Abstract
Background
We conducted single-marker, gene- and pathway-based analyses to examine the association between renin–angiotensin–aldosterone system (RAAS) variants and chronic kidney disease (CKD) progression among Chronic Renal Insufficiency Cohort study participants.
Methods
A total of 1523 white and 1490 black subjects were genotyped for 490 single nucleotide polymorphisms (SNPs) in 12 RAAS genes as part of the ITMAT-Broad-CARe array. CKD progression phenotypes included decline in estimated glomerular filtration rate (eGFR) over time and the occurrence of a renal disease event, defined as incident end-stage renal disease or halving of eGFR from baseline. Mixed-effects models were used to examine SNP associations with eGFR decline, while Cox proportional hazards models tested SNP associations with renal events. Gene- and pathway-based analyses were conducted using the truncated product method. All analyses were stratified by race, and a Bonferroni correction was applied to adjust for multiple testing.
Results
Among white and black participants, eGFR declined an average of 1.2 and 2.3 mL/min/1.73 m2/year, respectively, while renal events occurred in a respective 11.5 and 24.9% of participants. We identified strong gene- and pathway-based associations with CKD progression. The AGT and RENBP genes were consistently associated with risk of renal events in separate analyses of white and black participants (both P < 1.00 × 10−6). Driven by the significant gene-based findings, the entire RAAS pathway was also associated with renal events in both groups (both P < 1.00 × 10−6). No single-marker associations with CKD progression were observed.
Conclusions
The current study provides strong evidence for a role of the RAAS in CKD progression.
Keywords: chronic kidney disease, genetics, renin–angiotensin–aldosterone system
INTRODUCTION
The prevalence of end-stage renal disease (ESRD) has risen steadily over the past few decades [1]. Recent data indicate that over 600 000 individuals are now living with ESRD in the USA [1]. The rising prevalence of ESRD combined with its associated increases in morbidity and mortality make ESRD a quickly emerging public health challenge [1, 2]. However, ESRD makes up only a small portion of the total number of chronic kidney disease (CKD) cases nationwide [1]. Although CKD frequently worsens in severity over time, declines in renal function can vary substantially between individuals [3]. Well-known factors influencing progression of CKD to ESRD include proteinuria [4], diabetes [5], hypertension [6] and race [7]. While a genetic component to CKD progression has also been established [8], genomic factors underlying this complex phenotype remain largely unknown.
Physiological studies have implicated the renin–angiotensin–aldosterone system (RAAS) in the progression of CKD [9, 10]. While the RAAS likely contributes to CKD progression in part via blood pressure-mediated kidney damage, non-hemodynamic effects of the RAAS on this complex phenotype have also been described [9, 10]. In addition to its vasoconstrictive properties, angiotensin II (the main effector protein of the RAAS) has been shown to increase production of pro-inflammatory cytokines leading to inflammation and renal fibrosis [10]. The known physiological relevance of the RAAS makes genes in this pathway logical candidates for genomic study of CKD progression. While some studies have examined the role of select variants in RAAS genes on CKD-related phenotypes [11–18], a comprehensive exploration of common genetic variation in these genes for their association with CKD progression has not been carried out. Furthermore, no gene or pathway-based analyses have been conducted. Such analyses may increase statistical power to detect the likely modest effects of common RAAS variants on CKD progression phenotypes.
In the current study, we conducted single-marker, gene- and pathway-based analyses to examine the associations of common variants from 12 RAAS candidate genes [renin (REN); hydroxysteroid (11-beta) dehydrogenase 1 (HSD11B1); angiotensinogen (AGT); angiotensin II Type 1 receptor (AGTR1); nuclear receptor subfamily 3, group C, member 2 (NR3C2); cytochrome P450, family 11, subfamily B, polypeptide 1 (CYP11B1); cytochrome P450, family 11, subfamily B, polypeptide 2 (CYP11B2); hydroxysteroid (11-beta) dehydrogenase 2 (HSD11B2); angiotensin converting enzyme (ACE); angiotensin-converting enzyme 2 (ACE2); angiotensin II Type 1 receptor 2 (AGTR2) and renin-binding protein (RENBP)] with CKD progression phenotypes among white and black participants from the Chronic Renal Insufficiency Cohort (CRIC) study.
MATERIALS AND METHODS
Study population
Between June 2003 and August 2008, the CRIC study enrolled 3939 adult patients with CKD who are followed-up bi-annually for clinical and subclinical outcomes of kidney and cardiovascular diseases. A detailed description of the CRIC study design and participants has been reported previously [19, 20]. Briefly, the CRIC study recruited a racially and ethnically diverse group of adults aged 21–74 with a broad spectrum of renal disease severity [estimated glomerular filtration rate (eGFR) of 20–70 mL/min/1.73 m2] from seven clinical centers in the USA. Among the 3288 non-Hispanic white and black CRIC participants, 3013 (91.6%) with adequate phenotype and genotype data were eligible for the current analysis.
Data collection
At the baseline examination information was ascertained on demographic characteristics, medical history and medication use. Blood samples were collected and DNA extracted for genomic study. Anthropometric measurements were also obtained and blood pressure measured using a standard protocol [21]. Serum creatinine was measured from fasting blood samples at the baseline examination and during annual in-person study visits. The eGFR was calculated according to a CRIC-specific equation, which included serum creatinine, cystatin C, age, gender and race [22].
During the follow-up, incident ESRD was defined as receipt of chronic dialysis or kidney transplant. Information on the initiation and maintenance of dialysis and kidney transplant was obtained by annual clinical follow-up visits and interim telephone interviews and confirmed by a dialysis unit or hospital chart review. Ascertainment of ESRD in the CRIC study was supplemented by information from the US Renal Data System.
Genotyping and genotype quality control
A total of 1523 white and 1490 black CRIC participants were genotyped using the ITMAT-Broad-CARe (IBC) chip [23], which includes ∼50 000 SNPs from cardiovascular disease-related loci across the genome. To examine the association between RAAS variants and CKD progression, all 490 SNPs from 12 RAAS genes (REN, HSD11B1, AGT, AGTR1, NR3C2, CYP11B1, CYP11B2, HSD11B2, ACE, ACE2, AGTR2 and RENBP) were selected from the IBC chip for possible inclusion in the current study. Characteristics of these SNPs are shown in Supplementary Table S1. Quality control excluded RAAS variants with low minor allele frequency (<0.01), low genotyping call rate (<95%) or significant deviation from Hardy–Weinberg Equilibrium (Bonferroni-adjusted, <1.02 × 10−4). After SNP filtering, 254 SNPs and 375 SNPs remained among white and black participants, respectively. Individual quality control was conducted using genome-wide genotype data. Assessment of cryptic relatedness removed two white and four black participants who shared at least 12.5% of alleles identically by descent with another CRIC participant. In addition, race-stratified principal components analysis excluded a further 11 whites and 1 black with genomic ancestry at least 6 SDs from the mean [24]. After excluding cryptically related participants and participants with divergent ancestry, 1510 white and 1485 black participants remained for the analysis. Significant principal components were retained for ancestry adjustment in multivariable analysis.
Study outcomes
The current analysis examined two CKD progression phenotypes, which included rate of decline in kidney function (slope of eGFR over time) and the occurrence of a renal disease event (incident ESRD or halving of eGFR from baseline). For time-to-event analyses, time until halving of eGFR was imputed assuming a linear decline in kidney function between in-person annual follow-up visits.
Statistical analysis
Baseline characteristics and CKD progression outcomes were calculated separately among white and black CRIC participants as mean ± SD for continuous variables and as percentages for categorical variables.
Race-stratified mixed-effects regression models were used to test the additive associations between single SNPs and eGFR decline over time. In the mixed-effects models, eGFR was included as the dependent variable while each SNP, follow-up time and an SNP by follow-up time interaction term were included as independent variables (along with other covariables). The P-value for the interaction term was used to identify SNPs influencing eGFR decline over time. Autoregressive variance–covariance matrices were used to accommodate the correlations of repeated measurements within individuals. A race-stratified Cox proportional hazards model was used to examine the additive association between each SNP and time to renal event. In the main analysis of CKD progression phenotypes, models were adjusted for age, gender and ancestry (Model 1). Additional models were developed to explore the association of each SNP with CKD progression outcomes after adjustment for variables in the main model plus potential mediators such as baseline eGFR (Model 2) and both baseline eGFR and systolic blood pressure (Model 3). For analysis of variants on the X-chromosome, SNPs were encoded as [0, 2] for men and [0, 1, 2] for women, which assumes one of the two X chromosomes in women is fully inactivated. All single-marker analyses were carried out using SAS statistical software (version 9.3; SAS Institute, Cary, NC, USA).
While the influence of single SNPs on complex phenotypes may be modest, the joint effects of multiple SNPs at a single gene locus may be larger, increasing statistical power to identify genic associations [25]. In the current analysis, the truncated product method (TPM) was used to determine the overall association of each RAAS gene (with at least two genotyped SNPs) as well as the entire RAAS pathway with the CKD progression outcomes in each race group [26]. TPM combines P-values from a set of L hypothesis tests by leveraging features of the Fisher's product method and Wilkinson's truncation method. This procedure takes the product of P-values less than a specified cut-point (denoted as τ) and evaluates the probability of such a product, or smaller, under the overall hypothesis that all L hypotheses are true [26]. TPM has been evaluated extensively through simulation [27]. Furthermore, because the correlations between adjacent markers are taken into account in the permutation testing used to derive P-values, TPM allows for the statistical dependence of single-marker tests arising from linkage disequilibrium [26]. Similar to previous studies, TPM was used to estimate gene-based P-values by combining P-values from SNPs within each RAAS gene [28, 29]. TPM was also used to conduct pathway-based analyses, combining the TPM-generated gene-based P-values across all RAAS genes [28, 29]. The truncation point was set as τ = 0.10 for all analyses, and the P-value for TPM was estimated through simulation (1 000 000 replications). Sensitivity analyses were performed to determine whether significant variants from single-marker analyses could explain gene-based findings and whether significant genes from gene-based analyses could explain pathway-based findings. All gene- and pathway-based analyses were performed using R software (Version 3.0.1; http://www.r-project.org).
For all analyses, statistical significance was determined after Bonferroni adjustment for multiple testing.
RESULTS
Table 1 summarizes the baseline characteristics and CKD progression among 2995 CRIC study participants. On average, whites were 59 years of age, had systolic blood pressure of 122 mmHg and baseline eGFR of 44 mL/min/1.73 m2. Blacks, on average, were 58 years of age, had systolic blood pressure of 133 mmHg and baseline eGFR of 44 mL/min/1.73 m2. Sixty percent of whites and 49% of blacks were male. Over ∼3.7 and 3.5 years follow-up in whites and blacks, respectively, renal events occurred in 12% of whites and 25% of blacks. The eGFR declined an average of 1.2 and 2.3 mL/min/1.73 m2/year in whites and blacks, respectively.
Table 1.
Baseline characteristics and CKD progression among 2995 CRIC study participants
| Whites (n = 1510) | Blacks (n = 1485) | |
|---|---|---|
| Baseline characteristic | ||
| Age, years, mean (SD) | 59.0 (10.8) | 58.1 (10.6) |
| Male (%) | 59.9 | 48.7 |
| eGFR (mL/min/1.73 m2), mean (SD) | 43.6 (12.8) | 43.7 (13.9) |
| Systolic BP (mmHg), mean (SD) | 121.9 (18.6) | 132.9 (23.1) |
| CKD progression | ||
| eGFR slope (mL/min/1.73 m2/year), mean (SD) | −1.2 (3.7) | −2.3 (5.0) |
| Renal eventa (%b) | 11.5 | 24.9 |
BP, blood pressure; CKD, chronic kidney disease; CRIC, Chronic Renal Insufficiency Cohort; eGFR, estimated glomerular filtration rate; SD, standard deviation.
aDiagnosis of ESRD or a reduction of 50% in eGFR since baseline.
bPercent of participants with renal events over an average of 3.7 and 3.5 years follow-up in whites and blacks, respectively.
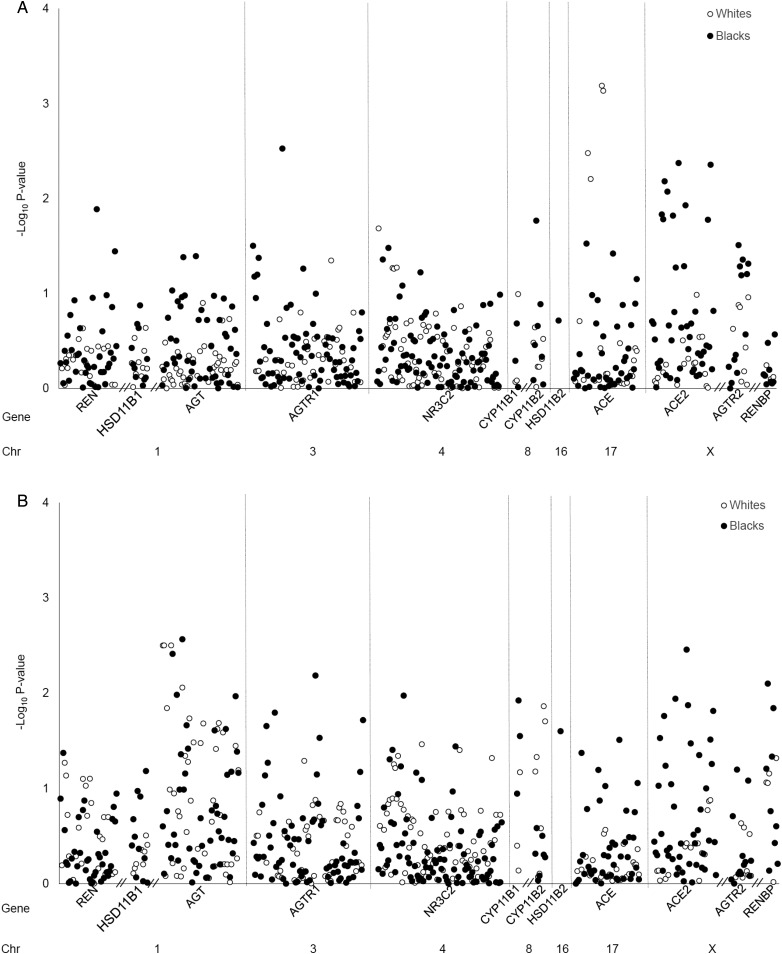
Results from the main multivariable model (adjusted for age, gender and ancestry) examining the association of each SNP with eGFR decline over time and time to renal event are shown in Figure 1A and B, respectively, and in Supplementary Table S2. After adjustment for multiple testing, no associations between RAAS variants and the CKD progression outcomes were identified in either white or black study participants. Results of Model 2 (Model 1 plus additional adjustment for baseline eGFR) and Model 3 (Model 1 plus additional adjustment for baseline eGFR and systolic blood pressure) were similar to those of the main analysis (data not shown).
FIGURE 1:
Association of RAAS variants with eGFR decline (A) and renal events (B) among participants of the CRIC study. Results represent findings from the main multivariable model (adjusted for age, gender and ancestry). No SNPs were significant after Bonferroni correction for multiple testing (P < 1.97 × 10−4 in whites and P < 1.33 × 10−4 in blacks). eGFR, estimated glomerular filtration rate; RAAS, renin–angiotensin–aldosterone system.
Table 2 summarizes gene- and pathway-based findings for eGFR decline. Among whites only, the ACE gene was significantly associated with eGFR decline. Results remained statistically significant after additional multivariable adjustments in Models 2 and 3. In blacks only, the ACE2 and AGTR2 genes as well as the entire RAAS pathway were significantly associated with eGFR decline in the main analysis. These associations remained in Models 2 and 3 for the ACE2 gene and the entire RAAS pathway. The AGTR2 gene remained associated with eGFR decline in Model 2 but not in Model 3.
Table 2.
Pathway- and gene-based associations of RAAS variants and eGFR decline among CRIC study participants
| HGNC symbol | k | Model 1a | Model 2b | Model 3c |
|---|---|---|---|---|
| Whites | ||||
| RAAS pathway | 254 | 0.22 | 0.23 | 0.26 |
| REN | 23 | 0.52 | 0.77 | 0.68 |
| HSD11B1 | 10 | 0.41 | 0.41 | 0.44 |
| AGT | 42 | 0.68 | 0.45 | 0.51 |
| AGTR1 | 44 | 0.63 | 0.78 | 0.72 |
| NR3C2 | 71 | 0.94 | 0.86 | 0.89 |
| CYP11B1 | 3 | 0.30 | 0.30 | 0.26 |
| CYP11B2 | 7 | 0.42 | 0.52 | 0.47 |
| ACE | 24 | 8.31 × 10−4 d | 9.93 × 10−4 d | 0.001d |
| ACE2 | 17 | 0.67 | 0.55 | 0.54 |
| AGTR2 | 8 | 0.58 | 0.59 | 0.40 |
| RENBP | 5 | 0.16 | 0.21 | 0.21 |
| Blacks | ||||
| RAAS pathway | 375 | 7.00 × 10−6 d | 3.79 × 10−4 d | 5.18 × 10−4 d |
| REN | 38 | 0.69 | 0.66 | 0.61 |
| HSD11B1 | 11 | 0.49 | 0.53 | 0.59 |
| AGT | 47 | 0.88 | 0.73 | 0.71 |
| AGTR1 | 73 | 0.59 | 0.26 | 0.49 |
| NR3C2 | 88 | 0.73 | 0.60 | 0.51 |
| CYP11B1 | 3 | 0.30 | 0.27 | 0.28 |
| CYP11B2 | 7 | 0.20 | 0.15 | 0.17 |
| ACE | 42 | 0.77 | 0.95 | 0.85 |
| ACE2 | 43 | <1.00 × 10−6 d | <1.00 × 10−6 d | <1.00 × 10−6 d |
| AGTR2 | 13 | 6.00 × 10−6 d | 0.003 d | 0.004 |
| RENBP | 9 | 0.38 | 0.51 | 0.51 |
eGFR, estimated glomerular filtration rate; HGNC, HUGO Gene Nomenclature Committee; k, number of SNPs; RAAS, renin–angiotensin–aldosterone system.
aMain multivariable model (adjusted for age, gender and genetic ancestry).
bMain multivariable model with additional adjustment for baseline eGFR.
cMain multivariable model with additional adjustment for baseline eGFR and systolic blood pressure.
dSignificant after Bonferroni correction for 12 gene- and pathway-based tests (P < 0.004).
The results of gene- and pathway-based analyses of renal events are displayed in Table 3. In the main multivariable model, genes AGT and RENBP as well as the entire RAAS pathway were significantly associated with renal events among both white and black study participants. The AGT gene and the entire RAAS pathway remained significantly associated with renal events in multivariable Models 2 and 3 in both groups. In contrast, the RENBP gene association was completely attenuated by additional adjustments in both Models 2 and 3 in whites and blacks. Among whites only, CYP11B2 appeared to be associated with renal events in Models 1 and 2 with a complete attenuation of the association in Model 3. Among blacks only, ACE2 was significantly associated with renal events in all models, a finding that was consistent with results from the gene-based analysis of eGFR decline.
Table 3.
Pathway- and gene-based associations of RAAS variants and renal event among CRIC study participants
| HGNC symbol | k | Model 1a | Model 2b | Model 3c |
|---|---|---|---|---|
| Whites | ||||
| RAAS pathway | 254 | <1.00 × 10−6 d | 1.10 × 10−5 d | 5.16 × 10−4 d |
| REN | 23 | 0.07 | 0.59 | 0.77 |
| HSD11B1 | 10 | 0.24 | 0.34 | 0.56 |
| AGT | 42 | <1.00 × 10−6 d | <1.00 × 10−6 d | <1.00 × 10−6 d |
| AGTR1 | 44 | 0.87 | 0.56 | 0.73 |
| NR3C2 | 71 | 0.53 | 0.47 | 0.72 |
| CYP11B1 | 3 | 0.20 | 0.15 | 0.26 |
| CYP11B2 | 7 | 1.00 × 10−6 d | 1.60 × 10−5 d | 0.33 |
| ACE | 24 | 0.45 | 0.77 | 0.55 |
| ACE2 | 17 | 0.61 | 0.33 | 0.60 |
| AGTR2 | 8 | 0.43 | 0.64 | 0.004 |
| RENBP | 5 | 1.70 × 10−5 d | 0.31 | 0.28 |
| Blacks | ||||
| RAAS pathway | 375 | <1.00 × 10−6 d | <1.00 × 10−6 d | <1.00 × 10−6 d |
| REN | 38 | 0.97 | 0.78 | 0.63 |
| HSD11B1 | 11 | 0.61 | 0.22 | 0.03 |
| AGT | 47 | <1.00 × 10−6 d | 0.001d | 1.00 × 10−6 d |
| AGTR1 | 73 | 0.15 | 0.45 | 0.60 |
| NR3C2 | 88 | 0.80 | 0.59 | 0.64 |
| CYP11B1 | 3 | 0.04 | 0.03 | 0.20 |
| CYP11B2 | 7 | 0.37 | 0.35 | 0.48 |
| ACE | 42 | 0.43 | 0.06 | 0.49 |
| ACE2 | 43 | <1.00 × 10−6 d | <1.00 × 10−6 d | <1.00 × 10−6 d |
| AGTR2 | 13 | 0.36 | 0.33 | 0.58 |
| RENBP | 9 | 2.20 × 10−5 d | 0.05 | 0.006 |
CRIC, Chronic Renal Insufficiency Cohort; HGNC, HUGO Gene Nomenclature Committee; k, number of SNPs; RAAS, renin–angiotensin–aldosterone system.
aMain multivariable model (adjusted for age, gender and genetic ancestry).
bMain multivariable model with additional adjustment for baseline eGFR.
cMain multivariable model with additional adjustment for baseline eGFR and systolic blood pressure.
dSignificant after Bonferroni correction for 12 gene- and pathway-based tests (P < 0.004).
After simultaneous removal of the genes identified by gene-based analysis, sensitivity analyses revealed a complete attenuation of the relationship between the RAAS pathway and eGFR progression that was observed in blacks. Similarly, removal of significantly identified genes for renal events explained the influence of the RAAS pathway with this CKD progression phenotype, attenuating the association in both whites and blacks. Because there were no single-marker findings, the influence of individual SNPs on gene-based findings was not examined.
DISCUSSION
In the first study, to examine the joint contribution of RAAS variants to CKD progression, we identified strong gene- and pathway-based associations with this complex phenotype. Both the AGT and RENBP genes were consistently associated with risk of renal events in independent subsamples of white and black participants of the CRIC study. Driven by significant gene-based findings, the entire RAAS pathway was also associated with renal events in both white and black CRIC participants. Race-specific findings included the associations of ACE and CYP11B2 with CKD progression phenotypes in whites and the associations of ACE2 and AGTR2 with CKD progression in blacks. These results are promising but should be interpreted with caution until consistency in other samples with similar ancestry is demonstrated. Since no individual SNP associations were identified by the current study, our results highlight the utility of gene- and pathway-based approaches to better understand the genomic mechanisms underlying CKD progression.
We observed a strong association of the AGT gene with renal events in both white and black CRIC study participants. This association persisted after adjustment for systolic blood pressure, suggesting that the influence of AGT on CKD progression may also be mediated by non-hemodynamic mechanisms of renal injury. The protein encoded by AGT is a precursor to angiotensin II, which may be involved in inflammation and renal fibrosis through its activation of mononuclear cells and by increasing pro-inflammatory mediators such as cytokines, chemokines, adhesion molecules and nuclear factor kB [10]. While we are the first to identify a gene-based association of AGT, individual AGT variants have previously been linked to CKD progression [11, 30]. Hsu et al. [11] identified an association of the AGT variant rs5051 (or the A-6G polymorphism) with renal events in black participants, while Lovati et al. [30] identified an association of rs699 (or the M235T polymorphism) with progression to ESRD in Europeans. These two markers are in high linkage disequilibrium in populations of European and African ancestry (r2 = 0.96 and 0.91, respectively). Interestingly, in the current study, rs5051 and rs699 were nominally or marginally associated with renal events among both whites (P = 0.06 and 0.03, respectively) and blacks (P = 0.05 and 0.17, respectively) after adjustment for systolic blood pressure. In aggregate, these findings strongly support a role of AGT in CKD progression.
A consistent association of the RENBP gene with renal events was also identified among white and black CRIC study participants. RENBP encodes a protein that inhibits renin activity [31]. Although this gene has not been implicated previously in CKD progression, its potential influence on blood pressure phenotypes has been reported in past epidemiologic studies [31, 32]. In the current study, findings were attenuated in multivariable models which included baseline eGFR and systolic blood pressure, suggesting that these factors may mediate the observed association. Since the multivariable models used could not disentangle the influence of systolic blood pressure from that of eGFR, a post hoc analysis was conducted to determine whether systolic blood pressure alone may have mediated the findings observed in the main analysis (the post hoc analysis included covariables in the main analysis plus baseline systolic blood pressure alone). Results showed a complete attenuation of the relation in whites (P = 0.19) but only a very slight attenuation in blacks (P = 2.50 × 10−5). While these inconsistent findings could be due to varying genomic mechanisms in whites and blacks, it could also be due to the different SNPs included in the gene-based analyses across race groups. The latter would suggest that blood pressure may only partly explain the observed association of RENBP with CKD progression.
Our analysis also identified race-specific gene-based findings, showing associations of ACE and CYP11B2 with CKD progression phenotypes in whites and associations of ACE2 and AGTR2 with CKD progression in blacks. Of particular interest was the consistent association of ACE2 with both eGFR decline and incident renal events in blacks. ACE2 is expressed in kidney endothelium and has renoprotective properties, converting angiotensin I to the inactive nonapeptide, angiotensin [1–9], and angiotensin II to the potent vasodilator, angiotensin [1–7, 33, 34]. Although several studies have identified relations of ACE2 with blood pressure phenotypes [35, 36], we are the first to report an association with a CKD-related trait. Interestingly, the association identified in the current study was independent of blood pressure, suggesting that the observed ACE2 association is not blood pressure mediated. While these results are promising, replication is needed to confirm our findings.
Our study has several strengths. We are the first to conduct gene- and pathway-based analyses to examine the influence of RAAS variants on CKD progression phenotypes. In addition, the CRIC study is a rigorously designed and conducted NIH-funded study, providing high-quality genomic, covariable and phenotypic data for genetic research. Furthermore, the large sample sizes of both white and black participants allowed us to explore the consistency of identified associations in independent CRIC subsamples employing stringent Bonferroni correction to adjust for multiple testing. Several limitations should also be acknowledged. Although our sample size was sufficient to identify aggregate effects of variants within RAAS genes and the entire RAAS pathway, we may have been underpowered to detect individual RAAS markers associated with CKD progression. In addition, although race-specific findings identified by our study are promising, research will be needed to explore whether they can be replicated in independent samples of similar ancestry. Furthermore, due to the limited number of SNPs in HSD11B2, gene-based analyses could not be performed on this RAAS gene. Future research will be needed to explore the gene-based association of HSD11B2 with CKD progression.
The current study provides strong evidence for a role of the RAAS in CKD progression. AGT and RENBP independently associated with renal events in both white and black participants. Since blood pressure did not completely explain the gene-based findings, our data provide further support for a non-hemodynamic role of the RAAS in renal disease progression. In aggregate, our findings emphasize the potential importance of gene- and pathway-based analyses to better understand the biological pathways underlying renal disease progression. Furthermore, our work contributes new insights into the cumulative understanding of the genomic mechanisms influencing this complex phenotype. Such information may one day be used to better predict renal disease progression in CKD patients, which could enable early prevention and intervention strategies for those at highest risk of ESRD.
SUPPLEMENTARY DATA
Supplementary data are available online at http://ndt.oxfordjournals.org.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the participants, investigators and staff of the CRIC study for their time and commitment. The CRIC Study Investigators are Lawrence J. Appel, MD, MPH, Harold I. Feldman, MD, MSCE, Alan S. Go, MD, John W. Kusek, PhD, James P. Lash, MD, Akinlolu Ojo, MD, PhD and Raymond R. Townsend, MD. Funding for the CRIC study was obtained under a cooperative agreement from National Institute of Diabetes and Digestive and Kidney Diseases (U01DK060990, U01DK060984, U01DK061022, U01DK061021, U01DK061028, U01DK060980, U01DK060963 and U01DK060902). In addition, this work was supported in part by the Perelman School of Medicine at the University of Pennsylvania Clinical and Translational Science Award NIH/NCATS UL1TR000003, Johns Hopkins University UL1 TR-000424, University of Maryland GCRC M01 RR-16500, Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, Michigan Institute for Clinical and Health Research (MICHR) V 2014.07.28 UL1TR000433, University of Illinois at Chicago CTSA UL1RR029879, Tulane University Translational Research in Hypertension and Renal Biology P30GM103337, Kaiser Permanente NIH/NCRR UCSF-CTSI UL1 RR-024131. Dr Raj is supported by R01 DK073665-01A1.
REFERENCES
- 1.US Renal Data System. USRDS 2013 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD, 2013 [Google Scholar]
- 2.Go AS, Chertow GM, Fan D, et al. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med 2004; 351: 1296–1305 [DOI] [PubMed] [Google Scholar]
- 3.McClellan WM, Flanders WD. Risk factors for progressive chronic kidney disease. J Am Soc Nephrol 2003; 14: S65–S70 [DOI] [PubMed] [Google Scholar]
- 4.Peterson JC, Adler S, Burkart JM, et al. Blood pressure control, proteinuria, and the progression of renal disease. The Modification of Diet in Renal Disease Study. Ann Intern Med 1995; 123: 754–762 [DOI] [PubMed] [Google Scholar]
- 5.Brancati FL, Whelton PK, Randall BL, et al. Risk of end-stage renal disease in diabetes mellitus: a prospective cohort study of men screened for MRFIT. Multiple Risk Factor Intervention Trial. JAMA 1997; 278: 2069–2074 [PubMed] [Google Scholar]
- 6.Klag MJ, Whelton PK, Randall BL, et al. Blood pressure and end-stage renal disease in men. N Engl J Med 1996; 334: 13–18 [DOI] [PubMed] [Google Scholar]
- 7.Cowie CC, Port FK, Wolfe RA, et al. Disparities in incidence of diabetic end-stage renal disease according to race and type of diabetes. N Engl J Med 1989; 321: 1074–1079 [DOI] [PubMed] [Google Scholar]
- 8.Parsa A, Kao WHL, Xie D, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 2013; 369: 2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remuzzi G, Perico N, Macia M, et al. The role of renin-angiotensin-aldosterone system in the progression of chronic kidney disease. Kidney Int Suppl 2005; 99: S57–S65 [DOI] [PubMed] [Google Scholar]
- 10.Mezzano SA, Ruiz-Ortega M, Egido J. Angiotensin II and renal fibrosis. Hypertension 2001; 38: 635–638 [DOI] [PubMed] [Google Scholar]
- 11.Hsu CC-C, Bray MS, Kao WHL, et al. Genetic variation of the renin-angiotensin system and chronic kidney disease progression in black individuals in the atherosclerosis risk in communities study. J Am Soc Nephrol 2006; 17: 504–512 [DOI] [PubMed] [Google Scholar]
- 12.Anbazhagan K, Sampathkumar K, Ramakrishnan M, et al. Analysis of polymorphism in renin angiotensin system and other related genes in South Indian chronic kidney disease patients. Clin Chim Acta 2009; 406: 108–112 [DOI] [PubMed] [Google Scholar]
- 13.Buraczynska M, Ksiazek P, Drop A, et al. Genetic polymorphisms of the renin-angiotensin system in end-stage renal disease. Nephrol Dial Transplant 2006; 21: 979–983 [DOI] [PubMed] [Google Scholar]
- 14.Campbell C, Fang B, Guo X, et al. Associations between genetic variants in the ACE, AGT, AGTR1 and AGTR2 genes and renal function in the Multi-ethnic Study of Atherosclerosis. Am J Nephrol 2010; 32: 156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad P, Tiwari A, Kumar K, et al. Chronic renal insufficiency among Asian Indians with type 2 diabetes: I. Role of RAAS gene polymorphisms. BMC Med Genet 2006; 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudnicki M, Mayer G. Significance of genetic polymorphisms of the renin-angiotensin-aldosterone system in cardiovascular and renal disease. Pharmacogenomics 2009; 10: 463–476 [DOI] [PubMed] [Google Scholar]
- 17.Tripathi G, Dharmani P, Khan F, et al. High prevalence of ACE DD genotype among north Indian end stage renal disease patients. BMC Nephrol 2006; 7: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong C, Kanetsky P, Raj D. Genetic polymorphisms of the RAS-cytokine pathway and chronic kidney disease. Pediatr Nephrol 2008; 23: 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldman HI, Appel LJ, Chertow GM, et al. The Chronic Renal Insufficiency Cohort (CRIC) Study: design and methods. J Am Soc Nephrol 2003; 14: S148–S153 [DOI] [PubMed] [Google Scholar]
- 20.Lash JP, Go AS, Appel LJ, et al. Chronic Renal Insufficiency Cohort (CRIC) Study: baseline characteristics and associations with kidney function. Clin J Am Soc Nephrol 2009; 4: 1302–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perloff D, Grim C, Flack J, et al. Human blood pressure determination by sphygmomanometry. Circulation 1993; 88: 2460–2470 [DOI] [PubMed] [Google Scholar]
- 22.Anderson AH, Yang W, Hsu C-Y, et al. Estimating GFR among participants in the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis 2012; 60: 250–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE 2008; 3: e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909 [DOI] [PubMed] [Google Scholar]
- 25.Ma L, Clark AG, Keinan A. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet 2013; 9: e1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaykin D, Zhivotovsky L, Westfall P, et al. Truncated product method for combining p-values. Genet Epidemiol 2002; 22: 170–185 [DOI] [PubMed] [Google Scholar]
- 27.Sheng X, Yang J. Truncated product methods for panel unit root tests. Oxf Bull Econ Stat 2013; 75: 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Zhu Y, Cole SA, et al. A gene-family analysis of 61 genetic variants in the nicotinic acetylcholine receptor genes for insulin resistance and type 2 diabetes in American Indians. Diabetes 2012; 61: 1888–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Zhu Y, Lee ET, et al. Joint associations of 61 genetic variants in the nicotinic acetylcholine receptor genes with subclinical atherosclerosis in American Indians: a gene-family analysis. Circ Cardiovasc Genet 2013; 6: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovati E, Richard A, Frey BM, et al. Genetic polymorphisms of the renin-angiotensin-aldosterone system in end-stage renal disease. Kidney Int 2001; 60: 46–54 [DOI] [PubMed] [Google Scholar]
- 31.Knoll A, Schunkert H, Reichwald K, et al. Human renin binding protein: complete genomic sequence and association of an intronic T/C polymorphism with the prorenin level in males. Hum Mol Genet 1997; 6: 1527–1534 [DOI] [PubMed] [Google Scholar]
- 32.Gu D, Kelly TN, Hixson JE, et al. Genetic polymorphisms in the renin-angiotensin-aldosterone system and salt-sensitivity of blood pressure. J Hypertens 2010; 28: 1210–1220 [PMC free article] [PubMed] [Google Scholar]
- 33.Donaghue M, Hsieh F, Baronas E, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res 2000; 87: e1–e9 [DOI] [PubMed] [Google Scholar]
- 34.Santos RA. Angiotensin-(1-7). Hypertension 2014; 63: 1138–1147 [DOI] [PubMed] [Google Scholar]
- 35.Yang M, Zhao J, Xing L, et al. The association between angiotensin-converting enzyme 2 polymorphisms and essential hypertension risk: a meta-analysis involving 14,122 patients. J Renin-Angiotensin-Aldosterone Syst 2014; doi:10.1177/1470320314549221 [DOI] [PubMed] [Google Scholar]
- 36.Malard L, Kakinami L, O'Loughlin J, et al. The association between the angiotensin-converting enzyme-2 gene and blood pressure in a cohort of adolescents. BMC Med Genet 2013; 14: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.