SUMMARY
Activation of NF-kB induces mesenchymal (MES) trans-differentiation and radio-resistance in glioma stem cells (GSCs), but molecular mechanisms for NF-kB activation in GSCs are currently unknown. Here we report that mixed lineage kinase 4 (MLK4) is overexpressed in MES but not proneural (PN) GSCs. Silencing MLK4 suppresses self-renewal, motility, tumorigenesis, and radio-resistance of MES GSCs via a loss of the MES signature. MLK4 binds and phosphorylates the NF-κB regulator IKKα, leading to activation of NF-κB signaling in GSCs. MLK4 expression is inversely correlated with patient prognosis in MES, but not PN high-grade gliomas. Collectively, our results uncover MLK4 as an upstream regulator of NF-kB signaling and a potential molecular target for the MES subtype of GBMs.
Keywords: glioblastoma, cancer stem cell, epithelial-to-mesenchymal transition, proneural-Mesenchymal transition
INTRODUCTION
Death in patients afflicted with cancer is mainly due to uncontrollable recurrence of the primary tumor following failure of current therapies. Glioblastoma (GBM) is one such cancer type wherein aggressive treatment strategies including surgery, ionizing radiation (IR) and chemotherapy provide only palliation (Chaichana et al., 2010; Goodwin et al., 2011; Kaper et al., 2006; Omuro and DeAngelis, 2013). Recent investigations have uncovered substantial phenotypic changes that occur in post-therapeutic cancers (Candolfi et al., 2012; Locatelli et al., 2013; Oh et al., 2012; Tafani et al., 2011).
Each GBM tumor is composed of heterogeneous tumor cell populations including those with stem cell properties, termed glioma initiating cells or glioma stem cells (GSCs)(Hemmati et al., 2003; Singh et al., 2003; Singh et al., 2004). Accumulating evidence suggests that stem cell properties in cancers contribute to therapeutic resistance and cancer initiation (Bao et al., 2006; Beier et al., 2011; Capper et al., 2009; Vescovi et al., 2006). Thus, development of therapeutics targeting GSCs may provide considerable benefit to cancer patients.
In various cancers, transition from epithelial to mesenchymal (MES) subtype is associated with advanced malignancy (Epithelial-MES Transition; EMT) (Nurwidya et al., 2012; Sanchez-Tillo et al., 2012; Shirkoohi, 2013). We and others have identified an EMT-like phenotypic shift that occurs in GBM in response to factors in the microenvironment or cytotoxic treatments that is now termed proneural (PN) to MES Transition (PMT) (Bhat et al., 2013; Halliday et al., 2014; Mao et al., 2013; Phillips et al., 2006). Of note, the MES identity is a hallmark of glioma aggressiveness and strongly associated with the poor outcome of patients (Bhat et al., 2013; Carro et al., 2010). Despite the central role of PMT in brain tumor recurrence, little is known about the molecular mechanisms that control this phenotypic shift in GSCs. Advances in genome-wide, genetic and genomic profiling have uncovered the aberrant signaling pathways that are essential for various phenotypes of cancers such as tumorigenesis, cellular proliferation, motility, and therapy resistance. Such oncogenic pathways include NF-κB and its downstream targets (Fan et al., 2005; Fan et al., 2010; Ferris and Grandis, 2007; Yamamoto et al., 2013). In particular, we have recently demonstrated that NF-κB is a master regulator that causes MES trans-differentiation with an associated radio-resistance in PN GSCs (Bhat et al., 2013).
Mixed lineage kinase 4 (MLK4) is a relatively poorly-characterized serine/threonine kinase. While the genetic mutation of MLK4 is frequent in microsatellite stable subtype of colorectal cancers (CRCs) that exhibit extremely poor prognosis compared to other subtypes, this genetic event is substantially rare in GBM (Martini et al., 2013). In CRCs, the role of MLK4 in intracellular signaling is associated with KRAS- and MAPK-mediated pathways (Marusiak et al., 2014). To date, it remains undetermined whether MLK4 plays a pathophysiological role in GBM and/or cancer stem cells. In this study, driven by the fact that MLK4 is a lead candidate from kinase screens, we seek to examine its role in gliomagenesis, MES trans-differentiation and radio-resistance in GSCs.
RESULTS
MLK4 is overexpressed in MES GSCs
To identify protein kinases that are associated with the MES signature, we compared the genome-wide expression levels of 349 kinase-encoding genes in the patient-derived PN and MES GSC-containing primary cultures grown as neurospheres (hereafter designated as glioma spheres) using 18 PN and 12 MES high-grade glioma (HGG) patient-derived tumor specimens. We found 6 genes (MLK4, LYN, MST4, VRK2, PRKCH, and MAPK9) that were significantly upregulated in MES glioma spheres by 4-fold or more compared to PN spheres and/or somatic cell types including normal neural progenitors, and astrocytes (Figure 1A). Upon silencing with short hairpin RNA (shRNA) technique, MLK4 was the only gene that was required for survival of MES, but not PN, glioma spheres as judged by increased number of cells in the subG1 phase of the cell cycle (Figure 1B and 1C). MLK4 was also the most highly expressed kinase in the MES glioma spheres when compared to normal neural progenitors or astrocytes (Figure 1D; Figure S1A). By contrast, the other members of the MLK family did not show any noticeable difference in expression between PN and MES glioma spheres (Figure S1B). We validated the elevated MLK4 levels in MES glioma spheres by real-time PCR (Figure S1C), Western blotting (Figure 1E), and immunofluorescence in HGG patient-derived glioma spheres (Figure 1F; Figure S1D, Table S1 and S2). Moreover, co-expression of MLK4 with a MES marker CD44 was confirmed using immunofluorescence and FACS analysis in tumor spheres (Figure 1G and 1H; Figure S1E and S1F). Interestingly, MLK4 promoter was hypermethylated in PN tumors with IDH1 mutation suggesting that epigenetic silencing could be one mechanism by which MLK4 is expressed at lower levels in PN GBMs (Figure S1G). Finally, we observed that MLK4 expression is enriched in the stemness associated ALDEFLUOR-positive subpopulations in MES 83 glioma spheres (Figure 1I and 1J). We compared MLK4 expression between self-renewing undifferentiated MES glioma spheres and their sister cultures that underwent differentiation. At 2 days after differentiation induction, MLK4 mRNA levels rapidly declined with a sharp contrast with the marked induction of an osteocyte differentiation marker Runx2 (Figure 1K). Taken together, these data indicate that MLK4 is over-expressed in the MES subtype of GSCs.
Figure 1. MLK4 is highly expressed in MES GSCs.
(A) Heatmap showing differentially expressed kinase-encoding genes in Proneural (PN), Mesenchymal (MES) glioma sphere cells, neural progenitors, and normal astrocytes.
(B) Average fold change of cell number in SubG1 phase upon target gene knockdown in MES 83 and PN 528 glioma spheres. Error bar represents standard deviation (SD) from 5 independent shRNAs for each gene. *p < 0.05. shNT, Non-Targeting control shRNA.
(C) Cell cycle analysis in glioma spheres with MLK4 knockdown (shMLK4; red) by PI staining using FACS. shNT as control (blue).
(D) Ranking of MLK4 among 349 kinase genes according to the fold difference of expression in MES glioma spheres relative to neural progenitors or normal astrocytes.
(E) Immunoblot (IB) analysis of MLK4 in glioma spheres. β-actin as internal control.
(F) Immunofluorescent (IF) staining for MLK4 expression in glioma spheres. MLK4 was labeled in green. Nuclei were counterstained with DAPI (blue). Scale bar represents 20 μm.
(G) IF staining of MLK4 and MES marker CD44 in glioma spheres. MLK4 was labeled in red and CD44 in green. Nuclei were counterstained with DAPI (blue). Scale bar represents 20 μm.
(H) FACS plots of MLK4 and CD44 in PN and MES glioma spheres.
(I) Quantitative RT-PCR (qRT-PCR) analysis of mRNA expression of MLK4 and ALDH1A3 in ALDEFLUOR-positive and ALDEFLUOR-negative subpopulations of MES 83 glioma spheres. Data are means ± SD (n = 3). ***p < 0.001.
(J) MLK4 protein expression in ALDEFLUOR-positive and ALDEFLUOR-negative subpopulations of MES 83 glioma spheres using immunoblotting. β-actin as internal control.
(K) qRT-PCR analysis of mRNA expression of MLK4 and an osteogenic differentiation marker Runx2 in MES 83 glioma spheres during osteogenic differentiation induction.
Data are means ± SD (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001. See also Figure S1, Table S1 and S2.
Silencing MLK4 induces apoptosis, reduces sphere formation and tumorigenesis in MES tumor spheres
Next, we investigated the physiological role of MLK4 in MES GSCs using two shRNA lentiviral vectors targeting independent regions of the MLK4 gene in two well-characterized glioma sphere samples (MES 83 and 267) (Bhat et al., 2013; Jeon et al., 2014; Mao et al., 2013). Western blotting showed more than 80% reduction of MLK4 by one of the MLK4 shRNA (shMLK4_2) in both MES 83 and 267 spheres (Figure S2A and S4C). In parallel, we generated MLK4 knockout (KO) cells by utilizing clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (CRISPR-Cas9) system (Figure S2B). Both shMLK4 silencing and MLK4 KO significantly attenuated the ability of in vitro clonal growth of MES 83 and 267 cells (Figure 2A; Figure S2C). Reduction of sphere growth could be only partially attributed to diminished proliferation, since silencing MLK4 caused weak, yet significant, reduction of the proportion of 5-ethynyl-2’-deoxyuridine (EdU)-incorporated cells (Figure 2B). To the contrary, shMLK4-infected MES spheres, but not PN 528 glioma spheres, showed a substantial accumulation of apoptotic cells as judged by Annexin V/ PI staining (Figure 2C; Figure S2D). In addition, there was no significant difference in the effects of shRNA targeting for the other 3 MLK genes on sphere formation between PN and MES glioma cells (Figure S2E), suggesting MLK4 is unique in that it is preferentially required for the self-renewal, and proliferation of MES glioma spheres in vitro while MLK1-3 may have a general role for self-renewal and proliferation of multiple cell types. Consequently, limiting dilution sphere formation assay exhibited that the proportion of self-renewing sphere-forming unit was largely reduced by both shMLK4 and MLK4 KO in these MES glioma cells (Figure 2D; Figure S2F).
Figure 2. Depletion of MLK4 attenuates a set of MES GSC phenotypes.
(A) Effects of MLK4 knockdown by shRNA (shMLK_1 or shMLK_2) and MLK4 knockout (KO) by CRISPR/Cas9 system on cell growth in MES 83 glioma spheres analyzed by AlamarBlue staining. Results were expressed as relative fluorescence units (RFU). Data are means ± SD (n = 6). ***p < 0.001.
(B) Representative images of EdU incorporation assays (left) and quantification of EdU positive cells (right) in MES 83 glioma spheres expressing shNT or shMLK4. Cells in green represent EdU positive cells. Nuclei were counterstained with DAPI (blue). Scale bar represents 20 μm. *p < 0.05.
(C) FACS plots of Annexin V and PI staining in MES 83 and PN 528 glioma spheres expressing shNT, shMLK4_1, or shMLK_2.
(D) Effects of or shMLK4_2 and MLK4 KO on sphere forming frequency of MES 83 glioma cells determined by limiting dilution assays. Stem cell frequency was calculated by ELDA analysis. Data are means ± SD (n = 18) ***p < 0.001. See also Figure S2.
To test the in vivo consequence of silencing MLK4 in MES 83 and 267 glioma spheres, we used the intracranial mouse models. MLK4 silencing significantly decreased in vivo tumor growth and/or extended median survival of mice without neurological deficit due to tumor burden compared to the control shNT-infected MES 83 glioma spheres (p = 0.0021 for shMLK4_1; p = 0.0003 for shMLK4_2, Log-rank p-value) and MES 267 glioma spheres (p = 0.0551 for shMLK4_1; p = 0.0013 for shMLK4_2, Log-rank p-value). Of note, the more effective shRNA clone (shMLK4_2 vs. shMLK4_1) had more impact on prolonged survival of mice (Figure 3A and 3B). These data indicate that MLK4 is required for both in vitro and in vivo growth of MES tumor spheres.
Figure 3. Silencing MLK4 in MES GSCs suppressed tumor growth and increased mouse survival.
(A) Representative hematoxylin and eosin (H&E) staining of mouse brains harvested on day 15 (MES 83) or day 58 (MES 267) after transplantation of MES glioma spheres expressing shNT, shMLK_1, or shMLK_2. Scale bars represent 2 mm. (B) Kaplan-Meier survival curves of mice intracranially transplanted with MES 83 or 267 glioma spheres that were infected with indicated shRNAs (n = 7). Tables show median survival of mice.
MLK4 is required for the MES phenotype in GSCs
Aberrant activation of the MES phenotype in GBM is linked to increased cellular motility, glycolysis, and genes associated with wound healing and inflammatory properties (Zhong et al., 2010). Therefore, we investigated whether MLK4 inhibition influences these traits in MES glioma spheres from the single-cell resolution measurement. We performed time-lapse monitoring of individual cells from dissociated shMLK4 lentivirus infected MES 83 spheres on micropatterned polydimethylsiolxane surfaces that mimic the fiber-like and/or conduit-like structures of in vivo parenchyma or stroma (Gallego-Perez et al., 2012). Travelling paths (tracks) of individual clones in each group displayed the clear difference of cellular motility in MES glioma sphere with shNT and shMLK4 infection (Figure 4A). Silencing MLK4 resulted in a significantly attenuated clonal motility in comparison to the control, determined by video time-lapse microscopy (Figure 4A; Movie S1 and S2).
Figure 4. Mesenchymal phenotypes in glioma spheres are dependent on MLK4.
(A) Effects of MLK4 knockdown (shMLK4_1 or shMLK4_2) on the motility of MES 83 glioma spheres. Representative track pattern images of individual MES 83 cells are shown (upper). Quantification shows the effect of shMLK_1 or shMLK4_2 on single-clone migration velocity (lower). Scale bar represents 200 μm. Data are means ± SD. ***p < 0.001.
(B) Effects of MLK4 knockdown (shMLK4_1 or shMLK4_2) on the glycolysis of MES 83 spheres were measured by recording the extracellular acidification rate (ECAR) in a Seahorse Bioanalyzer. Data are means ± SD. **p < 0.01.
(C) qRT- PCR analysis for gene expression of MES markers including MET, WT-1, BCL2A1, Vimentin, and Snail in MES 83 glioma spheres expressing shNT or shMLK4 and in PN 528 glioma spheres expressing control vector or kinase active mutant of MLK4 (R470C). Data are means ± SD (n = 3). **p < 0.01; ***p < 0.001.
(D) GSEA plot of TCGA MES gene signatures in shNT or shMLK4 of glioma spheres. Gene expression profile data are obtained by cDNA microarray using with MES glioma spheres expressing shNT or shMLK4. Gene sets for MES GBM defined by the TCGA MES signature were used for this analysis (Verhaak et al., 2010). The normalized enrichment scores (NES) is shown in the plot.
(E) Effects of MLK4 KO and MLK4 (R470C) overexpression in glioma spheres on CD44 expression by FACS analysis. The basal levels were determined by the results with the vector control.
(F) IF analysis of Vimentin expression in GBM xenografts derived from MES 83 and 267 glioma spheres with or without MLK4 knockdown. Vimentin was labeled in red. Nuclei were counterstained with DAPI (blue). Scale bar represents 50 μm.
We next investigated the glycolytic activity of shMLK4-infected MES glioma spheres. The extracellular acidification rate (ECAR) was measured as a glycolysis indicator. Diminished basal ECAR was observed in MES 83 spheres, when they were infected with either one of the 2 shMLK4 lentivirus but not shNT virus, and when mitochondrial respiration was blocked with Oligomycin, compensatory glycolysis was even further diminished in these shMLK4 infected MES 83 spheres (Figure 4B).
We then asked whether MLK4 is required and/or sufficient for the expression of MES markers in GSCs. qRT-PCR showed that MLK4 knockdown markedly decreased the expression of MES genes (MET, WT1, BCL2A1, Vimentin, and Snail), whereas kinase active MLK4 (R470C) overexpression in PN 528 spheres resulted in the opposite effect (Figure 4C; Martini et al., 2013). A similar reduction of MES genes was also observed in MLK4 KO cells compared to controls (Figure S3B). As expected, gene-set enrichment analysis (GSEA) demonstrated signature global reduction of MES signature (Figure 4D). Interestingly, depletion of MLK4 in these MES glioma spheres did not induce a shift toward a PN gene signature (Figure S3C). Consistent with these mRNA expression data, the MES cell surface antigen CD44 was decreased by MLK4 knockdown or KO in MES 83 and 267 spheres, and in turn, MLK4 overexpression increased CD44 protein expression in PN 528 spheres (Figure 4E; Figure S3D). The reduction of the MES markers was observed in xenograft tumors generated from shMLK4 lentivirus infected MES 83 and 267 spheres as evidenced by Vimentin immunoreactivity, accompanied by retained MLK4 elimination (Figure 4F; Figure S3E). Collectively, these results indicate that MLK4 could be a global regulator of the MES signature and traits associated with this subtype of tumors.
MLK4 regulates NF-κB signaling axis in MES GSCs via binding and phosphorylation of IKKa
To gain insight into the mechanisms by which MLK4 control the MES signature, we first examined downstream pathways that MLK4 could potentially regulate. It has been reported that four MLK genes (MLK1- 4) belong to the mitogen-activated-protein kinase (MAPK) family that activate c-Jun amino-terminal kinase (JNK) and p38 as downstream targets (Gallo and Johnson, 2002). However, we observed no noticeable difference in ERK, JNK, and phospho-p38 levels in glioma spheres of either PN or MES subtype (Figure S4A and S4B). Given the molecular link between NF-kB pathway activation and MES differentiation (Bhat et al., 2013), we examined if this pathway is altered in response to modulation of MLK4. Western blotting showed MLK4 silencing reduces phosphorylated form, but not the total form, of the upstream regulators of the NF-κB pathway, IKKα and IκB (Figure 5A). Luciferase assay using a NF-κB responsive element demonstrated that MLK4 knockdown considerably reduces the NF-κB transcriptional activity in MES 83 spheres (Figure 5B). Moreover, reduced NF-κB DNA binding activity was observed in the shMLK4 lysates compared to controls (Figure 5C). Immunoprecipitation and in vitro kinase analysis revealed that MLK4 physically interacts with both IKKα/β, but preferentially phosphorylates IKKα over IKKβ (Figure 5D–5E; Figure S4D). To test the function of IKKα on MLK4 mediated NF-κB reporter activity, we used siRNA targeting IKKα and kinase dead IKKα (S176A) to repress their expression levels. MLK4-mediated NF-κB luciferase activity was strongly suppressed by knockdown of IKKα wild-type, but not IKKα mutant (S176A) (Figure 5F; Figure S4E). Analysis of the microarray data with 30 glioma sphere samples exhibited that MLK4 expression is strongly correlated with the genes involved in the NF-κB pathway (Figure S4F and S4G). Furthermore, GSEA showed that genes affected by MLK4 silencing in MES glioma spheres were enriched for the NF-κB pathway signature (Figure 5G). These data implicate MLK4 as an upstream regulator of NF-kB signaling.
Figure 5. MLK4 binds and phosphorylates IKKa resulting in increased NF- B activity in MES glioma spheres.
(A) IB analysis of p-IKKα/β (S176/180), IKKα, and p-IκB (S32) expression in MES 83 glioma spheres expressing shNT or shMLK4.
(B) Effects of MLK4 knockdown on NF-κB response element driven Gaussia luciferase(NF-Gluc) activity in MES 83 glioma spheres. Data are means ± SD (n = 3). **p < 0.01.
(C) Gel shift assay (EMSA) with the nuclear extracts from MES 83 glioma spheres expressing shNT or shMLK4. Nuclear extracts were prepared and EMSA was performed with a radiolabeled oligonucleotide containing an NFκB-binding site.
(D) Immunoprecipitation (IP) analysis with IKKα antibody followed by immunoblot for MLK4 showing the IKKα-MLK4 protein complex in MES 83 glioma spheres. Whole cell lysates as input positive control and pull-down with IgG as negative control.
(E) In vitro kinase assay showing MLK4 phosphorylates IKKα, but not IKKβ in a dose dependent manner. After incubation with purified baculovirus-expressed IKKα or IKKβ and MLK4 recombinant proteins including ATP, immunoblot blot was performed with anti p-Serine antibody.
(F) Effects of IKKα siRNA knockdown in PN 528 glioma spheres on MLK4–mediated NF-Gluc activity. Data are means ± SD (n = 3). **p < 0.01; ##p < 0.01. (G) GSEA showing NF-κB gene signature is reduced in MLK4 depleted MES glioma spheres. The normalized enrichment scores (NES) is shown in the plot.
(H-I) IB analysis of p-IKKα/β (H) and NF-Gluc reporter activity (I) in vector or kinase-inactive MLK4 (K151A) in MES 83 glioma spheres. Data are means ± SD. *p < 0.05.
(J) Limiting dilution neurosphere forming assay in MES 83 glioma spheres with control vector or MLK4 (K151A) overexpression. Stem cell frequency was calculated by ELDA analysis. ***p < 0.001.
(K-L) Kaplan-Meier survival curves (K) and representative H&E staining images (L) of mice intracranially injected MES 83 glioma spheres with vector or MLK4 (K151A). Arrows indicate tumors. Scale bar represents 2 mm (upper) and 100 μm (lower).
(M) Representative IHC imaging for CD44 and Vimentin in vector- or MLK4 (K151A)-overexpressing MES 83 derived tumors. Scale bar represents 20 μm. See also Figure S4.
In order to evaluate whether the function of MLK4 in MES GSCs is dependent on its kinase activity, we carried out site directed mutagenesis to create MLK4 (K151A) mutant clone (Martini et al., 2013). Overexpression of MLK4 (K151A) in MES 83 spheres inhibited cell proliferation and self-renewal activity, accompanied by a reduction of p-IKKα/β expression and NF-κB reporter activity (Figure 5H–5J; Figure S4H). Furthermore, we found that IKKα exhibits strong association with MLK4-WT, whereas the association with MLK4 (K151A) was relatively weak, indicating MLK4-IKKα interaction is, at least partially, dependent on MLK4 phosphorylation (Figure S4I). We then investigated whether overexpression of this catalytically-inactive MLK4 affects in vivo tumorigenic potential using the orthotopic xenograft models (Figure 5L and 5K). MLK4 (K151A)-overexpression in MES 83 spheres significantly decreased in vivo tumor growth, thereby prolonged median survival of the tumor-bearing mice. Strikingly, central necrosis of tumors - one criteria for diagnosis of Grade IV GBM – was not observed in xenografted tumors derived from MES glioma spheres overexpressing K151A mutant form of MLK4, accompanied by reduced immunoreactivity to CD44 and Vimentin (Figure 5M). Together, these data suggest that MLK4 activates the NF-κB pathway via interaction and phosphorylation of IKKα in MES glioma spheres, and the catalytic activity of MLK4 is essential for maintenance of high grade glioma and the MES phenotype.
Following IR therapy, PN GSC-derived brain tumors develop sensitivity to MLK4 elimination
We previously reported that ionizing radiation (IR) to glioma spheres increases MES- marker expression with a concomitant decrease in PN markers, raising a possibility that IR induces PMT in GSCs in vitro (Mao et al., 2013). To determine whether MLK4 plays a role in this process, we first examined the change of MLK4 expression in IR-treated PN glioma spheres (PN 157 and 84) by qRT-PCR. MLK4 was dramatically upregulated 24 hr after IR treatment, with an accompanied increase in MES marker CD44 at later time-points, in these 2 PN glioma sphere samples (Figure 6A). In turn, when we combined MLK4 knockdown and IR treatment in MES 83 spheres, MLK4 silencing by shRNA almost completely abolished their IR-induced NF-κB activity (Figure 6B). Given that radioresistance of GBM tumors is linked to the MES subtype of GSCs (Bhat et al., 2013) and our data indicates the elevation of MLK4 by IR, we investigated the combined effect of MLK4 knockdown and IR treatment in mouse xenograft tumors using PN glioma spheres (GSC 23). Whole brain radiation was performed with four cycles of 2.5 Gy on consecutive days for these tumor-burden mice to mimic the clinical regimen of radiation therapy for GBM. As expected and consistent with our in vitro findings (Figure 2), silencing MLK4 alone did not noticeably affect the growth of these PN tumors, whereas IR prolonged survival of these mice (Figure 6C and 6D). In these tumors, expression levels of nuclear p-p65, CD44, and Vimentin were strongly induced by IR, which, in turn, were markedly suppressed by shMLK4 (Figure S5A and S5B). As expected, combination of MLK4 inhibition with IR treatment showed a further reduction in their growth kinetics, and significantly prolonged mouse survival (median survival of shNT vs. shNT+IR: 44 vs. 60.5 days and shMLK4 vs. shMLK4+IR: 44 vs. 70 days in GSC 23; Figure 6C and 6D). In the MES glioma sphere-derived mouse xenograft models, the combined IR treatment and shMLK4 also prolonged mouse survival (median survival of shMLK4_1 vs. shMLK4_1+IR in MES 83: 27 vs. 37 days; Figure S5C).
Figure 6. The effects of MLK4 silencing together with irradiation in PN GSC-derived xenograft tumors.
(A) qRT-PCR analysis for MLK4 and CD44 expression in response to 5 Gy irradiation in PN 157 and PN 84 glioma spheres in vitro. **p < 0.01.
(B) NF-κB reporter activities in MES 83 glioma spheres with lentiviral infection of shNT or shMLK4 in response to irradiation in vitro. **p < 0.01 in shNT vs. shNT + irradiation; ##p < 0.01 or #p < 0.05 in shNT vs. shMLK4 + Irradiation.
(C) Representative bio-luminescence images of shNT or shMLK4 infected GSC 23 transduced with pCignal lenti-CMV-luc injected intracranially into Foxn1nu mice. Mice were imaged at 2–3 weeks after implantation, after which the radiation group received four cycles of 2.5 Gy IR on consecutive days (n = 6 in each group).
(D) Kaplan-Meier survival curves of mice intracranially transplanted GSC 23 with shNT or shMLK4 in response to irradiation. Arrows indicate the time of irradiation.
(E) Kaplan-Meier survival curves of tumor-burden mice intracranially transplanted GSC 8-11 with empty or MLK4 overexpression in response to irradiation. Irradiation was performed on week 3 after implantation for 4 consecutive days at 2.5 Gy/day. See also Figure S5.
To investigate the role of MLK4 in promoting GBM radioresistance, we injected either the control- or MLK4 (R470C)-overexpressing PN 8-11 spheres intracranially into immunocompromised mouse brains. IR treatment extended the survival of the control vector-overexpressing PN sphere-derived tumor-bearing mice by 51.5 days (Figure 6E). In contrast, in MLK4 (R470C)-overexpressing tumor bearing mice, IR extended the mouse survival for only 21 days (Figure 6E). These results indicate that expression of MLK4 is not only required, but sufficient to promote radioresistance in GBM.
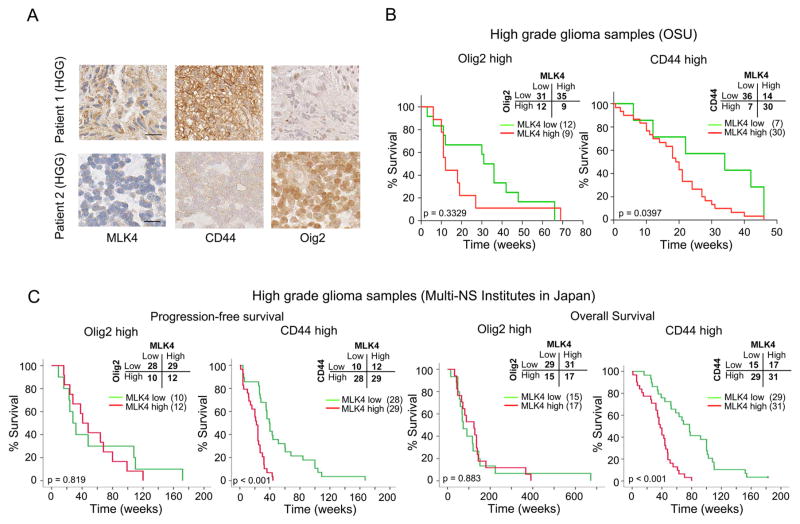
MLK4 expression is associated with poor survival of MES but not PN GBM
Lastly, we investigated the clinical relevance of our findings using tissues obtained from GBM patients. We performed immunohistochemical staining of MLK4, Olig2 (PN GBM marker), and CD44 (MES GBM marker) in 87 HGG specimens. As shown in Figure 7A, MLK4 expression strongly correlated with CD44 expression, whereas the staining of MLK4 and Olig2 were mutually exclusive. Intriguingly, in the Olig2high HGG patients, the MLK4 expression level was not informative for post-surgical patient survival. In contrast, high MLK4 patients displayed significantly shorter survival in the CD44high patient group, indicating MLK4 expression predicts survival of patients within the MES subgroup (Figure 7B). Following this exploratory study, we validated these findings in a larger cohort of cases (n = 108 cases). Consistent with the results observed with the initial datasets, both the progression-free survival and overall survival of patients with CD44high GBMs were significantly correlated with MLK4 expression, whereas those patients with Olig2high GBMs did not show any noticeable correlation (Figure 7C). Taken together, MLK4 is specifically upregulated in MES HGGs with CD44high expression and is negatively linked with post-surgical survival of patients with MES tumors.
Figure 7. MLK4 expression correlates with post-surgical poor survival of patients with MES GBM.
(A) Immunohistochemistry of human high grade glioma (HGG) samples for MLK4, CD44, and Olig2. Scale bar represents 20 μm.
(B) Kaplan Meier curve showing overall survival of HGG patients divided based on MLK4 expression in Olig2high PN-type (left) or CD44high MES-type (right) tumors with the exploratory cohort treated in the Department of Neurosurgery at OSU. (C) Kaplan Meier curve showing progression-free survival and overall survival of GBM patients divided based on MLK4 expression in Olig2high PN-type (left) or CD44high MES-type (right) tumors with the validation cohort treated in the Department of Neurosurgery in 4 Universities in Japan (designated as Multi-NS institutes).
DISCUSSION
In this study, we demonstrate that (1) MLK4 is highly expressed in glioma spheres derived from MES but not PN GBM, (2) overexpression of MLK4 promotes, while MLK4 knockdown attenuates a set of the MES glioma stem cell phenotypes, (3) MLK4 shRNA inhibits tumor initiation and propagation of MES GSCs in the mouse xenograft models, (4) radiation-induced MES transition in PN glioma spheres is associated with substantial upregulation of MLK4, (5) unlike the results of MLK4 knockdown alone, its combined treatment with IR in PN glioma sphere-derived tumors significantly attenuates tumor growth, thereby prolongs survival of tumor burden mice, (6) MLK4 binds and phosphorylates the NF-κB regulator IKKα, thereby activating the DNA binding of NF-κB in MES glioma spheres, and (7) MLK4 expression is highly elevated in CD44high MES subtype HGGs and is informative for the prognosis of patients with MES, but not PN HGG/GBM.
One major finding in this study is the differential role of MLK4 in PN and MES GSCs. Glioma spheres derived from Olig2high tumors, that were enriched with PN GSCs (Bhat et al., 2013; Mao et al., 2013), expressed substantially lower levels of MLK4 than MES tumor-derived cells and MLK4 silencing by shRNA did not induce apoptosis in vitro or affect tumor growth. In contrast, CD44high glioma spheres with MES GSC identity (Bhat et al., 2013; Mao et al., 2013) expressed substantially higher MLK4 and as a result, MLK4 knockdown reduced a set of stem cell traits (e.g. in vitro clonogenicity, in vivo tumorigenicity, increased tumor cell motility, and increased glycolysis) specifically in MES glioma sphere samples, making them selectively vulnerable to MLK4 inhibition.
Overexpression of MLK4 promoted whereas its knockdown reduced the expression of the MES-markers, suggesting that MLK4 is not only required but most likely sufficient to induce the MES phenotype. In orthotopic models, although mice bearing shMLK4 GSCs showed improved survival, they eventually died due to tumor burden despite retaining MLK4 downregulation (Figure S3E). It is not clear whether the MES glioma spheres initially contain de novo MLK4 resistant cell population or they acquire resistance during tumor formation in vivo. Molecular and phenotypic characterization of these tumors is crucial to determine whether persistent MES identity independent of MLK4-mediated signaling mechanisms promotes tumor growth or a shift of the MES phenotype to another subtype by shMLK4 subsequently leads to tumor growth despite MLK4 targeting. The frequency of MLK4 mutation in GBM is not very clear yet. One recent report by Martini et al. showed that 2 cases out of 9 had missense/nonsense mutation (Martini et al., 2013). Our study revealed the transcriptional and protein function of MLK4 in GBM and GSCs. Future studies will elucidate the genomic contribution of MLK4 on GBM.
The second finding in this study is the identification of a regulatory role for MLK4 in NF-κB signaling in GBM cells. We found that MLK4 interacts and phosphorylates IKKα in MES glioma spheres, thereby regulating its kinase activity. Through this interaction with IKKα, MLK4 controls NF-κB’s DNA binding activity (Figure 5D). Coupled with our previous study that NF-κB drives the MES signature by inducing master regulatory transcription factors (Bhat et al., 2013), the MLK4-NF-kB axis could be a key node in fine tuning the MES phenotype in GSCs. The current study also uncovers MLK4 as a potential link between tumor radioresistance and PMT in GSCs. IR-induced NF-κB activation was dependent on MLK4, and therefore silencing MLK4 presumably attenuates PMT following IR treatment. To date, there are no effective treatment options for recurrent GBMs after failure of the first-line therapies, and the identification of MLK4 presents an attractive target, inhibition of which could potentially improve patient outcome. Given the broad significance of the NF-κB pathway in a variety of cancers, future studies are required to explore the pathophysiological role of MLK4 in other cancers that depend on NF-κB for their growth and therapy resistance.
Unlike some other cancers (e.g. colon), virtually all GBMs are detected without prior pre- cancerous stage in the clinic, except for the minor population of the IDH mutant secondary GBMs. Therefore, whether GBMs initiate as PN tumors and evolve to gain more MES identity is an important question, but still remains unknown. Recent studies including our own indicate that PMT is likely associated with post-treatment reinitiation of tumors as recurrence (Chen et al., 2012). Indeed, some of newly-diagnosed GBM tumors that arise as PN tumors come back as MES tumors at recurrence (Phillips et al., 2006). Nonetheless, it is still debatable whether or not post-therapeutic PMT of GBM tumors is a universal phenomenon. In breast cancers, therapeutic insult appears to develop MES phenotypes, termed de novo-recurrence evolution (Mani et al., 2008; Moody et al., 2005). However, intratumoral sampling variations may mislead the data interpretation and are merely indicative of tumor heterogeneity. This open question aside, our data indicated that stimulation of PN glioma spheres with TNF-α upregulated MLK4 expression (data not shown). Previously we reported that TNF-α is derived from macrophages/microglia and is associated with the transcriptomic plasticity of the PN and MES states. It is known that the MES GBMs exhibit a high degree of macrophages/microglial infiltration. Possibly, macrophages and microglia may provide extrinsic signals including TNF-α to cause MLK4 activation thereby promoting PMT of tumors and/or GSCs via NF-κB activation (Bhat et al., 2013). Alternatively or on top of these extrinsic signals, MLK4 activation in MES GSCs could be driven by intrinsic signals in GSCs. In recent reports, a set of core transcriptome signatures was identified to determine tumor evolutionary dynamics and subsequent cellular identities in cancers and GSCs (Hnisz et al., 2013; Suva et al., 2014). It is important to further elucidate how intrinsic signaling mechanisms and extrinsic microenvironment factors cooperate / compete to sustain the MES identity in GSCs and GBM tumors.
In the current neuro-oncology clinic, irradiation is a mainstay of post-surgical GBM treatment and recent studies including ours elucidated that NF-κB is activated in post-IR tumors (Bhat et al., 2013; Brach et al., 1991). NF-κB is an essential molecule in various cancers to regulate cell proliferation, motility, therapy resistance, and metastasis (Bhat et al., 2013; Bivona et al., 2011; Bonavia et al., 2012; Fan et al., 2005; Fan et al., 2010; Ferris and Grandis, 2007; Gan et al., 2009; Tanaka et al., 2011; Yamamoto et al., 2013). Nonetheless, development of targeted therapies for transcription factors (TFs) such as NF-κB is notoriously challenging. To date NF-κB targeting cancer therapeutics has not been established. In addition, although NF-κB is potentially one of the promising candidates in MES GSCs, we should take into consideration that excessive and prolonged NF-κB inhibition is possibly detrimental because of its suppression in innate immunity (Baud and Karin, 2009; Greten et al., 2007). In this context, this study provides an alternative mode of NF-κB activation that could be exclusive to MES GSCs. MLK4-driven IKKα / NF-κB signaling axis therefore is a therapeutic target for the MES subtypes of GBMs. Moreover, the toxicity of specific MLK4 inhibitors may be modest because MLK4 is not expressed in normal progenitors and normal brain.
In conclusion, this study reveals a critical role of MLK4 in controlling the MES identity in GSCs and suggests MLK4 as an attractive target for treatment in aggressive cancers including MES GBM. The poorly characterized serine/threonine kinase MLK4 is abundantly expressed in various cancers (Abi Saab et al., 2012; Martini et al., 2013). Therefore, our data is applicable to other cancer types beyond GBM and provides a rationale for the therapeutic targeting of MLK4 and/or the MLK4/ NF-κB complex in a broader context. Given the importance of NF-κB signaling in cancers in general, our findings not only provide a better understanding of the molecular mechanisms underlying maintenance of stem cell characteristics in GBM but also identify a molecular target for therapeutic intervention. In addition, identification of MLK4 specific inhibitors could potentially create a new paradigm in the discovery and development of molecular targeted therapeutics for cancers including GBM.
EXPERIMENTAL PROCEDURES
Ethics
All of the work related to human tissues was exempt from requiring consent by Institutional Review Board (IRB) at University of Alabama at Birmingham (IRB no. #N151013001 and #N151014002), The Ohio State University (IRB no. #2005C0075), and M.D. Anderson Cancer Center (IRB no.#LAB04-0001). All mouse studies were conducted under the approved protocols by Institutional Animal Care and Use Committee (IACUC) and in accordance with NIH guidelines. The unique identity of all patient-derived glioma cell line was confirmed by short tandem repeats (STR) analysis as described in the methods section.
Establishment of GSC cultures
High grade glioma patient-derived neurospheres were molecularly characterized as previously described (Bhat et al., 2013; Gu et al., 2013; Joshi et al., 2013; Mao et al., 2013). Freshly resected glioma tumor samples were dissociated established GSC cells were cultured in defined medium containing Dulbecco’s modified Eagle’s medium (DMEM)/F12/Glutamax (Invitrogen) supplemented with B27 (Miltenyi Biotec), heparin (2.5 ug/ml), bFGF (20 ng/ml), and EGF (20 ng/ml). Growth factors (bFGF and EGF) were added twice a week.
Tissue Microarray (TMA)
TMA consisting of three to six representative 0.6-mm cores from formalin-fixed, paraffin-embedded tissue blocks was generated in the Department of Pathology and Laboratory Medicine at The Ohio State University as described previously (Guvenc et al., 2013; Miyazaki et al., 2012).
Gaussia NF-κB promoter luciferase assay
Gaussia Luciferase reporter lentiviral vector containing NF-κB binding sites (NF-Gluc-reporter) was provided by Dr. Christian Badr (Badr et al., 2009). After infection with NF-Gluc reporter lentivirus in glioma spheres, 15 μl aliquots of the cell-free conditioned medium were collected. Gluc activity was assayed by adding 20 μM coelenterazine, the Gluc substrate (Nanolight) to the supernatant and measuring photon counts in a 96-well plate FLUOstar luminometer (BMG labtech) over 10 sec.
Plasmids and lentiviral transduction
Lentiviral vectors expressing non-target shRNA, two shRNA constructs targeting MLK4, shMLK4_1 (Clone name: NM_032435.x-4199s1c1) and shMLK4_2 (Clone name: NM_032435.x-1413s1c1), used to silence MLK4 expression were obtained from Sigma. Full-length MLK4 cDNA in pCMV-Entry vector is obtained from Origene and transferred into pLenti-C-Myc-DDK lentiviral overexpression vector with RapidShuttling kit (Origene). 293FT (Invitrogen) cells were transfected using calcium phosphate (Clontech) for lentivirus production. Lentivirus was harvested at 72 hr after transfection and concentrated 100-fold using Lenti-X concentrator (Clontech). Infection of lentivirus was performed according to the manufacturer’s protocol.
Single cell motility assay
Single cell motility was monitored under guided migration conditions for approximately 16 hr (Gallego-Perez et al., 2012; Irimia and Toner, 2009; Johnson et al., 2009; Petrie et al., 2009). Glioma spheres were dissociated into single cells, plated, and allowed to adhere and spread on micropatterned polydimethylsiloxane surfaces in serum-containing, heparin-free CO2-independent medium (Invitrogen). Such surfaces were fabricated through a simple replica-molding process from a photolithographically-patterned Si master (Gallego-Perez et al., 2012). These substrates were sterilized in 70% ethanol prior to cell seeding. Cell motility was traced via time-lapse microscopy. Images were collected every 10 min, and analyzed using the manual tracker plugin in Fiji.
Limiting dilution neurosphere forming assay
Limiting dilution assay was performed in 96-well plates as described previously (Flavahan et al., 2013). Briefly, dissociated cells from glioma spheres on the 0.5 μg/cm2 laminin (Sigma) pre-coated flasks were seeded in 96-well plates containing GSCs culture medium (1 to 50 or 1 to 100 cells per well). After 7 days for MES 83 and 14 days for MES 267, each well was examined for formation of tumor spheres. Stem cell frequency was calculated using extreme limiting dilution analysis (ELDA) (http://bioinf.wehi.edu.au/software/elda/).
Statistics
Statistical analysis and graphing were performed using Microsoft Excel 2010, SPSS statistical package version 19 for Macintosh, and Graphpad Prism 6.0 software. Log-rank analysis was used to determine statistical significance of Kaplan-Meier survival curve.
Supplementary Material
SIGNIFICANCE.
Proneural (PN) and mesenchymal (MES) glioma stem cells (GSCs) represent two mutually-exclusive, and biologically distinct GSC subtypes. GBM patients with the MES GSC signature belong to the poorer prognosis subclass and are resistant to irradiation. Identification of regulatory mechanisms that regulate MES trans-differentiation is therefore critical for developing GSC-targeted therapy. Here we find that silencing MLK4 inhibits de novo and acquired (radiation-induced) MES GSCs both in vitro and in vivo. In addition, we present evidence of IKKα as a direct molecular target of MLK4 that drives NF-κB pathway activation, thereby promoting MES trans-differentiation of GSCs. Targeting the MLK4-driven NF-κB signaling axis could be a therapeutic strategy for GBM patients with a MES signature.
HIGHLIGHTS.
Silencing MLK4 attenuates Mesenchymal identity in glioma stem cells
MLK4 activates NF-κB signaling by directly phosphorylation of IKKα
MLK4 promotes radioresistance in GBM
MLK4 expression negatively correlates with patient survival of MES but not PN GBM
Acknowledgments
We are grateful to the members in the Nakano Laboratory for constructive discussion for this study. We thank Junfeng Shi (OSU), Drs. Joji Ishida (OU), and Seigo Kimura (OMC) for technical help and Dr. James Walker (MGH) for collaborative effort. We also appreciate the support from the Comparative Pathology and Mouse Phenotyping Shared Resource of The Ohio State University (P30 CA016058). This work was supported by the American Cancer Society MRSG-08-108-01 and NIH/NCI P01 CA163205, R21 CA175875, NIH/NINDS R01 NS083767, and R01 NS087913 (to I.N.); Grant-in-Aid for Scientific Research (B-26293322) from the Japan Society for the Promotion of Science and Takeda Science Foundation (to M.N.); Career Enhancement Project grant (2P50CA1270011) from the M.D. Anderson Brain Tumor SPORE (to K.B.). The authors declare no conflict of interest.
Footnotes
Author Contributions
S.H.K. and I.N. designed the experiments; S.H.K., R.E., E.P., D.G.P., A.S., D.T., K.L., T.F., H.S., R.C., J.H.C., A.M., S.B., K.K., T.K., R.I., and A.A. conducted the experiments; S.H.K., A.M., S.B., J.K., E.P.S., S.C., L.J.L., M.N., D.G., B.D., V.G., K.B., and I.N. analyzed data; S.H.K., K.B., and I.N. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abi Saab WF, Brown MS, Chadee DN. MLK4beta functions as a negative regulator of MAPK signaling and cell invasion. Oncogenesis. 2012;1:e6. doi: 10.1038/oncsis.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr CE, Niers JM, Tjon-Kon-Fat LA, Noske DP, Wurdinger T, Tannous BA. Real-time monitoring of nuclear factor kappaB activity in cultured cells and in animal models. Mol Imaging. 2009;8:278–290. [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Schulz JB, Beier CP. Chemoresistance of glioblastoma cancer stem cells--much more complex than expected. Mol Cancer. 2011;10:128. doi: 10.1186/1476-4598-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. Mesenchymal Differentiation Mediated by NF-kappaB Promotes Radiation Resistance in Glioblastoma. Cancer cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, Moonsamy P, Dahlman K, Miller VA, Costa C, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia R, Inda MM, Vandenberg S, Cheng SY, Nagane M, Hadwiger P, Tan P, Sah DW, Cavenee WK, Furnari FB. EGFRvIII promotes glioma angiogenesis and growth through the NF-kappaB, interleukin-8 pathway. Oncogene. 2012;31:4054–4066. doi: 10.1038/onc.2011.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brach MA, Hass R, Sherman ML, Gunji H, Weichselbaum R, Kufe D. Ionizing radiation induces expression and binding activity of the nuclear factor kappa B. J Clin Invest. 1991;88:691–695. doi: 10.1172/JCI115354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candolfi M, King GD, Yagiz K, Curtin JF, Mineharu Y, Muhammad AK, Foulad D, Kroeger KM, Barnett N, Josien R, et al. Plasmacytoid dendritic cells in the tumor microenvironment: immune targets for glioma therapeutics. Neoplasia. 2012;14:757–770. doi: 10.1593/neo.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capper D, Gaiser T, Hartmann C, Habel A, Mueller W, Herold-Mende C, von Deimling A, Siegelin MD. Stem-cell-like glioma cells are resistant to TRAIL/Apo2L and exhibit down-regulation of caspase-8 by promoter methylation. Acta neuropathol. 2009;117:445–456. doi: 10.1007/s00401-009-0494-3. [DOI] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichana KL, McGirt MJ, Laterra J, Olivi A, Quinones-Hinojosa A. Recurrence and malignant degeneration after resection of adult hemispheric low-grade gliomas. J Neurosurg. 2010;112:10–17. doi: 10.3171/2008.10.JNS08608. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Gao M, Meng Q, Laterra JJ, Symons MH, Coniglio S, Pestell RG, Goldberg ID, Rosen EM. Role of NF-kappaB signaling in hepatocyte growth factor/scatter factor-mediated cell protection. Oncogene. 2005;24:1749–1766. doi: 10.1038/sj.onc.1208327. [DOI] [PubMed] [Google Scholar]
- Fan S, Meng Q, Laterra JJ, Rosen EM. Scatter factor protects tumor cells against apoptosis caused by TRAIL. Anti-cancer drugs. 2010;21:10–24. doi: 10.1097/CAD.0b013e32832afc3b. [DOI] [PubMed] [Google Scholar]
- Ferris RL, Grandis JR. NF-kappaB gene signatures and p53 mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2007;13:5663–5664. doi: 10.1158/1078-0432.CCR-07-1544. [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, Weil RJ, Nakano I, Sarkaria JN, Stringer BW, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Perez D, Higuita-Castro N, Denning L, DeJesus J, Dahl K, Sarkar A, Hansford DJ. Microfabricated mimics of in vivo structural cues for the study of guided tumor cell migration. Lab Chip. 2012;12:4424–4432. doi: 10.1039/c2lc40726d. [DOI] [PubMed] [Google Scholar]
- Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- Gan HK, Lappas M, Cao DX, Cvrljevdic A, Scott AM, Johns TG. Targeting a unique EGFR epitope with monoclonal antibody 806 activates NF-kappaB and initiates tumour vascular normalization. J Cell Mol Med. 2009;13:3993–4001. doi: 10.1111/j.1582-4934.2009.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin CR, Lal B, Ho S, Woodard CL, Zhou X, Taeger A, Xia S, Laterra J. PTEN reconstitution alters glioma responses to c-Met pathway inhibition. Anti-cancer drugs. 2011;22:905–912. doi: 10.1097/CAD.0b013e3283484750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten FR, Arkan MC, Bollrath J, Hsu LC, Goode J, Miething C, Goktuna SI, Neuenhahn M, Fierer J, Paxian S, et al. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130:918–931. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Banasavadi-Siddegowda YK, Joshi K, Nakamura Y, Kurt H, Gupta S, Nakano I. Tumor-specific activation of the C-JUN/MELK pathway regulates glioma stem cell growth in a p53-dependent manner. Stem cells. 2013;31:870–881. doi: 10.1002/stem.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvenc H, Pavlyukov MS, Joshi K, Kurt H, Banasavadi-Siddegowda YK, Mao P, Hong C, Yamada R, Kwon CH, Bhasin D, et al. Impairment of glioma stem cell survival and growth by a novel inhibitor for Survivin-Ran protein complex. Clin Cancer Res. 2013;19:631–642. doi: 10.1158/1078-0432.CCR-12-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T, Holland EC. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci USA. 2014;111:5248–5253. doi: 10.1073/pnas.1321014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol. 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HM, Kim SH, Jin X, Park JB, Kim SH, Joshi K, Nakano I, Kim H. Crosstalk between Glioma-Initiating Cells and Endothelial Cells Drives Tumor Progression. Cancer Res. 2014;74:4482–4492. doi: 10.1158/0008-5472.CAN-13-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J, Nowicki MO, Lee CH, Chiocca EA, Viapiano MS, Lawler SE, Lannutti JJ. Quantitative analysis of complex glioma cell migration on electrospun polycaprolactone using time-lapse microscopy. Tissue Eng Part C Methods. 2009;15:531–540. doi: 10.1089/ten.tec.2008.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K, Banasavadi-Siddegowda Y, Mo X, Kim SH, Mao P, Kig C, Nardini D, Sobol RW, Chow LM, Kornblum HI, et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem cells. 2013;31:1051–1063. doi: 10.1002/stem.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper F, Dornhoefer N, Giaccia AJ. Mutations in the PI3K/PTEN/TSC2 pathway contribute to mammalian target of rapamycin activity and increased translation under hypoxic conditions. Cancer Res. 2006;66:1561–1569. doi: 10.1158/0008-5472.CAN-05-3375. [DOI] [PubMed] [Google Scholar]
- Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli M, Ferrero S, Martinelli Boneschi F, Boiocchi L, Zavanone M, Maria Gaini S, Bello L, Valentino S, Barbati E, Nebuloni M, et al. The long pentraxin PTX3 as a correlate of cancer-related inflammation and prognosis of malignancy in gliomas. J Neuroimmunol. 2013;260:99–106. doi: 10.1016/j.jneuroim.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl AcadSci USA. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini M, Russo M, Lamba S, Vitiello E, Crowley EH, Sassi F, Romanelli D, Frattini M, Marchetti A, Bardelli A. Mixed lineage kinase MLK4 is activated in colorectal cancers where it synergistically cooperates with activated RAS signaling in driving tumorigenesis. Cancer Res. 2013;73:1912–1921. doi: 10.1158/0008-5472.CAN-12-3074. [DOI] [PubMed] [Google Scholar]
- Marusiak AA, Edwards ZC, Hugo W, Trotter EW, Girotti MR, Stephenson NL, Kong X, Gartside MG, Fawdar S, Hudson A, et al. Mixed lineage kinases activate MEK independently of RAF to mediate resistance to RAF inhibitors. Nat Commun. 2014;5:3901. doi: 10.1038/ncomms4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Pan Y, Joshi K, Purohit D, Hu B, Demir H, Mazumder S, Okabe S, Yamori T, Viapiano M, et al. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb. Clin Cancer Res. 2012;18:1268–1280. doi: 10.1158/1078-0432.CCR-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Nurwidya F, Takahashi F, Murakami A, Takahashi K. Epithelial mesenchymal transition in drug resistance and metastasis of lung cancer. Cancer Res Treat. 2012;44:151–156. doi: 10.4143/crt.2012.44.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MC, Kim JM, Safaee M, Kaur G, Sun MZ, Kaur R, Celli A, Mauro TM, Parsa AT. Overexpression of calcium-permeable glutamate receptors in glioblastoma derived brain tumor initiating cells. PloS one. 2012;7:e47846. doi: 10.1371/journal.pone.0047846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Sanchez-Tillo E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A, Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirkoohi R. Epithelial mesenchymal transition from a natural gestational orchestration to a bizarre cancer disturbance. Cancer Sci. 2013;104:28–35. doi: 10.1111/cas.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafani M, Di Vito M, Frati A, Pellegrini L, De Santis E, Sette G, Eramo A, Sale P, Mari E, Santoro A, et al. Pro-inflammatory gene expression in solid glioblastoma microenvironment and in hypoxic stem cells from human glioblastoma. J Neuroinflammation. 2011;8:32. doi: 10.1186/1742-2094-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, Dang J, Zhu S, Yang H, De Jesus J, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Taguchi Y, Ito-Kureha T, Semba K, Yamaguchi N, Inoue J. NF-kappaB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun. 2013;4:2299. doi: 10.1038/ncomms3299. [DOI] [PubMed] [Google Scholar]
- Zhong J, Paul A, Kellie SJ, O'Neill GM. Mesenchymal migration as a therapeutic target in glioblastoma. J Oncol. 2010;2010:430142. doi: 10.1155/2010/430142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.