Abstract
Cell-cell communication between cardiac and vascular cells and from stem and progenitor cells to differentiated cardiovascular cells is both an important and complex process, achieved through a diversity of mechanisms that have an impact on cardiovascular biology, disease and therapeutics. In recent years, evidence has accumulated suggesting that extracellular vesicles (EVs) are a new system of intercellular communication. EVs of different sizes are produced via different biogenesis pathways and have been shown to be released and taken up by most of known cell types, including heart and vascular cells, and stem and progenitor cells. This review will focus on exosomes, the smallest EVs (up to 100 nm in diameter) identified so far. Cells can package cargoes consisting of selective lipids, proteins and RNA in exosomes and such cargoes can be shipped to recipient cells, inducing expressional and functional changes. This review focuses on exosomes and microRNAs in the context of cardiovascular disease and repair. We will describe exosome biogenesis and cargo formation and discuss the available information on in vitro and in vivo exosomes-based cell-to-cell communication relevant to cardiovascular science. The methods used in exosome research will be also described. Finally, we will address the promise of exosomes as clinical biomarkers and their impact as a biomedical tool in stem cell-based cardiovascular therapeutics.
Keywords: exosomes, microRNAs, stem cells, angiogenesis, ischemia, ischemia/reperfusion injury, cardiac protection
Graphical Abstract
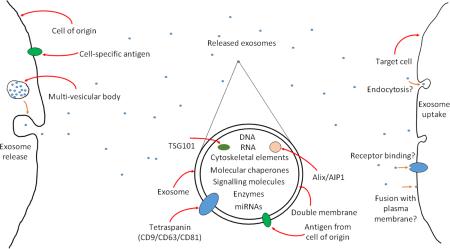
An introduction to exosomes. Exosomes are released from cells via multi-vesicular bodies. They have a double membrane and carry markers of their cell of origin. They also carry markers of their endosomal pathway of origin, for example tetraspanins, ALG-2 interacting protein X (Alix)/ALG-2 interacting protein 1 (AIP1) and tumour susceptibility 101 (TSG101). They are able to carry a cargo, which can include DNA, RNA cytoskeletal elements, molecular chaperones, signalling molecules, enzymes and microRNAs. Exosomes are currently of considerable interest as they are now known to participate in a novel form of cellular signalling by interacting with target cells, including transporting microRNAs from one cell to another. The exact methods by which exosomes interact with target cells is not fully understood – they may be taken up by endocytosis, by fusion with the plasma membrane, or bind to receptors on the cell surface, triggering intracellular signalling pathways.
1. Introduction
It has been known for some considerable time that when cells undergo apoptosis small vesicles, known as apoptotic bodies, form [1]. It was initially thought that all particles smaller than 4μm fell into this category and were, essentially, debris that was just a means of packaging the remnants of the dead cells in a way that would not cause any collateral damage to other cells in the vicinity, a form of “rubbish collection” [2]. However, evidence has emerged that the smallest of these extracellular vesicles (EVs), generally those in the size range of 30nm to 1μm, are not necessarily released during cell death and have a biological function. Exosomes are some of the smallest of these EVs and are often described as having a size of the order of 30-100nm, while microparticles (MPs) are generally between 100 and 1μm. These size ranges, however, are not considered absolute [3]. The mechanism of release of these different particles is also different, in that exosomes are produced through the endosomal pathway, whereas MPs are released through budding from the cell membrane. Exosomes carry on their surface some of the cell surface markers of their cell of origin and evidence is mounting that they are able to interact with the cell surface receptors on neighbouring and possibly also distant cells, in an almost hormonal fashion [4]. In addition, the vesicular nature of exosomes means that they are able to carry a cargo, which includes proteins[5], messenger RNAs (mRNAs) and microRNAs (miRNAs) [6], and to transfer these cargos to recipient cells, thus contributing to cell-to-cell communication. The EV cargo might considerably vary in function of the producing cell type and its “health status”, thus producing very different functional results in the incorporating cells.
This review will focus on exosomes and exosomal miRNAs and discuss their roles in cardiovascular protection and regeneration.
2. Exosome Biogenesis, Release and Uptake
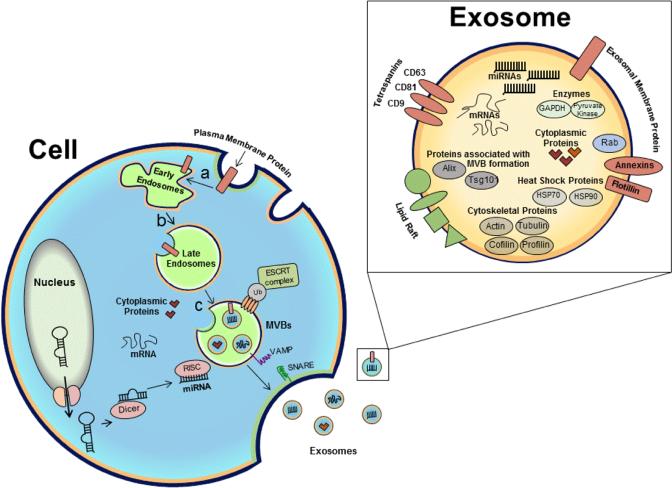
Exosomes are a subtype of membrane vesicles released from the endocytic compartment of live cells. Exosome biogenesis is exemplified in Figure 1. The endocytic vesicles originate from highly dynamic membrane compartments involved in internalization of extracellular ligands following the invagination of plasma membrane. Surface proteins found on the plasma membrane may be transferred to the inner membrane (towards the lumen) of these endocytic vesicles during the process. Inward budding of the limiting membrane of the endocytic vesicles generates intraluminal vesicles or exosomes with cytoplasmic components within them. Accumulation of exosomes in the lumens of endosomes results in multivesicular bodies (MVBs). The sorting mechanisms present on the membrane of MVBs ensure that cargoes are selectively loaded into the exosomes [7]. Exosomes from many cell types may contain similar surface proteins to the cell from which they are derived [8, 9]. The exosomal membranes contain lipid raft microdomains, which are enriched in cholesterol, sphingomyelin, glycolipids and ceramide. Phosphatidylserine is externalized on some exosomes membrane [10]. Exosomes retain the membrane typology of the parent cell. The cytosolic side of the lipid bilayer remains inside the vesicle, isolating signalling domains of membrane receptors from the cytosol, restricting their function and dampening signal transduction, while the luminal part is exposed. Different mechanisms are known to exist in the specific sorting of proteins into exosomes, including ESCRT (endosomal sorting complexes required for transport), tetraspanins and lipid-dependent mechanisms. The ESCRT complex is composed of several subcomplexes (0, I and III) that function in concert to produce intraluminal vesicles that bud into multivesicular bodies and sort monoubiquitinated proteins into them. Tetraspanins such as CD81, CD9 and CD63 play a key role in the composition of ESCRT-independent loading into exosomes. Through their interaction with other transmembrane proteins, cytosolic proteins and lipids, tetraspanins organize the membrane into tetraspanin-enriched domains. The existence of ESCRT-dependent and - independent mechanisms for loading of proteins into exosomes indicates the presence of heterogeneous populations of MVBs and exosomes. It has been shown that breast cancer cells secrete several types of exosomes, which differ in size and miRNA composition [11]. Additionally, B-cell lymphoma [9] and colon carcinoma cells [12] are shown to release different subpopulations of exosomes which are segregated based on their different surface antigens that may have different protein content and function. The existence of different populations of exosomes released from apical and basolateral cell surfaces not only demonstrates the heterogeneity of vesicles but also strongly suggests the existence of very specialized mechanisms to control the selective sorting of cargo into these vesicles.
Figure 1. Exosome Biogenesis and Cargo formation.
Early endosomes form by the fusion of small vesicles that originate from invaginations of the plasma membrane. Plasma membrane proteins may also be transferred to the surfaces of the small vesicles. Early endosomes mature into late endosomes by changes in their protein composition and acidification (a, b). Inward budding of the limiting membrane of endocytic vesicles generates exosomes with cytoplasmic content within them. Accumulation of exosomes within the late endosome results in a multivesicular body (MVB) (c). MiRNAs originate from the nucleus as pre-miRNAs, processed by Dicer and RISC complex in the cytoplasm and selectively loaded into exosomes. Sorting mechanisms including ESCRT and tetraspanins present on the membrane of MVBs ensure the selectivity of the exosomal cargo. SNARE and VAMP proteins either by direct or indirect involvement facilitate MVB fusion to the plasma membrane and release of exosomes outside of the cell. A wide range of cargo is transported within exosomes, including mRNA, miRNA, cytoskeletal elements, proteins, enzymes, molecular chaperones and signalling molecules. (Figure adapted from O'Loughlin, Wood et. al. 2012).
Upon movement of MVBs to the plasma membrane and subsequent fusion, the internal vesicles are released into the extracellular space as exosomes. Exosome release involves contributions from several Rab proteins, a subfamily of small GTPases with more than 60 known members, involved in the regulation of intracellular vesicle transportation. Further, once MVBs are docked to the plasma membrane, SNARE (Soluble N-ethylmaleimide-sensitive-fusion-protein-attachment-protein-receptor) and VAMP (v-SNAREs such as VAMP7 and VAMP2) proteins participate directly or indirectly in the docking and fusion of MVB to the plasma membrane and subsequently their release outside of the cells [13]. Different lipids and lipid-related enzymes may also control the secretion of these exosomes [7]. The release of exosomes may be constitutive or induced by stimuli including calcium, mitogens, cytokines or stress [14]. Following release, exosomes may travel to near or distant cells for uptake.
Exosomes may be internalized into recipient cells via a number of potential mechanisms: i) exosomes could be endocytosed or internalized into an endocytic compartment or MVB, from which they may undergo back-fusion, fusing with the limiting membrane and releasing their cargo into the cytoplasm of the recipient cell [15]; ii) The exosome membrane may fuse with the plasma membrane, releasing the cargo directly into the cytoplasm of the recipient cell [16]; iii) receptor-ligand mediated interactions could result in either signal transduction or exosomal internalization [14, 17]. Exosomes are increasingly recognized as major players in both local and distant cell-cell communications and often travel long distances in biological fluids.
3. Exosome cargos
As shown in Figure 1, a wide range of cargo is transported within exosomes including mRNA, miRNA (further described below), cytoskeletal elements (e.g. actins), proteins, enzymes, molecular chaperones and signalling molecules. In fact, Valadi et al identified 1,300 mRNAs and 120 miRNAs in exosomes from mast cells, many of which were not expressed in the donor cell cytoplasm, indicating that the RNA was targeted to exosomes via a selective mechanism [18]. They also found that the RNA from mast cell exosomes was transferable to other mouse and human mast cells. After transfer of mouse exosomal RNA to human mast cells, new mouse proteins were found in the recipient cells, indicating that transferred exosomal mRNA can be translated after entering another cell [18]. Indeed, one of the most interesting discoveries regarding exosomes is that they are a natural carrier system, transporting mRNA, miRNA and proteins between cells [6]. Moreover, circulating exosomes are highly regarded in biomarker studies because they allow for the protected circulation of biological material originating from the donor cells. Thus, measuring putative clinical biomarkers in the exosome fraction rather than in total biological fluids could improve the sensitivity of the analyses.
Recent studies provide evidence of the importance of exosome-encased miRNAs in cell-to-cell communication within the cardiovascular system [19, 20], from stem cells to cardiovascular cells [21-23] and from the heart to bone marrow (BM) stem cells [24]. MiRNAs are small, non-coding RNA molecules. Their primitive forms (pri-miRNAs) are either encoded within introns of protein-coding genes or autonomous miRNA genes, often in polycistronic units, and are transcriptionally regulated similarly to any protein coding genes. Pri-miRNAs are matured in the nucleus to smaller precursor forms (pre-miRNAs) and finally exported to the cytosol or the endoplasmic reticulum to be finally cut to their mature form of around 22 nucleotide by the ribonucleases Dicer. In its mature form, the miRNA is a single RNA strand of approximately 22 nucleotides in length. It can be recruited to the RNA-induced silencing complex (RISC), which contains Dicer, Argonaute (Ago) proteins and other RNA binding proteins. It is this complex that carries out the repression of the target genes of the miRNA targets. One miRNA can repress several genes, which it recognizes primarily using a “seed sequence” of 8 nucleotides, which is located in its 5’ untranslated region (5’UTR). This seed sequence is complimentary (or semi-complimentary) to one or more miRNA binding sites in the 3’ UTR of the target mRNA. The result is the repression of the targeted mRNAs, through degradation, transcript deadenylation, translation inhibition or sequestration of the mRNAs in the processing body (P-body). Being carried either within exosomes or other EVs, or being released conjugated to either lipoproteins or Ago proteins affords the extracellular miRNAs protection against degradation.
In the context of this review, it is also particularly important that stem cell-derived exosomes contain cardioprotective enzymes, which show beneficial effect in a rat myocardial infarct/reperfusion model (further described below) [25].
4. Methods for Exosome Characterization
Characterizing and observing exosomes is challenging, given their small size. The upper limit of their size range is below the threshold that instruments that would be classically used for their characterization, such as a flow cytometer, can accurately distinguish from noise, or individual particles from each other. This means that the flow cytometer cannot be used to observe individual exosomes. One therefore needs to be more creative in characterizing exosomes and use several different techniques in order to do so.
4.1. Antibody-based exosome identification
Given that exosomes are produced in the endosomal pathway, antibodies targeting markers associated with this pathway can be used to identify exosomes. These include the tetraspanins (CD9, CD63 and CD81), AIP1/Alix, TSG101 and CD326/EPCAM. In addition, exosomes also express phosphatidylserine [26]. These can be used in conjunction with some of the techniques described below to aid in the identification of exosome populations.
4.2. Electron Microscopy
The visualization of exosomes by electron microscopy (EM) is somewhat straightforward and an important step in their characterization. Purified exosome populations in suspension can be placed on to electron microscopy (EM) sample grids. Following negative staining using uranyl acetate and methylcellulose, exosomes can be clearly visualised using transmission EM. The exosomes can be seen as double-membrane bound vesicles of the correct size range. They are often described as having a “cup-shaped” morphology, although while this is often true, it is the result of an artefact caused by the drying of the sample during preparation for EM [26]. Therefore, while this is something which may well be seen, it should not be taken as a definitive feature of exosomes, nor should it be relied upon for their identification.
EM can also be used in conjunction with the exosome-specific antigens mentioned above in order to further strengthen the evidence of their identification. Rather than simply using a negative stain in the EM preparation, one can carry out a single or double staining against the antigens mentioned above. It is then possible to do a secondary staining using gold nanoparticles of different sizes (for example 6nm for one antibody and 10nm for the other) specific for the primary antibody. These nanoparticles can be clearly seen and distinguished for their sized on an electron micrograph and can be used to aid confirmation of the presence of exosomes within the preparation [27]. Moreover, exosomes can be identified on tissue electron micrographs using standard preparation techniques [27], and it is possible to get an even more accurate assessment of the presence of exosomes using a combination of both antibody and tissue sectioning methods [28].
4.3. Approaches for particle identification by size
One of the most common methods used to count particles of exosome size is Nanoparticle Tracking Analysis (NTA), produced by NanoSight (now part of Malvern, developers of dynamic light scattering). This is a light microscope on which is mounted a high-definition video camera. NTA utilizes the properties of both light scattering and Brownian motion in order to obtain the particle size distribution of samples in liquid suspension and it allows particles in the range of 50-1000 nm in liquid suspension to be directly and individually visualized and counted in real-time. Simultaneously, NTA provides high-resolution particle size distribution profiles and concentration measurements. The sample is passed through a flow cell placed on the microscope stage at an accurate and constant rate of flow. A laser is shone through the sample, allowing the camera to record video of the particles flowing through the flow cell. The NTA software then uses the Stokes-Einstein equation to calculate the size of the particles in the sample based on their Brownian motion [29, 30]. The method allows for use of fluorescent markers with the laser, so, in theory, it could be used to determine how many of the particles in the exosome size range were positive for a particular exosome marker. With the caveat that not all of the particles that this machine identifies will be exosomes, this technology can be used to semi-quantify the exosome/microvesicle preparations.
An alternative to NTA is Dynamic Light Scattering (DLS) [8]. Both techniques work on the principle of calculating the Brownian motion of the particles. However, DLS does this by determining the scattering of laser light passed through a sample, and thereby calculating the speed of the particles, rather than measuring the distance they travel over a given time [31]. Finally, IZON qNano can measure the size and concentration based on the Tunable Resistive Pulse Sensing (TRPS) technique.
The above approaches are reliable, semi-quantitative and quick to use. Moreover, they require very small samples for analyses. However, their weakness is that they do not specifically identify exosomes, but every particle of a given size (vesicles above ~10nm size). This could include fragments of membrane and other cellular debris, or lipoprotein complexes. Consequently, they should be integrated with other more “qualitative” techniques enabling the identification of exosomes, especially EM.
4.4. Flow cytometry
Exosomes currently fall below the size threshold where routine flow cytometer analyses can accurately distinguish particles from noise [30]. Exosomes tagged with fluorophores can be identified by flow cytometry machines, but the exosome numbers cannot be quantified because of swarming effects [32] and the inability of the machine to accurately separate them from noise. Alternative approaches based on the use of latex beads to immobilize exosomes have also been proposed. Exosomes can be bound to latex beads either directly or using exosome-specific antibodies. The antibody-conjugated beads can either be made in-house or bought commercially. The bound exosomes can then be stained for other exosome-specific markers using fluorophore-bound antibodies. While it is not possible to determine absolute numbers of exosomes using this technique, as one cannot determine how many exosomes are bound to each bead, it is possible to analyse their surface antigens [33]. All this said, however, this problem is under investigation and new techniques, such as those developed by Marca Wauben, are beginning to address it [34].
4.5. Fluorescence and confocal microscopy
It is possible to mark exosomes using a membrane-binding dye, such as PKH67 [35]. While one cannot actually visualise each individual exosome using this technique, is possible to determine if marked exosomes have been taken up into cells [35].
4.6. Other approaches
Other methods to detect single exosomes are atomic force microscopy [36], field emission scanning electron microscopy (FESEM)[36], Raman microspectroscopy [37], micro nuclear magnetic resonance, small-angle X-ray scattering (SAXS), and anomalous SAXS [38] and resistive pulse sensing [39].
5. Exosomes in Cardiovascular Biology, Disease and Repair
5.1. Exosomes in cardiovascular cell-to-cell communication
Evidence that exosomes are secreted by cardiac and vascular cells and stem cells in culture have emerged [22, 40-42]. Moreover, exosomes have been shown to mediate communication between endothelial cells (ECs) and smooth muscle cells (SMCs) [43], ECs and pericytes (A Caporali and C Emanueli, unpublished data, 2014), cardiac myocytes and ECs [44] and fibroblasts and cardiac myocytes [20]. Figure 2 summarized the role of exosomes and exosomal miRNAs in cardiovascular cell-to-cell communication.
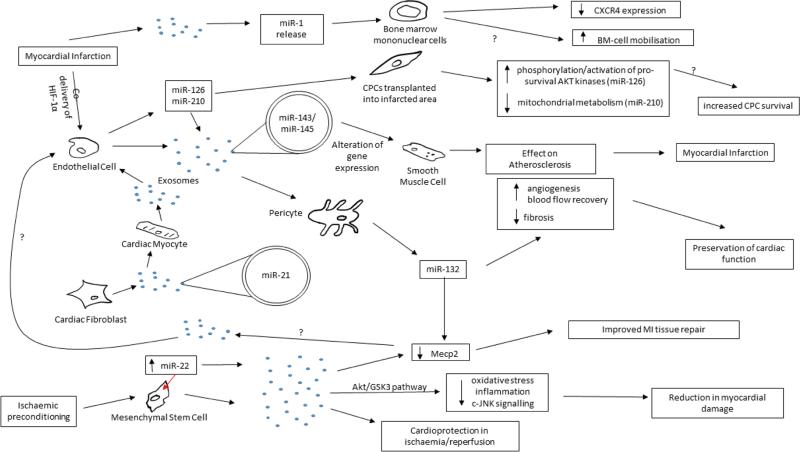
Figure 2. Exosomes in cardiovascular cell-cell communication.
An outline of the known cellular communication pathways that exosomes are known, or thought, to participate in. Those marked with a “?” are hypothesised but not yet demonstrated. For a detailed explanation of the pathways see text in section 5. Abbreviations – CPC: cardiac progenitor cell; HIF: Hypoxia-inducible factor; Akt: Protein Kinase B; GSK: Glycogen synthase kinase 3; Mecp2: methyl CpG binding protein 2; c-JNK: C-Jun N-terminal kinase; CXCR4: C-X-C chemokine receptor type 4; BM: bone marrow.
EVs have been shown to play a role in cell-cell communication during atherosclerotic protection [43]. SMCs have an important role in the formation of atherosclerotic plaques within arteries, which can lead to myocardial infarction (MI). The study demonstrates that ECs release microvesicles enriched with miR-143/145, which are taken up by SMCs and regulate gene expression in the recipient cells. Injection of miR-143/145-containing EVs in an atherosclerotic mouse model resulted in reduction in atherosclerotic lesion formation. Further, it has been demonstrated that atherosclerotic plaques contain large amounts of MPs, mostly of platelets or leukocyte origin [45]. In a recent investigation, the Thum laboratory demonstrated a cross talk between cardiac fibroblasts and cardiomyocytes [20]. Cardiac fibroblasts were shown to secrete exosomes enriched with miR-21* as a crucial paracrine signalling mediator of cardiac hypertrophy. MiR-21* was shuttled to cardiomyocytes affecting the expression of the miR-21* target genes within them and leading to cellular hypertrophy. Pharmacological inhibition of miR-21* obtained by injection of an “antagomiR-21*” attenuated the development of cardiac hypertrophy in mice infused with Angiotensin II (AngII). These findings illustrate that investigations on exosome-mediated communication systems may lead to the discovery of novel mechanisms contributing to cardiac failure. Exosomes can also exert beneficial actions. For example, Giricz et al. reported that EVs released from the rat heart after ischemic preconditioning are important and responsible for the transmission of remote conditioning signals for cardioprotection [46]. The aforementioned studies consistently implicate the transfer of one or more miRNAs in the action elicited by the exosomes. Nonetheless, it is possible than other components of the exosome cargo have also contributed to the effects. Moreover, additional reports have shown miRNA transfer between cells without identifying the method of transportation from one cell type to the other. For example, we have previously shown that the pro-angiogenic activity of pericytes partly depends on miR-132, which they produce and release, especially in response to hypoxia. Pericyte-derived miR-132 is taken up by ECs, resulting in a higher pro-angiogenic capacity [47]. Interestingly, there is evidence that both ischemic and healthy human and mouse cardiomyocytes may release exosomes-like vesicles in vivo [48]. Further, a recent report indicates that circulating miRNA-1, which in mice with an acute MI is released to the blood stream via exosomes, supresses the expression of the SDF-1 receptor CXCR-4 in BM mononuclear cells. This might contribute to the increased BM-cell mobilization elicited by an acute ischemic episode, such as MI [24]. These studies implicate exosomes-mediated communication mechanisms may play a significant role in disease dissemination, cardiac repair and regeneration.
5.2. Exosomes in stem and progenitor cell actions
There is mounting evidence to suggest that exosomes have a role in ischemic tissue repair. Early experiments which attempted to transplant stem cells to sites of injury following myocardial infarction suggested that many of the transplanted cells did not integrate into the damaged tissue. However, functional improvement were observed. This led to the hypothesis that the transplanted stem cells might work through a paracrine manner. It has been recently described that stem cells can also release exosomes or other EVs that may contain both autocrine and paracrine angiogenic factors [8, 42, 49, 50]. Figure 3 provides examples of stem and progenitor cell-derived exosomes actions in the setting of cardiovascular diseases. Lim and colleagues have detected RNAs and miRNAs in the supernatant of human mesenchymal stem cells (MSCs) that may have been associated with exosomes [51]. They identified a cardio-protective effect of these exosomes when injected into a rat model of ischemia/reperfusion [52]. Moreover, human CD34+ peripheral blood-derived hematopoietic stem cells were found to secrete exosomes, which can promote angiogenesis both in vitro and in vivo (mouse cornea model). These exosomes, which were found to carry the CD34+ protein on their surface, were enriched with pro-angiogenic miRNAs such as miR-126, miR-130a [8], which were most likely responsible for the exosome pro-angiogenic function [50]. Cardiac progenitor cells (CPCs) have been shown to exert potentially anti-fibrotic effects by transferring exosomes to fibroblasts, and by promoting angiogenesis and cardiac myocyte survival in vitro [22]. Moreover, Ibrahim et al. identified exosomes as critical agents of cardiac regeneration triggered by cardiosphere-mediated cell therapies [53]. The cardiosphere-derived exosomes were found to be enriched with miR-146a, which mediated their regenerative function. Similarly, Chen and co-workers demonstrated that cardiac progenitor cells produced exosomes and that these conferred a protective preconditioning effect when injected into mouse hearts immediately after induction of ischemia [54]. They also demonstrated that these EVs contained a high level of miR-451, a miRNA already known to improve the clinical outcome when transfected into ischaemic rodent hearts [54]. The authors inferred that it was the exosomes that caused the improvement in outcome when CPCs were introduced into the ischaemic heart [54]. MiR-22 has also been implicated in the repair of injury following MI. A study that demonstrated the upregulation of miR-22 in MSCs subjected to ischaemic preconditioning also showed that this and other miRNAs were packaged into exosomes released from these cells. When injected into infarcted mouse hearts, these exosomes appeared to confer an improvement in the repair of tissue damage resulting from MI via a mechanism involving the downregulation of Mecp2 (methyl CpG binding protein 2) [23]. Interestingly, the profibrotic Mecp2 is also targeted by miR-132, which is released by human perivascular pericytes (the “Bristol pericytes”) [55] and transferred to ECs, possibly via an exosome-mediated mechanism [47]. The Bristol pericytes proved to be therapeutic in a mouse MI model, where they induced angiogenesis and blood flow recovery, and inhibited fibrosis, thus preserving cardiac function. These therapeutic actions were negated when miR-132 was inhibited in the cells before transplantation into mouse hearts. Further studies will clarify the importance of exosomes in the paracrine actions elicited by Bristol pericytes. Another study investigating the effects of MSC-derived exosomes on myocardial ischemia/reperfusion (I/R) injury used a combination of in vivo and ex vivo techniques to demonstrate that these isolated exosomes reduced myocardial damage [25]. This study did not investigate the role of miRNAs, although it did demonstrate that the Akt/GSK3 pathway was activated by exosome delivery. The authors also showed that there was a reduction in inflammation, oxidative stress and c-JNK signalling. A more recent study by the Wu group looked at the potential role of hypoxia-inducible factor-1α (HIF-1α) in improving the environment around damaged tissue following an acute MI [21]. They have demonstrated that co-delivery of CPCs with a nonviral minicircle plasmid carrying HIF1 (MC-HIF1) into the ischemic myocardium can improve the survival of transplanted CPCs. Complementary in vitro studies showed that cardiac ECs produced exosomes that were internalized by recipient CPC and that exosomes from EC overexpressing HIF1 had higher contents of miR-126 and miR-210. The exosomes from ECs with increased HIF1 could be taken up by transplanted CPCs into the infarcted area and contributed to CPC's increasing tolerance under hypoxic stress. The investigators showed that uptake of miR-126 caused increased phosphorylation/activation of the pro-survival AKT kinases, and that uptake of miR-210 caused a reduction in mitochondrial metabolism. The authors suggested that these effects could be the cause of the increased survival of the transplanted CPCs that was seen when either HIF-1 or the extracted exosomes were administered at the same time as the transplanted cells [21].
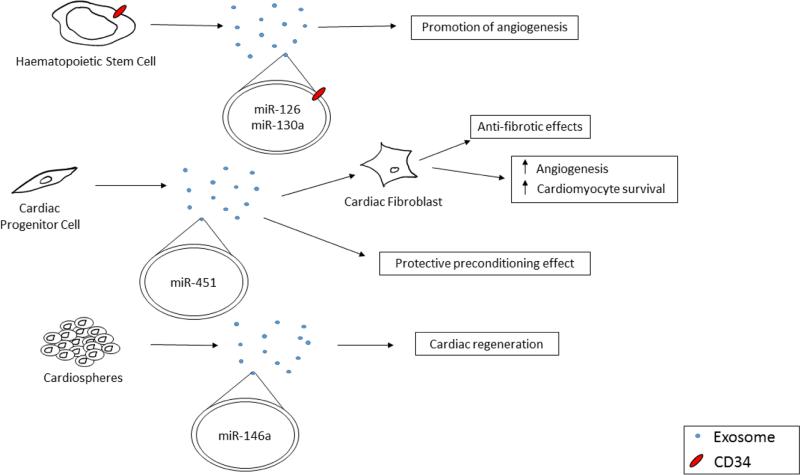
Figure 3. Examples of exosomes in stem and progenitor cell actions in cardiovascular disease.
An outline of the ways in which exosomes are thought to have a role in the actions of stem and progenitor cells. CD34+ haematopoietic stem cells are thought to promote angiogenesis via miR-126 and miR-130a packaged in exosomes. Cardiac progenitor cells release exosomes that can interact with cardiac fibroblasts and exert anti-fibrotic effects, along with increasing angiogenesis and cardiomyocyte survival. These exosomes have been demonstrated to contain miR-451, which can exert a protective preconditioning effect. Cardiospheres have been shown to release exosomes containing miR-146a, and these exosomes promote cardiac regeneration.
Collectively, the above evidence indicates that exosomes from several different sources can modulate post-ischemic angiogenesis, tissue protection and repair in animal models of heart and kidney ischemia, or ischemia/reperfusion. These cumulative data strongly suggest that exosomes can be a suitable target for the development of novel therapeutics.
6. Translational perspectives: Exosomes as new therapeutics and clinical biomarkers
Exosomes are naturally adapted for the transport and intercellular delivery of proteins and nucleic acids. This makes them particularly attractive as pharmaceutical delivery agents. Moreover, due to their biophysical properties, exosomes are easy to isolate and their RNA and protein contents manipulated [56, 57]. Therefore, in addition to being used as therapeutic entities themselves, both natural exosomes and exosome-mimetic nanovesicles are regarded as possible therapeutic Trojan Horses to deliver biological therapeutics, including short interfering RNAs and recombinant proteins, which if released free in the circulation would be prone to degradation and have limited ability to cross biological membranes. The use of exosomes as carriers of biological therapeutics is a promising strategy to overcome these issues and to achieve efficient delivery of drugs across different biological barriers to the cytosol of target cells. The first proof that exosomes can be used as delivery vehicle for nucleic acid cargoes was exhibited by the group of MJ Wood [56, 57], demonstrating that modified exosomes expressing a neuron-specific protein on their surface can target siRNA-carrying exosomes to mouse brains [57]. Furthermore, exosomes carrying recombinant proteins and tumour antigens expressed by cancer vaccines [58] were demonstrated to have a therapeutic potential in two Phase I clinical trials on patients with stage III/IV melanoma. Beneficial effects of stem cell-derived exosomes for ischemic tissue repair and regeneration have also been shown by several preclinical studies [21, 49, 52, 53]. Despite these interesting possibilities for cardiovascular exosome treatment, these novel therapeutic approaches still need thorough investigation in order to translate them into cardiovascular clinical applications. Some of the major challenges to overcome before endogenous exosomes may be used in clinical setting are the complex structure of naturally-derived exosomes, which are difficult to characterize pharmaceutically. In addition, they have complex roles in health and disease and their potential off-target effects have not been well characterized. Given that it is quite possible that not all components of exosomes are required for their proper function, an alternative strategy would be to mimic these vesicles and to choose their cargoes synthetically. Exosomal components that would be required for the assembly of functional exosome mimetic remain to be identified. Nevertheless, lessons from the ongoing exosome-based preliminary clinical trials in the field of cancer would positively encourage such ideas.
Another avenue where disease-related exosomes can be used clinically is their potential to be used as biomarkers. Exosomes contain a wide variety of proteins, lipids, mRNAs, non-transcribed RNAs, miRNAs and small RNAs that are representative to their cellular origin and reflective of the pathological condition of the cell of its origin. The detection of exosomes in the serum of glioblastoma [59] or ovarian cancer patients [60] emphasizes the potential utility of exosomes and their contents as biomarkers for cancer and other pathological conditions. The possibility to isolate and characterize exosomes from bodily fluids makes them very attractive diagnostic markers. Nevertheless, despite the abundance of exosomes in biological fluids, the actual use of exosome-derived proteins or miRNAs as biomarkers has not yet been implemented in the clinical practice, primarily due to a lack of a fast and efficient method to process a large number of biological samples. Several commercial enterprises have initiated development of exosome-based can.cer diagnostics recently, including Caris Life Sciences, Exosomes Diagnostics and Hansa BioMed. In the field of cardiovascular medicine and surgery the exosomes are still an unexplored world which we are committed to pioneer.
Funding and Acknowledgement
The work was funded by the Leducq Transatlantic Network in Vascular microRNAs (MIRVAD) and the British Heart Foundation (BHF) Regenerative Medicine Centres (both to CE); the National Health Research Institute (NIHR) Bristol Cardiovascular Biomedical Research Unit (BRU) to GDA and by National Institute of Health (NIH) and American Heart Association (AHA) funded grants to SS. CE is a BHF Senior Research Fellow; GDA in a BHF Chair of Cardiac Surgery and a NIHR Senior Investigator. The views expressed are those of the Authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26(4):239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elmore S. Apoptosis: a review of programmed cell death. Toxicologic pathology. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of extracellular vesicles. 2013;2:1–25. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skokos D, et al. Mast Cell-Derived Exosomes Induce Phenotypic and Functional Maturation of Dendritic Cells and Elicit Specific Immune Responses In Vivo. The Journal of Immunology. 2003;170:3037–3045. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 5.Malik ZA, et al. Cardiac myocyte exosomes: stability, HSP60, and proteomics. Am J Physiol Heart Circ Physiol. 2013;304:H954–65. doi: 10.1152/ajpheart.00835.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Villarroya-Beltri C, et al. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahoo S, et al. Exosomes from human CD34(+) stem cells mediate their proangiogenic paracrine activity. Circ Res. 2011;109(7):724–8. doi: 10.1161/CIRCRESAHA.111.253286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oksvold MP, et al. Expression of B-cell surface antigens in subpopulations of exosomes released from B-cell lymphoma cells. Clin Ther. 2014;36(6):847–862. e1. doi: 10.1016/j.clinthera.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Zakharova L, Svetlova M, Fomina AF. T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J Cell Physiol. 2007;212(1):174–81. doi: 10.1002/jcp.21013. [DOI] [PubMed] [Google Scholar]
- 11.Palma J, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40(18):9125–38. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tauro BJ, et al. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol Cell Proteomics. 2013;12(3):587–98. doi: 10.1074/mcp.M112.021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fader CM, et al. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793(12):1901–16. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 14.O'Loughlin AJ, Woffindale CA, Wood MJ. Exosomes and the emerging field of exosome-based gene therapy. Curr Gene Ther. 2012;12(4):262–74. doi: 10.2174/156652312802083594. [DOI] [PubMed] [Google Scholar]
- 15.Fitzner D, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci. 2011;124(Pt 3):447–58. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 16.Parolini I, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284(49):34211–22. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segura E, et al. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J Immunol. 2007;179(3):1489–96. doi: 10.4049/jimmunol.179.3.1489. [DOI] [PubMed] [Google Scholar]
- 18.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shantikumar S, Angelini GD, Emanueli C. Diabetes, microRNAs and exosomes: Les liaisons dangereuses. J Mol Cell Cardiol. 2014;74:196–8. doi: 10.1016/j.yjmcc.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Bang C, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124(5):2136–46. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong SG, et al. Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation. 2014;130(11 Suppl 1):S60–9. doi: 10.1161/CIRCULATIONAHA.113.007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray WD, et al. Identification of Therapeutic Covariant microRNA Clusters in Hypoxia Treated Cardiac Progenitor Cell Exosomes using Systems Biology. Circ Res. 2014 doi: 10.1161/CIRCRESAHA.116.304360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y, et al. Ischemic preconditioning potentiates the protective effect of stem cells through secretion of exosomes by targeting Mecp2 via miR-22. PloS one. 2014;9:e88685. doi: 10.1371/journal.pone.0088685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng M, et al. American Heart Association Scientific Sessions. Chicago, IL, USA: 2014. Circulating Exosomal miR-1a is Markedly Induced by Myocardial Infarction and Downregulates CXCR4 Expression in the Bone Marrow Mononuclear Cells. [Google Scholar]
- 25.Arslan F, et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem cell research. 2013;10:301–12. doi: 10.1016/j.scr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature reviews. Immunology. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 27.van Weering JRT, et al. Intracellular membrane traffic at high resolution. Methods in cell biology. 2010;96:619–48. doi: 10.1016/S0091-679X(10)96026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Current opinion in cell biology. 2004;16:415–21. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Gardiner C, et al. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dragovic RA, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine : nanotechnology, biology, and medicine. 2011;7:780–8. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malvern [29/07/2014];Dynamic Light Scattering: An Introduction in 30 Minutes. 2014 Available from: http://www.malvern.com/en/support/resource-center/technical-notes/TN101104DynamicLightScatteringIntroduction.aspx.
- 32.van der Pol E, et al. Single vs. swarm detection of microparticles and exosomes by flow cytometry. J Thromb Haemost. 2012;10(5):919–30. doi: 10.1111/j.1538-7836.2012.04683.x. [DOI] [PubMed] [Google Scholar]
- 33.Ostrowski M, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature cell biology. 2010;12:19–30. 1–13. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 34.van der Vlist EJ, et al. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311–26. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- 35.Lugini L, et al. Immune surveillance properties of human NK cell-derived exosomes. Journal of immunology (Baltimore, Md. : 1950) 2012;189:2833–42. doi: 10.4049/jimmunol.1101988. [DOI] [PubMed] [Google Scholar]
- 36.Sharma S, et al. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4(4):1921–6. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatischeff I, et al. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J Extracell Vesicles. 2012;1:19179. doi: 10.3402/jev.v1i0.19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Pol E, et al. Innovation in detection of microparticles and exosomes. J Thromb Haemost. 2013;11(Suppl 1):36–45. doi: 10.1111/jth.12254. [DOI] [PubMed] [Google Scholar]
- 39.van der Pol E, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12(7):1182–92. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 40.Akyurekli C, et al. A Systematic Review of Preclinical Studies on the Therapeutic Potential of Mesenchymal Stromal Cell-Derived Microvesicles. Stem Cell Rev. 2014 doi: 10.1007/s12015-014-9545-9. [DOI] [PubMed] [Google Scholar]
- 41.Lamichhane TN, et al. Emerging Roles for Extracellular Vesicles in Tissue Engineering and Regenerative Medicine. Tissue Eng Part B Rev. 2014 doi: 10.1089/ten.teb.2014.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barile L, et al. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovascular Research. 2014;103(4):530–541. doi: 10.1093/cvr/cvu167. [DOI] [PubMed] [Google Scholar]
- 43.Hergenreider E, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14(3):249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, et al. Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. J Mol Cell Cardiol. 2014;74:139–50. doi: 10.1016/j.yjmcc.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leroyer AS, et al. Cellular origins and thrombogenic activity of microparticles isolated from human atherosclerotic plaques. J Am Coll Cardiol. 2007;49(7):772–7. doi: 10.1016/j.jacc.2006.10.053. [DOI] [PubMed] [Google Scholar]
- 46.Giricz Z, et al. Cardioprotection by remote ischemic preconditioning of the rat heart is mediated by extracellular vesicles. J Mol Cell Cardiol. 2014;68:75–8. doi: 10.1016/j.yjmcc.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 47.Katare R, et al. Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132. Circ Res. 2011;109(8):894–906. doi: 10.1161/CIRCRESAHA.111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahoo S, Losordo DW. Exosomes and Cardiac Repair After Myocardial Infarction. Circulation Research. 2014;114(2):333–344. doi: 10.1161/CIRCRESAHA.114.300639. [DOI] [PubMed] [Google Scholar]
- 49.Mackie AR, et al. Sonic hedgehog-modified human CD34+ cells preserve cardiac function after acute myocardial infarction. Circ Res. 2012;111(3):312–21. doi: 10.1161/CIRCRESAHA.112.266015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mocharla P, et al. AngiomiR-126 expression and secretion from circulating CD34(+) and CD14(+) PBMCs: role for proangiogenic effects and alterations in type 2 diabetics. Blood. 2013;121(1):226–36. doi: 10.1182/blood-2012-01-407106. [DOI] [PubMed] [Google Scholar]
- 51.Chen TS, et al. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38(1):215–24. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai RC, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4(3):214–22. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Ibrahim AG, Cheng K, Marban E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports. 2014;2(5):606–19. doi: 10.1016/j.stemcr.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, et al. Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochemical and biophysical research communications. 2013;431:566–71. doi: 10.1016/j.bbrc.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campagnolo P, et al. Human adult vena saphena contains perivascular progenitor cells endowed with clonogenic and proangiogenic potential. Circulation. 2010;121(15):1735–45. doi: 10.1161/CIRCULATIONAHA.109.899252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 57.El-Andaloussi S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc. 2012;7(12):2112–26. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 58.Rountree RB, et al. Exosome targeting of tumor antigens expressed by cancer vaccines can improve antigen immunogenicity and therapeutic efficacy. Cancer Res. 2011;71(15):5235–44. doi: 10.1158/0008-5472.CAN-10-4076. [DOI] [PubMed] [Google Scholar]
- 59.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor DD, Homesley HD, Doellgast GJ. “Membrane-associated” immunoglobulins in cyst and ascites fluids of ovarian cancer patients. Am J Reprod Immunol. 1983;3(1):7–11. doi: 10.1111/j.1600-0897.1983.tb00204.x. [DOI] [PubMed] [Google Scholar]