Abstract
Nonsense-mediated mRNA decay (NMD) is a eukaryotic surveillance mechanism that monitors cytoplasmic mRNA translation and targets mRNAs undergoing premature translation termination for rapid degradation. From yeasts to humans, activation of NMD requires the function of the three conserved Upf factors: Upf1, Upf2, and Upf3. Here, we summarize the progress in our understanding of the molecular mechanisms of NMD in several model systems and discuss recent experiments that address the roles of Upf1, the principal regulator of NMD, in the initial targeting and final degradation of NMD-susceptible mRNAs. We propose a unified model for NMD in which the Upf factors provide several functions during premature termination, including the stimulation of release factor activity and the dissociation and recycling of ribosomal subunits. In this model, the ultimate degradation of the mRNA is the last step in a complex premature termination process.
Keywords: quality control, Upf proteins, translation termination, ribosome dissociation
INTRODUCTION
Nonsense-mediated mRNA decay (NMD), one of several cytoplasmic surveillance mechanisms that monitor mRNA translation in eukaryotes, targets mRNAs containing a premature termination codon (PTC) for rapid degradation (109, 140, 201). The pathway was initially recognized in Saccharomyces cerevisiae and Caenorhabditis elegans (126, 174, 180), but NMD exists in all eukaryotic cells examined and the core NMD machinery is conserved from yeast to humans (18, 109, 193, 194). NMD’s function was originally thought to be limited to quality control, i.e., the elimination of mRNAs derived from genes harboring nonsense mutations to prevent the accumulation of potentially deleterious truncated polypeptides (84, 180). However, the pathway’s substrates include not only PTC-containing mRNAs but also a significant fraction of apparently normal and physiologically functional wild-type mRNAs (194). NMD thus serves as both a surveillance system and a fundamental post-transcriptional regulatory mechanism for eukaryotic gene expression. In the latter mode, NMD function is linked to diverse cellular processes, including cell growth and proliferation (12, 138, 221), development and differentiation (24, 70, 147, 148, 151, 231), innate immunity (68), antiviral or stress responses (13, 60, 189), and neuronal activity or behavior (39, 66).
Multiple aspects of NMD have substantial clinical significance. Mutations in human genes that regulate NMD can cause serious neurodevelopmental disorders, predispose patients to such disorders, or associate with specific tumor types (101, 135, 160, 161, 212, 232). Further, nonsense mutations are responsible for ~15% of the single base-pair mutations that cause inherited disorders, and some disease genes have considerably higher nonsense mutation frequencies (156). Patients with nonsense mutations tend to have more serious disease than those with missense mutations, presumably because of marked reductions in specific gene expression (115). Variations in NMD efficiency also appear to lead to significant differences in disease pathology and outcome (15, 57, 87, 111, 152). In addition, because all diseases caused by nonsense mutations generally share two key gene expression problems, premature translation termination and mRNA reduction through NMD, therapeutic approaches to both problems are being investigated, with the understanding that a single drug has the potential to treat a large number of different disorders (107, 175).
The onset of NMD for a particular transcript is linked to premature translation termination (109), but NMD activation by a PTC stands in striking contrast to events occurring during stop codon recognition by the ribosome at the end of each round of normal translation. Accordingly, a mechanistic understanding of NMD depends on elucidating the differences between premature and normal termination, determining how premature termination is coupled to accelerated mRNA decay, and defining the roles and functional order for factors that activate and execute these processes.
SUBSTRATES OF NONSENSE-MEDIATED DECAY
NMD targets both PTC-containing and apparently wild-type transcripts in eukaryotic cells. Depending on the organism or cell type, approximately 5% to 20% of the transcripts in a typical transcriptome are substrates of the NMD pathway (83, 131, 150, 151, 183–185, 210, 221, 222, 231, 237). Transcripts degraded by the NMD pathway can be classified into several general categories. One category, exemplifying typical NMD substrates, includes mRNAs with a destabilizing PTC in their coding region. These transcripts are generated from endogenous genes harboring nonsense or frameshift mutations (83), pseudogenes (83, 146), nonproductively rearranged genetic loci (134), or alternative splicing events that lead to intron retention or inclusion of a PTC-containing exon (62, 97, 122, 143, 162, 222). A second category contains mRNA-like transcripts with limited or no apparent coding potential, e.g., long noncoding RNAs (117, 143, 211), small RNAs derived from intragenic regions (206, 215), and transcripts of inactivated transposable elements (83). A third category contains a subset of physiologically relevant transcripts that appear to be normal, such as mRNAs with upstream open reading frames (uORFs) (10, 59, 83, 167, 185), atypically long 3′-UTRs (untranslated regions) (106, 108, 204, 238), and mRNAs coding for selenoproteins (155, 196). Only some mRNAs with uORFs are degraded by the NMD pathway (59, 83, 167, 185). Those that are resistant to NMD appear to have diminished nonsense codon occupancy by the ribosome (59) or contain downstream sequences that inactivate NMD (187, 188). Termination codons flanked by atypically long 3′-UTRs also elicit NMD (106, 108, 204, 238), an observation implying that termination context can influence mRNA susceptibility to NMD (4) (see below).
The broad range of NMD substrates indicates that NMD has substantial biological impact on eukaryotic gene expression; not only does NMD ensure removal of junk (e.g., some by-products of transcription and alternate splicing or pseudogene mRNAs), but it also effectively renders most nonsense alleles as null alleles (42, 152) and has been co-opted to regulate the levels and locations of specific proteins (39, 66, 99, 125).
THE NONSENSE-MEDIATED DECAY MACHINERY
NMD is a translation-dependent process triggered by the presence of a premature stop codon in the A site of an elongating ribosome (109, 140, 193). The NMD machinery identifies unique features in the prematurely terminating messenger ribonucleoprotein (mRNP), targets PTC-containing transcripts for rapid degradation, and includes the conserved Upf factors, the regulators and effectors of the Upf factors, and, most likely, the factors directly involved in translation termination.
Core Upf Proteins
From yeast to humans, activation of NMD requires a set of conserved core regulatory factors, the Upf1-3/Smg2-4 proteins (41, 61, 78, 80, 109, 126, 141, 163, 165, 176). Upf1 and Upf2 are localized primarily in the cytoplasm, whereas Upf3 shuttles between the nucleus and the cytoplasm (141, 195). Upf1, Upf2, and Upf3 interact with each other, the ribosome, and multiple mRNA decay and translation factors, but their exact roles in NMD, as well as their mechanism of association with a premature termination complex that includes an mRNA, an 80S ribosome, and numerous factors, remain to be clarified (109).
Upf1 is a superfamily I RNA helicase with two major domains: an N-terminal cysteine- and histidine-rich (CH) domain and a helicase domain toward its C terminus (3, 29, 38, 114, 127). Purified Upf1 binds ATP and RNA and manifests RNA-dependent ATPase and 5′-to-3′ RNA helicase activities (20, 44). Upf1’s ATP-binding and hydrolysis activities are critical for activation of NMD, as mutations that eliminate these activities abolish NMD activity in vivo (226–228), and Upf1 proteins that lack these activities are defective in association with the 40S ribosomal subunit (153), disassembly of a terminating mRNP (56), and recycling of components of the protein synthesis and NMD machineries (65). Maximal in vitro activation of Upf1’s ATPase and helicase activities requires both Upf2 and Upf3 (30). The CH domain of yeast Upf1 binds to the C-terminal region of Upf2 (78), the ribosomal protein Rps26 (153), and the decapping enzyme subunit Dcp2 (80, 82), and also self-associates (79), suggesting that it may play a role in sequential molecular interactions during execution of NMD. Metazoan Upf1 contains extra conserved extensions at both its N and C termini. The N-terminal extension is rich in proline/glycine residues, and the C-terminal extension is rich in serine/glutamine residues (8, 176). In human Upf1, these N- and C-terminal extensions contain multiple phosphorylated serine or threonine residues (169, 236) and exhibit both phosphorylation-dependent and phosphorylation-independent interactions with Smg5, Smg6, and Smg7 (28, 164, 168, 169). Human Upf1 also interacts with Dcp2 (54, 139). In contrast to yeast Upf1:Dcp2 interaction, human Upf1:Dcp2 is mediated primarily by Upf1’s N-and C-terminal extensions (136).
Upf2 is commonly thought to be a molecular bridge between Upf1 and Upf3, but its function has additional complexity. The N-terminal two-thirds of Upf2 contains three conserved eIF4G-like (MIF4G) domains (9, 37, 103, 179), the most C terminal of which (MIF4G-3) interacts with the central RRM (RNA recognition motif) in Upf3 (Upf3b in metazoans) (78, 103) and the C-terminal domain of Smg1. In vitro, the Upf2 MIF4G-1 and -3 domains both bind RNA (55, 103). The C terminus of Upf2 binds to the CH domain of Upf1 (38, 77), and Upf2 binding to Upf1 switches Upf1 from RNA clamping to RNA unwinding and promotes Upf1’s RNA helicase activity (29, 30).
Upf3 is a basic protein. A single isoform of Upf3 exists in Saccharomyces cerevisiae and Caenorhabditis elegans, but two isoforms (Upf3a and Upf3b) are present in human cells (141, 195). The two Upf3 isoforms exhibit different expression and regulation patterns (31), as well as different NMD-inducing activities (116). For both Upf3a and Upf3b, the central RRM domain binds to Upf2 (103, 195) and the C-terminal domain binds to a composite surface on the exon junction complex (EJC) (25, 64).
Additional Smg Proteins
In addition to the core Upf factors, activation of NMD in multicellular organisms also requires the function of additional proteins, Smg1 and Smg 5–9 (109, 194, 233). Collectively, these Smg proteins control the phosphorylation and dephosphorylation of Upf1 and in some cases also function as effectors of Upf1 by activating or recruiting specific mRNA decay activities.
Smg1, Smg8, and Smg9 form a kinase complex that catalyzes Upf1 phosphorylation, a rate-limiting step of NMD in metazoans (76, 104, 235, 236). Smg1-mediated Upf1 phosphorylation occurs in the decay-inducing complex (DECID), and this process also requires Upf2, Upf3, and components of the EJC (104). Smg5, Smg6, and Smg7 are structurally related proteins (61) that function in Upf1 dephosphorylation (170, 233) and also act as effectors of Upf1 (51, 90, 136, 216). Each of these proteins contains a phosphopeptide-binding 14-3-3-like domain and binds to phosphorylated Upf1 (28, 58, 102, 168, 169). Smg5 and Smg6 each contain a C-terminal PilT N-terminus (PIN) domain, which is related to the RNase H family (67). Smg5 and Smg7 form a heterodimer with their 14-3-3-like domains (7, 102), and the resulting complex acts through Smg7 to recruit the CCR4-NOT deadenylase complex and promote mRNA deadenylation and decapping (136, 216). Smg5 interacts with the structural and catalytic subunits of the PP2A phosphatase (7, 168). Smg6 is monomeric in solution and can also bind the Upf1 helicase domain and C-terminal extension in a phosphorylation-independent manner (28, 164). The C-terminal PIN domain of Smg6 exhibits nuclease activity in vitro (67), and, in vivo, Smg6’s endonuclease activity cleaves NMD substrates (22, 51, 90, 143, 192). Upf1 phosphorylation is also observed in yeast (47, 123, 218), but these cells lack an ortholog of the Smg1 kinase, and yeast Upf1 also lacks the regions targeted by Smg1. Currently, there is no direct evidence that links Upf1 phosphorylation to NMD regulation in yeast (123, 218).
Release Factors
NMD is functionally and physically linked to the activities of the eukaryotic release factors eRF1 and eRF3 (Sup45 and Sup35, respectively, in yeast). All three Upf proteins interact with the release factors, and these interactions may function to recruit the NMD machinery to prematurely terminate mRNPs and launch the NMD process. Upf1 interacts with both eRF1 and eRF3 (43, 95, 104, 204), and in human cells this leads to formation of the SURF (Smg-1-Upf1-release factor) complex (104, 235). Yeast Upf2 and Upf3 interact with eRF3 and may compete with eRF1 for a specific eRF3 interaction domain (219). Upf1 binding to eRF1 and eRF3 inhibits Upf1’s ATPase activity (43), a result supporting the notion that Upf1 is initially recruited to a prematurely terminating ribosome in an inactive form and is subsequently activated by interaction with a Upf2:Upf3 complex (30).
Exon Junction Complex
In mammalian cells, nuclear pre-mRNA splicing deposits a protein complex 20–24 nt upstream of an exon-exon junction (128). The core of this complex is formed by four conserved proteins: eIF4AIII, Y14, Magoh, and MLN51 (23). Dubbed the EJC, this group of proteins associates with spliced mRNA, travels with it to the cytoplasm, and controls several aspects of mRNA function, including nuclear export and cytoplasmic localization, translation, and decay (129). A role for the EJC in the activation of NMD was suggested by several observations, including those noting that (a) mammalian NMD is often enhanced by an exon-exon junction 50–55 nt downstream of a PTC (159, 214, 239); (b) the NMD factor Upf3 associates with the components of the EJC (112, 142); (c) tethering of EJC factors downstream of a termination codon can trigger mRNA degradation by NMD (63, 64, 142); and (d) elimination of core EJC components can selectively stabilize the PTC-containing mRNAs (64, 172, 198). In spite of these tantalizing observations, the exact role of the EJC in NMD is still unclear. Because EJCs promote the translational efficiency of spliced mRNAs (32, 144, 166), and because NMD is absolutely dependent on recognition of a PTC by the ribosome (19, 59, 73, 137, 224, 240), the observed effects of EJC components on NMD may be indirect, reflecting the consequences of EJCs on mRNA translation. Further blurring this issue and the interpretation models for EJC function in NMD (93, 193), recent genome-wide analyses revealed that EJC occupancy on spliced mRNAs exhibits significant heterogeneity, with a large fraction (20%) of splicing events not resulting in EJC deposition and approximately 40–50% of EJCs in human cells binding to noncanonical regions of spliced mRNAs (191, 203).
NONSENSE-MEDIATED DECAY AND TRANSLATION TERMINATION
As noted above, NMD and translation termination are linked because nonsense codon recognition by the translation apparatus is a requirement for NMD (19, 59, 73, 137, 224, 240). Although both premature and normal translation termination occur in response to a stop codon in the ribosomal A site, at least some aspects of the ensuing events are different, as PTCs, but not NTCs, activate NMD (Figure 1). At a minimum, premature termination is considerably less efficient than normal termination (4, 173), a difference that may be a key to understanding NMD.
Figure 1.
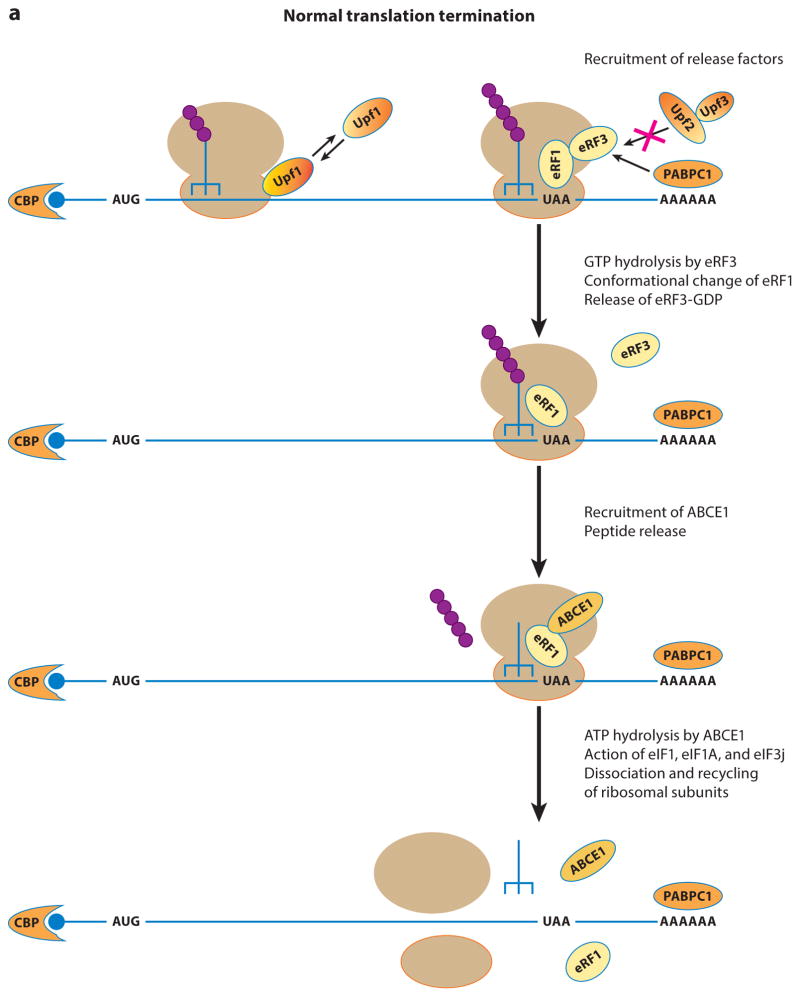
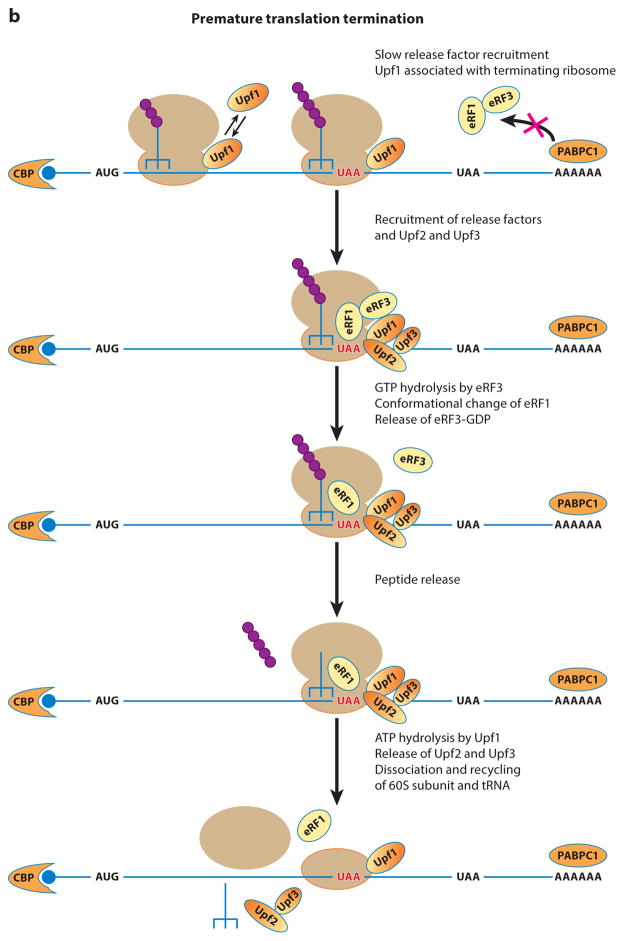
Possible mechanistic differences between normal translation termination and premature translation termination. (a) An overview of normal translation termination. Here, the release factors eRF1 and eRF3 are thought to be efficiently recruited to the A site of the terminating ribosome, thereby preventing Upf2 and Upf3 from associating with the termination complex and precluding the stable association of Upf1 with the 40S subunit. The ordered and coordinated actions of eRF1, eRF3, and ABCE1 promote efficient peptide release, after which ATP hydrolysis by ABCE1 and the combined actions of initiation factors eIF1, eIF1A, and eIF3j promote the dissociation and recycling of the ribosomal subunits. (b) An overview of premature translation termination. Owing to the lack of a 3′-UTR-based stimulatory activity, the release factors eRF1 and eRF3 are inefficiently recruited to the A site of the prematurely terminating ribosome. The delayed recruitment of the release factors or a particular structural feature of a premature termination complex triggers the binding of Upf2 and Upf3 to the terminating ribosome. The joining of Upf2 and Upf3 stabilizes Upf1’s binding to the premature terminating ribosome and, collectively, the three Upf factors control several activities of the termination process. At the early stage, they function in recruiting the release factors or stimulating release factor activities to promote peptide release. At the next stage, they promote the dissociation and recycling of ribosomal subunits. The function of the Upf factors at this stage of premature termination is similar to that of ABCE in normal termination, but mechanistic differences must exist. For example, ABCE1 activity at normal termination leads to complete dissociation of the ribosomal subunits from the mRNA, but the activity of the Upf factors at premature termination may only lead to dissociation of the 60S subunit. Abbreviations: CBP, cap-binding protein; PABPC1, poly(A)-binding protein.
Normal Translation Termination
In eukaryotes, termination at UAA, UAG, or UGA codons requires the coordinated functions of the release factors eRF1 and eRF3 (2) (Figure 1a). eRF1 recognizes nonsense codons in the ribosomal A site and induces hydrolysis of peptidyl-tRNA (2, 33, 208). eRF3 is a GTPase that stimulates eRF1 activity on the ribosome and links GTP hydrolysis to efficient peptide release (96, 190). After peptide release, eRF1 remains bound to the post-termination complex, recruits the ATP-binding cassette protein ABCE1 (Rli1 in yeast), and together with ABCE1 dissociates the complex into free 60S subunits and tRNA- and mRNA-associated 40S subunits (14, 16, 178). The subsequent dissociation of tRNA and mRNA from the 40S subunits is carried out by the combined actions of translation initiation factors eIF3, eIF1, eIF1A, and eIF3j (177, 178), and all components are thus rendered ready to be recycled for new rounds of translation. In vitro, Rli1 can also stimulate the peptide hydrolysis activity of eRF1, and this stimulatory activity is inhibited by eRF3, suggesting that efficient peptide release in vivo may involve the sequential actions of both the release factor eRF3 and the recycling factor ABCE1 (200). Using nonsense codon read-through as an assay, several other proteins, including poly(A)-binding protein (Pab1 in yeast; PABPC1 in humans), have been shown to influence translation termination. Pab1-eRF3 interaction is thought to play a role in the formation of an mRNP complex favorable to normal translation termination (6, 40, 85, 88), and overexpressing or deleting Pab1, respectively, enhances or reduces termination efficiency in yeast or human cells (40, 88, 95). Recent Pab1 tethering experiments suggest that the Pab1-eRF3 interaction functions mainly to promote the recycling of ribosomal subunits to the 5′ end of a translating mRNA (53).
Premature Translation Termination Is Aberrant
Although all termination events (and NMD) begin with a nonsense codon in the ribosomal A site, several observations indicate that the subsequent process appears to be mechanistically different for normal and premature translation termination (Figure 1b): (a) normal termination events do not trigger NMD; (b) yeast ribosomes recognizing normal termination codons (NTCs) in vitro do not yield toe prints unless eRF1 is inactivated by a temperature-sensitive lesion (4), but ribosomes at PTCs readily yield toe-print signals without eRF1 inactivation (4), and comparable results have been observed for termination at a PTC versus NTC of human β-globin mRNA (173); and (c) although some NTCs allow nonsense codon read-through (49, 72), PTCs are, in general, much more susceptible to read-through than NTCs (21, 48, 69, 75, 107, 120, 132, 133, 175, 223, 241). Thus, although translation termination at NTCs and PTCs are both triggered by the presence of a stop codon in the A site, the kinetics and the efficiency of the termination events at NTCs and PTCs are markedly different. The diminished efficiency of premature termination suggests that at least some aspects of termination events at PTCs may have fundamentally different kinetics than those at NTCs. The potential differences at PTCs may include the delayed recruitment and action of the release factors due to lack of the stimulatory activity of Pab1. As a consequence of such potential differences in kinetics, premature termination may use a different mechanism to dissociate and recycle the components of terminating mRNPs (Figure 1b).
MOLECULAR MECHANISMS OF NONSENSE-MEDIATED DECAY
NMD selectively targets mRNAs undergoing premature translation termination or the contextual equivalent of premature termination. An understanding of the molecular mechanisms of NMD requires answers to four key questions. First, what are the unique molecular features or events that distinguish premature termination from normal termination and make the premature termination process susceptible to NMD? Second, when and how are the key NMD factors targeted to prematurely terminating mRNPs? Third, how are the targeted mRNAs degraded? And, finally, in addition to their decay-promoting activities, what are the ancillary roles of the NMD factors and what insights do these roles provide about the differences between premature and normal termination?
Defining Features of Prematurely Terminating mRNPs
Three principal models describe unique or defining features of mRNPs undergoing premature translation termination and propose the ways in which these features might trigger NMD. These include the EJC model, the Upf1 3′-UTR sensing and potentiation model, and the faux 3′-UTR model. All three models invoke the importance of an mRNA’s 3′-UTR in initiating NMD but propose significantly different roles for the 3′-UTR. The EJC and the Upf1 3′-UTR sensing and potentiation models propose that the long 3′-UTR of a PTC-containing mRNA contains specific mRNP structures that can induce or potentiate NMD. In contrast, the faux 3′-UTR model proposes that a long 3′-UTR creates an mRNP structure that has diminished stimulatory activity for translation termination, leading to aberrant termination.
Exon Junction Complex Model
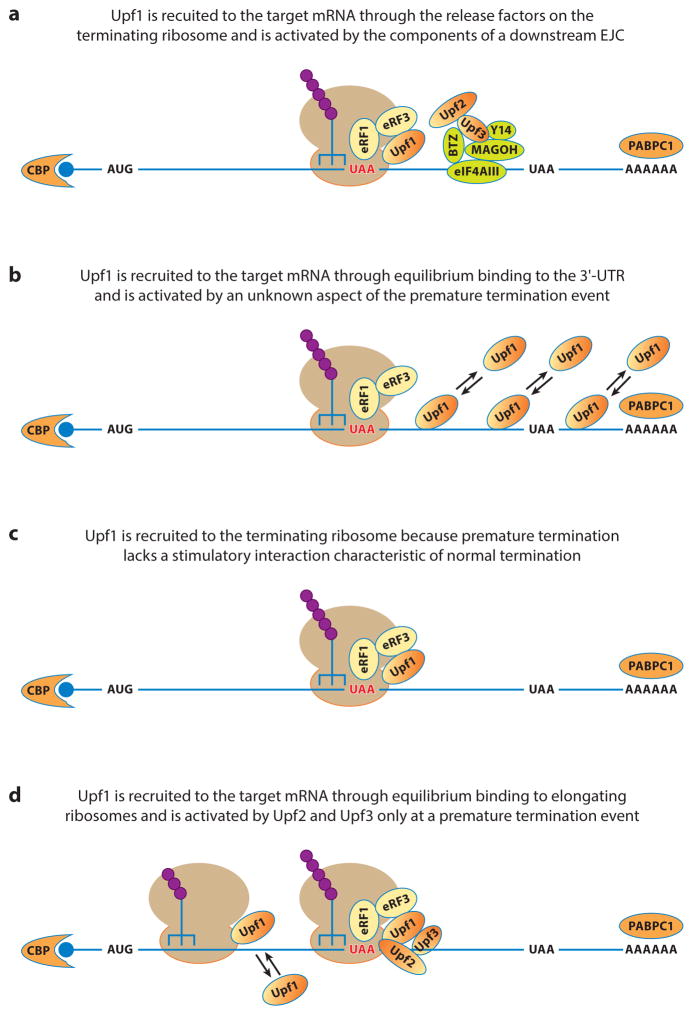
The EJC model is principally applicable to the mechanism of NMD in mammalian cells, yet it follows from an initial observation in yeast. The latter indicated that degradation of PTC-containing PGK1 mRNA requires both a downstream cis-acting element and a trans-acting factor (Hrp1) that binds to this element (71). Analogous principles in mammalian cells followed from several important observations pertaining to the role of pre-mRNA splicing in NMD, including those showing that (a) the vast majority of mammalian genes contain introns, and nuclear pre-mRNA splicing deposits EJCs on spliced mRNAs (128); (b) the normal stop codons for the vast majority of mammalian genes are located in the last exon, and mammalian NMD is often enhanced by a splicing event downstream of the PTC (159, 239); and (c) EJCs travel with spliced mRNAs to the cytoplasm and associate with the NMD factor Upf3 (112, 142). These and other observations led to a model proposing that during the initial or pioneer round of translation, elongating ribosomes displace EJCs from an mRNA unless the mRNA contains a PTC, in which case EJCs downstream of the PTC remain mRNA-associated. A PTC would then promote NMD because Upf1 is thought to be recruited to the prematurely terminating ribosome by the release factors eRF1 and eRF3, and then activated by Upf2 and Upf3 bound to the downstream EJC (Figure 2a). Activated Upf1, either by itself or through additional effectors, is then thought to recruit deadenylation, decapping, or endonucleolytic enzymes to the mRNA to promote transcript degradation (93, 94, 194). However, a role for a retained EJC in Upf1 activation fails to explain NMD of mRNAs derived from intronless precursors (182), spliced mRNAs without EJC components (225) or proper EJC spacing (217, 239), or viral unspliced mRNAs (124, 181). In addition, the EJC model is also challenged by recent experimental evidence showing that in human cells, NMD targets not only cap-binding protein (CBP)-bound mRNAs during the pioneer round of translation but also eIF4E-bound mRNAs during subsequent rounds of translation (50, 186).
Figure 2.
Possible modes of Upf1 recruitment and activation during nonsense-mediated mRNA decay (NMD). Four different modes of Upf1 recruitment and activation during NMD are described. (a) Upf1 recruitment and activation based on the EJC (exon junction complex) model. (b) Upf1 recruitment and activation based on the Upf1 3′-UTR (untranslated region) sensing and potentiation model. (c) Upf1 recruitment and activation based on the faux 3′-UTR model. (d) Upf1 recruitment and activation based on stochastic binding of Upf1 and activation by Upf2 and Upf3 binding only at a premature termination codon. Abbreviations: CBP, cap-binding protein; PABPC1, poly(A)-binding protein.
Upf1 3′-UTR Sensing and Potentiation Model
PTCs usually create long 3′-UTRs, and some wild-type mRNAs with long 3′-UTRs also trigger NMD. Recognizing the NMD-promoting role of such UTRs, Hogg & Goff (86) used an RNA-based affinity tag to isolate and then characterize the proteins associated with several reporter mRNAs with long 3′-UTRs. They observed that (a) Upf1 preferentially associates with transcripts containing NMD-inducing 3′-UTRs; (b) the site of Upf1 association with such transcripts was the 3′-UTR, and this binding was dependent on the 3′-UTR length but not the sequence; (c) Upf1’s association with NMD-inducing long 3′-UTR transcripts was independent of translation but disrupted by frequent ribosome read-through events; and (d) infrequent ribosome read-through events preserved Upf1 binding to the mRNA but prevented the activation of NMD. In the Upf1 3′-UTR sensing and potentiation model that is based on these observations, NMD is thought to be activated through a two-step mechanism (Figure 2b). In the first step, Upf1 is proposed to bind to 3′-UTRs and sense their lengths. Upf1 binding to long 3′-UTRs is then hypothesized to prime transcripts for decay, with another yet unidentified mRNP structure or termination event bearing responsibility for triggering Upf1 activation and promoting decay. This model and the general notion of Upf1 association with 3′-UTRs are supported by several genome-wide Upf1 CLIP (cross-linking and immunoprecipitation)-seq experiments (74, 91, 118, 242). However, length-dependent but sequence-independent Upf1 binding is difficult to reconcile with preferential binding of Upf1 to NMD-inducing, but not to non-NMD-inducing, long 3′-UTRs (86). Further, the translation-independence of Upf1 binding to NMD-inducing 3′-UTRs conflicts with earlier observations that, at steady state, the majority of yeast and human Upf1 is associated with polyribosomes (11, 171). The key experimental evidence for length-dependent Upf1 binding to NMD-inducing 3′-UTRs was based on quantitative coimmunoprecipitation of 3′-UTR fragments by Upf1 antibodies after RNase H-mediated cleavage (86). However, given the extensive molecular interactions thought to occur between mRNA 5′ and 3′ ends during translation (5, 207), a single cleavage site in an mRNP may not generate two independent mRNP domains, i.e., it is possible that coprecipitation of 3′-UTR fragments is mediated through the 5′-mRNP domain and its translating ribosomes. Consistent with this interpretation, both general translation factors and ribosomal proteins were present in the purified mRNP fraction (86). In addition, the DNA probe used to assess the efficiency of Upf1 association with different 3′-UTR fragments was capable of more extensive hybridization to longer 3′ cleavage products than to shorter ones (86), thus complicating quantitative interpretations of the relationship between UTR length and Upf1 binding.
Faux–Untranslated Region Model
The faux-UTR model was originally proposed to explain NMD in yeast cells (4), but its principal tenets were subsequently supported by results in other systems, including fly, plant, and human cells (17, 108, 204). This model integrated several important observations pertinent to both NMD and translation termination, namely that (a) PTCs that trigger NMD usually create long 3′-UTRs; (b) artificial extension of an mRNA’s 3′-UTR can trigger NMD (158); and (c) compared with termination at NTCs, termination at PTCs is intrinsically inefficient (4). The faux-UTR model posits that efficient translation termination depends on interactions between the release factors and proteins bound to a normal 3′-UTR. Accordingly, the mRNP structure associated with a 3′-UTR created by a PTC is thought to lack at least one critical factor that normally enhances termination (4, 6, 109). Upf1 involvement follows from the hypothesis that its association with a prematurely terminating ribosome could be favored in the faux 3′-UTR mRNP context, owing either to the loss of effective interaction between one of the release factors and a protein normally associated with the 3′-UTR, or to at least one kinetically delayed step during termination (109) (Figure 2c). In support of this model, artificial tethering of poly(A)-binding protein Pab1 (or PABPC1) downstream of a PTC, or shortening the distance between a PTC and the poly(A) tail by restructuring the 3′-UTR, can antagonize NMD (4, 17, 52, 95, 110, 202). In contrast to the model, some mRNAs that lack poly(A) are still subject to NMD regulation (197) and some mRNAs with long 3′-UTRs can evade NMD (124, 181, 204). In at least one instance, an mRNA in the latter class appears to contain a specific cis-acting element downstream of the termination codon that counteracts NMD activity (220, 230). If the faux-UTR model is correct, such elements may inhibit NMD by promoting efficient translation termination.
Upf1 Recruitment to Targeted mRNAs
Upf1 is the central regulator of NMD from yeasts to humans, and an understanding of the timing and means by which it is recruited to its target mRNAs is critically important for deciphering the molecular mechanisms of NMD. As might be expected, this is a contentious issue (118, 243). In the EJC and faux-UTR models, Upf1 is thought to be selectively recruited to NMD-targeted mRNAs during translation, most likely through interactions with the release factors eRF1 and eRF3 located on prematurely terminating ribosomes. This mode of Upf1 targeting is supported by substantial experimental evidence, including observations that (a) NMD requires mRNA translation (109); (b) the bulk of yeast and human Upf1 is associated with polyribosomes at steady state (11, 171) and yeast Upf1 copurifies with ribosomal 40S subunits (153); (c) Upf1 interacts with eRF1 and eRF3 (43, 95, 104, 204); and (d) Upf1 preferentially associates with NMD-targeted mRNAs (45, 98, 100, 118, 119). Nevertheless, translation-dependent Upf1 targeting of NMD substrate mRNAs has been challenged by the results of mRNP purification (86) and Upf1 CLIP-seq experiments (74, 91, 242), both of which imply that Upf1 associates with mRNA 3′-UTRs independently of mRNA translation. Although the mRNP purification experiments indicated selective Upf1 binding to NMD-inducing long 3′-UTRs, the CLIP experiments suggested that Upf1 targets the 3′-UTRs of both NMD-sensitive and NMD-insensitive transcripts (74, 91, 242). In addition, because inhibiting translation with cycloheximide or puromycin enhanced Upf1 binding to mRNA coding regions in the CLIP experiments, it has also been suggested that Upf1 may target most mRNAs prior to their translation, only to be displaced from mRNA coding regions by elongating ribosomes (242, 243). Complicating the matter further, an indiscriminate, translation-independent mode of Upf1 targeting is inconsistent with results obtained in Upf1 RNA immunoprecipitation (RIP) experiments in multiple experimental systems (98, 100, 118, 119, 202). All of these RIP experiments demonstrated selective Upf1 binding to NMD-targeted mRNAs, in some cases showing that Upf1 binding to the target mRNAs is dependent on translation (118, 119).
These discrepancies are substantial and complicate the likelihood of finding a common model, thus warranting a careful examination for possible technical differences that may explain the inconsistencies. Upf1 CLIP-seq and Upf1 RIP experiments differ significantly in the method of final RNP sample preparation and the CLIP approach appears to underestimate Upf1 association with polyribosomes. The CLIP experiments involve size selection of RNP complexes by gel electrophoresis and thus are biased against large RNP complexes that would include ribosomes. The RIP experiments [followed by either microarray analysis (98) or deep sequencing (118)] largely overcame this limitation and appear to have captured ribosome-associated mRNAs. Concerns about the recovery of polysome-associated mRNAs extend as well to data analysis. Current analysis pipelines for deep sequencing data include a potential bias against ribosome-associated proteins that may have cross-linked to rRNA. In most data analyses, an early step is the elimination of rRNA reads. For example, more than 75% of the sequencing reads from one CLIP-seq experiment were discarded (242). The RIP-seq and mRNP purification experiments overcome this limitation, with both protocols likely to encompass ribosomal complexes. In fact, the purified mRNP fraction contains ribosomal proteins and translation factors (86). The indiscriminate Upf1 binding to 3′-UTRs seen in CLIP experiments may thus reflect a direct interaction with mRNA that is independent of NMD. It is worth noting that the CLIP-seq experiments also reveal Upf1 association with some noncoding RNAs (MALAT1 and GAS5). Interestingly, these RNAs appear to contain cis-acting, translation-enhancing elements (229), are associated with translating ribosomes (92), and are subject to NMD (143). In contrast to an earlier proposition (242), these noncoding RNAs are also likely targeted by Upf1 through a translation-dependent mechanism. Collectively, these considerations lead us to speculate that Upf1 targets PTC-containing mRNAs through terminating ribosomes. One likely scenario is that Upf1 associates transiently with elongating ribosomes of all translating mRNAs but is activated only by the unusual termination events occurring at PTCs, such as the delayed recruitment and action of eRF1 and eRF3, or lack of ABCE1 activity (Figure 2d).
Roles of Upf1’s ATPase/Helicase Activities in Nonsense-Mediated Decay
Yeast and human Upf1 manifest RNA-dependent ATPase activity in vitro (20, 29, 30, 44). This ATPase activity is essential for NMD, as mutations that eliminate Upf1’s RNA binding, ATP-binding, or ATP hydrolysis functions all abolish NMD in vivo (104, 226–228). However, the function, site of action, and mechanism of activation of the Upf1 ATPase activity remain unclear. Genetic analyses in yeast suggest that the Upf1 ATPase activity likely promotes some steps during premature translation termination, including peptide release and ribosomal subunit dissociation or recycling (109). This conclusion is supported by several lines of evidence. First, deletion of UPF1 causes high levels of translation read-through at PTCs (145, 219, 227), only some of which is attributable to indirect effects (79, 99). Second, although upf1 mutants deficient in ATP-binding or ATP hydrolysis fail to promote NMD, these mutants prevent nonsense codon read-through as efficiently as the wild-type UPF1 allele (79, 226). Third, deletion of UPF1 also results in defects in translation reinitiation in vivo and ribosome recycling in vitro (65). Recent functional analyses of ATPase-deficient Upf1 mutants in human cells suggest that Upf1 is also required for the final step of NMD, i.e., Xrn1-mediated exonucleolytic decay, and that the ATPase activity promotes mRNP disassembly, including the dissociation of NMD factors (56). Several important observations support this conclusion; for example, (a) ATPase-deficient hUpf1 causes the accumulation of 3′ decay intermediates of NMD-targeted mRNAs in vivo; (b) ATPase-deficient hUpf1 and additional NMD factors accumulate on 3′ decay intermediates; and (c) the 3′ decay intermediates that accumulate in vivo are resistant to Xrn1 digestion. Although the timing and function of the yeast and human Upf1 ATPase activities in NMD appear to differ, these differences can be accommodated by a unified model and a single site of Upf1 action. In this model, Upf1 functions at prematurely terminating ribosomes, and its ATPase activity promotes conformational and compositional transitions in the terminating mRNPs that are essential for both the initiation and completion of NMD. ATPase-deficient Upf1 is unable to promote these transitions, causing the terminating mRNPs to stall at an intermediate step. ATPase-deficient Upf1 associated with the stalled mRNP could still recruit decay factors to initiate NMD but is unable to promote key transitions in the terminating mRNPs, thus causing the accumulation of 3′ decay intermediates. Alternative models for the function of Upf1 ATPase activity propose that Upf1 functions mainly as an RNA helicase after translation termination, using its ATPase activity to scan the RNP components in mRNA 3′-UTRs (149, 199). The experimental evidence for these models is still very limited.
Roles of Upf1 Phosphorylation Cycle in Nonsense-Mediated Decay
In metazoans, Upf1 function in NMD is also controlled by a cycle of phosphorylation and dephosphorylation (168, 170). Upf1 phosphorylation occurs after its targeting to PTC-containing mRNAs and while it is associated with prematurely terminating ribosomes. The modification is carried out by the Smg1 kinase and also requires the functions of Upf2, Upf3, and some EJC components (46, 76, 104, 236). Upf1 dephosphorylation most likely occurs after Upf1 dissociation from targeted mRNAs, and this step is dependent on the functions of Smg5, Smg6, and Smg7, and also requires the activity of the protein phosphatase PP2A (7, 168–170). Upf1 phosphorylation creates a set of binding sites for its downstream effector molecules. The new molecular interactions created by Upf1 phosphorylation probably coordinate mRNP remodeling events during NMD and may also control the mechanisms of degradation for different PTC-containing mRNAs. For example, phosphorylation of threonine 28 at the N-terminal extension of hUpf1 enhances Upf1 interaction with the endonuclease Smg6 (169). In contrast, phosphorylation of serine 1096 and 1116 at the C-terminal extension of hUpf1 promotes Upf1 binding of the Smg5-Smg7 heterodimer (28, 102, 169). In addition, hyperphosphorylated Upf1 also exhibits preferential binding to the decapping factors Dcp1 and human proline-rich nuclear receptor PNRC2 (36). Yeast Upf1 also contains several phosphorylated residues. However, there is no evidence that links these Upf1 phosphorylation events to functions in NMD (123, 218), suggesting that the function of Upf1 phosphorylation in NMD evolved in metazoans to deal with the increased complexity in mRNP and ribosomal structures, or the translation termination process.
Mechanisms of Nonsense-Mediated Decay–Targeted mRNA Degradation
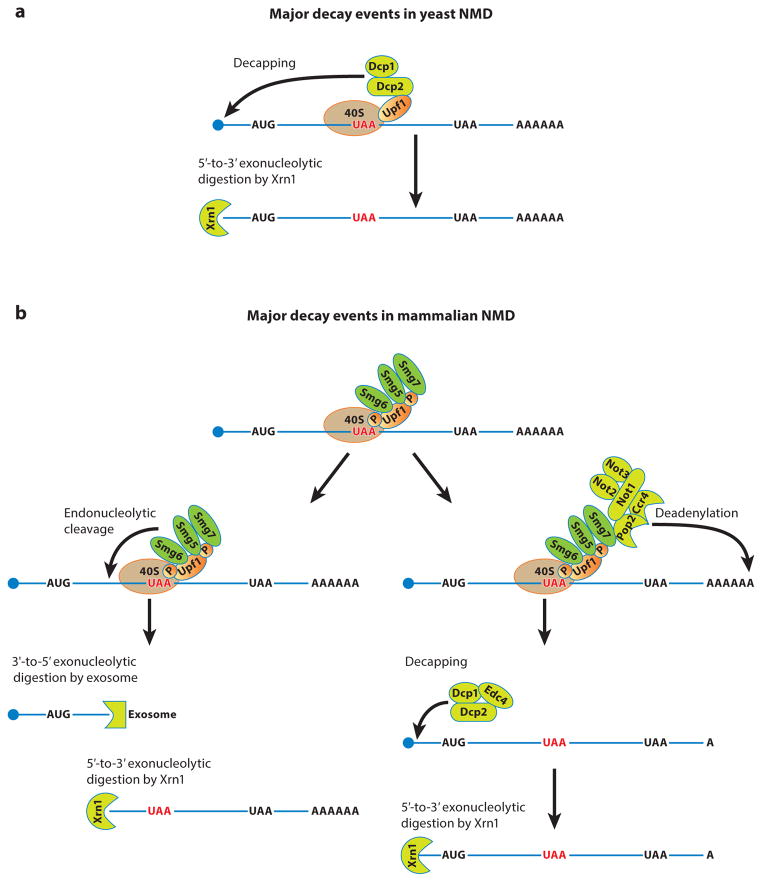
In yeast or human cells, the actual degradation of NMD-targeted mRNAs occurs through multiple mechanisms. In yeast cells, NMD-targeted mRNAs are degraded predominantly through a deadenylation-independent decapping mechanism involving decapping by the Dcp1/Dcp2 decapping enzyme and 5′-to-3′ exonucleolytic digestion by Xrn1 (81, 157) (Figure 3a). In this 5′-to-3′ decay pathway, the decapping enzyme is recruited to the targeted mRNAs by direct Upf1:Dcp2 interaction (80), possibly when the mRNA is still associated with the terminating ribosome (89). NMD-targeted mRNAs can also be degraded by a 3′-to-5′ decay mechanism involving deadenylation and by 3′-to-5′ digestion by the cytoplasmic exosome. This pathway appears to be preferred in cells lacking decapping activity (26, 154, 209). In human cells, NMD-targeted mRNAs are degraded by endonucleolytic cleavage (51, 90), deadenylation-dependent decapping (130, 136, 139, 216, 234), and exosome-mediated 3′-to-5′ decay (130) (Figure 3b). Initiation of both the endonucleolytic decay and deadenylation-dependent decapping is mediated by Upf1. In the endonucleolytic decay pathway, Upf1 recruits the endonuclease Smg6 to targeted mRNAs through both phosphorylation-dependent and phosphorylation-independent interactions (28, 164, 169). Smg6 cleaves its substrate mRNAs in the vicinity of PTCs and the resulting 5′ and 3′ fragments are degraded by the exosome and Xrn1, respectively (22, 51, 90). In the deadenylation-dependent decay pathway, phosphorylated Upf1 recruits the Smg5-Smg7 heterodimer to the targeted mRNAs. Subsequently, Smg7, via an interaction between its C-terminal domain and Pop2, recruits the Ccr4-Not deadenylase. After deadenylation, mRNAs are decapped by the Dcp2 decapping enzyme and finally degraded 5′ to 3′ by Xrn1 (136, 216). Human Upf1, through its N-and C-terminal extensions, also interacts directly with Dcp2, and this Upf1:Dcp2 interaction may promote both deadenylation-dependent and deadenylation-independent decapping of targeted mRNAs (136, 216). The question of how the cytoplasmic exosome is recruited to NMD-targeted mRNAs remains unanswered (130). Recent genome-wide 5′ end-seq [or PARE (parallel analysis of RNA ends)] analyses reveal that Smg6-mediated endonucleolytic decay targets thousands of transcripts in the human transcriptome and is the major decay-initiating event in mammalian NMD (143, 192). Deadenylation-dependent or independent decapping serves mostly as a backup for the Smg6 endonucleolytic decay pathway but also preferentially targets a specific set of mRNAs (143, 192). Further, although Smg6 usually cleaves targeted mRNAs at several locations in the vicinity of PTCs, the actual cleavage sites exhibit significant sequence specificity (192).
Figure 3.
Major decay events in yeast and mammalian nonsense-mediated mRNA decay (NMD). (a) Major decay events in yeast NMD. At the end of the premature termination process, Upf1 is still associated with the 40S subunit, which remains attached to the mRNA. Upf1 on the 40S subunit then interacts with the Dcp1/Dcp2 decapping enzyme to trigger decapping of the mRNA. After decapping, the mRNA is digested by the Xrn1 5′-to-3′ exonuclease. (b) Major decay events in mammalian NMD. At the end of the premature termination process, phosphorylated Upf1 is still associated with the 40S subunit, which remains attached to the mRNA. Phosphorylated Upf1 on the 40S subunit then interacts with the Smg6 endonuclease and the effector Smg5-Smg7 heterodimer to trigger either endonucleolytic cleavage in the vicinity of the premature termination codon or deadenylation from the 3′ end of the mRNA. In the endonucleolytic cleavage pathway, the 5′ cleavage product is digested by the cytoplasmic exosome and the 3′ cleavage product is digested by Xrn1. In the deadenylation pathway, the Ccr4-Not deadenylase is recruited to the mRNA through an interaction between Smg7 and Pop2. After deadenylation, the mRNA is decapped by the Dcp1/Dcp2/Edc4 complex and then digested by Xrn1.
PNCR2 was isolated based on its interaction with human Upf1 in a yeast–two hybrid screen; it also interacts with Dcp1 (36). On the basis of these observations and other results, it was suggested that PNCR2 plays an essential role in mammalian NMD (34, 36), but that notion has recently been challenged (136). PNCR2 is a small proline-rich polypeptide that interacts strongly with Dcp1 (121) but is, at best, a very weak interactor with Upf1 (136). Tests for direct interaction between PNCR2 and Upf1 have been attempted but never demonstrated, and the proposed Dcp1 and Upf1 binding sites on PNCR2 are close to each other (<20 amino acids apart) (121). Thus, it is possible that the originally observed two-hybrid interaction between hUpf1 and PNCR2 may be indirect and bridged through the yeast decapping enzyme, i.e., PNCR2 may have a more general function in mRNA decapping but probably does not function as a NMD factor. Consistent with this interpretation, PNCR2 is also involved in Staufen1-mediated mRNA decay (35).
UPF1 FUNCTIONS OUTSIDE OF NONSENSE-MEDIATED DECAY
In addition to its central role in NMD, Upf1 is also involved in several seemingly unrelated cellular processes. Upf1 is recruited by mammalian Staufen1 to specific mRNA 3′-UTRs to elicit mRNA decay (113), and it is also recruited by the stem-loop binding protein (SLBP) to replication-dependent histone mRNA 3′-UTRs to promote degradation of those mRNAs (105). Upf1 is a component of HIV-1 RNPs and depletion of Upf1 attenuates HIV-1 genomic RNA levels and protein expression (1). Upf1 is also a component of L1-RNPs generated from human active LINE-1 retrotransposable elements that limits L1 mRNA expression (213). All these Upf1 functions appear to be independent of the other NMD factors. Because human Upf1 also forms complexes with the Dcp2 decapping enzyme (54, 139), it is possible that these Upf1 activities are mediated by the presence of Upf1 in decapping complexes, i.e., they are independent of Upf1’s functions at terminating ribosomes during NMD.
A NEW MODEL FOR NONSENSE-MEDIATED DECAY
NMD is triggered by a PTC in the A site of the ribosome. From yeast to humans, NMD requires a set of conserved Upf factors, Upf1, Upf2, and Upf3, indicating that the fundamental mechanism of NMD has been conserved during eukaryotic evolution. Although the eventual outcome of NMD is the accelerated degradation of a targeted mRNA, much of the NMD mechanism is intimately entangled with the process of premature translation termination. This conclusion follows most easily from several observations in yeast. First, deletion of the genes encoding Upf1, Upf2, or Upf3 promotes translation read-through at PTCs (145, 219). Although complicated to some extent by being partially attributable to an indirect effect on intracellular Mg2+ levels (99), this observation indicates that Upf1, Upf2, and Upf3 may act at an early stage of the premature termination process, possibly having a role in enhancing the recruitment of the release factors to the terminating ribosome or stimulating release factor activities. Second, deletion of the genes encoding Upf1, Upf2, or Upf3 also causes a deficiency in translation reinitiation in vivo, a defect in ribosome recycling, and loss of aberrant toe prints associated with premature termination in vitro (4, 65). These observations indicate that Upf1, Upf2, and Upf3 also act at a late stage of the premature termination process, possibly in the dissociation and recycling of ribosomal subunits. Finally, the functions of the Upf factors in preventing translation read-through and promoting mRNA decay appear to be distinct. The former function can bypass Upf2 and Upf3, and is independent of Upf1’s ATPase activity (79, 145). In contrast, the latter function requires Upf2 and Upf3, and is dependent on Upf1’s ATPase activity (79, 226). Based largely on these observations, we propose a new model for NMD (Figure 1b). In this model, Upf1 normally associates transiently with elongating ribosomes on all translating mRNAs via 40S subunit interaction (153). Upf1 association with a prematurely terminating ribosome is thought to be stabilized if Upf2 and Upf3 also associate, with the latter event occurring because of the delayed recruitment of the release factors (eRF1 and eRF3) or an atypical conformation for a terminating ribosome. Ribosome-associated Upf1, Upf2, and Upf3 would then control several aspects of the premature termination process. At the early stage, these factors could either recruit the release factors to the terminating ribosome or stimulate release factor activities to promote peptide release. After peptide release, the Upfs would promote the dissociation and recycling of the terminating ribosome by activating the Upf1’s ATPase activity. After ATP hydrolysis by Upf1, the 60S subunit and the deacylated tRNA are thought to be released from the terminating mRNP, but Upf1 continues to be associated with the 40S subunit still bound to the mRNA. Upf1 on the 40S ribosomal subunit would then, directly or via effector molecules, recruit mRNA decay activities to the targeted mRNA to promote mRNA degradation (Figure 3).
OPEN QUESTIONS
Work over the past two-and-a-half decades has provided substantial insights into the multifaceted aspects of NMD. The core NMD factors and their major regulators or effectors were identified and characterized in several model organisms, substrates of the pathway were delineated, and decay-initiating events were illuminated in diverse cell types. In addition, we also learned a considerable amount about Upf1’s biochemical activities, structure, and regulation. To move to the next step and gain a better mechanistic understanding of NMD, several key questions remain to be addressed. First, NMD is linked to translation termination, yet the termination events that discriminate premature termination from normal termination are still unknown. Second, NMD requires the functions of the core Upf factors, yet the timing and the location of the association of these factors with terminating ribosomes remain obscure. Third, Upf1’s ATPase/helicase activity is essential for NMD and, in metazoans, Upf1’s function in NMD is also controlled by a cycle of phosphorylation and dephosphorylation. However, the target of Upf1’s ATPase/helicase activity and the relationship between this Upf1 activity and its phosphorylation status are also not clear. Finally, we have little understanding of the ways that NMD activity appears to be dictated by the length of translated regions (45) or is dependent on the specific positions of PTCs (27, 174, 217). Answers to all of these questions are required and our understanding of this process would be enhanced by structures of the Upfs in association with each other and their interactors (including the ribosome), as well as development of in vitro systems capable of carrying out at least some of the multiple steps in NMD.
Acknowledgments
Research in the authors’ laboratory was supported by National Institutes of Health grant R37GM27757 to A.J.
Footnotes
DISCLOSURE STATEMENT
A.J. is cofounder, scientific advisory board chair, and a director and shareholder of PTC Therapeutics, Inc., a company that markets and develops drugs that promote therapeutic nonsense suppression. F.H. is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Feng He, Email: feng.he@umassmed.edu.
Allan Jacobson, Email: allan.jacobson@umassmed.edu.
LITERATURE CITED
- 1.Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, et al. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA. 2008;14:914–27. doi: 10.1261/rna.829208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–36. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 3.Altamura N, Groudinsky O, Dujardin G, Slonimski PP. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–87. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- 4.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–18. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 5.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–80. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–25. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 7.Anders KR, Grimson A, Anderson P. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–50. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Applequist SE, Selg M, Raman C, Jack HM. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–21. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravind L, Koonin EV. Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res. 2000;10:1172–84. doi: 10.1101/gr.10.8.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arribere JA, Gilbert WV. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013;23:977–87. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkin AL, Altamura N, Leeds P, Culbertson MR. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–25. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA. 2011;17:624–38. doi: 10.1261/rna.2404211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balistreri G, Horvath P, Schweingruber C, Zund D, McInerney G, et al. The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe. 2014;16:403–11. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. PNAS. 2011;108:3228–33. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashyam MD. Nonsense-mediated decay: linking a basic cellular process to human disease. Expert Rev Mol Diagn. 2009;9:299–303. doi: 10.1586/erm.09.18. [DOI] [PubMed] [Google Scholar]
- 16.Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–6. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–53. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 19.Belgrader P, Cheng J, Maquat LE. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. PNAS. 1993;90:482–86. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–35. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bidou L, Allamand V, Rousset JP, Namy O. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol Med. 2012;18:679–88. doi: 10.1016/j.molmed.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Boehm V, Haberman N, Ottens F, Ule J, Gehring NH. 3′ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep. 2014;9:555–68. doi: 10.1016/j.celrep.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Bono F, Gehring NH. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 2011;8:24–29. doi: 10.4161/rna.8.1.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–10. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, et al. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. PNAS. 2010;107:10050–55. doi: 10.1073/pnas.1000993107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao D, Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–45. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 28.Chakrabarti S, Bonneau F, Schussler S, Eppinger E, Conti E. Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res. 2014;42:9447–60. doi: 10.1093/nar/gku578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 31.Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–53. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 32.Chazal PE, Daguenet E, Wendling C, Ulryck N, Tomasetto C, et al. EJC core component MLN51 interacts with eIF3 and activates translation. PNAS. 2013;110:5903–8. doi: 10.1073/pnas.1218732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–18. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK. SMG5-PNRC2 is functionally dominant compared with SMG5-SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res. 2013;41:1319–28. doi: 10.1093/nar/gks1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho H, Kim KM, Han S, Choe J, Park SG, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 37.Clerici M, Deniaud A, Boehm V, Gehring NH, Schaffitzel C, Cusack S. Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res. 2014;42:2673–86. doi: 10.1093/nar/gkt1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, et al. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–65. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, et al. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI+] propagation. Mol Cell Biol. 2002;22:3301–15. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–36. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 42.Culbertson MR. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 43.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–77. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–23. [PMC free article] [PubMed] [Google Scholar]
- 45.Decourty L, Doyen A, Malabat C, Frachon E, Rispal D, et al. Long open reading frame transcripts escape nonsense-mediated mRNA decay in yeast. Cell Rep. 2014;6:593–98. doi: 10.1016/j.celrep.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 46.Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. Cloning of a novel phosphatidyl-inositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001;276:22709–14. doi: 10.1074/jbc.C100144200. [DOI] [PubMed] [Google Scholar]
- 47.de Pinto B, Lippolis R, Castaldo R, Altamura N. Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance. Mol Microbiol. 2004;51:1129–42. doi: 10.1046/j.1365-2958.2003.03889.x. [DOI] [PubMed] [Google Scholar]
- 48.Drake KM, Dunmore BJ, McNelly LN, Morrell NW, Aldred MA. Correction of nonsense BMPR2 and SMAD9 mutations by ataluren in pulmonary arterial hypertension. Am J Respir Cell Mol Biol. 2013;49:403–9. doi: 10.1165/rcmb.2013-0100OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand S, Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol. 2013;20:702–9. doi: 10.1038/nsmb.2575. [DOI] [PubMed] [Google Scholar]
- 51.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 52.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLOS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fatscher T, Boehm V, Weiche B, Gehring NH. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA. 2014;20:1579–92. doi: 10.1261/rna.044933.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–15. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Fourati Z, Roy B, Millan C, Coureux P-D, Kervestin S, et al. A highly conserved region essential for NMD in the Upf2 N-terminal domain. J Mol Biol. 2014;426:3689–702. doi: 10.1016/j.jmb.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–50. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 58.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–47. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 59.Gaba A, Jacobson A, Sachs MS. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell. 2005;20:449–60. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 60.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–70. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ge Y, Porse BT. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. BioEssays. 2014;36:236–43. doi: 10.1002/bies.201300156. [DOI] [PubMed] [Google Scholar]
- 63.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 64.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–49. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 65.Ghosh S, Ganesan R, Amrani N, Jacobson A. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA. 2010;16:1832–47. doi: 10.1261/rna.1987710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–91. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 67.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–25. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gloggnitzer J, Akimcheva S, Srinivasan A, Kusenda B, Riehs N, et al. Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe. 2014;16:376–90. doi: 10.1016/j.chom.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 69.Goldmann T, Overlack N, Wolfrum U, Nagel-Wolfrum K. PTC124-mediated translational readthrough of a nonsense mutation causing Usher syndrome type 1C. Hum Gene Ther. 2011;22:537–47. doi: 10.1089/hum.2010.067. [DOI] [PubMed] [Google Scholar]
- 70.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gonzalez CI, Ruiz-Echevarria MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–99. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- 72.Goodenough E, Robinson TM, Zook MB, Flanigan KM, Atkins JF, et al. Cryptic MHC class I–binding peptides are revealed by aminoglycoside-induced stop codon read-through into the 3′ UTR. PNAS. 2014;111:5670–75. doi: 10.1073/pnas.1402670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gozalbo D, Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr Genet. 1990;17:77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- 74.Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, et al. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell. 2014;54:573–85. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 75.Gregory-Evans CY, Wang X, Wasan KM, Zhao J, Metcalfe AL, Gregory-Evans K. Postnatal manipulation of Pax6 dosage reverses congenital tissue malformation defects. J Clin Investig. 2014;124:111–16. doi: 10.1172/JCI70462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grimson A, O’Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7483–90. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He F, Brown AH, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–70. [PMC free article] [PubMed] [Google Scholar]
- 78.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–94. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He F, Ganesan R, Jacobson A. Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast. Mol Cell Biol. 2013;33:4672–84. doi: 10.1128/MCB.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–54. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 81.He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–30. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He F, Jacobson A. Control of mRNA decapping by positive and negative regulatory elements in the C-terminal domain of Dcp2. RNA. 2015;21:1633–47. doi: 10.1261/rna.052449.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–52. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 84.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. PNAS. 1993;90:7034–38. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hilleren P, Parker R. mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA. 1999;5:711–19. doi: 10.1017/s1355838299990519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hogg JR, Goff SP. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 2010;143:379–89. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–8. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 88.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–80. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 89.Hu W, Petzold C, Coller J, Baker KE. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–47. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–17. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–50. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–17. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 94.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–56. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 95.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–47. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 97.Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–62. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- 98.Johansson MJ, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. PNAS. 2007;104:20872–77. doi: 10.1073/pnas.0709257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johansson MJ, Jacobson A. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 2010;24:1491–95. doi: 10.1101/gad.1930710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol Cell Biol. 2007;27:5630–38. doi: 10.1128/MCB.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22:4673–87. doi: 10.1093/hmg/ddt315. [DOI] [PubMed] [Google Scholar]
- 102.Jonas S, Weichenrieder O, Izaurralde E. An unusual arrangement of two 14-3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev. 2013;27:211–25. doi: 10.1101/gad.206672.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–37. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 104.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–67. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 106.Kebaara BW, Atkin AL. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2771–78. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–94. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, et al. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–57. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–12. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kervestin S, Li C, Buckingham R, Jacobson A. Testing the faux-UTR model for NMD: analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie. 2012;94:1560–71. doi: 10.1016/j.biochi.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–81. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 112.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–36. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 113.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 114.Koonin E. A new group of putative RNA helicases. Trends Biochem Sci. 1992;17:495–97. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- 115.Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 1998;63:474–88. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–22. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. PNAS. 2009;106:2453–58. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, et al. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28:1900–16. doi: 10.1101/gad.245506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kurosaki T, Maquat LE. Rules that govern UPF1 binding to mRNA 3′ UTRs. PNAS. 2013;110:3357–62. doi: 10.1073/pnas.1219908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kuschal C, DiGiovanna JJ, Khan SG, Gatti RA, Kraemer KH. Repair of UV photolesions in xeroderma pigmentosum group C cells induced by translational readthrough of premature termination codons. PNAS. 2013;110:19483–88. doi: 10.1073/pnas.1312088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lai T, Cho H, Liu Z, Bowler MW, Piao S, et al. Structural basis of the PNRC2-mediated link between mRNA surveillance and decapping. Structure. 2012;20:2025–37. doi: 10.1016/j.str.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 122.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–29. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 123.Lasalde C, Rivera AV, Leon AJ, Gonzalez-Feliciano JA, Estrella LA, et al. Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1. Nucleic Acids Res. 2014;42:1916–29. doi: 10.1093/nar/gkt1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.LeBlanc JJ, Beemon KL. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J Virol. 2004;78:5139–46. doi: 10.1128/JVI.78.10.5139-5146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee MH, Schedl T. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 2004;18:1047–59. doi: 10.1101/gad.1188404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–14. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 127.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–77. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–69. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–16. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 130.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–87. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 131.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–19. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lentini L, Melfi R, Di Leonardo A, Spinello A, Barone G, et al. Toward a rationale for the PTC124 (ataluren) promoted readthrough of premature stop codons: a computational approach and GFP-reporter cell-based assay. Mol Pharm. 2014;11:653–64. doi: 10.1021/mp400230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li M, Andersson-Lendahl M, Sejersen T, Arner A. Muscle dysfunction and structural defects of dystrophin-null sapje mutant zebrafish larvae are rescued by ataluren treatment. FASEB J. 2014;28:1593–99. doi: 10.1096/fj.13-240044. [DOI] [PubMed] [Google Scholar]
- 134.Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–41. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 135.Liu C, Karam R, Zhou Y, Su F, Ji Y, et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med. 2014;20:596–98. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Loh B, Jonas S, Izaurralde E. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013;27:2125–38. doi: 10.1101/gad.226951.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. PNAS. 1979;76:5134–37. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, et al. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep. 2014;6:748–64. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–21. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lykke-Andersen J, Bennett EJ. Protecting the proteome: eukaryotic cotranslational quality control pathways. J Cell Biol. 2014;204:467–76. doi: 10.1083/jcb.201311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–31. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 142.Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–39. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 143.Lykke-Andersen S, Chen Y, Ardal BR, Lilje B, Waage J, et al. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014;28:2498–517. doi: 10.1101/gad.246538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–13. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 145.Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–93. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 147.McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. PNAS. 2010;107:12186–91. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 149.Melero R, Buchwald G, Castano R, Raabe M, Gil D, et al. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol. 2012;19:498–505. S1–2. doi: 10.1038/nsmb.2287. [DOI] [PubMed] [Google Scholar]
- 150.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–78. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 151.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLOS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Miller JN, Pearce DA. Nonsense-mediated decay in genetic disease: friend or foe? Mutat Res. 2014;762:52–64. doi: 10.1016/j.mrrev.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Min EE, Roy B, Amrani N, He F, Jacobson A. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA. 2013;19:1105–15. doi: 10.1261/rna.039396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′ → 5′ degradation. Mol Cell. 2003;11:1405–13. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 155.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–39. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–347. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 157.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–81. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 158.Muhlrad D, Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–99. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 160.Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry. 2012;17:1103–15. doi: 10.1038/mp.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neurodevelopmental disorders. Hum Mol Genet. 2013;22:1816–125. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- 162.Ni J, Grate L, Donohue J, Preston C, Nobida N, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nicholson P, Joncourt R, Muhlemann O. Analysis of nonsense-mediated mRNA decay in mammalian cells. Curr Protoc Cell Biol. 2012 doi: 10.1002/0471143030.cb2704s55. [DOI] [PubMed] [Google Scholar]
- 164.Nicholson P, Josi C, Kurosawa H, Yamashita A, Muhlemann O. A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res. 2014;42:9217–35. doi: 10.1093/nar/gku645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–22. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nyiko T, Sonkoly B, Merai Z, Benkovics AH, Silhavy D. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol Biol. 2009;71:367–78. doi: 10.1007/s11103-009-9528-4. [DOI] [PubMed] [Google Scholar]
- 168.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 169.Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–66. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–51. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–57. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 173.Peixeiro I, Inacio A, Barbosa C, Silva AL, Liebhaber SA, Romao L. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 2012;40:1160–73. doi: 10.1093/nar/gkr820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–54. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 175.Peltz SW, Morsy M, Welch EM, Jacobson A. Ataluren as an agent for therapeutic nonsense suppression. Annu Rev Med. 2013;64:407–25. doi: 10.1146/annurev-med-120611-144851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Jr, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. PNAS. 1996;93:10928–32. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–99. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ponting CP. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem Sci. 2000;25:423–26. doi: 10.1016/s0968-0004(00)01628-5. [DOI] [PubMed] [Google Scholar]
- 180.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–97. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 181.Quek BL, Beemon K. Retroviral strategy to stabilize viral RNA. Curr Opin Microbiol. 2014;18:78–82. doi: 10.1016/j.mib.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rajavel KS, Neufeld EF. Nonsense-mediated decay of human HEXA mRNA. Mol Cell Biol. 2001;21:5512–19. doi: 10.1128/MCB.21.16.5512-5519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 2009;10:R101. doi: 10.1186/gb-2009-10-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rayson S, Arciga-Reyes L, Wootton L, De Torres Zabala M, Truman W, et al. A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLOS ONE. 2012;7:e31917. doi: 10.1371/journal.pone.0031917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–44. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rufener SC, Muhlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2013;20:710–17. doi: 10.1038/nsmb.2576. [DOI] [PubMed] [Google Scholar]
- 187.Ruiz-Echevarria MJ, Peltz SW. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–19. [PMC free article] [PubMed] [Google Scholar]
- 188.Ruiz-Echevarria MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–51. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 189.Sakaki K, Yoshina S, Shen X, Han J, DeSantis MR, et al. RNA surveillance is required for endoplasmic reticulum homeostasis. PNAS. 2012;109:8079–84. doi: 10.1073/pnas.1110589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24:7769–78. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–31. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 192.Schmidt SA, Foley PL, Jeong DH, Rymarquis LA, Doyle F, et al. Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res. 2014;43:309–23. doi: 10.1093/nar/gku1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–59. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense-mediated mRNA decay: mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–23. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 195.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–23. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Seyedali A, Berry MJ. Nonsense-mediated decay factors are involved in the regulation of selenoprotein mRNA levels during selenium deficiency. RNA. 2014;20:1248–56. doi: 10.1261/rna.043463.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shen PS, Park J, Qin Y, Li X, Parsawar K, et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347:75–78. doi: 10.1126/science.1259724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–51. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 199.Shigeoka T, Kato S, Kawaichi M, Ishida Y. Evidence that the Upf1-related molecular motor scans the 3′-UTR to ensure mRNA integrity. Nucleic Acids Res. 2012;40:6887–97. doi: 10.1093/nar/gks344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. PNAS. 2011;108:E1392–98. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14:563–76. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–64. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLOS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell. 2013;51:249–64. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, et al. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014;7:1858–66. doi: 10.1016/j.celrep.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–29. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 208.Song H, Mugnier P, Das AK, Webb HM, Evans DR, et al. The crystal structure of human eukaryotic release factor eRF1: mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–21. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 209.Takahashi S, Araki Y, Sakuno T, Katada T. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 2003;22:3951–59. doi: 10.1093/emboj/cdg374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, et al. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 2012;9:1370–79. doi: 10.4161/rna.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLOS ONE. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–33. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Taylor MS, Lacava J, Mita P, Molloy KR, Huang CR, et al. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell. 2013;155:1034–48. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–94. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–96. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 217.Wang J, Gudikote JP, Olivas OR, Wilkinson MF. Boundary-independent polar nonsense-mediated decay. EMBO Rep. 2002;3:274–79. doi: 10.1093/embo-reports/kvf036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, Gonzalez CI. A role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:3390–4000. doi: 10.1128/MCB.26.9.3390-3400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–90. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Weil JE, Beemon KL. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA. 2006;12:102–10. doi: 10.1261/rna.2129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–96. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, et al. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13:R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 224.Welch EM, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–45. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wen J, Brogna S. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J. 2010;29:1537–51. doi: 10.1038/emboj.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–90. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Weng Y, Czaplinski K, Peltz SW. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–14. [PMC free article] [PubMed] [Google Scholar]
- 229.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Withers JB, Beemon KL. Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon. Retrovirology. 2010;7:65. doi: 10.1186/1742-4690-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, et al. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29:3517–28. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xu X, Zhang L, Tong P, Xun G, Su W, et al. Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin Genet. 2013;83:560–64. doi: 10.1111/cge.12014. [DOI] [PubMed] [Google Scholar]
- 233.Yamashita A. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells. 2013;18:161–75. doi: 10.1111/gtc.12033. [DOI] [PubMed] [Google Scholar]
- 234.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–63. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 235.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–28. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–18. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zaborske JM, Zeitler B, Culbertson MR. Multiple transcripts from a 3′-UTR reporter vary in sensitivity to nonsense-mediated mRNA decay in Saccharomyces cerevisiae. PLOS ONE. 2013;8:e80981. doi: 10.1371/journal.pone.0080981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–15. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang S, Welch EM, Hogan K, Brown AH, Peltz SW, Jacobson A. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–44. [PMC free article] [PubMed] [Google Scholar]
- 241.Zhou Y, Jiang Q, Takahagi S, Shao C, Uitto J. Premature termination codon read-through in the ABCC6 gene: potential treatment for pseudoxanthoma elasticum. J Investig Dermatol. 2013;133:2672–77. doi: 10.1038/jid.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zünd D, Gruber AR, Zavolan M, Mühlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol. 2013;20:936–43. doi: 10.1038/nsmb.2635. [DOI] [PubMed] [Google Scholar]
- 243.Zünd D, Mühlemann O. Recent transcriptome-wide mapping of UPF1 binding sites reveals evidence for its recruitment to mRNA before translation. Translation. 2013;1:e26977. doi: 10.4161/trla.26977. [DOI] [PMC free article] [PubMed] [Google Scholar]