Introduction
Interest in and knowledge of the gut microbiome has increased exponentially in the past decade. An internet search using key words such as “gut microbiome” or “gut microbiota” generates between 1 and 2.2 million results. Once overlooked, this component of the gastrointestinal (GI) tract is now gaining appreciation for its importance in optimal health. The food industry has taken this to full heights and a plethora of food and supplement products touting “probiotic” or “fermented foods” populate store shelves; and advertisements for them can be found in magazines, the internet and television commercials. Never before has the discussion of one’s bowel habits and GI symptoms been more mainstream. Interestingly, this rapid growth has left both consumers and many clinicians confused as the data, while growing, still leave many unanswered questions.
Dating back to the early 1900’s Eli Metchnikoff, a Russian scientist of the Pasteur Institute in Paris, associated longevity of rural Bulgarians to their consumption of fermented milk products. He postulated that the lactic acid bacteria in the fermented milk products ingested by these peasants living in harsh climate and poverty provided an anti-aging effect which contributed to their greatly outliving wealthier Europeans. He named the organism “Lactobacillus bulgaricus.” As Metchnikoff researched, he hypothesized that seeding the gut with healthy bacteria by drinking fermented milk products could fight off harmful bacteria and prolong life. He was the first scientist to suggest that it was possible to modify the gut microbiome by replacing bad bacteria with good bacteria, and he earned a Nobel Prize in 1908 for his work in immunity. The discovery of penicillin in 1928 by Sir Alexander Fleming, a Scottish biologist, turned the attention of researchers away from using bacteria to assist with healing to that of using soil fungi derivatives to kill bacteria. During the golden age of antibiotic discovery from 1940–1960, most antibiotics were originally isolated by screening soil-derived actinomycetes (1). However as returns diminished on this discovery platform, new discoveries were based upon synthetic compounds.
However this target-focused screening process of large libraries also failed, partly due to the inability of these synthetic compounds to penetrate the bacterial envelope (1). Unfortunately the deceleration of antibiotic discovery is accompanied with the spread of resistant bacteria, and a major public health threat of untreatable infections. With advances in micro- and molecular biology techniques, the culture is poised to revisit past successes with bacterial therapy for gut health and immunity in order to develop future treatments.
One of several international efforts, the Human Microbiome Project (HMP) utilizes high through multi ‘omics analyses to identify and study the microbiome in human health (2,3). Funded by the NIH common fund in 2008, the HMP has resulted in the isolation and sequencing of over 1,300 reference strains thus far from the human body (2,3). The HMP Consortium has reported the structure and function of the human microbiome in 300 healthy adults (18–40 yrs) at 18 body sites from a single time point. This has led to an unprecedented amount of data about the complexity of the human microbiome allowing for a baseline for further research into the impacts of the microbiome on health and disease. As a precursor to the Human Microbiome Project, the Human Gut Microbiome Project has widened our appreciation for the bacterial ecosystem that resides within the human intestinal tract. This system is comprised of microorganisms such as bacteria, archaea, fungus and viruses that are distributed throughout the entire gastrointestinal tract (4). Ongoing investigations are revealing the importance of the gut microorganisms in exerting beneficial effects on human health.
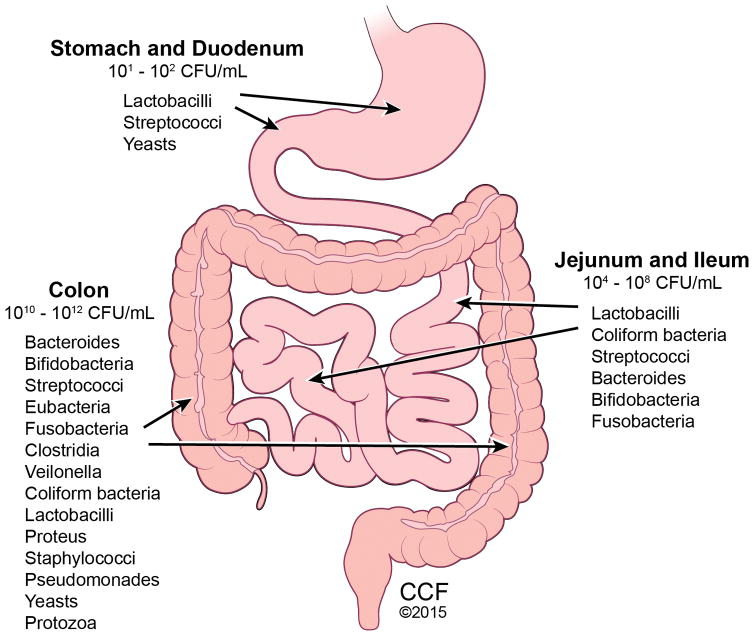
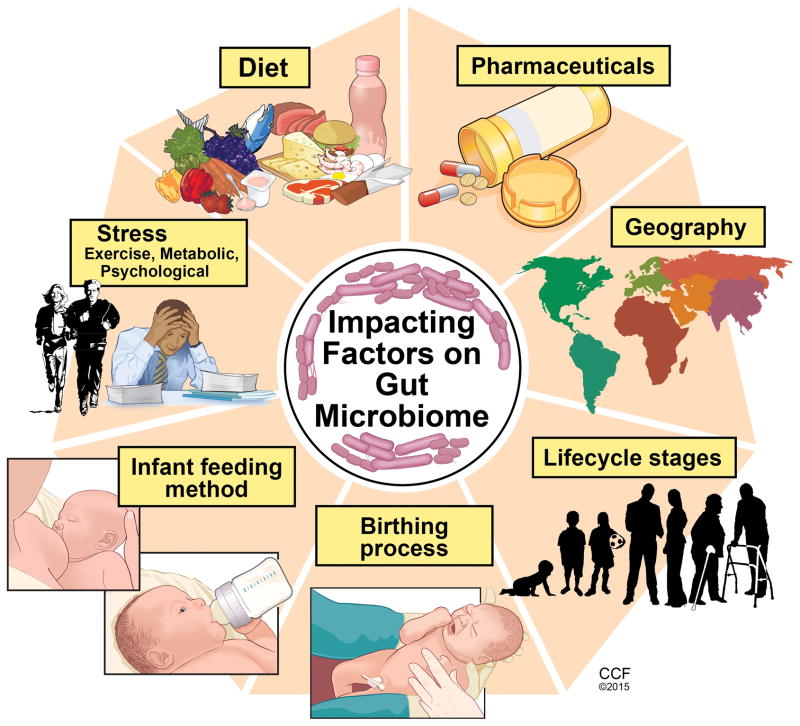
The typical healthy person is inhabited with trillions of microbes (Figure 1). But to confuse matters, two healthy people may have very different microbiomes. Analysis of HMP samples with lifestyle and history questionnaires has revealed associations between life-history characteristics and microbiome composition (5). Ding and Schloss analyzed a 16S rRNA gene sequence data set from the HMP Consortium. As a significant difficulty in analyzing microbiome data has been the considerable intra- and interpersonal variation in the composition of the human microbiome, this study took the approach of clustering samples into bins based on their taxonomic similarity. Using bacterial community structures collected from18 body sites and up to three time points, they applied community typing analysis to better understand the factors that affect the structure of the microbiome and contribute to human health. Using Dirichlet multinomial mixture models to partition the data into community types for each body site, their analysis of simulated data and the HMP data suggest that the community types represent clusters of relative abundance profiles. Three important observations were made with this approach. Firstly, there were strong associations between whether someone was breastfed, their gender and education level with their community types at several body sites. Secondly, the community types of the oral cavity and gut microbiome were predictive of each other even though the specific taxonomic compositions were different. Lastly, stability of community types was least stable in the oral cavity and most stable in the vagina and gut communities over the course of the sampling period. Therefore the authors concluded that despite substantial intra- and interpersonal variation in the human microbiome, by partitioning this variation intocommunity types, the types became predictive of each other and are likely the result of life-history characteristics (5). The gut microbiome is influenced by multiple factors including mode of infant delivery and feeding, aging process, diet composition, geography, medications, and stress (Figure 2). The following sections provide a brief overview of current knowledge regarding variances in the gut microbiota related to these factors.
Figure 1.
Figure 2.
Our first colonization
In utero
It was previously believed that the GI tract was sterile in utero, and that during the birthing process was when first colonization and initiation of the gut microbiome occurred (6). However, recent studies have challenged this belief by demonstrating the presence of microorganisms in the placenta, amniotic fluid and umbilical cord (7–9). This work is still preliminary but deserves further study as it highlights the importance of maternal gut microbiota health. It is hypothesized that by swallowing amniotic fluid and its bacteria in utero, the fetus begins to colonize the developing GI tract (10). The meconium, the first infant stool sample, has also been shown to contain microorganisms (11). Notably, the meconium from preterm infants shows a different microbiota composition from that seen in a sample acquired after the first week of life, demonstrating how normal gut colonization takes place throughout the course of fetal development (10). Absence of this progressive colonization could place preterm infants at risk for various gastrointestinal infections, including necrotizing enterocolitis (NEC) (10). As a means to mimic the natural inoculation of the gut, multiple studies have evaluated probiotic supplementation as a means to influence the incidence and severity of NEC. A recent Cochrane review, which included 24 randomized controlled trials (n=5529), assessed the role of probiotic supplementation in preterm neonates. The review confirmed previous findings that probiotic supplementation significantly reduces the risk of stage II or greater necrotizing enterocolitis and all cause mortality in this patient population (12). As probiotics are known to have potentially beneficial effects on gut function and maturity, Athalye-Jape et al conducted a meta-analysis that included 25 randomized controlled trials (n=5895) to determine if probiotic supplementation influenced the time to establish first full enteral feeds in preterm neonates (13). Overall, the subjects supplemented with probiotics took less time to achieve full feeds (mean difference: −1.54 d; 95% CI: −2.75, −0.32d; p=0.01; I2 = 93%). The type and/or number of probiotic strains (e.g., Bifidobacterium or non-Bifidobacterium) did not influence this improvement. Additionally, probiotic supplementation reduced hospital length of stay, feeding intolerance, and duration of indirect hyperbilirubinemia as well as increased weight gain and growth velocity. None of the trials reported adverse effects of probiotics on outcomes.
Delivery Mode
The birthing process exposes newborns to a wide range of microorganisms that also contribute to the colonization of the gut microbiome. The mode of delivery affects the composition of the infant’s gut microbiota, and interestingly, the gut microbiota of the newborn will closely resemble the microbiota with which they encountered during birth. During a vaginal birth, the infant is inoculated with vaginal microbiota, which differs from skin microbiota encountered during a caesarian section (14). At the phylum level, studies have shown that infants delivered vaginally have a larger population of Bacteroidetes than Firmicutes when compared to infants delivered via cesarean section (15).
Despite exposure in utero, the majority of microbes that will colonize the infant gut are acquired postpartum. The initial colonization pattern is thought to be chaotic, and increasing evidence suggests that environmental exposures early in life, including diet, are responsible for these variations (16). Gut colonization in an infant occurs in a succession of stages. Early on, the gut becomes colonized by primarily aerobic organisms, such as enterobacteria, staphylococci, and streptococci, many of which have the potential to be pathogenic. These early colonizers begin to change the gut environment, paving the way for colonization by an increasingly anaerobic community of microbes (17). The structure of the gut community continues to change over the first year of life and thereafter in response to external factors such as diet and antibiotic use (18, 19). Weaning, breastfeeding status, and the successive introduction of different types of food all correspondingly affect the infant gut microbiome and immune system (20).
Infant Feeding Method
Breast milk is the optimal food for infants as it meets all their nutritional and physiologic requirements. Human milk contains protein, fat and carbohydrate, as well as immunoglobulins and endocannabinoids (21). Breast milk is not sterile as it contains as many as 600 different species of bacteria including beneficial Bifidobacterium breve, B. adolescentis, B. longum, B. bifidum, and B. dentium (22). In addition to lactose, the carbohydrate component of human milk also contains oligosaccharides which comprise the third largest solid component (23). Human milk oligosaccharides are indigestible polymers formed by a small number of different monosacchrides that serve as prebiotics by selectively stimulating growth of members of the genus Bifidobacterium(23). Some of the beneficial effects of breast feeding over formula feeding are attributed to the effects of oligosaccharides on the beneficial bacteria as an increased proportion of Bifidobacteria is noted in breast-fed infants compared to those fed formula (24). Bifidobacteria have been linked with strengthening gut mucosal protection through activities against pathogens (25). Bifidobacteria have been shown to increase the production of immunoglobulin A which is correlated with modulation of the intestinal immune system (26).
Schwartz et al studied breast-fed and formula-fed infants and their mothers. Stool samples from each infant were collected, and microbial DNA was extracted and sequenced; and mRNA was isolated from stool containing host gut exfoliated epithelial cells and processed for microarray analysis (20). They found that the microbiome of breast-fed infants to be significantly enriched in genes associated with virulence functionality, and demonstrated a multivariate correlation between the gut flora genes associated with bacterial pathogenicity and the expression of host genes associated with immune and defense mechanisms. Interestingly, the operational taxonomic unit (OTU) composition and genetic potential of the microbiota differed between breast-fed and formula-fed infants. The researchers suggested their findings indicate human milk promotes the mutualistic crosstalk between the mucosal immune system and the microbiome in the maintenance of intestinal homeostasis. More research is needed to better determine the mechanisms by which Bifidobacteria produce these effects.
Aerotolerance of the intestinal microbiota also seems to differ between breast-fed and formula-fed infants. Aerobic organisms are more common in the feces of breast-fed infants, whereas anaerobic and facultatively anaerobic organisms, which preferentially use anaerobic glycolysis, are more frequently identified in feces of formula-fed infants (27). Bacteroides and Clostridia colonization differs between the two types of feeding, with breast-fed infants characterized with lower concentrations of both (28–30).
With a better understanding of the composition of human breast milk, developments of complex infant formulas have attempted to mimic its nutritional value making them an acceptable alternative for mothers unable to breast-feed. Infant formulas are not a perfect substitute for human milk since bioactive compounds contained in breast milk known to affect nutrient absorption and digestion, immune protection, and defense against potentially pathogenic microbes are lacking (21). Unfortunately it is difficult to mimic the actions of these bioactive compounds. While infants fed a formula enriched with oligosaccharides have been shown to harbor more Bididobacteria in the feces, more evidence is needed for affirmation that infant formulas designed to mimic breast milk are beneficial (31).
Changes in the Gut Microbiome with Age
Revealed by metagenomic analysis, gut microbiota composition transforms throughout early stages of human development and is influenced by the diet (32). As an infant’s diet comprises breast milk and formula, this is reflective in that the microbiome has minimal diversity and is enriched in genes to facilitate lactate utilization (33). A shift in the functional capacity to preferentially utilize plant-derived glycans occurs prior to the introduction of solid foods. Around 3-years of age, the bacterial composition resembles that of an adult and remains stable until old age when variability in community composition increases (34). In terms of ecological succession, the Bifidobacterium-dominated microbiota of the infant changes over time into the Bacteroidetes- and Firmicutes-dominated microbiota of the adult (35). This distribution remains fairly stable throughout adulthood in the absence of perturbations, such as long-term dietary changes or repeated antibiotic usage.
Declines in dentition, salivary function, digestion and intestinal transit time may affect the gut microbiota upon ageing (36). There are notable differences in the microbiota in elderly people compared to young adults, with relative proportions of Bacteroidetes predominating in the elderly compared to higher proportions of Firmicutes in young adults (37). The elderly are also noted to have significant decreases in Bifidobacteria, Bacteriodes, and Clostridium cluster IV (38). Variability varies greatly among individuals ranging from 3 to 92% for Bacteroidetes and 7 to 94% for Firmicutes (34). However the microbiota are less variable within individual subjects (34).
Alterations in gut microbiome are associated with health concerns pertaining to the elderly, such as frailty. A significant reduction in microbial diversity with reduced composition of lactobacilli, Bacteroides/Prevotella and Faecalibacterium prausnitzii, and increased proportions of Ruminococcus, Atopobium and Enterobacteriaceae was observed in people with high frailty scores (39). Claesson et al investigated the links between diet, environment, health and microbiota in 178 older people (>65 years) subjects and found an association between gut microbiome diversity and functional independence (39). Decreased microbial diversity was noted in individuals living in short- or long-term residential care compared to those living in the community, and this difference was associated with increased frailty, decreased diet diversity and health parameters as well as increased inflammatory markers (serum TNF-α, IL-6, IL-8 and C-reactive protein) (38). Dietary patterns in residential location correlated with separations based on microbiota composition with the most discriminating food types being vegetables, fruit and meats. Complete linkage clusterings revealed four dietary groupings: low fat/high fiber and moderate fat/high fiber included 98% of the community and day hospital subjects; and moderate fat/low fiber and high fat/low fiber included 83% of long-stay subjects (39).Significant associations between several health/frailty measurements were found with minimum variability amongst community dwellers, but within the long-stay subjects the most significant associations were related to functional independence, Barthel index (functional assessment) and nutrition, followed by blood pressure and calf circumference. The authors speculated that the later may be attributable to the influence of diet and/or the microbiota on muscle mass and sarcopenia and hence frailty.
Influences on the Gut Microbiome
Geography
Geographical location and ethnicity have been shown to be determinants of the diversity and overall composition of microbiota. A 2013 study performed by Prideaux (40) et al looked at Caucasian and Chinese subjects in the United States and Hong Kong and found that microbial composition differed between countries and between ethnicities within the same country. In an elegant study, Yatsunenko et al characterized bacterial species in fecal samples from 531 individuals comprised of healthy children and adults from the Amazonas of Venezuela, rural Malawi and US metropolitan areas and included mono- and dizygotic twins (41). All populations studied showed shared features of gut microbiome development during the first three years of life including age-associated changes in the genes involved in vitamin biosynthesis and metabolism. Phylogenetic composition of fecal microbiota was significantly altered between individuals living in the different countries, with pronounced separation occurring between the US and the Malawian and Amerindian gut communities; this was true for individuals aged 0–3 years, 3–17 years, and for adults. However there was also separation between the non-US populations. Bacterial diversity increased with age in all three populations with the microbiome of the US population having the least diversity. Microbiome datasets from breastfed children (n=110: (24 babies (0.6–5 months old), 60 children and adolescents (6 months to 17 years old) and 26 adults) were analyzed to determine which bacterial taxa changed monotonically with increasing age within and between the three sampled populations. In all babies 16S rRNA sequences mapped to members of the Bifidobacterium genus, and while Bifidobacteria dominated fecal communities during the first year of life, they proportionally declined during this period. Also noted were age and population-related changes in metabolism. A total of 476 enzymes were identified as being significantly different in US versus Malawian and Amerindian breastfed babies (p<.0.001). The most prominent differences involved pathways related to vitamin biosynthesis and carbohydrate metabolism. Malawian and Amerindian babies, but not adults, had higher representation of enzymes that were components of the vitamin B2 (riboflavin) biosynthetic pathway. Compared with adults, baby microbiomes were enriched in enzymes involved in the scavenging of glycans represented in breastmilk and the intestinal mucosa (mannans, sialylated glycans, galactose and fucosyloligosaccharides), with several of these genes significantly overrepresented in Amerindian and Malawian baby microbiomes compared with US. Interestingly, urease gene representation was significantly higher but decreased with age in Malawian and Amerindian baby microbiomes, as opposed to the United States, where it was low from infancy to adulthood. Urea, which comprises up to 15% of the nitrogen present in human breast milk, is broken down by urease to ammonia which can then be used for microbial biosynthesis of essential and nonessential amino acids. Urease also has a crucial role in nitrogen recycling, particularly when diets are deficient in protein making urease potentially advantageous to both the microbiome and host when dietary nitrogen supply is suboptimal (42). A typical US diet is rich in protein, whereas diets in Malawian and Amerindian populations are dominated by corn and cassava. The differences between US and Malawian/Amerindian microbiomes can be related to these differences in diet. The enzymes that were the most significantly enriched in US fecal microbiomes parallel differences observed in carnivorous versus herbivorous mammals (43). Interestingly, several enzymes involved in the degradation of amino acids were overrepresented in adult US fecal microbiomes including aspartate, proline, ornithine and lysine as were enzymes involved in the catabolism of simple sugars (glucose-6-phosphate dehydrogenase and 6-phosphofructokinase), sugar substitutes (L-iditol 2-dehydrogenase, which degrades sorbitol), and host glycans (α-mannosidase, β-mannosidase and α-fucosidase). By contrast Malawian and Amerindian microbiomes had an over-representation of α-amylase which participates in the degradation of starch reflecting their corn-rich diet. Perhaps reflecting a fat-rich diet, the US microbiomes had an over-representation of enzymes involved with vitamin biosynthesis (cobalamin, biotin and lipoic acid), in the metabolism of xenobiotics (phenylacetate CoA ligase), which participates in the metabolism of aromatic compounds, and mercury reductase, and in bile salt metabolism (choloylglycine hydrolase).
To obtain insights into commonalities and differences between gut microbiomes across different populations, Arumugam et al sequenced 22 European metagenomes from Danish, French, Italian and Spanish individuals that were selected for diversity and combined them with previous sequencing results from Japanese and Americans to total 39 individuals (44). They found that despite the vast number of species residing in the gut and their inter-individual variability, the microbiota composition can be classified into at least 3 distinct groups, or enterotypes. The enterotypes contain functional markers that correlate with individual features such as age and body mass index (BMI). For example, starch degradation enzymes such as glycosidases and glucan phosphorylases increase with age, which could be a reaction to decreased efficiency of host breakdown of dietary carbohydrates with age (45). Three marker modules, two of which were ATPase complexes, correlated strongly with the hosts’ BMI supporting the link found between the gut microbiota’s capacity for energy harvest and obesity in the host (46). The authors concluded that these functional markers might be utilizable for diagnostic and perhaps even prognostic tools for numerous human disorders, for instance colorectal cancer and obesity-linked co-morbidities such as metabolic syndrome, diabetes and cardiovascular pathologies.
Food Supply
Diet has emerged as one of the most relevant factors in influencing the gut microbiome. Significant and meaningful changes in the gut microbiota have been associated with dietary alterations, primarily consumption of dietary fiber from fruits, vegetables and other plants. A diet that is varied and complex is associated with a more diversified microbiome. Globally the microbiome composition is noted to be different amongst different populations and cultures. De Filippo et al compared the gut microbiota of children aged 1–6 years living in a village of rural Africa in an environment that still resembles that of Neolithic subsistence farmers with the gut microbiota of western European children (Florence, Italy) of the same age, with dietary habits and living conditions typical of the developed world (47). Traditionally the African children are breast-fed until the age of 2 after which their diet is predominantly vegetarian; low in fat and animal protein and rich in starch, plant polysaccharides and fiber. All food resources are completely produced locally, cultivated and harvested nearby the village. The dietary content of carbohydrate, fiber and non-animal protein is very high. Animal protein intake is low and consists of mainly chicken and termites during the rainy season. The European children were breast-fed for up to 1 y of age after which they ate a typical Western diet high in animal protein, sugar, starch, and fat and low in fiber. Average caloric intake differed between the two populations (African children: 1–2 y old, 672 kcal/d; 2–6 y old, 996 kcal/d; European children: 1–2 y old, 1,068 kcal/d; 2–6 y old, 1,512 kcal/d). Multiplex pyrosequencing of the 16S rRNA gene revealed that 94.2% of the sequences in all of the African and European samples were found to belong to the four most populated bacterial phyla, namely Actinobacteria, Bacteroidetes, Firmicutes,and Proteobacteria, in agreement with previous studies describing such phyla as those contributing to the majority of human gut microbiota (48). Relevant differences were found in the proportions of four phyla: Actinobacteria and Bacteroidetes were more represented in African than in European children’s microbiota (10.1% versus 6.7% and 57.7% versus 22.4%, respectively), whereas Firmicutes and Proteobacteria were more abundant in European than in African children (63.7% versus 27.3% and 6.7% versus 0.8%, respectively). Firmicutes were twice as abundant in the European children as evidenced by the different ratio between Firmicutes and Bacteroidetes (F/B ratio ± SD, 2.8 ± 0.06 in European and 0.47 ± 0.05 in African). While short-chain fatty acid (SCFA)-producing bacteria were found in both populations, bacteria (Xylanibacter, Prevotella, Butyrivibrio, and Treponema) which use xylane, xylose, and carboxymethylcellulose to produce high levels of SCFAs and have a protective role against gut inflammation (49) were found exclusively in the African children. Notably, the African children had higher levels of total SCFA than the European children, with 4 times higher levels of butyrate and propionate. The authors concluded a correlation exists between polysaccharide-degrading microbiota and the calories that the host can extract from his/her diet, potentially influencing the survival and fitness of the host, suggesting that the microbiome of the African children co-evolved with their diet to assist with energy harvest by producing high levels of SCFA.
Microbial enrichment has been associated with diets high in fruits, vegetables and fiber compared to a Western diet rich in fat, sugars and animal protein and depleted of fiber. Zimmer et al analyzed the fecal flora of a large group of healthy volunteers on a strict vegetarian or vegan diet with classical microbiological culture and compared them with age and gender matched subjects consuming an omnivorous diet (50). The faecal microbiota of vegetarian and vegan subjects showed significantly lower microbial counts of Bifidobacterium, Bacteroides, E. coli and Enterobacteriaceae species and lower stool pH compared with omnivores. Compared to an omnivore diet, a vegetarian/vegan diet is associated with a higher carbohydrate and fiber content in which the undigestible polysaccharides can be fermented into SCFA by the gut microbiota. Production of SCFA is associated with decreasing luminal pH. The fact that E. coli and Enterobacteriacea do not thrive in lower pH ranges (5.5–6.5) and that they prefer proteins as their energy source, may explain the lower counts in the subjects consuming a vegan/vegetarian diet. Depleted microbial biodiversity of the gut microbiota in people consuming a Western diet is associated with increasing incidence of obesity, coronary vascular disease, metabolic syndrome and certain malignancies. Detailed discussion of each of these associations is beyond the scope of this paper. At a minimum, the diversity of the gut microbiome may be a future biomarker of long-term consumption of a “healthy” versus “unhealthy” diet which may be linked to potential for disease development.
Stress
Stress is defined as an organism’s total response to environmental demands or pressures. There are several different types of stressors, such as acute or chronic, acute on chronic, or repetitive acute. Stress can be predictable and controllable as well as unpredictable and uncontrollable, mild or severe, and occur in or out of context (51). Both the perception of stress and the persistence of its consequences vary between people. Stress contributes to susceptibility to disease and disabilities, therefore represents a severe economic burden.
Psychological and physical stressors activate the hypothalamic-pituitary-adrenal (HPA) axis. This results in a series or hormonal responses including release of corticotrophin-releasing hormone which induces the release of adrenocorticotrophic hormone systemically with then stimulates glucocorticoid synthesis (cortisol) in the adrenal cortex (52). Additionally, catecholamines (noradrenaline and adrenaline) are also released following psychological and physical stressors. The GI tract, and more recently known, the gut microbiota, is sensitive to stress and stress mediators. Enteric bacteria respond to the release of stress-related neurochemical mediators by the host which can influence the response to a bacterial infection (53). Recent theory suggests that bacteria act as delivery vehicles for neuroactive compounds, and therefore can affect host physiology by providing neurochemicals (54).
Physiological Stress-Exercise
High intensity exercise is a physiological stressor that can lead to gastrointestinal distress. There are some reports that between 30–90% of distance runners have experienced intestinal problems related to exercise (55). The degree of intestinal distress can range from mild to severe, and symptoms include nausea, vomiting, abdominal angina, and bloody diarrhea. High intensity training has been associated with reduced GI blood flow, tissue hyperthermia and hypoxia leading to possible alterations of microbiota and gut barrier. Athletes often have different dietary pattern from non-athletes depending upon the type of activity they in which they participate. In a study of professional rugby players during a regulated environment of preseason training, Clarke et al demonstrated the impact of exercise and associated dietary changes on the gut microbiota (56). Enhanced gut microbial diversity was significantly higher and positively correlated with increased exercise and dietary protein intake in athletes as compared with size matched (high Body Mass Index – BMI ~30 kg/m2) and age/gender matched (lower BMI <25) non-athletic control groups, with fewer differences seen between the two control groups. Interestingly, when compared to controls, the athletes also exhibited lower inflammatory markers and improved metabolic markers. Athletes consumed more calories, with a higher percentage of protein, and grazed eating throughout the day compared to non-athletes. Microbiota diversity measures were positively correlated with protein intake and plasma creatine kinase levels, a marker of extreme exercise, suggesting that both diet and exercise were influencing changes in microbial diversity. The athlete’s increased microbial diversity was reflected by the presence of representatives of 22 phyla of bacteria in contrast to 11 and 9 phyla in the low and high BMI controls, respectively.
There are several potential mechanisms by which physical activity and fitness might modify the microbiota. Abrupt exercise produces multiple metabolites and inflammatory mediators whereas habitual exercise and fitness leads to suppression of basal inflammatory cytokines suggesting a regulatory loop between exercise biology and host immunity (57). Regular exercise has an anti-inflammatory effect which improves the immunological profile in type 2 diabetes mellitus, coronary artery disease, peripheral arterial disease and obesity (58–61). In animal models, repeated exercise results in reduced pro-inflammatory cytokine expression and increased anti-inflammatory IL-10 expression (62, 63). Regular exercise also decreases colonic oxidative insult in a rat model of colitis (64).
Contrary to the benefits of regular exercise, prolonged excessive exercise can negatively affect intestinal function. As high-intensity exercise can lead to prolonged intestinal hypo-perfusion, intestinal ischemia may result. Increased intestinal permeability can result making the gut susceptible to endotoxin translocation (65–67). Probiotic supplements, in conjunction with other dietary strategies, have been studied in athletes as a means to improve gut health, as characterized by decreased symptoms of nausea, cramping, bloating and diarrhea,as well as improved immunity. Primarily supplements containing Lactobacillus and/or Bifidobacterium species have been utilized. Products have been provided from 1 to 6 months, before and/or after exercise or a competition, and at varying doses (109–1012 CFU/day). Some studies report clinical outcomes of improved upper respiratory tract illness (64–69), gastrointestinal illness (70–72)), as well as immunological measures and outcomes (67, 68, 73–76)).
Physiological Stress-Critical Illness
As seen in high intensity exercise, intestinal hypoperfusion resulting from redistribution of splanchnic circulation severe enough to cause gut ischemia and mucosal injury, often occurs in the critically ill. In the critically ill, the gut has an important role in promoting infectious complications and multiple organ dysfunction syndrome (MODS). This is due to deteriorated intestinal epithelium, altered gut immune system, and dysfunctional metabolic activities of commensal bacteria (77). Shimizu et al, evaluated the gut microbiota and gut environment (fecal pH and presence of organic acids) in patients with systemic inflammatory response syndrome (SIRS) (78). In comparison to healthy controls, patients with severe SIRS had significantly lower total anaerobic bacterial counts (especially 2–4 log fewer commensal Bifidobacterium and Lactobacillus) and 2 log higher potentially pathogenic Staphylococcus and Pseudomonas group counts. Concentrations of total organic acids, in particular the short-chain fatty acids (SCFA) acetate, propionate and butyrate, were significantly decreased in the patients, whereas pH was markedly increased. This group of researchers further investigated the impact of fecal pH in critical illness in 138 trauma patients (79). Patients with acidic or alkaline feces were noted to have decreased Bacteroide and Bifidobacterium species. The incidence of bacteremia in patients with an acidic or alkaline fecal pH was significantly higher than those with a fecal pH in the normal range (P < 0.05 versus normal range). The incidence of both bacteremia and mortality were associated with an increased pH of 6.6. When the pH level was increased or decreased by one, the incidence of bacteremia more than tripled and mortality more than doubled. Total SCFA (propionate, butyrate) concentrations decreased with pH >6.6; lactic, succinic, and formic acids were increased in acidic feces which is notable as these are produced by Enterobacteriaceae (80). Whether these changes are a cause or consequence of SIRS is yet to be determined. While this study lends potential that fecal pH could be a risk factor marker, it does have limitations because these data were made with the use of culture-based interrogation of the microbiota and because the pH was not specified by gastrointestinal region. Alterations in the gut microbiome have been shown to occur within 6 hours of a metabolic insult and the microbiome fails to return to microbial patterns seen in healthy controls (81).
Numerous studies attempting to modify the gut microbiome and improve clinical outcomes in the critically ill by providing probiotics, prebiotics or synbiotics have been performed (82–89). Details of these studies are beyond the scope of this review. Methods of dosing, supplement duration and supplements utilized vary amongst studies making general recommendations difficult. Several meta-analyses and systematic reviews have been performed evaluating probiotics in the critically ill (89–91)). The choice of the studies included in the analysis will determine the overall outcome of the study. Generally, it seems probiotic supplementation in the critically ill favorably improves outcomes. However, the issue of heterogeneity and lack of good quality studies is a highlight of systematic reviews of probiotic use in critically ill patients. Knowing that unrecoverable gut dysbiosis occurs early after metabolic insult and knowing the potential mechanisms of action of certain probiotic strains, further rigorous study on probiotic provision in the critically ill deserves attention. Future studies should focus on 1) clarification of viable probiotic strain(s) provided, and their dosage, route and timing of delivery; 2) clear definitions for the outcomes attempting to modify (ventilator associated pneumonia, diarrhea); and 3) focus effect of probiotic supplementation on meaningful clinical endpoints (mortality, duration of mechanical ventilation, other infections, length of stay).
Psychological Stress
The discovery of hormonal regulation of digestion initiated the concept of the gut-brain axis (92). The interaction of psychological factors and altered gut physiology via the gut-brain axis, where brain and gut symptoms are reciprocally influencing each other’s expression, is a conceptual framework of functional GI disorders (irritable bowel syndrome, functional dyspepsia). Early life stressors (e.g., psychological, sexual and/or physical abuse) have been suggested as important contributors to the pathogenesis of functional GI disorders (93–96)). This is a crucial developmental period when the gut microbiota are diversifying, thus making it particularly vulnerable to these stressors.
Animal models of maternal separation, a model commonly used for early life stress, induces prolonged HPA-axis hyperactivity (97–103), anxiety-like behavior (100,,104–106), visceral hypersensitivity (107, 108–110), and altered cholinergic activity in the gut (111, 112), along with increased intestinal permeability (107, 113–116). This model results in a dysfunctional gut-brain axis and produces phenotypes found in irritable bowel syndrome patients (106, 116– 118). This animal model also yields gut dysbiosis in addition to other physiological and behavioral features of irritable bowel syndrome (118–120)). Various means to restore gut dysbiosis with probiotics have been investigated and appear to improve the negative stress-induced effects (107, 111, 120– 122).). Morphologic changes also result from maternal separation with increased goblet cells in the crypts of proximal colon with a subsequent increase in mucus secretion and a thinner mucosal layer (112). Thus changes in the gut microbiota in maternally separated animals could be resultant from changes to gut physiology (111– 114)) and morphology (112).
Although the exact mechanisms are unknown, data thus suggest that stress, whether acute or chronic, creates a dysbiotic gut microbiome which then may induce anxiety and depression (52). It seems that metabolites produced by the gut microbiota might modulate brain biochemistry and behavior (54, 123–125). Through regulation of the vagus nerve, behavior can be altered by the gut microbiota which then affects neurotransmitter metabolism (126, 127)) or other undefined pathways (128)). Thus futuristic therapy for patients with irritable bowel syndrome with co-morbid depression or anxiety may be targeted probiotic and/or synbiotic either alone or as an adjuvant to traditional therapy.
Pharmaceuticals
There is substantial variation in the pharmacokinetics of most commonly used drugs, and large differences in the responses of ill persons to therapeutics. The gut microbiota assists in the conversion of inactive therapeutics (e.g., prodrugs) and dietary bioactives into their active forms (129–131). For example, a medication prescribed for ulcerative colitis, sulfasalazine remains inactive until it reaches the distal gut. This prodrug consists of an ant-iinflammatory 5-aminosalicylic acid (5-ASA) molecule connected to a sulfapyridine molecule through an N-N double bond. Gut microbiota encoded azoreductases cleave the N-N double bond to release active 5-ASA (131). Likewise, foods such as fruits, vegetables, cereals and coffee contain conjugated hydroxycinnamates, antioxidant and anti-inflammatory compounds which are activated following microbial biotransformation (132). Therefore, a question arises whether variation in medication response amongst people is due to alterations in their gut microbiome?
The host possesses many protective mechanisms to protect against ingested pathogens including an acidic gastric environment, optimal bile flow, peristalsis and the gut microbiota. Current data suggest that the gut microbiota protects the host from pathogens by competing for binding sites, competing for requirements, and by direct inhibition through release of inhibitory molecules (133). When these protective mechanisms are disrupted, an imbalance in the gut microbiota can occur.
Gastric Acid Suppression
Several conditions such as intestinal dysmotility, altered GI anatomy, immune deficiencies, and hypochlorhydria (134) are predisposing factors for the development of small intestinal bacterial overgrowth (SIBO).
While hypochlorhydria can be a result of Helicobacter pylori colonization and aging, many people also take medications to reduce their gastric acidity for stress ulcer prophylaxis or gastric esophageal reflux disease. The pH of gastric acid in the stomach lumen is normally 1.5–3.5; maintaining intragastric pH above 3.5–5.0 prevents gastric mucosal injury and may be facilitated with histamine 2 receptor antagonists (H2RAs), sucralfate, and proton pump inhibitors (PPIs) (135). Previous studies show that PPIs are known to alter the GI bacterial population in 50% of patients on long-term treatment with any type of effective antisecretory drug (135, 136). Lombardo et al, conducted a study in 200 patients with gastroesophageal reflux disease (GERD) receiving PPI treatment, 200 patients with irritable bowel syndrome (IBS) not receiving PPI treatment, and 50 healthy control subjects not receiving PPI treatment greater than 10 years and showed that SIBO occurs more frequently in long-term PPI users than patients with IBS or control subjects (137). They found the incidence of SIBO was significantly different (P< .001) in patients using PPIs (50%) as compared to patients with IBS not using PPIs (24.5%), and healthy control subjects (6%). According to a multivariate analysis including 5,387 elderly subjects, the presence of diarrhea was significantly associated with use of PPIs (odds ratio [OR] 2.97, 95% confidence interval [CI] 2.03–4.35) and antibiotics (OR 4.58, 95% CI 1.95–10.73) (138). Theisen et al, found that the suppression of gastric acid with omeprazole led to a high prevalence of SIBO, leading to a markedly increased concentration of unconjugated bile acids (139). Conversely a retrospective chart review completed in 1,191 patients with glucose hydrogen breath testing (GHBT) evaluated whether this value positively correlated with patients on PPI therapy (134). Interestingly, GHBT was positively associated with older age and antidiarrheal use, but not with PPI use. Therefore, the authors concluded that PPI use was not associated with the presence of SIBO as measured by GHBT. However, Lombardo (140) challenged these results making a point that PPI use has become very common, both prescription and over-the-counter, and many patients may not consider non-prescription use as a medication and not disclose it to their physician. The authors were unable to confirm actual use of PPI due to the nature of a retrospective chart review. Additionally, as PPI duration is related to incidence of SIBO, the lack of knowledge regarding the duration the patients were taking a PPI leads one to speculate that if they only took it for a short time would it have an influence in GHBT results? Knowing the protective role gastric acidity has in regards to protecting the host against ingested pathogens, it is plausible that prolonged use of gastric acid suppressants contributes to the incidence of SIBO and that clinicians should be judicious in recommending their use.
Antibiotics
Antibiotic therapies not only target pathogenic microorganisms, but also the host-associated microbial communities in the gut. Most antibiotics have broad-spectrum activity so they can be used to treat many diseases. Thus, although antibiotics are designed to target pathogenic organisms, related members of the microbiota are also affected, leaving a lasting negative effect on the gut microbial community long after the antibiotics have ceased (141). Antibiotics can also promote the expansion of antibiotic-resistant strains which can act as a reservoir for resistance genes in the gut microenvironment (142). Decreased diversity in the microbiome typically follows antibiotic treatment, and even though most of the microbiota return to pretreatment levels, some members are lost from the community indefinitely (142). As microorganisms can be dependent upon other colonizers for the provision of nutrition, secondary metabolites or removal of toxic waste products, alterations in microbial co-dependence can leave detrimental effects (143). The antibiotic spectrum of activity will influence the shift in gut microbiota composition. Additionally, the administration dose of the antibiotic is also important in determining the ecological impact it will have on the microbiota. While the sub-therapeutic antibiotic dosing often used by the agriculture industry prophylactically and to promote animal growth does not reduce the total bacterial mass, this use is criticized as it shifts the composition of the microbiota and promotes dangerous levels of antibiotic resistance (144). Similarly in humans, antibiotic therapeutic doses are designed to minimize these effects, but despite these efforts, subsets of the microbiota can shift to that of increased colonization by opportunistic pathogens such as Clostridium difficile and Candida albicans. Noteworthy is that these effects of antibiotic delivery are not limited to oral delivery. Intravenously delivered antibiotics can have an effect on the gut microbiota as they become incorporated into bile and secreted into the intestine via the biliary system (144).
Gut microbial community alterations can result in dysregulation of host immune homeostasis and an increased susceptibility to disease. Host-microbial interactions are very specific and antibiotic therapy causes alterations or loss of highly co-evolved processes (145, 146). Antibiotic-induced changes that are important to microbial regulation of host immunity include loss of bacterial ligands that are recognized by the host, loss of specific bacterial signals, and alterations in the metabolites produced by the microbiota such as short-chain fatty acids (147). Short-chain fatty acids (SCFA) are beneficial for gut health as they serve as a primary fuel source for the colonocyte, are involved with water and electrolyte absorption, help to maintain the intestinal barrier, and modulate cell proliferation, differentiation, growth and apoptosis (147). Provision of butyrate during broad-spectrum antibiotic therapy mitigated the negative effects of antibiotics on gut health, notably the mRNA and protein expression for several anion exchangers, butyrate receptor and transports as well as protein expression of intestinal tight junction proteins (148).
Changes in gut immunity increase host susceptibility to infection by pathogens. For instance, treatment with metronidazole, an antibiotic that targets anaerobic bacteria, reduces the integrity of the mucus layer and accelerates mucosal attachment of C. rodentium (149). Changes in gut microbiota structure and function after antibiotic treatment create a metabolic environment that favors C. difficile germination and colonization and associated infectious diarrhea (150). While antibiotic therapy is the major risk factor for C. difficile colonization, the accepted treatment for C. difficile infection is further antibiotic therapy, initially with metronidazole and then vancomycin, if C. difficile persists (151). Repeated antibiotic use is often necessary in the treatment of recurrent Clostridium difficile infections which unfortunately causes further disruption of an imbalanced microbiota (152). Chang et al, identified that patients with recurrent C. difficile infection possessed a microbiota characterized by markedly reduced overall diversity compared to controls and to patients with an initial episode of the infection (152). C. difficile infection is debilitating to patients and extremely costly, with symptoms ranging from diarrhea to fulminant colitis, toxic megacolon and death (153). Fecal Microbiota Transplantation (FMT) is the process by which a homogenized stool sample from a healthy donor is administered into the GI tract of an individual. This therapy is now becoming a sought-out treatment to eradicate C. difficile colonization and correct gut dysbiosis. FMT has been shown to be a successful treatment option to eliminate recurrent and refractory C. difficile infection and to re-establish a healthy colony in certain patients (154). With its 91% primary and 98% secondary cure rates, FMT is gaining increased interest among researchers as a treatment option for other gastrointestinal disorders (154).
Perspective and future directions
With technological advances, knowledge regarding the gut microbiome has expanded from earlier beliefs (155, 156). The topics covered in this review are examples of what is currently understood regarding how the human gut microbiome is first established and how certain life encounters alter the composition, but they are by no means inclusive. The influence of the gut microbiome on health and disease is currently one of the most exciting areas of science. Learning more about how breaches upon the gut microbiome early in life coupled with further insults throughout life influences health may provide new insights for treatments of many challenging syndromes and diseases (e.g., fibromyalgia, irritable bowel disease, IBD, autism). We are just beginning to uncover the impact of how things we ingest can influence pathology. Future research should direct its focus to the gut microbiome as a biomarker for dietary intake and disease development, the effect of probiotic supplementation on clinical endpoints, and bacterial therapy for gut health, immunity and therapeutic treatments. New available and developing technologies to uncover the metabolon opens the path of exploration to this exciting frontier. The horizons are broad with exciting developments sure to come.
Supplementary Material
Contributor Information
Gail A. Cresci, Email: crescig@ccf.org, Department of Gastroenterology/Hepatology, Cleveland Clinic, Cleveland, OH 44195, Ph: 216-445-8317, Fax: 216-444-9401.
Emmy Bawden, Dietetic Intern, Cleveland Clinic, Cleveland, OH 44195.
References
- 1.Lewis K. Platforms for antibiotic discovery. Nature Reviews. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;86:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Nature. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–60. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salazar N, Arboleya S, Valdés L, et al. The human intestinal microbiome at extreme ages of life. Frontiers in Genetics. 2014;5:1–9. doi: 10.3389/fgene.2014.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiGiulio DB. Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med. 2012;17:2–11. doi: 10.1016/j.siny.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez E, Fernándes L, Marín M, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. 2005;51:270–274. doi: 10.1007/s00284-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 9.Aagard K, Ma J, Antony K, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237–265. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, Fernández L, Rodríguez JM, Jiménez E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first months of life. PLoS ONE. 2013;8:1–13. doi: 10.1371/journal.pone.0066986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jimenez E, Marín ML, Martín R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Alfaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolotis in preterm infants. The Cochrane Database Syst Rev. 2014;4:CD005496. doi: 10.1002/14651858.CD005496.pub4. [DOI] [PubMed] [Google Scholar]
- 13.Athalye-Jape G, Deshpande G, Rao S, Patole S. Benefits of probiotics on enteral nutrition in preterm neonates: a systematic review. Am J Clin Nutr. 2014;100:1508–19. doi: 10.3945/ajcn.114.092551. [DOI] [PubMed] [Google Scholar]
- 14.Clemente JC, Ursell LK, Parfrey LW, et al. The impact of the gut microbiota on human health: An integrated view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobsson HE, Abrahamsson TR, Jenmalm MC, et al. Decreased gut microbiota diversity, delayed Bacteriodetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 16.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pop M. We are what we eat: how the diet of infants affects their gut microbiome. Genome Biology. 2012;13:152. doi: 10.1186/gb-2012-13-4-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, Relman DA, Banfield JF. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci U S A. 2011;108:1128–1133. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S, Friedberg I, Ivanov I, Davidson LA, Goldsby JS, Dahl DB, et al. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biology. 2012;13:r32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Huerou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23:23–36. doi: 10.1017/S0954422410000065. [DOI] [PubMed] [Google Scholar]
- 22.Martín R, Jiménez E, Heilig H, Fernández L, Marín ML, Zoetendal EG, Rodríguez JM. Isolation of Bifidobacteria from breast milk and assessment of the bifidobacterial population by pcr-denaturing gradient gel electrophoresis and quantitative real-time pcr. Applied and Environmental Microbiology. 2009;75:965–969. doi: 10.1128/AEM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.German JB, Freeman SL, Lebrilla CB, Mills DA. Human Milk Oligosaccharides: Evolution, Structures and Bioselectivity as Substrates for Intestinal Bacteria. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:205–222. doi: 10.1159/000146322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balmer SE, Wharton BA. Diet and faecal flora in the newborn: breast milk and infant formula. Arch Dis Child. 1989;64:1672–7. doi: 10.1136/adc.64.12.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 26.Ouwehand A, Isolauri E, Salminen S. The role of the intestinal microflora for the development of the immune system in early childhood. Eur J Nutr. 2002;41(Suppl 1):I32– 7. doi: 10.1007/s00394-002-1105-4. [DOI] [PubMed] [Google Scholar]
- 27.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15:189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- 28.Harmsen HK, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–7. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Stark PL, Lee A, Parsonage BD. Colonization of the large bowel by Clostridium difficile in healthy infants: quantitative study. Infect Immun. 1982;35:895–9. doi: 10.1128/iai.35.3.895-899.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcobal A, Barboza M, Sonnenburg ED, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beereman-Wauters G, Staelens S, Van de Broek H, et al. Physiological and bifidogenic effects of prebiotic supplements in infant formulae. J Pediatr Gastroenterol Nutr. 2011;52:763–71. doi: 10.1097/MPG.0b013e3182139f39. [DOI] [PubMed] [Google Scholar]
- 32.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salazar N, Arboleya S, Valdés L, et al. The human intestinal microbiome at extreme ages of life. Frontiers in Genetics. 2014;5:1–9. doi: 10.3389/fgene.2014.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4586– 4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ottman N, Smidt H, de Vos WM, Belzer C. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol. 2012;2:104. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovat LG. Age related changes in gut physiology and nutritional status. Gut. 1996;38:306– 309. doi: 10.1136/gut.38.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwielehner J, Liszt K, Handschur M, Lassl C, Lapin A, Haslberger AG. Combined PCR-DGGE fingerprinting and quantitative PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp Gerontol. 2009;44:440–446. doi: 10.1016/j.exger.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Van Tongeren SP, Slaets JPJ, Harmsen HKM, Welling GW. Fecal microbiota composition and frailty. Appl Environ Microbiol. 2005;71:6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 40.Prideaux L, Kang S, Wagner J, et al. Impact of ethnicity, geography, and disease on the microbiota in health and inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:2906– 2918. doi: 10.1097/01.MIB.0000435759.05577.12. [DOI] [PubMed] [Google Scholar]
- 41.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meakins TS, Jackson AA. Salvage of exogenous urea nitrogen enhances nitrogen balance in normal men consuming marginally inadequate protein diets. Clin Sci (Lond) 1996;90:215–2. doi: 10.1042/cs0900215. [DOI] [PubMed] [Google Scholar]
- 43.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodmansey EJ. Intestinal bacteria and ageing. J Appl Microbiol. 2007;102:1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- 46.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 47.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JP, Massart S, Collini S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 50.Zimmer J, Lange B, Frick JS, Sauer H, Zimmermann K, Schwiertz A, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. European Journal of Clinical Nutrition. 2012;66:53–60. doi: 10.1038/ejcn.2011.141. [DOI] [PubMed] [Google Scholar]
- 51.Lucassen PJ, Pruessner J, Sousa N, Almeida OF, Van Dam AM, Rajkowska G, et al. Neuropathology of stress. Acta Neuropathol. 2014;127:109–135. doi: 10.1007/s00401-013-1223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Palma G, Collins SM, Bercik P, Verdu EF. The microbiota-gut-brain axis in gastrointestinal disorders: stressed bugs, stressed brain or both? J Physiol. 2014;14:2989– 2997. doi: 10.1113/jphysiol.2014.273995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyte M, Vulchanova L, Brown DR. Stress at the intestinal surface: catecholamines and mucosa-bacteria interactions. Cell Tissue Res. 2011;343:23–32. doi: 10.1007/s00441-010-1050-0. [DOI] [PubMed] [Google Scholar]
- 54.Lyte M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: microbial endocrinology in the design and use of probiotics. Bioessays. 2011;33:574–581. doi: 10.1002/bies.201100024. [DOI] [PubMed] [Google Scholar]
- 55.de Oliveira EP, Burini RC, Jeukendrup A. Gastrointestinal complaints during exercise: Prevalence, etiology, and nutritional recommendations. Sports Med. 2014;44(Suppl 1):S79– 85. doi: 10.1007/s40279-014-0153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clarke SF, Murphy EF, O’Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;64:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 57.O’Sullivan O, Cornin W, Clarke SF, Murphy EF, Molloy MG, Shanahan F, et al. Exercise and the microbiota. Gut Microbes. 2015;6(2):131–136. doi: 10.1080/19490976.2015.1011875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balducci S, Zanuso S, Nicolucci A, Fernando F, Cavalli S, Cardelli P, Fallucca S, et al. Anti- inflammatory effect of exercise training in subjects with type 2 diabetes and the metabolic syndrome is dependent on exercise modalities and independent of weight loss. Nutr Metab Cardiovasc Dis. 2010;20:608–17. doi: 10.1016/j.numecd.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 59.Lara Fernandes J, Serrano CV, Toledo F, Hunziker MM, Zamperini A, Teo FH, et al. Acute and chronic effects of exercise on inflammatory markers and B type natriuretic peptide in patients with coronary artery disease. Clin Res Cardiol. 2011;100:77–84. doi: 10.1007/s00392-010-0215-x. [DOI] [PubMed] [Google Scholar]
- 60.Tisi PV, Hulse M, Chulakadabba A, Gosling P, Shearman CP. Exercise training for intermittent claudication: does it adversely affect biochemical markers of exercise- induced inflammatory response? Eur J Vasc Endovasc Surg. 1997;14:344–50. doi: 10.1016/s1078-5884(97)80283-3. [DOI] [PubMed] [Google Scholar]
- 61.Ho SS, Dhaliwal SS, Hills AP, Pal S. Effects of chronic exercise training on inflammatory markers in Australian overweight and obese individuals in a randomized controlled trial. Inflammation. 2013;36:625–32. doi: 10.1007/s10753-012-9584-9. [DOI] [PubMed] [Google Scholar]
- 62.Hoffman-Goetz L, Spangnuolo PA, Guan J. Repeated exercise in mice alters expression of IL-10 and TNF-alpha in intestinal lymphocytes. Brain, Behav Immun. 2008;22:195–9. doi: 10.1016/j.bbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 63.Hoffman-Goetz L, Pervaiz N, Packer N, Guan J. Free-wheel training decreases pro- and increases anti-inflammatory cytokine expression in mouse intestinal lymphocytes. Brain, Behav Immun. 2010;24:1105–15. doi: 10.1016/j.bbi.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Kasimay O, Guzel E, Gemici A, Abdyli A, Sulovari A, Ercan F, Yegen BC. Colitis-induced oxidative damage of the colon and skeletal muscle is ameliorated by regular exercise in rats: the anxiolytic role of exercise. Exp Physiol. 2006;91:897–906. doi: 10.1113/expphysiol.2006.034439. [DOI] [PubMed] [Google Scholar]
- 65.Oktedalen O, Lunde OC, Opstad PK, Aabakken L, Kvernebo K. Changes in the gastrointestinal mucosa after long-distance running. Scand J Gastroenterol. 1992;27(4):270–4. doi: 10.3109/00365529209000073. [DOI] [PubMed] [Google Scholar]
- 66.Pals KL, Chang RT, Ryan AJ, Gisolfi CV. Effect of running intensity on intestinal permeability. J Appl Physiol. 1997;82(2):571–6. doi: 10.1152/jappl.1997.82.2.571. [DOI] [PubMed] [Google Scholar]
- 67.Brock-Utne JG, Gaffin SL, Wells MT, Gathiram P, Sohar E, James MF, Morrell DF, Normal RJ. Endotoxaemia in exhausted runners after a long-distance race. S Afr Med J. 1988;73:533–6. [PubMed] [Google Scholar]
- 68.Tiollier E, Chennaoui M, Gomez-Merino D, Drogou C, Filaire E, Guezennec CY. Effect of a probiotics supplementation on respiratory infections and immune and hormonal parameters during intense military training. Military Medicine. 2007;172:1006–1011. doi: 10.7205/milmed.172.9.1006. [DOI] [PubMed] [Google Scholar]
- 69.Gleeson M. Intense exercise training and immune function. Nestlé Nutrition Institute Workshop series. 2013;76:39–50. doi: 10.1159/000350254. [DOI] [PubMed] [Google Scholar]
- 70.Gleeson M, Bishop N, Oliveira M, McCauley T, Tauler P, Lawrence C. Effects of a Lactobacillus salivarius probiotic intervention on infection, cold symptom duration and severity, and mucosal immunity in endurance athletes. International Journal of Sports Nutrition and Exercise Metabolism. 2012;22:235–242. doi: 10.1123/ijsnem.22.4.235. [DOI] [PubMed] [Google Scholar]
- 71.Haywood B, Black K, Baker D, McGarvey J, Healey P, Brown R. Probiotic supplementation reduces the duration and incidence of infections but not severity in elite rugby union players. Journal of Science and Medicine in Sport. 2014;17:356–360. doi: 10.1016/j.jsams.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 72.Cox AJ, Pyne DB, Saunders PU, Fricker PA. Oral administration of the probiotic Lactobacilus fermentum VRI-003 and mucosal immunity in endurance athletes. British Journal of Sports Medicine. 2010;44:222–226. doi: 10.1136/bjsm.2007.044628. [DOI] [PubMed] [Google Scholar]
- 73.West N, Pyne D, Cripps A, Hopkins W, Eskesen D, Jairath A, et al. Lactobacillus fermentum (PCC®) supplementation and gastrointestinal and respiratory tract illness symptoms: A randomized control trial in athletes. Nutrition Journal. 2011;10:30. doi: 10.1186/1475-2891-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.West N, Horn P, Pyne D, Gebski V, Lahtinen S, Fricker P, Cripps A. Probiotic supplementation for respiratory and gastrointestinal illness symptoms in healthy physically active individuals. Clinical Nutrition. 2014;33:581–587. doi: 10.1016/j.clnu.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 75.Kekkonen RA, Vasankari TJ, Vuorimaa T, Haahtela T, Julkunen I, Korpela R. The effect of probiotics on respiratory infections and gastrointestinal symptoms during training in marathon runners. International Journal of Sports Nutrition and Exercise Metabolism. 2007;17:352–363. doi: 10.1123/ijsnem.17.4.352. [DOI] [PubMed] [Google Scholar]
- 76.Martarelli D, Verdenelli MC, Scuri S, Cocchioni M, Silvi S, Cecchini C, Pompei P. Effect of a probiotic intake on oxidant and antioxidant parameters in plasma of athletes during intense exercise training. Current Microbiology. 2011;62:1689–1696. doi: 10.1007/s00284-011-9915-3. [DOI] [PubMed] [Google Scholar]
- 77.Mittal R, Coopersmith CM. Redefining the gut as the motor of critical illness. Trends in Mol Med. 2014;20:214–223. doi: 10.1016/j.molmed.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu K, Ogura H, Goto M, et al. Altered gut flora and environment in patients with severe SIRS. J Trauma. 2006;60:126–133. doi: 10.1097/01.ta.0000197374.99755.fe. [DOI] [PubMed] [Google Scholar]
- 79.Shimizu K, Ogura H, Hamasaki T, Goto M, Tasaki O, Asahara T, Nomoto K, et al. Altered gut flora are associated with septic complications and death in critically ill patients with systemic inflammatory response syndrome. Dig Dis Sci. 2010;56:1171–1177. doi: 10.1007/s10620-010-1418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osuka A, Shimizu K, Ogura H, et al. Prognostic impact of fecal pH in critically ill patients. Critical Care. 2012;16:R119. doi: 10.1186/cc11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayakawa M, Asahara T, Henzan N, Murakami H, Yamamoto H, et al. Dramatic changes of the gut flora immediately after severe and sudden insults. Dig Dis Sci. 2011;56:2361–2365. doi: 10.1007/s10620-011-1649-3. [DOI] [PubMed] [Google Scholar]; Junko W. Carbohydrate fermentation in the colon. J Intest Microbiol. 2005;19:169–177. [Google Scholar]
- 82.Siempos II, Ntaidou TK, Falagas ME. Impact of the administration of probiotics on the incidence of ventilator-associated pneumonia: A meta-analysis of randomized controlled trials. Crit Care Med. 2010;38:954–962. doi: 10.1097/CCM.0b013e3181c8fe4b. [DOI] [PubMed] [Google Scholar]
- 83.Jain P, McNaught CE, Anderson ADG, MacFie J, Mitchell CJ. Influence of synbiotic containing Lactobacillus acidophilus La5, Bifidobacterium lactis Bb 12, Streptococcus thermophilis, Lactobacillus bulgaricus and oligofructose on gut barrier function and sepsis in crically ill patients: a randomized controlled trial. Clin Nutr. 2004;23:467–475. doi: 10.1016/j.clnu.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 84.Klarin B, Johansson ML, Molin G, Larsson A, Jeppsson B. Adhesion of the probiotic bacterium Lactobacillus plantarum 299v onto the gut mucosa in critically ill patients: a randomised open trial. Crit Care. 2005;9:R285–R93. doi: 10.1186/cc3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kotzampassi K, Giamarellos-Bourboulis EJ, Voudouris A, Kazamias P, Eleftheriadis E. Benefits of a synbiotic formula (Synbiotic 2000Forte) in critically ill trauma patients: early results of a randomized controlled trial. W J Surg. 2006;30:1848–55. doi: 10.1007/s00268-005-0653-1. [DOI] [PubMed] [Google Scholar]
- 86.McNaught C, Woodcock NP, Anderson ADG, MacFie J. A propsective randomized trial of probiotics in critically ill patients. Clin Nutr. 2005;24:211–9. doi: 10.1016/j.clnu.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 87.Besselink MG, van Santvoort HC, Buskens E, Boermeester MA, van Goor H, Timmerman HM, Nieuwenhuijs VB, Bollen TL, van Ramshorst B, Witteman BJ, Rosman C, Ploeg RJ, Boermeester MA, et al. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet. 2008 Feb 23;371(9613):651–9. doi: 10.1016/S0140-6736(08)60207-X. [DOI] [PubMed] [Google Scholar]
- 88.Spindler-Vesel A, Bengmark S, Vovk I, Cerovic O, Kompan L. Synbiotics, prebiotics, glutamine, or peptide in early enteral nturition: a randomized study in trauma patients. JPEN J Parenter Enteral Nutr. 2007;31:119–26. doi: 10.1177/0148607107031002119. [DOI] [PubMed] [Google Scholar]
- 89.Gu WJ, Wei CY, Yin RX. Lack of Efficacy of Probiotics in Preventing Ventilator-Associated Pneumonia: A Systematic Review and Meta-analysis of Randomized Controlled Trials. Chest. 2012;142(4):859–868. doi: 10.1378/chest.12-0679. [DOI] [PubMed] [Google Scholar]
- 90.Petrof EO, Dhaliwal R, Manzanares W, Johnstone J, Cook D, Heyland DK. Probiotics in the critically ill: a systematic review of the randomized trial evidence. Crit Care Med. 2012;40:3290–302. doi: 10.1097/CCM.0b013e318260cc33. [DOI] [PubMed] [Google Scholar]
- 91.Watkinson PJ, Barber VS, Dark P, et al. The use of pre-pro-and synbiotics in adult intensive care unit patients: Systematic review. Clin Nutr. 2007;26:182–192. doi: 10.1016/j.clnu.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 92.Track NS. The gastrointestinal endocrine system. Can Med Assoc J. 1980;122:287–292. [PMC free article] [PubMed] [Google Scholar]
- 93.Heitkemper MM, Cain KC, Burr RL, Jun SE, Jarrett ME. Is childhood abuse or neglect associated with symptom reports and physiological measures in women with irritable bowel syndrome? Biol Res Nurs. 2011;13:399–408. doi: 10.1177/1099800410393274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu JC. Physchological co-morbidity in functional gastrointestinal disorders: epidemiology, mechanisms, and management. J Neurogastroenterol Motil. 2012;18:13–18. doi: 10.5056/jnm.2012.18.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Tilburg MA, Palsson OS, Whitehead WE. Which psychological factors exacerbate irritable bowel syndrome? Development of a comprehensive model. J Psychosom Res. 2013;74:486–492. doi: 10.1016/j.jpsychores.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ladd CO, Huot RL, Thrivikraman KV, Nemeroff CB, Meaney MJ, Plotsky PM. Long-term behavioral and neuroendocrine adaptations to adverse early experience. Prog Brain Res. 2000;122:81–103. doi: 10.1016/s0079-6123(08)62132-9. [DOI] [PubMed] [Google Scholar]
- 97.Barreau F, Ferrier L, Fioramonti J, Bueno L. Neonatal maternal deprivation triggers long term alterations in colonic epithelial barrier and mucosal immunity in rats. Gut. 2004;53:501–506. doi: 10.1136/gut.2003.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Daniels WM, Pietersen CY, Carstens ME, Stein DJ. Maternal separation in rats leads to anxiety-like behavior and a blunted ACTH response and altered neurotransmitter levels in response to a subsequent stressor. Metab Brain Dis. 2004;19:3–14. doi: 10.1023/b:mebr.0000027412.19664.b3. [DOI] [PubMed] [Google Scholar]
- 99.Lippmann M, Bress A, Nemeroff CB, Plotsky PM, Monteggia LM. Long-term behavioral and molecular alterations associated with maternal separation in rats. Eur J Neurosci. 2007;25:3091–3098. doi: 10.1111/j.1460-9568.2007.05522.x. [DOI] [PubMed] [Google Scholar]
- 100.Aisa B, Tordera R, Lasheras B, Del Río J, Ramírez MJ. Effects of maternal separation on hypothalamic–pituitary–adrenal responses, cognition and vulnerability to stress in adult female rats. Neuroscience. 2008;154:1218–1226. doi: 10.1016/j.neuroscience.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 101.Gareau MG, Silva MA, Perdue MH. Pathophysiological mechanisms of stress-induced intestinal damage. Curr Mol Med. 2008;8:274–281. doi: 10.2174/156652408784533760. [DOI] [PubMed] [Google Scholar]
- 102.Oines E, Murison R, Mrdalj J, Grønli J, Milde AM. Neonatal maternal separation in male rats increases intestinal permeability and affects behavior after chronic social stress. Physiol Behav. 2012;105:1058–1066. doi: 10.1016/j.physbeh.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 103.Varghese AK, Verdu EF, Bercik P, Khan WI, Blennerhassett PA, Szechtman H, Collins SM. Antidepressants attenuate increased susceptibility to colitis in a murine model of depression. Gastroenterology. 2006;130:1743–1753. doi: 10.1053/j.gastro.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 104.Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience. 2010;170:1179–1188. doi: 10.1016/j.neuroscience.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 105.O’Mahony SM, Hyland NP, Dinan TG, Cryan JF. Maternal separation as a model of brain gut axis dysfunction. Psychopharmacology (Berl) 2011;214:71–88. doi: 10.1007/s00213-010-2010-9. [DOI] [PubMed] [Google Scholar]
- 106.Li M, Xue X, Shao S, Shao F, Wang W. Cognitive, emotional and neurochemical effects of repeated maternal separation in adolescent rats. Brain Res. 2013;1518:82–90. doi: 10.1016/j.brainres.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 107.Eutamene H, Lamine F, Chabo C, Theodorou V, Rochat F, Bergonzelli GE, Corthesy-Theulaz I, Fioramonti J, Bueno L. Synergy between Lactobacillus paracasei and its bacterial products to counteract stress-induced gut permeability and sensitivity increase in rats. J Nutr. 2007;137:1901–1907. doi: 10.1093/jn/137.8.1901. [DOI] [PubMed] [Google Scholar]
- 108.Felice VD, Gibney SM, Gosselin RD, Dinan TG, O’Mahony SM, Cryan JF. Differential activation of the prefrontal cortex following psychological stress and colorectal distension in the maternally separated rat. Neuroscience. 2014;267:252–262. doi: 10.1016/j.neuroscience.2014.01.064. [DOI] [PubMed] [Google Scholar]
- 109.Moloney RD, O’Leary OF, Felice D, Bettler B, Dinan TG, Cryan JF. Early-life stress induces visceral hypersensitivity in mice. Neurosci Lett. 2012;512:99–102. doi: 10.1016/j.neulet.2012.01.066. [DOI] [PubMed] [Google Scholar]
- 110.O’Malley D, Julio-Pieper M, Gibney SM, Dinan TG, Cryan JF. Distinct alterations in colonic morphology and physiology in two rat models of enhanced stress-induced anxiety and depression-like behavior. Stress. 2010;13:114–122. doi: 10.3109/10253890903067418. [DOI] [PubMed] [Google Scholar]
- 111.Gareau MG, Jury J, MacQueen G, Sherman PM, Perdue MH. Probiotic treatment of rat pups normalizes corticosterone release and ameliorates colonic dysfunction induced by maternal separation. Gut. 2010;56:1522–1528. doi: 10.1136/gut.2006.117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Soderholm JD, Yates DA, Gareau MG, Yang PC, Macqueen G, Perdue MH. Neonatal maternal separation predisposes adult rats to colonic barrier dysfunction in response to mild stress. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1257–G1263. doi: 10.1152/ajpgi.00314.2002. [DOI] [PubMed] [Google Scholar]
- 113.Barreau F, Cartier C, Ferrier L, Fioramonti J, Bueno L. Nerve growth factor mediates alterations of colonic sensitivity and mucosal barrier induced by neonatal stress in rats. Gastroenterology. 2004;127:524–534. doi: 10.1053/j.gastro.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 114.Gareau MG, Jury J, Perdue MH. Neonatal maternal separation of rat pups results in abnormal cholinergic regulation of epithelial permeability. Am J Physiol Gastrointest Liver Physiol. 2007;293:G198–G203. doi: 10.1152/ajpgi.00392.2006. [DOI] [PubMed] [Google Scholar]
- 115.Oines E, Murison R, Mrdalj J, Grønli J, Milde AM. Neonatal maternal separation in male rats increases intestinal permeability and affects behavior after chronic social stress. Physiol Behav. 2012;105:1058–1066. doi: 10.1016/j.physbeh.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 116.Barreau F, Ferrier L, Fioramonti J, Bueno L. New insights in the etiology and pathophysiology of irritable bowel syndrome: contribution of neonatal stress models. Pediatr Res. 2007;62:240–245. doi: 10.1203/PDR.0b013e3180db2949. [DOI] [PubMed] [Google Scholar]
- 117.O’Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EM, Cryan JF, Dinan TG. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65:263–267. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 118.Barouei J, Moussavi M, Hodgson DM. Effect of maternal probiotic intervention on HPA axis, immunity and gut microbiota in a rat model of irritable bowel syndrome. PLoS One. 2012;7:e46051. doi: 10.1371/journal.pone.0046051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garcıa-Rodenas CL, Bergonzelli GE, Nutten S, Schumann A, Cherbut C, Turini M, Ornstein K, Rochat F, Corthésy-Theulaz I. Nutritional approach to restore impaired intestinal barrier function and growth after neonatal stress in rats. J Pediatr Gastroenterol Nutr. 2006;43:16–24. doi: 10.1097/01.mpg.0000226376.95623.9f. [DOI] [PubMed] [Google Scholar]
- 120.Eutamene H, Bueno L. Role of probiotics in correcting abnormalities of colonic flora induced by stress. Gut. 2007;56:1495–1497. doi: 10.1136/gut.2007.124040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Distrutti E, Cipriani S, Mencarelli A, Renga B, Fiorucci S. Probiotics VSL#3 protect against development ofvisceral pain in murine model of irritable bowel syndrome. PLoSOne. 2013;8:e63893. doi: 10.1371/journal.pone.0063893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barrett E, Ross RP, O’Toole PW, Fitzgerald GF, Stanton C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 124.Barrett E, Fitzgerald P, Dinan TG, Cryan JF, Ross RP, Quigley EM, Shanahan F, Kiely B, Fitzgerald GF, O’Toole PW, Stanton C. Bifidobacterium breve with α-linolenic acid and linoleic acid alters fatty acid metabolism in the maternal separation model of irritable bowel syndrome. PLoS One. 2012;7:e48159. doi: 10.1371/journal.pone.0048159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bercik P, Park AJ, Sinclair D, Khoshdel A, Lu J, Huang X, Deng Y, Blennerhassett PA, Fahnestock M, Moine D, Berger B, Huizinga JD, Kunze W, McLean PG, Bergonzelli GE, Collins SM, Verdu EF. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut –brain communication. Neurogastroenterol Motil. 2011;23:1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bravo JA, Dinan TG, Cryan JF. Alterations in the central CRF system of two different rat models of comorbid depression and functional gastrointestinal disorders. Int J Neuropsychopharmacol. 2011;14:666–683. doi: 10.1017/S1461145710000994. [DOI] [PubMed] [Google Scholar]
- 127.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. 609e.1–3. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 128.Bercik P, Verdu EF, Foster JA, Macri J, Potter M, Huang X, Malinowski P, Jackson W, Blennerhassett P, Neufeld KA, Lu J, Khan WI, Corthesy-Theulaz I, Cherbut C, Bergonzelli GE, Collins SM. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology. 2010;139:2102–2112. doi: 10.1053/j.gastro.2010.06.063. [DOI] [PubMed] [Google Scholar]
- 129.Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest. 2014;124:4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 131.Peppercorn MA, Goldman P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharm Exp Ther. 1972;181:555–562. [PubMed] [Google Scholar]
- 132.Laparra JM, Sanz Y. Interactions of gut mcirobiota with functional food components and nutraceuticals. Pharmacol Res. 2010;61:219–225. doi: 10.1016/j.phrs.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 133.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 134.Rataupli SK, Ellington TG, O’Neill MT, Umar SB, Harris LA, Foxx-Orenstein AE, et al. Proton pump inhibitor therapy use does not predispose to small intestinal bacterial overgrowth. Am J Gastroenterol. 2012;107:730–735. doi: 10.1038/ajg.2012.4. [DOI] [PubMed] [Google Scholar]
- 135.Alhazzani W, Alenezi F, Jaeschke RZ, Moayyedi P, Cook DJ. 2 Proton Pump Inhibitors Versus Histamine 2 Receptor Antagonists for Stress Ulcer Prophylaxis in Critically Ill Patients: A Systematic Review and Meta-Analysis. Crit Care Med. 2013;41:693–705. doi: 10.1097/CCM.0b013e3182758734. [DOI] [PubMed] [Google Scholar]
- 136.Sanduleanu S, Jonkers D, De Bruine A, Hameeteman W, Stockbrugger RW. Non- Helicobacter pylori bacterial flora during acid-suppressive therapy: differential findings in gastric juice and gastric mucosa. Aliment Pharmacol Ther. 2001;15:379–388. doi: 10.1046/j.1365-2036.2001.00888.x. [DOI] [PubMed] [Google Scholar]
- 137.Lombardo L, Foti M, Ruggia O, Chiecchio A. Increased incidence of small intestinal bacterial overgrowth during proton pump inhibitor therapy. Clin Gastroenterol and Hepat. 2010;8:504–508. doi: 10.1016/j.cgh.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 138.Pilotto A, Franceschi M, Vitale D, Zaninelli A, Di Mario F, Seripa D, Rengo F. The Prevalence of Diarrhea and Its Association With Drug Use in Elderly Outpatients: A Multicenter Study. The American Journal of Gastro. 2008;103:2816–2823. doi: 10.1111/j.1572-0241.2008.02107.x. [DOI] [PubMed] [Google Scholar]
- 139.Theisen J, Nehra D, Citron D, et al. Suppression of gastric acid secretion in patients with GERD results in gastric bacterial overgrowth and deconjugation of bile acids. J Gastrointest Surg. 2000;4:50–4. doi: 10.1016/s1091-255x(00)80032-3. [DOI] [PubMed] [Google Scholar]
- 140.Lombardo L. PPI Use and SIBO: Predisposition or Cause? Am J Gastroenterol. 2012;107:1923. doi: 10.1038/ajg.2012.257. [DOI] [PubMed] [Google Scholar]
- 141.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 142.Lofmark S, Jernberg C, Jansson JK, Edlund C. Clindamycin-induced enrichment and long- term persistence of resistant Bacteroides spp. And resistance genes. J Antimicrob Chemother. 2006;58:1160–1167. doi: 10.1093/jac/dkl420. [DOI] [PubMed] [Google Scholar]
- 143.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Smith DL, Harris AD, Johnson JA, Silbergeld EK, Morris JG., Jr Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc Natl Acad Sci USA. 2002;99:6434–6439. doi: 10.1073/pnas.082188899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nature Reviews. 2011;9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 146.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–64. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 148.Cresci G, Nagy LE, Ganapathy V. Lactobacillus GG and tributyrin supplementation reduce antibiotic-induced intestinal injury. JPEN J Parenter Enteral Nutr. 2013;37:763–74. doi: 10.1177/0148607113486809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–45. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Theriot CM, Koenigsknecht MJ, Carlson PE, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature Comm. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hamilton MJ, Weingarden AR, Sadowsky MJ, et al. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:761–767. doi: 10.1038/ajg.2011.482. [DOI] [PubMed] [Google Scholar]
- 152.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–8. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 153.Brandt LJ, Aroniadis OC, Mellow M, et al. Long-Term Follow-Up of Colonoscopic Fecal Microbiota Transplant for Recurrent Clostridium difficile Infection. Am J Gastroenterol. 2012;107:1079–1087. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- 154.Wage ZK, Yang YS, Chen Ye, et al. Intestinal microbiota pathogenesis and fecal microbiota transplantation for inflammatory bowel disease. World J Gastroenteral. 2014;20:14805–14820. doi: 10.3748/wjg.v20.i40.14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ouwehand A, Isolauri E, Salminen The role of the intestinal microflora for the development of the immune system in early childhood. Eur J Nutr. 2002;41(Suppl 1):I32–I37. doi: 10.1007/s00394-002-1105-4. [DOI] [PubMed] [Google Scholar]
- 156.Young VB, Schmidt TM. Overview of the gastrointestinal microbiota. Adv Exp Med Biol. 2008;635:29–40. doi: 10.1007/978-0-387-09550-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.