad range amplification protocol previously described (Supplementary Table A1) [25, 26]. To adapt the earlier approach to be compatible with results of MiSeq (Illumina, San Diego, California) 150–base pair paired end sequencing (150 cycles), the previously described 432–base pair 18S ribosomal DNA (rDNA) region of the Entamoeba genus was amplified in 2 sections, using the primers in (Supplementary Table A1) [25, 26]. The primers also included Illumina adaptors to permit the preparation of multiplex next-generation sequencing libraries as used in previously described protocols [8, 27, 28]. No new E. histolytica–positive samples were identified in the sample subset, verifying the utility of the E. histolytica–specific qPCR assay used in this study [20]. To expand the characterization of Entamoeba strains beyond what has been previously published, a second set of broad range primers was used to amplify the conserved transmembrane sequences of the E. histolytica heavy subunit of the Gal/GalNAc lectin hgl gene (Supplementary Figure A2A). The hgl genes of E. histolytica are a 4-member gene family and were not as highly amplified as the 18S Entamoeba target, which is contained within multicopy rDNA episomes [29, 30]. Therefore, a nested priming strategy was used that involved an initial amplification with the E_GALNAC primers and a second amplification with the internal E_illumina_n_GalNAc primer set (Supplementary Table A1). Sequence reads were assembled de novo, using Geneious 7 software (Biomatters, Auckland, New Zealand), and aligned with the E. histolytica rDNA and Gal/GalNAc lectin sequences, using ClustalW and Mega 6 software (Supplementary Figure A2C) [31, 32]. The hgl sequences of Entamoeba bangladeshi were confirmed by amplification with the primers described in Supplementary Table A1 and Sanger sequencing.
E. bangladeshi Surveillance
A qPCR assay was designed specific for E. bangladeshi, using the primers described in Supplementary Table A1 and the SensiFAST SYBR PCR mix (Supplementary Figure A2B) (Bioline, Taunton, Massachusetts). Reaction conditions involved a 3-minute hot start at 95°C, followed by a DNA amplification for 40 cycles, which consisted of a denaturation step at 95°C for 10 seconds followed by an annealing/extension step at 63°C for 15 seconds. The companion E. histolytica–specific assay used similar reaction conditions except that the annealing/extension step took place at 70°C; the level of fluorescent double-stranded DNA was measured using a CFX96 machine (Bio-Rad). Melting curve analysis was then used to confirm the amplification of the correct target.
Statistical Methods
Demographic information, surveillance data, and clinical and laboratory findings were computed in data files, using Fox-Pro (Microsoft, Redmond, Washington). Double data entry was used for all data sets. Differences in categorical data were tested by the χ2 test and the Fisher exact test, and differences in continuous data were tested by either a 2-sample t test or a nonparametric test, where applicable, using the Prism program (GraphPad Software, La Jolla, California). The time to the first E. histolytica–positive event was analyzed as time-to-event data under the survival analysis framework. The survival probabilities from E. histolytica were estimated using the Kaplan–Meier method, and differences in the time to infection were evaluated with the log-rank test.
Ethical Considerations
The study was approved by the ethical and research review committees of the International Centre for Diarrhoeal Disease Research, Bangladesh, and the Institutional Review Board of the University of Virginia. Informed written consent was obtained from the parents or guardians for the participation of their child in the study.
RESULTS
Study Characteristics
Children enrolled in the prospective study came from households where the median income was less than $100 per month and <40% of the mothers had formal education. However, almost all households reported the use of safe food-handling practices and had access to municipal water (Table 1) [2, 10].
E. histolytica Infection Was Prevalent in the Study Population
By the end of the second year of life, approximately 80% of infants in the study population had experienced at least 1 E. histolytica–positive infection, of which 17.3% were associated with diarrhea (Tables 2–4). To place this in context, in our cohort there were a record of 951 instances of diarrheal disease (mean no. [±SD] per child, 2.93 ± 1.856) in the first year of life and 1688 by the end of the second (4.66 ± 3.124). Diarrheal disease could be attributed to E. histolytica in 1 of every 20 cases (47 [5%] of cases in the first year and 48 (6.5%) in the second year).
Table 3.
Characteristics of 162 Children With a Second Entamoeba histolytica Infection
| Characteristic | Diarrhea | Colonizationa |
|---|---|---|
| Overall | 28/162 (17.3) | 134/162 (82.7) |
| Age at infection, d, median (range) | 430 (153–718) | 493 (94–719) |
| E. histolytica IgA positive–stool | 8/28 (28.6) | 17/134 (12.7) |
| Had third infection | ||
| Overall | 11/28 (39.3) | 43/134 (32.1) |
| Symptomatic, no. | 1 | 8 |
| Asymptomatic (colonization), no. | 10 | 35 |
Data are no. of children with the specified characteristic/no. of children in the cohort of interest (%), unless otherwise indicated. Children were considered to be infected with E. histolytica if they had a diarrhea or surveillance stool specimen that tested positive by quantitative polymerase chain reaction and were negative for E. histolytica during the previous 60 days.
Abbreviation: IgA, immunoglobulin A.
a Defined as an E. histolytica–positive stool specimen collected from an asymptomatic child during monthly surveillance.
Table 2.
Characteristics of 307 Children With an Initial Entamoeba histolytica Infection
| Characteristic | Diarrhea | Colonizationa |
|---|---|---|
| Overall | 54/307 (17.6) | 253/307 (82.4) |
| Age at infection, d, median (range) | 200 (4–701) | 214 (3–717) |
| E. histolytica IgA positive–stool | 8/54 (14.8) | 57/253 (22.5) |
| Had second infection | ||
| Overall | 30/54 (55.6) | 132/253 (52.2) |
| Symptomatic, no. | 5 | 23 |
| Asymptomatic (colonization), no. | 25 | 109 |
Data are no. of children with the specified characteristic/no. of children in the cohort of interest (%), unless otherwise indicated. Children were considered to be infected with E. histolytica if they had a diarrhea or surveillance stool specimen that tested positive by quantitative polymerase chain reaction and were negative for E. histolytica during the previous 60 days.
Abbreviation: IgA, immunoglobulin A.
a Defined as an E. histolytica–positive stool specimen collected from an asymptomatic child during monthly surveillance.
Table 4.
Characteristics of 54 Children With a Third Entamoeba histolytica Infection
| Characteristic | Diarrhea | Colonizationa |
|---|---|---|
| Overall | 9/54 (16.7) | 45/54 (83.3) |
| Age at infection, d, median (range) | 621 (332–720) | 614 (156–716) |
| E. histolytica IgA positive–stool | 2/9 (22) | 1/45 (2.2) |
| Had fourth infection | ||
| Overall | 3/9 (33.3) | 10/45 (22.2) |
| Symptomatic, no. | 1 | 2 |
| Asymptomatic (colonization), no. | 2 | 8 |
Data are no. of children with the specified characteristic/no. of children in the cohort of interest (%), unless otherwise indicated. Children were considered to be infected with E. histolytica if they had a diarrhea or surveillance stool specimen that tested positive by quantitative polymerase chain reaction and were negative for E. histolytica during the previous 60 days.
Abbreviation: IgA, immunoglobulin A.
a Defined as an E. histolytica–positive stool specimen collected from an asymptomatic child during monthly surveillance.
Anti-E. histolytica IgA Anti-Gal/GalNAc Lectin Was Associated With Protection From Infection
As previously shown, protection from E. histolytica in the first year of life was associated with the level of anti-E. histolytica IgA Gal/GalNAc lectin in the mother's breast milk (Supplementary Figure A4A) [10]. As expected, since exclusive breast-feeding did not take place in the second year of life, breast milk IgA was not associated with protection in infants during year 2 (Supplementary Figure A4B). In addition, a significantly longer E. histolytica–free period was observed if the child developed fecal anti-E. histolytica IgA at the time of the initial E. histolytica–positive sample: the median survival interval between the first and second infection in IgA-negative children was 373.3 days, compared with 526 days in IgA-positive children, and the log-rank (Mantel–Cox) test result indicated that the survival curves were significantly different (P = .02), with a log-rank hazard ratio of 1.586 (95% confidence interval, 1.078–2.140; Figure 1). We concluded that IgA antibodies against the Gal/GalNAc lectin, both in breast milk from the child's mother and from the child's gut, were associated with protection of the infant from amebiasis.
Figure 1.
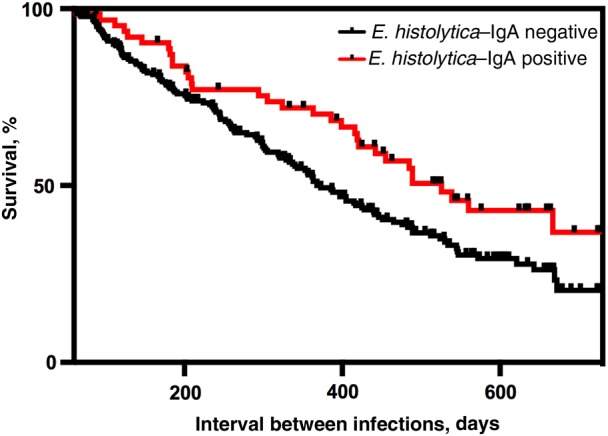
Children with fecal anti-Entamoeba histolytica Gal/GalNAc lectin immunoglobulin A (IgA) had a lower rate of reinfection. The y-axis denotes the percentage of children with an E. histolytica infection (n = 307), and the x-axis denotes the interval between the first and second E. histolytica infections. A statistically significant increase in the time to reinfection was observed for children who were fecal IgA anti-lectin positive (red line), compared with those who were negative (black line; P = .0175, by the log-rank [Mantel–Cox] test). A total 65 children tested positive and 242 tested negative for E. histolytica IgA.
A Higher Parasite Burden Was Associated With Diarrhea in the First but Not Second Year of Life
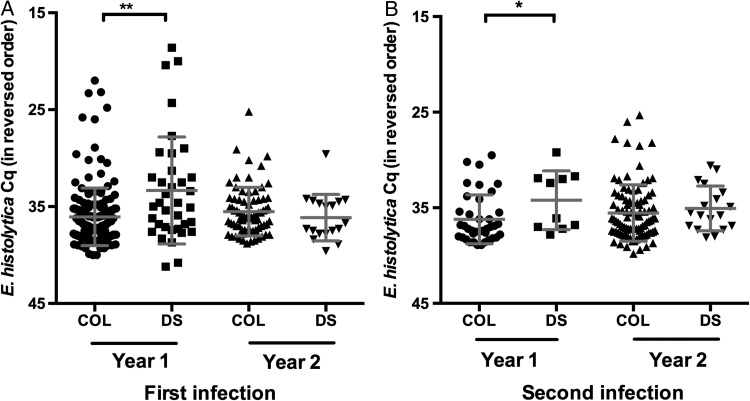
It has been hypothesized that symptomatic disease occurs when host defenses are overwhelmed by parasite numbers [33]. In E. histolytica events, this supposition was challenged by studies comparing the parasite load in older children and adults seeking care at the Dhaka Hospital with those in asymptomatic community controls [1]. In the current study, however, in a subset of the asymptomatic and diarrhea samples from children aged <1 year, a higher parasite burden was observed in symptomatic cases [3]. Using the entire data set, we confirmed that, in the first year of life, a statistically significant increase in parasite load occurred in symptomatic cases (Figure 2). However, in the second year of life, our findings were similar to the results from the earlier study: no difference in the parasite burden of symptomatic cases was observed [1]. This finding remained true even if it was repeated using only the children who experienced their initial E. histolytica event in the second year of life. This effect on parasite burden was independent of the number of preceding E. histolytica events, as determined by examining the qPCR Cq values (Supplementary Figure A6). These results suggested that parasite load was higher in diarrheal stool specimens in the first but not second year of life.
Figure 2.
Increased parasite burden with diarrhea in first but not second year of life. The amount of Entamoeba histolytica in stool specimens from children who had diarrhea and those who were asymptomatic was determined by quantitative polymerase chain reaction (qPCR). Samples collected from an E. histolytica–associated diarrheal sample (DS) or a sample in which E. histolytica was detected but colonization was not coincident with disease (COL) during the first infection (A) and second infection (B). The mean and standard error are shown. The y-axis indicates the threshold value of the qPCR assay specific for E. histolytica. As the quantification cycle distribution was not Gaussian, Kruskal–Wallis 1-way analysis of variance was used to determine that this data set contained significant differences. The Dunn multiple comparison test was used to determine that the year 1 data were significantly different from each other. *P ≤ .05 and **P ≤ .01.
Insights Into the Natural History of Human E. bangladeshi
During the in-depth MiSeq analysis of selected specimens sequences encoding the conserved transmembrane region of the Gal/GalNAc lectin family (Supplementary Table A1), data were obtained from an E. Bangladeshi–positive specimen (Supplementary Figure A1). A qPCR survey was conducted of cultured microscopically positive samples that could not be assigned to the common Entamoeba species of E. histolytica, E. dispar, or E. moshkovskii. Additional E. bangladeshi isolates were identified in 3 diarrheal samples and 8 surveillance samples derived from this study cohort [25]. Given the small number of E. bangladeshi–positive samples, it was not possible to come to any conclusions regarding the propensity of E. bangladeshi to cause diarrheal disease.
E. histolytica does not appear to confer cross-species protective immunity against E. moshkovskii or E. dispar colonization [34, 35]. In our study, we observed an E. histolytica colonization event prior to the collection 5 days later of an E. bangladeshi–positive sample. While this suggests that no barrier exists between colonization with both these Entamoeba species, a detectable anti-E. histolytica IgA response was not detected, suggesting that a robust mucosal E. histolytica immune defense was not mounted.
Environmental Factors Associated With Diarrhea Due to E. histolytica
One factor linked to E. histolytica–induced diarrhea in amebiasis is inflammation [36]. In regions where diarrheal disease is endemic, environmental enteropathy (blunted small-intestinal villi with lamina propria inflammation) is known to occur [37]. Cases of reported diarrhea from any cause in the cohort were examined to determine whether the number of days of diarrheal illness predicted whether a child experienced overt diarrheal disease or asymptomatic colonization upon an E. histolytica event: this was not significantly different in children aged 1–2 years (mean duration [±SD], 8.73 ± 6.2 days vs 7.18 ± 5.74 days [P = .1245]; Supplementary Figure A7).
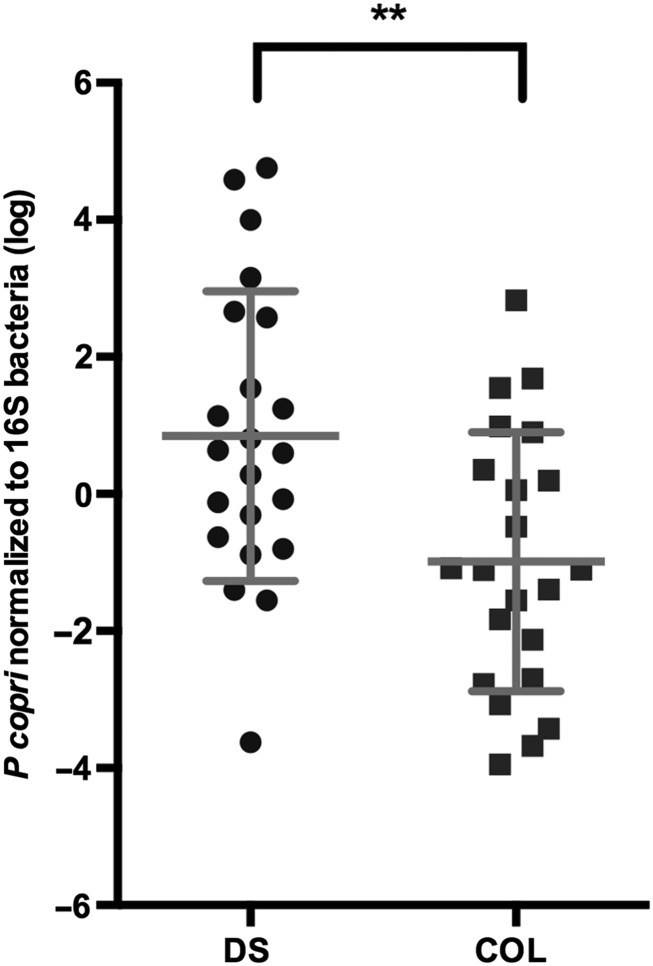
P. copri was associated with plant fiber consumption in humans [38], associated with colonic inflammation in mouse models [24], and found in E. histolytica–positive fecal samples (C. A. Gilchrist, unpublished observation). We therefore hypothesized that E. histolytica diarrhea would be more common in children with high P. copri levels. qPCR was used to examine the incidence of P. copri infection in symptomatic and asymptomatic children colonized with E. histolytica and Enterobacteriaceae rDNA used as an internal reference (Supplementary Figure A7) [15]. The P. copri level (normalized to Enterobacteriaceae values) was significantly increased in diarrhea, compared with asymptomatic infection (P = .0095, by the Mann–Whitney test; Supplementary Figure A8), in children between the ages of 600–800 days. An additional 18 samples from another cohort combined with 26 samples from the study cohort children to yield 44 samples (P = .0079, by the Mann–Whitney test; Figure 3). This larger data set allowed the effect of age to be scrutinized in conjunction with child symptoms and P. copri levels. As expected, in the regression analysis, a significant association with both age and child symptoms (model P = .001; R2 = 0.276) was observed with an estimated mean age effect (±SD) of 1.002 ± 0.431 per month (P = .025), indicating that each month increment in age was associated with a 1.002 increase in log P. copri levels [12, 13]. The P. copri levels, however, remained significantly lower in asymptomatic children, compared with those with diarrhea, even when age was taken into consideration (estimated mean effect [±SD], −5.220 ± 1.392; P < .001).
Figure 3.
Expansion of Prevotella copri was associated with diarrhea due to Entamoeba histolytica in children from 600 to 800 days of age. Samples collected between 600 and 800 days of age from an E. histolytica–associated diarrheal sample (DS) or a sample in which E. histolytica was detected but colonization was not coincident with disease (COL) were compared after normalization to Enterobacteriaceae levels. The y-axis denotes the P. copri qPCR signal normalized to the Enterobacteriaceae burden, as determined by 16S bacteria ribosomal DNA load (log), and the x-axis denotes the specimen type. **P ≤ .01.
Adults with new-onset untreated rheumatoid arthritis had an expansion of P. copri at the expense of a more beneficial member of the Bacteroides family, B. thetaiotaomicron [24]. We therefore examined the level of B. thetaiotaomicron in these children, but in contrast to the findings of Scher et al, Bacteroides levels did not significantly change (data not shown).
DISCUSSION
This study represents the first prospective longitudinal of E. histolytica infection from birth to 2 years of life. Infection occurred in 80% of the children from the Mirpur slum in Dhaka, with 17% of infections resulting in diarrhea. The most intriguing finding was the association between parasite burden and symptomatic amebiasis in the first year of life and in the second with increased levels the level of a pathobiont, P. copri. We had suspected that intestinal dysbiosis and the domination of the gut environment by a pathobiont could affect the phenotype of an E. histolytica infection, as the children in the study cohort had had intense exposure to enteric infections [3]. In the second year of life, when the association of P. copri with E. histolytica diarrhea was observed, only 8.4% of children (33 of 392) had not experienced diarrhea. In a study examining the potential role of the microbiome in older E. histolytica–positive patients, the Bacteroides species were in general downregulated [39]. We did not see a significant change in B. thetaiotaomicron levels in our population, but any effect may have been masked by the age of our study participants, as an increase in Bacteroides numbers occurs during weaning [12].
Environmental enteropathy is a condition caused by repeated diarrheal events and is often associated with an unfavorable outcome in enteric disease–associated infections [40]. Because of the association of P. copri with systemic gut inflammation in patients and in animal models [24], this work suggests that P. copri, already associated with excessive immune activation, may tip the balance to the development of amebic diarrhea upon E. histolytica infection and could be linked, at least in our population, with the syndrome of environmental enteropathy. Combined with recent work showing the importance of the microbiota in controlling susceptibility in the mouse model of amebiasis [41], the work in humans therefore provides insights into the factors that may control virulence expression [40].
Studies are needed not only to confirm the link between P. copri and amebiasis in human populations but also to investigate the interplay between this bacterium and host inflammation, as well as the role of other members of the gut microflora. In this population, therapies aimed at controlling the expansion of pathobionts may be particularly useful in protecting children from diarrheal disease [42]. A deeper analysis of the microbiome of these children may allow identification of additional potential deleterious or protective microbes, and future investigations are planned in this regard.
Fecal anti-E. histolytica IgA against the CRD region of the Gal/GalNAc lectin was associated with protection however this natural response was insufficient to ameliorate the burden of disease in this age group. There is a need to develop a vaccine against E. histolytica that can prevent disease and provide strong and long lasting protection.
Additional studies are also still needed to understand the virulence determinants of the Entamoeba species that infect humans. A key component of E. histolytica virulence is the Gal/GalNAc lectin, the subunits of which are encoded by a family of genes [43]. The genome of E. bangladeshi also appeared to encode a Gal/GalNAc lectin gene family (Supplementary Figure A2C), but the absence of cross-species immunity or the cross-reaction with E. histolytica Gal/GalNAc lectin–based immunodiagnostics used in this study suggest that the extracellular domain of the encoded proteins, like those of E. dispar, may be dissimilar [44]. Specific tools therefore will be needed to determine the true prevalence of this organism. This work indicated both that this protozoan organism was circulating within the Bangladesh community and that E. bangladeshi could be associated with diarrhea that was occurring at the time of isolation in 3 of the 11 E. bangladeshi–positive infants.
In summary, this work suggests that, in addition to the impact of host and parasite genetics in providing protection in amebiasis, the commensal microbiota and the development of protective immunity in regions of endemicity are important protective factors that should be examined to fully understand the risk of contracting symptomatic disease [7–9, 17]. In this study, severity of amebic colitis was influenced by different factors in the gut environment, depending on child age. Early in life, prenatal and perinatal factors, such as birth LAZ score and maternal breast milk IgA antibody level, were associated with E. histolytica infection incidence. However, important as these factors were in year 1 of life, they no longer played a significant role in older children. Instead, in children aged 1–2 years, a strong link was observed between an expansion of P. copri and diarrheal disease, suggesting that, in this age group, symptomatic outcome of E. histolytica is influenced by the microbiome and the inflammatory state of the gut.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank Dr B. Mann for careful reading of the manuscript.
C. A. G., W. A. P., and R. H. designed and R. H., W. A. P., M. A., and M. K. oversaw the work described in this article. C. A. G. wrote the manuscript. S. E. P., B. N. S., D. J. R., S. B., C. S. J., K. P. E., M. K., and M. A. acquired the data used in this article. C. A. G., J. Z. M., N. J., S. E. P., W. A. P., and R. H. reviewed and analyzed study data. K. W. and S. L. B. assisted in the interpretation of data.
Disclaimer. The funders had no role in study design, data collection and analysis, or decision to submit for publication.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grants AI103536-01 [to C. A. G.] and AI043596 [to W. A. P.]).
Potential conflicts of interest. The University of Virginia has licensed technology for amebiasis diagnosis to TechLab, and W. A. P. receives licensing income from TechLab for amebiasis diagnostics; this income is donated in its entirety to the American Society for Tropical Medicine and Hygiene, without benefit to W. A. P. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Haque R, Mondal D, Karim A et al. Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in Bangladesh. Clin Infect Dis 2009; 48:1191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mondal D, Minak J, Alam M et al. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in Bangladesh. Clin Infect Dis 2012; 54:185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taniuchi M, Sobuz SU, Begum S et al. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J Infect Dis 2013; 208:1794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotloff KL, Nataro JP, Blackwelder WC et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–22. [DOI] [PubMed] [Google Scholar]
- 5.Fischer Walker CL, Aryee MJ, Boschi-Pinto C, Black RE. Estimating diarrhea mortality among young children in low and middle income countries. PLoS One 2012; 7:e29151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haque R, Huston CD, Hughes M, Houpt E, Petri WAJ. Amebiasis. N Engl J Med 2003; 348:1565–73. [DOI] [PubMed] [Google Scholar]
- 7.Ali IKM, Haque R, Alam F, Kabir M, Siddique A, Petri WAJ. Evidence for a link between locus R-R sequence type and outcome of infection with Entamoeba histolytica. Clin Microbiol Infect 2012; 18:E235–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilchrist CA, Ali IKM, Kabir M et al. A multilocus sequence typing system (MLST) reveals a high level of diversity and a genetic component to Entamoeba histolytica virulence. BMC Microbiol 2012; 12:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggal P, Guo X, Haque R et al. A mutation in the leptin receptor is associated with Entamoeba histolytica infection in children. J Clin Invest 2011; 121:1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korpe PS, Liu Y, Siddique A et al. Breast milk parasite-specific antibodies and protection from amebiasis and cryptosporidiosis in Bangladeshi infants: a prospective cohort study. Clin Infect Dis 2013; 56:988–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haque R, Mondal D, Duggal P et al. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect Immun 2006; 74:904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koenig JE, Spor A, Scalfone N et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011; 108(suppl):4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian S, Huq S, Yatsunenko T et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014; 509:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandtzaeg P. The gut as communicator between environment and host: immunological consequences. Eur J Pharmacol 2011; 668(suppl):S16–32. [DOI] [PubMed] [Google Scholar]
- 15.Barman M, Unold D, Shifley K et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 2008; 76:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mondal D, Haque R, Sack RB, Kirkpatrick BD, Petri WA Jr. Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am J Trop Med Hyg 2009; 80:824–6. [PMC free article] [PubMed] [Google Scholar]
- 17.Haque R, Duggal P, Ali IM et al. Innate and acquired resistance to amebiasis in Bangladeshi children. J Infect Dis 2002; 186:547–52. [DOI] [PubMed] [Google Scholar]
- 18.Christy NCV, Hencke JD, Escueta-De Cadiz A et al. Multisite performance evaluation of an ELISA for the detection of Giardia, Cryptosporidium, and Entamoeba histolytica antigens in human stool. J Clin Microbiol 2012; 50:1762–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque R, Neville LM, Wood S, Petri WA. Short report: detection of Entamoeba histolytica and E. dispar directly in stool. Am J Trop Med Hyg 1994; 50:595–6. [DOI] [PubMed] [Google Scholar]
- 20.Haque R, Roy S, Siddique A et al. Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am J Trop Med Hyg 2007; 76:713–7. [PubMed] [Google Scholar]
- 21.Ali IKM, Hossain MB, Roy S et al. Entamoeba moshkovskii infections in children, Bangladesh. Emerg Infect Dis 2003; 9:580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haque R, Ali IK, Akther S, Petri WA. Comparison of PCR, isoenzyme analysis, and antigen detection for diagnosis of Entamoeba histolytica infection. J Clin Microbiol 1998; 36:449–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haque R, Ali IM, Petri WA. . Prevalence and immune response to Entamoeba histolytica infection in preschool children in Bangladesh. Am J Trop Med Hyg 1999; 60:1031–4. [DOI] [PubMed] [Google Scholar]
- 24.Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013; 2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Royer TL, Gilchrist C, Kabir M et al. Entamoeba bangladeshi nov. sp., Bangladesh. Emerg Infect Dis 2012; 18:1543–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stensvold C, Lebbad M, Victory E. Increased sampling reveals novel lineages of Entamoeba: consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist 2011; 162:525–41. [DOI] [PubMed] [Google Scholar]
- 27.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc 2010; 2010:pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 28.Caporaso JG, Lauber CL, Walters WA et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J [Internet]. Int Soc Microb Ecol 2012; 6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sehgal D, Mittal V, Ramachandran S, Dhar SK, Bhattacharya A, Bhattacharya S. Nucleotide sequence organisation and analysis of the nuclear ribosomal DNA circle of the protozoan parasite Entamoeba histolytica. Mol Biochem Parasitol 1994; 67:205–14. [DOI] [PubMed] [Google Scholar]
- 30.Ramakrishnan G, Ragland BD, Purdy JE, Mann BJ. Physical mapping and expression of gene families encoding the N-acetyl D-galactosamine adherence lectin of Entamoeba histolytica. Mol Microbiol 1996; 19:91–100. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2002; Chapter 2:Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 33.Janeway CA, Travers P, Walport M, Shlomchik MJ. Immunobiology: the immune system in health and disease. 5th ed New York, NY: Garland Science, 2001. http://www.ncbi.nlm.nih.gov/books/NBK10757/. Accessed 7 September 2014. [Google Scholar]
- 34.Shimokawa C, Kabir M, Taniuchi M et al. Entamoeba moshkovskii is associated with diarrhea in infants and causes diarrhea and colitis in mice. J Infect Dis 2012; 206:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ximénez C, Morán P, Rojas L, Valadez A, Gómez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect Genet Evol 2009; 9:1023–32. [DOI] [PubMed] [Google Scholar]
- 36.Moonah SN, Jiang NM, Petri WA. Host immune response to intestinal amebiasis. PLoS Pathog 2013; 9:e1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korpe PS, Petri WA. Environmental enteropathy: critical implications of a poorly understood condition. Trends Mol Med 2012; 18:328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruengsomwong S, Korenori Y, Sakamoto N, Wannissorn B, Nakayama J, Nitisinprasert S. Senior Thai fecal microbiota comparison between vegetarians and non-vegetarians using PCR-DGGE and real-time PCR. J Microbiol Biotechnol 2014; 24:1026–33. [DOI] [PubMed] [Google Scholar]
- 39.Verma AK, Verma R, Ahuja V, Paul J. Real-time analysis of gut flora in Entamoeba histolytica infected patients of Northern India. BMC Microbiol 2012; 12:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerrant RL, DeBoer MD, Moore SR, Scharf RJ, Lima AAM. The impoverished gut--a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 2013; 10:220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess SL, Buonomo E, Carey M et al. Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A-dependent protection against Entamoeba histolytica colitis. MBio 2014; 5:e01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barr JJ, Youle M, Rohwer F. Innate and acquired bacteriophage-mediated immunity. Bacteriophage 2014; 3:e25857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petri WAJ, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu Rev Microbiol 2002; 56:39–64. [DOI] [PubMed] [Google Scholar]
- 44.Weedall GD, Sherrington J, Paterson S, Hall N. Evidence of gene conversion in genes encoding the Gal/GalNac lectin complex of Entamoeba. PLoS Negl Trop Dis 2011; 5:e1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.