Abstract
The capacity of sex to modify behavior in health and illness may stem from biological differences between males and females. One such difference – fundamental to the biological definition of sex - is inequality of X chromosome dosage. Studies of Turner syndrome (TS) suggest that X-monosomy profoundly alters mammalian brain development. However, use of TS as a model for X chromosome haplonsufficiency is complicated by karyotypic mosaicism, background genetic heterogeneity and ovarian dysgenesis. Therefore, to better isolate X chromosome effects on brain development and identify how these overlap with normative sex differences, we used whole-brain structural imaging to study X-monosomic mice (free of mosaicism and ovarian dysgenesis) alongside their karyotypical normal male and female littermates. We demonstrate that murine X-monosomy (XO) causes (i) accentuation of XX vs XY differences in a set of sexually dimorphic structures including classical foci of sex-hormone action, such as the bed nucleus of the stria terminal and medial amygdala, (ii) parietal and striatal abnormalities that recapitulate those reported TS, and (iii) abnormal development of brain systems relevant for domains of altered cognition and emotion in both murine and human X-monosomy. Our findings suggest an unexpected role for X-linked genes in shaping sexually dimorphic brain development, and an evolutionarily conserved influence of X-linked genes on both cortical and subcortical development in mammals. Furthermore, our murine findings highlight the bed nucleus of the stria terminalis and periaqueductal gray matter as novel neuroanatomical candidates for closer study in TS. Integration of these data with existing genomic knowledge generates a set of novel, testable hypotheses regarding candidate mechanisms for each observed pattern of anatomical variation across XO, XX and XY groups.
Keywords: Sex Chromosomes, Aneuploidy, Sex Differences, Brain Development
1. INTRODUCTION
Since unequal X chromosome gene dosage is a fundamental genetic sex-difference in mammals, understanding X chromosome influences on brain development emerges as a priority in the search for biological contributions to sex-differences in mammalian brain and behavior (Arnold, 2004). The potential for X chromosome dosage to directly influence the mammalian brain is clearly demonstrated by recent experimental work in genetically modified mice allow sex chromosome effects to be dissociated from those of circulating sex steroids (Hughes et al., 2012). A complementary, naturally occurring model for X chromosome influences on mammalian brain development is Turner Syndrome (TS): this sex chromosome aneuploidy syndrome arises due to X chromosome monosomy in females and has been linked to alterations of both sub-cortical and cortical brain anatomy (Lepage et al., 2012; Raznahan et al., 2010).
However, a number of challenges complicate use of TS as a homogeneous model for X chromosome influences on brain development. First, karyotypic mosaicism (an assorted chromosome complement within the same individual) is evident in over 50% of TS cases within clinical cohorts (Wolff et al., 2010). Individuals with TS can also vary according to their X chromosome parent of origin - which may influence expression of X-linked genes (Davies et al., 2005) and brain function (Skuse et al., 1997). Finally, in TS, X haploinsufficiency is usually accompanied by complete ovarian dysgenesis (Sybert and McCauley, 2004). This lack of functional ovarian tissue effectively removes the possibility of intact sex-steroid influences on brain development in TS – particularly during perinatal (Garagorri et al., 2008) and pubertal (Garces et al., 2008) life when the hypothalamic-pituitary-gonadal axis undergoes robust up-regulation in typical development. Although some recent studies in TS have sought to mitigate against this confound by focusing on the pre-pubertal age range (Yamagata et al., 2012), X haploinsufficiency in TS cannot be uncoupled from the potential consequences of perinatal hormonal deficits arising from ovarian dysgenesis.
In order to examine X chromosome influences on brain development in a way that attenuates the potentially confounding variables above, we used high resolution (~30μm3) ex vivo neuroimaging (Lerch et al., 2008) to conduct a spatially fine-grained whole-brain anatomical survey in X-monosomic mice (XO) alongside their XX and XY littermates. Unlike humans with TS, the murine X-monosomy model we studied is necessarily free of karyotypic mosaicism and allows X chromosome parent-of-origin to be pre-specified (Davies et al., 2005). Moreover, ovarian dysgenesis is not a feature of murine X-monosomy, and the fact that XO mice are fertile suggests grossly intact ovarian function during early life (Probst et al., 2008; Trolle et al., 2012). Use of a murine model also means that neuroanatomical differences across XO, XX and XY groups can be characterized while providing a degree of homogeneity in genetic and environmental background that is hard to achieve in humans.
The neuroimaging methods we employed to compare XO, XX and XY groups provide an important complement to classical histochemical methods for the study of murine anatomy by allowing a highly efficient and technically homogenous survey of anatomy across the entire brain in a spatially un-biased manner. Thus, whole-brain neuroimaging makes it possible to simultaneously assay multiple classical foci of anatomical sex difference in the brain [e,g bed nucleus of the stria terminalis (BNST), amygdala, olfactory tubercle, and basal forebrain] (Morris et al., 2004; Shah et al., 2004), while also allowing for the discovery of novel foci of importance beyond these a priori regions of interest.
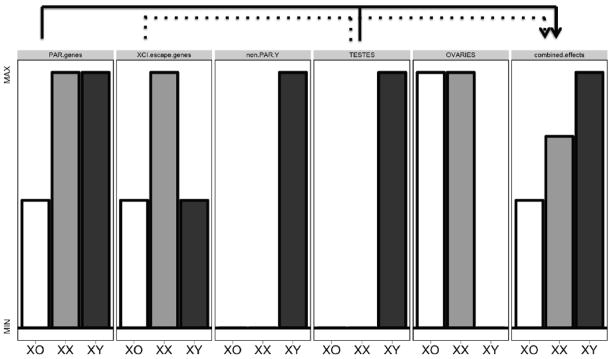
Our study design enabled us to first identify brain regions that vary by group, and then to sub-classify these regions according to the exact pattern of anatomical variation across XX, XY and XO groups. Each potential distribution of anatomical disparity between groups is best mirrored by a different set of known group inequalities in chromosomal and gonadal “dosage” (Figure 1). For example, the most parsimonious basis for XO-specific effects in a region of parity between XX and XY groups would be group differences in pseudoautosomal regions (PAR) dosage: stretches of shared sequence and obligate meiotic pairing between X and Y chromosomes (Otto et al., 2011). In contrast, an XX specific effect in a region of parity between XO and XY groups would best mirror dosage difference in non PAR genes that “escape” X chromosome inactivation (XCI) - the random “silencing” of one X chromosome in female XX cells (Lyon, 1961) that is hypothesized to compensate for male-female differences in X chromosome count. Finally, XY specific effects in a region of parity between XO and XX groups would be most parsimoniously accounted for by the well-documented masculinizing effects of testicular androgens (Morris et al., 2004) (although could also theoretically reflect the action of non-PAR Y genes). Using this conceptual framework, we specify which sets of chromosomal or gonadal factors constitute the strongest candidate mechanism for each observed focus of significant anatomical variation across XO, XX and XY mice.
Figure 1. Sex-Linked Genetic and Gonadal “Dosage” Differences Between XO, XX and XY Mice in this Study.
Simplified overview of XO, XX and XY profiles for sex-linked genetic and gonadal factors – separately, and in an illustrative combination. Abbreviations: PAR- pseudoautosomal regions, XCI – X chromosome inactivation. See main text for definition of PAR and XCI. Plots are intended to convey relative dosage differences between groups: y-axis scales are arbitrary and not comparable across plots. Plots provide a working conceptual framework for sources of observed anatomical variation across groups, and do not account for complexities such as (i) functional dissimilarity between X–Y homologs (Xu et al., 2002), (ii) “genomic conflict” between X–Y homologs (Cocquet et al., 2012), and (iii) regionally-specific (Gregg et al., 2010b) and sexually-dimorphic (Gregg et al., 2010a) expression of parentally-imprinted X chromosome genes in the mouse brain.
The informativeness of our anatomical data is also enhanced by the wealth of experimental evidence specifying regions of the rodent brain where male-female differences in anatomy are clearly partially driven by sex-differences in circulating androgens. These include the bed nucleus of the stria terminalis (BNST), medial amygdala, olfactory tubercle, and basal forebrain (Morris et al., 2004; Shah et al., 2004). By being able to assay for XO effects on anatomy within these classical foci of androgen dependent sexual dimorphism, our study effectively allows a direct test for spatial convergence between sex chromosome and sex hormone influences on brain development. Such convergence has never been demonstrated, but is predicted by newer theories that replace the classical “androgen-dominated” account for sexually dimorphic development (Phoenix et al., 1959), with models that posit an intimate interaction between sex-specific genes and hormones in shaping the sexually-dimorphic brain(Lenz et al., 2012).
2. MATERIALS AND METHODS
2.1 Mice
The mice used in this study were generated according to the following breeding scheme. In the grandparental generation, female XO mice carrying the tabby (EdaTa) coat-color marker (JAX, Bar Harbor, ME: JAX stock number 000314) (Probst et al., 2008) were crossed with B6CBACaF1/J-Aw-J/A males (JAX stock number 001201) to derive XO mice without the tabby coat-color marker. This step was taken because hemizygosity for the tabby coat-color marker in XO mice causes dental alterations (Wallace et al., 2013) which may confound study of the X-monosomic state through secondary nutritional problems. The XO females arising from this cross (the parental generation) were then bred with purebred C57BL/6J males (JAX stock number 000664) to generate the XO (n=10), XX (n=12), and XY (n=11) mice used for this study. All XO mice in this study bore an X -chromosome of paternal origin. X chromosome copy number was determined by quantitative PCR (qPCR) (see Figure 2). PCR primers and qPCR probes were designed to amplify a portion of the autosomal gene for adenine phosphoribosyl transferase (Aprt) and a portion of the X chromosome gene for hypoxanthine guanine phosphoribosyl transferase (Hprt) (Aprt.for: 5′-GACGTGAGTTTAGCGTGCTG-3′; Aprt.rev: 5′-ATTGGGAAGTCGGGGAAG-3′; Aprt.probe: 5′-VIC-TTCCGACATGGCCGCGTGC-TAMRA-3′; Hprt.for: 5′-CAGCGTTTCTGAGCCATTG-3′; Hprt.rev: 5′-AAAGCGGTCTGAGGAGGAAG-3′; and Hprt.probe: 5′-6FAM-TAGCACCTCCTCCGCC-MGBNFQ-3′)[TaqMan®, Applied Biosystems, now known as Life Technologies, Grand Island, NY]. qPCR was performed according to the manufacturer’s instructions and analyzed with their CopyCaller software.
Figure 2. X chromosome Copy Number from Mice Within Breeding Scheme as Determined by qPCR.
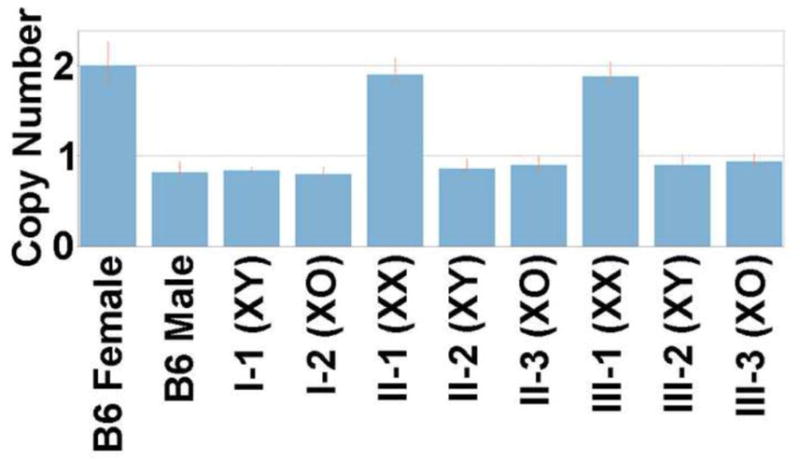
The results from the previously karyotyped animals from Probst et al, 2008, are shown. Error bars represent the range of results from the replicates for each sample. Abbreviations: B6 (Black 6). Roman numerals relate to generation within the breeding scheme (I - grandparental, II-parental, III-present). Arabic numerals relate to karyotype classes within each generation.
All mice were housed in a specific-pathogen free (SPF), Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited animal facility at Baylor College of Medicine. Food and water were provided ad libitum. This work was approved by the Institutional Animal Care and Use Committee (IACUC) of Baylor College of Medicine.
2.2 Brain Sample Preparation
Mice were sacrificed in young adulthood, at 80 days of age, following deep anesthesia by intraperitoneal injection with a combination of ketamine (Fort Dodge Animal Health, Fort Dodge, IA, 150mg/kg) and xylazine (MP Biomedicals, LLC, Solon, OH, 10mg/kg). Thoracic cavities were opened, and animals were perfused through the left ventricle with 30 mL of phosphate-buffered saline (PBS) (pH 7.4) plus heparin with 2 mM ProHance® (gadoteridol, Bracco Diagnostics Inc., Princeton, NJ) at room temperature (25 °C) at a rate of approximately 100 mL/h. This was followed by infusion with 30 mL of 4% paraformaldehyde (PFA) in PBS with 2 mM ProHance® at the same rate. Following perfusion, the heads were removed along with the skin, lower jaw, ears and the cartilaginous nose tip. The remaining skull structures were allowed to postfix in 4% PFA at 4 °C with 2 mM ProHance® for 12 h. Following an incubation period of 5 days in PBS plus 0.02% sodium azide with 2 mM ProHance® at 4 °C, the skulls were transferred to a PBS and 2 mM ProHance® solution for at least 7 days at 4 °C. MR imaging occurred 12 to 21 days post-mortem.
2.3 Imaging
A multi-channel 7.0 Tesla MRI scanner (Varian Inc., Palo Alto, CA) with a 6-cm inner bore diameter insert gradient was used to acquire anatomical images of brains within skulls. Prior to imaging, the samples were removed from the contrast agent solution, blotted and placed into plastic tubes (13 mm diam) filled with a proton-free susceptibility-matching fluid (Fluorinert FC-77, 3M Corp., St. Paul, MN). Three custom-built, solenoid coils (14 mm diam, 18.3 cm in length) with over wound ends were used to image three brains in parallel. Parameters used in the scans were optimized for gray/white matter contrast: a T2-weighted, 3D fast spin-echo sequence with 6 echoes, with TR/TE= 325/32 ms, four averages, field-of-view 14 × 14 × 25 mm3 and matrix size = 432 × 432 × 780 giving an image with 32 μm isotropic voxels. Geometric distortion due to position of the three coils inside the magnet was calibrated using a precision-machined MR phantom. Total imaging time was 11.3 h (Cahill et al., 2012).
2.4 Image Processing and Statistical Analyses
All 33 acquired anatomical MRIs were analyzed using an image registration pipeline described previously (Lerch et al., 2011). Briefly, after an initial affine registration to correct for postural differences, pair-wise 12-parameter registrations were computed to define a consensus space for all scans. Three generations of non-linear registrations using a diffeomorphic registration algorithm (Avants et al., 2008) then brought all scans into precise alignment. The Jacobian determinant of the deformations fields was used as the metric of local volume change. We first used voxel-wise F-tests, with False Discovery Rate (FDR) correction for multiple comparisons [with q (the expected proportion of falsely rejected nulls) set at 0.05 (Genovese et al., 2002)], to identify regions where the omnibus effects of all three groups (XO, XX, XY) in combination predicting a significant proportion of anatomical variance. Then, for the peak voxel within each anatomical structure highlighted by the omnibus F-test, we ran three post-hoc pairwise contrasts (XO vs XX, XO vs. XY, XX vs. XY), to distinguish regions of overlap and dissociation between XO effects and typical sex-differences.
3. RESULTS
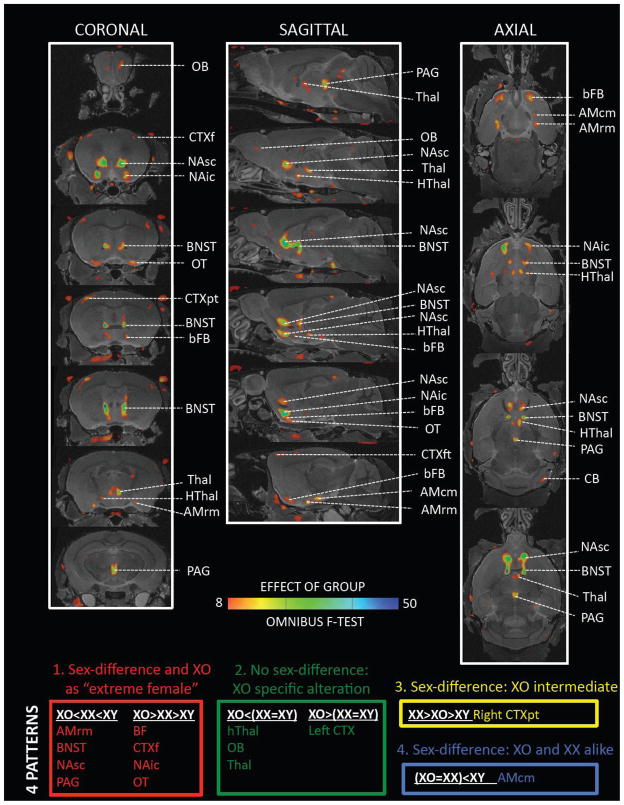
Brain regions for which karyotype grouping was a strong predictor of structural variation adopted a highly symmetric spatial distribution, respected known anatomical boundaries, and fell into four distinct groups based on the results of post-hoc pair-wise group contrasts (see Figures 3 and 4).
Figure 3. Coronal, Sagittal and Axial Maps Showing Regions Of Significant Anatomical Variation Across XO, XX and XY Mice, With Grouping of These Regions According to Pattern of Pairwise Group Differences.
Color-maps encode a FDR-thresholded F-statistic for the omnibus effect of group. The color-coding for each of the four observed patterns relates to the plots shown in Figure 4 Abbreviations: AMcm – caudomedial amygdala, AMrm – rostromedial amygdala, bFB – basal forebrain, BNST – bed nucleus of the stria terminalis, CB – cerebellum, CTX – cortex (f –frontal, p-parietal), HThal – hypothalamus, NAic – infracommisural nucleus accumbens, NAsc – supracommisural nucleus accumbens, OB – olfactory bulb, OT – olfactory tubercle, PAG – periaqueductal gray, Thal – thalamus.
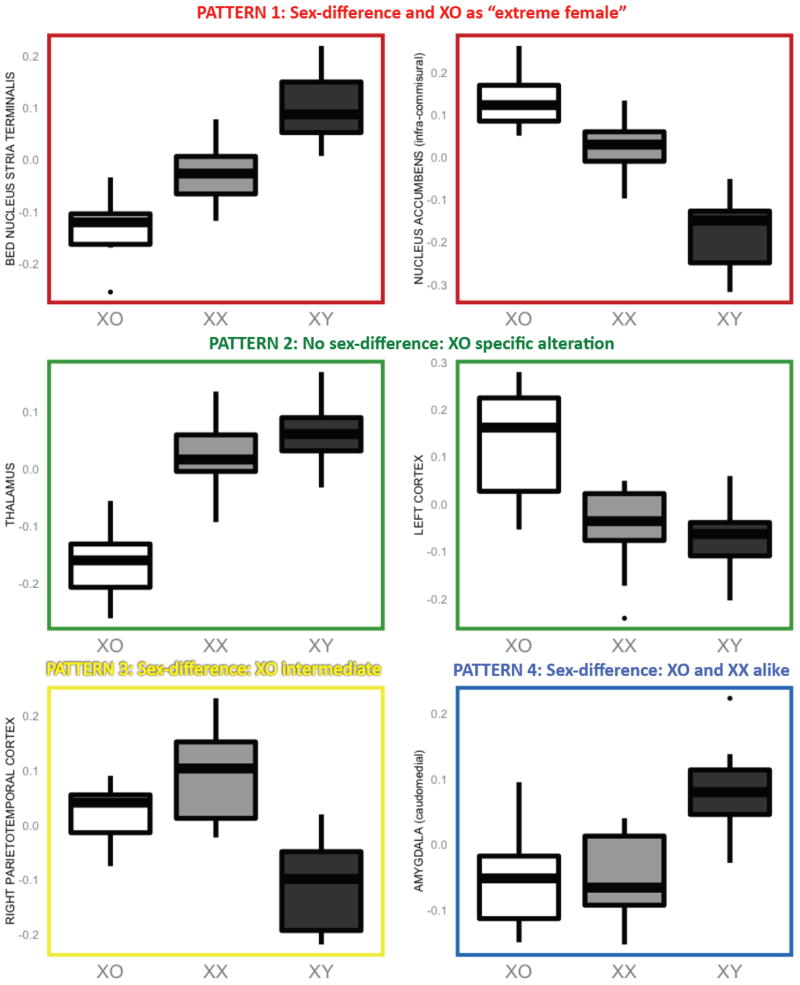
Figure 4. Illustrative Box-Plots for Each of the Four Patterns of Anatomical Variation Observes Across XO, XX and XY Mice.
Color codes link with those for different patterns of anatomical variation shown in Figure 3. These boxplots show local volume differences by group for selected structures. Y axis values are the deformation index for local volume change (in log Jacobians).
The dominant pattern of anatomical variation observed was that of a significant difference between XX and XY mice, accompanied by significant XO-XX differences in which XO mice carried an accentuation of the difference observed in XX females relative to males (“Pattern 1” in Figs 3 and 4). This pattern either took the form of stepwise volume increase moving from XO to XX to XY groups [BNST, supra-collicular nucleus accumbens, rostromedial amygdala, and periaqueductal gray (PAG)], or step-wise volume decrease [olfactory tubercle, basal forebrain, infra-collicular nucleus accumbens and right frontal cortex].
A contrasting set of regions showed XO effects in the absence of detectable sexual-dimorphism (“Pattern 2” in Figs 3 and 4). These regions either involved an abnormal volume reduction in XO relative to both XX and XY groups (olfactory bulb, thalamus and hypothalamus), or an abnormal volume increase (left fronto-parietal cortex).
We also observed two other patterns of anatomical variation which were each highly circumscribed in nature: (i) a sexually dimorphic region in the right parietal cortex of volume excess in XX relative to XY mice, with XO mice adopting an intermediate position of significantly lower volume than XX, but greater volume than XY (“Pattern 3”, Figs 3 and 4), and (ii) a bilateral caudomedial amygdala focus of volume excess in males relative to both XX and XO groups (who were in turn statistically indistinguishable from each other) (“Pattern 4”, Figs 3 and 4).
4. DISCUSSION
Our combined analysis of sex-differences and X chromosome dosage effects on murine neuroanatomy gave rise to several novel findings that could potentially advance understanding of sexually dimorphic brain development in health, and altered brain development in TS.
4.1 Relevance for Understanding of Sexually Dimorphic Brain Development
We find that the majority of brain regions which are sensitive to X monosomy also show significant anatomical differences between XX and XY mice (i.e Patterns 1 and 3 in Figs 3 and 4). This convergence most often takes the form of step-wise anatomical differences moving from XO to XX to XY groups, such that XO and XY mice show “mirrored” anatomical differences from XX mice – i.e XO < XX when XY > XX and visa-versa (Pattern 1 in Figs 3 and 4). XY-XX differences in the volume of many such regions – especially BNST, medial amygdala, olfactory tubercle, and basal forebrain - have been well described by prior histological work, and are believed to arise via masculinizing effects of testicular androgens in males (Morris et al., 2004; Shah et al., 2004; Zuloaga et al., 2008). However, the fact that many of these regions also show volume differences between XO and XX mice raises the possibility that differences in X-chromosome dosage between karyotypically normal males and females may be acting alongside hormonal factors in shaping these classical foci of sexual dimorphism within the brain. We explicate this idea below using the BNST as an example.
The difference in BNST volume between XO and XX mice could be accounted for by XO-XX dosage differences in two classes of X chromosome gene - PAR genes or non-PAR genes that escape X-inactivation (henceforth “XCIe” genes). If lowered BNST volume in XO vs. XX mice is caused by reduced PAR gene dosage in XO relative to XX mice, then the greater BNST volume in XY vs. XX mice - which both carry two copies of each PAR gene - can be wholly attributed to well described androgenic mechanisms for BNST volume enhancement in XY vs. XX mice (Zuloaga et al., 2008). In contrast, under the alternative hypothesis that lowered BNST volume in XO vs. XX mice is caused by reduced XCIe gene dosage in XO relative to XX mice, then this same tendency to lowered BNST volume relative to XX females should presumably also be conferred to XY males, because both XO and XY mice have one rather than two copies of XCIe genes. Thus, attributing BNST volume reduction in XO vs XX mice to a relative reduction of XCIe gene dosage, implies that BNST volume differences between XY and XX mice arise through opposing effects of sex-differences in XCIe gene dosage and sex differences in circulating androgen levels. Specifically, the influence of a sex-difference in testicular androgens to drive BNST volume excess in XY vs. XX mice (Zuloaga et al., 2008) would be operating in counterpoint to an opposing influence of sex-differences in XCIe gene dose to confer a BNST volume reduction in XY vs. XX mice. To create the step-wise volume differences seen in BNST and other “Pattern 1” regions (Figs 3 and 4), the set point for opposing effects of greater circulating androgens in males and greater XCIe dose in females would have to be one where effects of androgens predominate. Full compensation of one effect by the other would ablate observable volume differences between XX and XY mice, but leave an observable difference between XO and others: providing an alternative mechanism to group differences in PAR dosage as an explanation for the XO specific effects we see in thalamus, hypothalamus, olfactory bulb and left fronto-parietal cortex (Pattern 2 in Fig 3 and Fig 4).
The hypothesis that XY-XX differences in regional brain volume might reflect the sum of opposing sex-steroid and XCIe gene effects could be experimentally addressed using the “Four Core Genotypes” mouse model which allow sex-chromosome and gonadal profile to be dissociated (McCarthy et al., 2012), with important implications for our understanding of sexually-dimorphic neurobiology. For example, brain systems that achieve structural or functional equivalence in males and females through sexually-differentiated underlying mechanisms represent easily overlooked, but potentially critical biological substrates for well-documented sex-differences in human psychopathology (Giedd et al., 2012). The seemingly counterintuitive idea - that defining genetic and endocrine sex-differences might counteract each other to lessen what would otherwise be a much larger dissimilarity between males and female - runs against canonical models(Lenz et al., 2012), but has been lent recent empirical support in studies of murine immunological phenotypes (Palaszynski et al., 2005).
Our study also identifies a caudomedial amygdala focus where males are different from both XX and XO females, who are in turn equivalent to each other (Pattern 4 in Fig 3 and Fig 4). This finding can be fully accounted for by well-documented androgen-dependent increases in medial amygdala size in males relative to females(Morris et al., 2004).
4.2 Relevance for Understanding of Turner Syndrome Neurobiology
As already outlined, the XO mice examined in our study offer a number of advantages over TS as a model for X chromosome influences in mammalian brain development. Many of these advantages simultaneously lessen the power of XO mice as a model for TS. For example, unlike clinical TS cohorts, the XO mice we study (i) do not carry a combination of X-chromosome haploinsufficiency and ovarian dysgenesis, and (ii) all carry an X-chromosome of paternal origin, whereas maternal X chromosome parent of origin is more common than paternal in clinical cohorts of females with TS. The consequence of these differences for generalizability of our murine findings to humans is however hard to establish given limited knowledge regarding the degree to which interactions between gene-dosage and hormonal alterations influence the TS phenotype, or the comparability of X chromosome parent of origin effects in mice and humans.
Notwithstanding these considerations, and in spite of the obvious lack of straightforward homology between the brains of mice and humans, we find several anatomical differences between XO and XX mice that converge with neuroimaging reports in TS. Specifically, a recent systematic review (Raznahan et al., 2010) and subsequent MRI studies in TS (Lepage et al., 2012; Marzelli et al., 2011) have identified replicated foci of anatomical difference between XO and XX humans, which we now show to be significantly altered in XO mice relative to their XX littermates, including the parietal cortex and striatum. Since these shared anatomical alterations in human and murine X-monosomy occur despite the lack of overt ovarian dysgenesis in murine X-monosomy, they potentially point towards a regionally-specific and evolutionarily preserved role for X chromosome genes on brain development. Strong candidate genes in this regard are those that escape X inactivation both humans and mice: STS, MID1, KDM6A, EIF2S3, DDX3X, KDM5C, CA5B and CXorf38 (Yang et al., 2010). It is important to note, however, that specification of XCIe genes in both humans and mice is challenged by the recent observations that in mice alone XCIe genes can vary by organ (Arias-Vasquez et al., 2011; Lopes et al., 2010) and brain region (Gregg et al., 2010b).
Several of the anatomical differences we identify between XO and XX mice have not been yet reported in the existing TS neuroimaging literature: PAG, BNST, basal forebrain, olfactory tubercle, and OB. Clarifying whether these smaller structures are also altered in human X-monosomy will require targeted in-vivo sMRI studies with specialized approaches to image acquisition and segmentation. From a functional perspective, the recognized importance of PAG (Linnman et al., 2012; Silveira et al., 1993) and BNST (Dumont, 2009; Duvarci et al., 2009) for fear processing in the mammalian brain highlights these structures as being of potential interest with respect to reported associations between altered X chromosome dosage and anxiety-related phenotypes in X-monosomic humans (Kesler, 2007) and mice (Isles et al., 2004).
4.3 Next Steps
Our findings in XO mice highlight a number of important areas for future investigation.
First, closer study of endocrine function will be required to detail the relative contribution of gonadally-independent vs. sex-steroid mediated pathways to the anatomical alterations we see in XO mice. Although ovarian function in XO is sufficiently intact to support fertility, XO mice have smaller ovaries, fewer ovarian follicles, and earlier menopause than their XX counterparts(Deckers et al., 1981). These ovarian abnormalities could potentially be relevant for brain development in XO mice. For instance, compromised ovarian function could theoretically reduce androgen biosynthesis in XO mice below that of XX counterparts (Erickson et al., 1985) – which would represent a very parsimonious account for our observation of volume reductions in XO vs XX mice within several regions of androgen-dependent brain volume increase in XY vs XX mice. Presently, conducting an adequate test of this androgenic account is complicated by a lack of good normative models for androgen metabolism and central nervous system function in typically-developing female mice.
Second, the XO mice in our study all carried a paternally inherited X chromosome, and it will be important to test whether our findings generalize to XO mice with maternally inherited X chromosomes(Davies et al., 2005). Third, we studied XO mice post-pubertally, and longitudinal studies of live mice will be required to better characterize the developmental timing of anatomical differences among XX, XY, and XO mice. Fourth, future in-vivo studies will also make it possible to investigate the behavioral relevance of anatomical findings.
Notwithstanding the importance of these areas for future research, our findings on parallel analysis of anatomy in XO, XX, and XY (i) pose highly testable sets of competing hypotheses regarding the nature of sex-specific hormonal and chromosomal influences in brain development, (ii) support the notion that involvement of the parietal cortex and striatum in TS reflects an evolutionarily preserved role for X chromosome influences on mammalian brain development, and (iii) raise the PAG, BNST, basal forebrain and olfactory systems as novel targets for closer neuroanatomical study in TS.
Acknowledgments
This study was funded through the National Institutes of Health, National Institute of Health Intramural Research, and the Canadian Institute of Health Research. FJP holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. We thank Monica J Justice for the use of laboratory space and reagents; and Christine Laliberte, Shoshana Spring and Dr. Miriam Friedel for their assistance with data acquisition and processing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arias-Vasquez A, Altink ME, Rommelse NN, Slaats-Willemse DI, Buschgens CJ, Fliers EA, Faraone SV, Sergeant JA, Oosterlaan J, Franke B, Buitelaar JK. CDH13 is associated with working memory performance in attention deficit/hyperactivity disorder. Genes Brain Behav. 2011;10:844–851. doi: 10.1111/j.1601-183X.2011.00724.x. [DOI] [PubMed] [Google Scholar]
- Arnold AP. Sex chromosomes and brain gender. Nat Rev Neurosci. 2004;5:701–708. doi: 10.1038/nrn1494. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill LS, Laliberte CL, Ellegood J, Spring S, Gleave JA, Eede MC, Lerch JP, Henkelman RM. Preparation of fixed mouse brains for MRI. Neuroimage. 2012;60:933–939. doi: 10.1016/j.neuroimage.2012.01.100. [DOI] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. A genetic basis for a postmeiotic x versus y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8:e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, Wilkinson L. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nature Genetics. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- Deckers JFM, van der Kroon PHW, Douglas L. Some characteristics of the X0 mouse (Mus musculus L.) II. Reproduction: fertility and gametic segregation. Genetica. 1981;57:3–11. [Google Scholar]
- Dumont EC. What is the bed nucleus of the stria terminalis? Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2009;33:1289–1290. doi: 10.1016/j.pnpbp.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvarci S, Bauer EP, Pare D. The bed nucleus of the stria terminalis mediates inter-individual variations in anxiety and fear. Journal of Neuroscience. 2009;29:10357–10361. doi: 10.1523/JNEUROSCI.2119-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson GF, Magoffin DA, Dyer CA, Hofeditz C. The ovarian androgen producing cells: a review of structure/function relationships. Endocrine Reviews. 1985;6:371–399. doi: 10.1210/edrv-6-3-371. [DOI] [PubMed] [Google Scholar]
- Garagorri JM, Rodriguez G, Lario-Elboj AJ, Olivares JL, Lario-Munoz A, Orden I. Reference levels for 17-hydroxyprogesterone, 11-desoxycortisol, cortisol, testosterone, dehydroepiandrosterone sulfate and androstenedione in infants from birth to six months of age. European Journal of Pediatrics. 2008;167:647–653. doi: 10.1007/s00431-007-0565-1. [DOI] [PubMed] [Google Scholar]
- Garces C, de Oya I, Lopez-Simon L, Cano B, Schoppen S, Gil A, de Oya M. Hormone levels in 12- to 15-year-old boys and girls in Spain and their relationship with anthropometric variables. Clinical Biochemistry. 2008;41:621–624. doi: 10.1016/j.clinbiochem.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of Statistical Maps in Functional Neuroimaging Using the False Discovery Rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Raznahan A, Mills K, Lenroot RK. Review: magnetic resonance imaging of male/female differences in human adolescent brain anatomy. Biol Sex Differ. 2012;3:19. doi: 10.1186/2042-6410-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010b;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EJ, Bond J, Svrckova P, Makropoulos A, Ball G, Sharp DJ, Edwards AD, Hajnal JV, Counsell SJ. Regional changes in thalamic shape and volume with increasing age. Neuroimage. 2012;63:1134–1142. doi: 10.1016/j.neuroimage.2012.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isles AR, Davies W, Burrmann D, Burgoyne PS, Wilkinson LS. Effects on fear reactivity in XO mice are due to haploinsufficiency of a non-PAR X gene: implications for emotional function in Turner’s syndrome. Human Molecular Genetics. 2004;13:1849–1855. doi: 10.1093/hmg/ddh203. [DOI] [PubMed] [Google Scholar]
- Kesler SR. Turner syndrome. Child and Adolescent Psychiatric Clinics of North America. 2007;16:709–722. doi: 10.1016/j.chc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz KM, Nugent BM, McCarthy MM. Sexual differentiation of the rodent brain: dogma and beyond. Front Neurosci. 2012;6:26. doi: 10.3389/fnins.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage JF, Mazaika PK, Hong DS, Raman M, Reiss AL. Cortical Brain Morphology in Young, Estrogen-Naive, and Adolescent, Estrogen-Treated Girls with Turner Syndrome. Cerebral Cortex. 2012 doi: 10.1093/cercor/bhs195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Carroll JB, Dorr A, Spring S, Evans AC, Hayden MR, Sled JG, Henkelman RM. Cortical thickness measured from MRI in the YAC128 mouse model of Huntington’s disease. Neuroimage. 2008;41:243–251. doi: 10.1016/j.neuroimage.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Sled JG, Henkelman RM. MRI phenotyping of genetically altered mice. Methods in Molecular Biology. 2011;711:349–361. doi: 10.1007/978-1-61737-992-5_17. [DOI] [PubMed] [Google Scholar]
- Linnman C, Moulton EA, Barmettler G, Becerra L, Borsook D. Neuroimaging of the periaqueductal gray: state of the field. Neuroimage. 2012;60:505–522. doi: 10.1016/j.neuroimage.2011.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes AM, Burgoyne PS, Ojarikre A, Bauer J, Sargent CA, Amorim A, Affara NA. Transcriptional changes in response to X chromosome dosage in the mouse: implications for X inactivation and the molecular basis of Turner Syndrome. BMC Genomics. 2010;11:82. doi: 10.1186/1471-2164-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marzelli MJ, Hoeft F, Hong DS, Reiss AL. Neuroanatomical spatial patterns in Turner syndrome. Neuroimage. 2011;55:439–447. doi: 10.1016/j.neuroimage.2010.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Arnold AP, Ball GF, Blaustein JD, De Vries GJ. Sex differences in the brain: the not so inconvenient truth. Journal of Neuroscience. 2012;32:2241–2247. doi: 10.1523/JNEUROSCI.5372-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Jordan CL, Breedlove SM. Sexual differentiation of the vertebrate nervous system. Nature Neuroscience. 2004;7:1034–1039. doi: 10.1038/nn1325. [DOI] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman TL, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends in Genetics. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Palaszynski KM, Smith DL, Kamrava S, Burgoyne PS, Arnold AP, Voskuhl RR. A yin-yang effect between sex chromosome complement and sex hormones on the immune response. Endocrinology. 2005;146:3280–3285. doi: 10.1210/en.2005-0284. [DOI] [PubMed] [Google Scholar]
- Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology. 1959;65:369–382. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Cooper ML, Cheung SW, Justice MJ. Genotype, phenotype, and karyotype correlation in the XO mouse model of Turner Syndrome. Journal of Heredity. 2008;99:512–517. doi: 10.1093/jhered/esn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Cutter W, Lalonde F, Robertson D, Daly E, Conway GS, Skuse DH, Ross J, Lerch JP, Giedd JN, Murphy DD. Cortical anatomy in human X monosomy. Neuroimage. 2010;49:2915–2923. doi: 10.1016/j.neuroimage.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NM, Pisapia DJ, Maniatis S, Mendelsohn MM, Nemes A, Axel R. Visualizing sexual dimorphism in the brain. Neuron. 2004;43:313–319. doi: 10.1016/j.neuron.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Silveira MC, Sandner G, Graeff FG. Induction of Fos immunoreactivity in the brain by exposure to the elevated plus-maze. Behavioural Brain Research. 1993;56:115–118. doi: 10.1016/0166-4328(93)90028-o. [DOI] [PubMed] [Google Scholar]
- Skuse DH, James RS, Bishop DV, Coppin B, Dalton P, Aamodt-Leeper G, Bacarese-Hamilton M, Creswell C, McGurk R, Jacobs PA. Evidence from Turner’s syndrome of an imprinted X-linked locus affecting cognitive function. Nature. 1997;387:705–708. doi: 10.1038/42706. [DOI] [PubMed] [Google Scholar]
- Sybert VP, McCauley E. Turner’s syndrome. New England Journal of Medicine. 2004;351:1227–1238. doi: 10.1056/NEJMra030360. [DOI] [PubMed] [Google Scholar]
- Trolle C, Hjerrild B, Cleemann L, Mortensen KH, Gravholt CH. Sex hormone replacement in Turner syndrome. Endocrine. 2012;41:200–219. doi: 10.1007/s12020-011-9569-8. [DOI] [PubMed] [Google Scholar]
- Wallace GL, Robustelli B, Dankner N, Kenworthy L, Giedd JN, Martin A. Increased gyrification, but comparable surface area in adolescents with autism spectrum disorders. Brain. 2013;136:1956–1967. doi: 10.1093/brain/awt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff DJ, Van Dyke DL, Powell CM Working Group of the ALQAC. Laboratory guideline for Turner syndrome. Genet Med. 2010;12:52–55. doi: 10.1097/GIM.0b013e3181c684b2. [DOI] [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Human Molecular Genetics. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Yamagata B, Barnea-Goraly N, Marzelli MJ, Park Y, Hong DS, Mimura M, Reiss AL. White matter aberrations in prepubertal estrogen-naive girls with monosomic Turner syndrome. Cerebral Cortex. 2012;22:2761–2768. doi: 10.1093/cercor/bhr355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Research. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuloaga DG, Puts DA, Jordan CL, Breedlove SM. The role of androgen receptors in the masculinization of brain and behavior: what we’ve learned from the testicular feminization mutation. Hormones and Behavior. 2008;53:613–626. doi: 10.1016/j.yhbeh.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]