Abstract
Conjugation of biologically active proteins to polymeric materials is of great interest in the treatment of cancer and other diseases of protein deficiencies. The conjugation of such biomacromolecules is challenging both due to their hydrophilicity and propensity to denature under non-native conditions. We describe a novel reactive self-assembly approach to “wrap” a protein with polymers, simultaneously protecting its delicate folded state and silencing its enzymatic activity. This approach has been demonstrated using caspase-3, an apoptosis-inducing protein, as the first case study. The protein-polymer conjugation is designed to be reversed under the native conditions for caspase-3, i.e. the reducing environment found in the cytosol. The current strategy allowed release and recovery of up to 86% of caspase activity and nanogel-caspase-3 conjugates induced 70–80% apoptotic cell death shortly thereafter. This approach is widely generalizable and should be applicable to the intracellular delivery of a wide range of therapeutic proteins for treatment of complex and genetic diseases.
TOC Graphic
Proteins perform vital biological functions, ranging from gene regulation to catalysis of metabolic reactions and cell signaling to programmed cell death. Proteins are widely used as therapeutics (‘biologics’), because they exhibit higher specificity and offer more nuanced functions than can be achieved by small synthetic molecules.1–2 For example, small-molecule-based drugs often suffer from off-target activities, especially because of their inability to differentiate within protein sub-classes. These off-target effects can be avoided by using a deficient, repressed or down-regulated protein as a drug directly. However, in vivo stability of proteins has been a significant issue with this approach. PEGylation of proteins has been effective in the in vivo stabilization of protein-therapeutics,3–4 but complementary approaches are necessary as this approach can result in irreversible modification of the surfaces of the protein cargo. Many of the biologically important proteins also have inherent liabilities for manipulation and direct administration, including conformational flexibility, a metastable “folded” state, large size, propensity to aggregate and susceptibility to oxidation or degradation. As a result, the development of new protein-polymer nanoconjugates to stabilize and deliver proteins is gaining tremendous interest.5–8 Traditionally, conjugation of proteins to polymeric nanocarriers has been quite challenging due to the propensity of proteins to denature during polymer conjugation, which often requires using organic solvents and harsh conditions for synthesis. 9–10
The past decade has seen some brilliant contributions to address this need. Conjugation of proteins to telechelic, branched, star polymers and polymeric supramolecular molecules has been achieved without compromising the activity or integrity of certain proteins.11–15 Since most of the current approaches use irreversible covalent conjugation to reactive, surface exposed residues of proteins (such as lysines and cysteines), and since multiple copies of these amino acids might be present on the surface, there have been efforts to genetically modify proteins whereby reactive functional groups can be placed in specific locations,16–21 but not without risks to the inherent immunogenicity. We are interested in developing a strategy that provides for complete reversibility in the protein-polymer conjugation and utilizes native proteins. More specifically, we are also interested in an approach that allows for turning-off the protein activity, and then turning the activity ‘on’ when it reaches its target environment using a specific biological trigger.
A commonly used strategy for reversibility involves the utilization of electrostatic complementarity, which has advantages because of the simplicity in obtaining the formulation.22–25 Since there is a significant literature that suggests that charge-neutral nanoscopic systems are desired for long circulation times, we particularly focus on methods that can conveniently provide a charge-neutral surface. Inverse mini-emulsion methods use large volumes of organic solvents, where the aqueous phase is the dispersed phase. This dispersed phase can be utilized to trap water-soluble monomers, crosslinkers, and proteins and then encapsulate the protein using a polymerization reaction.9–10,26–29 We are interested in developing a reactive self-assembly of a precursor polymer to conjugate proteins, where: (i) the conjugation is covalent, but reversible; (ii) the process does not use organic solvents; (iii) the protein is in its native form; (iv) the protein is encapsulated to be protected from proteases in order to avoid premature degradation; (v) the protein activity is turned-off in the encapsulated form; and (vi) the protein activity is recovered in its native environment. In this manuscript, we disclose a reactive self-assembly strategy that provide a polymer-protein nanoassembly with all these characteristics.
We use a caspase protein to demonstrate the utility of the approach outlined here. Caspases are cysteine proteases that are known for their exquisite specificity for cleaving after particular aspartic acid residues and rapidly inducing apoptotic cell death.30–31 Their ability to trigger apoptosis makes them attractive cargo for selective cell killing, especially in cancer therapeutics. However, caspases are inherently fragile because they contain dynamic loops, reactive cysteines poised for chemistry at their active site, and are prone to aggregation due to their tetrameric composition. These qualities therefore necessitate a delivery system that will not damage the protein and protect it simultaneously. Among the family of apoptotic caspases, caspase-3 is of special interest due to its major role in cleaving substrates during apoptosis32 as well as its high catalytic rate.33 Reversible conjugation with silenced activity, targeted in this study, is also ideal to study with this protein, because accidental release of active caspases can lead to irreversible proteolytic damage or even unwanted apoptotic death. Thus, the therapeutic potential of caspases for inducing cell death using intrinsic biological pathways could be ultimately harnessed, if it were possible to silence caspase activity through polymer conjugation and then recover its activity in response to a specific trigger.
EXPERIMENTAL SECTION
Methods and Materials
Polyethylene glycol monomethyl ether methacrylate (PEGMA) (MW 475), 2,2’-dithiodipyridine, 2,2’-azobis(2-methylpropionitrile) (AIBN), 4-cyano-4-(phenylcarbonothioylthio) pentanoic acid (chain transfer agent), D,L-dithiothreitol (DTT), and other conventional reagents were obtained from commercial sources and were used without further purification, except for AIBN which was purified by recrystallization. Pyridyl disulfide ethyl methacrylate (PDSMA) was prepared using the previously reported procedure (Macromolecules 2006, 39, 5595–5597). 1H-NMR spectra were recorded on a 400 MHz Bruker NMR spectrometer using the residual proton resonance of the solvent as the internal standard. Chemical shifts are reported in parts per million (ppm). Molecular weight of the polymer was estimated by gel permeation chromatography (GPC) in THF using the poly(methyl methacrylate) (PMMA) standard with a refractive index detector. Dynamic light scattering (DLS) measurements were performed using a Malvern Nanozetasizer. UV-Visible absorption spectra were recorded on a Varian (Model EL 01125047) spectrophotometer. Size exclusion chromatography was performed on Amersham Biosciences chromatography system equipped with a GE health care life sciences superdex 75 10/300 GL column. Activity assay was performed utilizing Spectramax M5 spectrophotometer. MALDI-MS analyses were performed on a Bruker Autoflex III time-of-flight mass spectrometer. All mass spectra were acquired in the reflectron mode, and an average of 200 laser shots at an optimized power (60%) was used. Cell imaging was performed using Zeiss 510 META confocal microscope.
Synthesis of p(PEGMA-co-PDSMA) (P1)
A mixture of PDSMA (536.8 mg, 2.1 mmol), PEGMA (1 g, 2.1 mmol), 4-Cyano-4-(phenylcarbonothioylthio) pentanoic acid (21 mg, 0.0756 mmol) and AIBN (1.2 mg, 0.00756 mmol) were dissolved in 3 mL THF in a 10 mL Schlenk flask and degassed by performing three freeze-pump-thaw cycles with an argon inflow into the reaction. The reaction vessel was sealed and placed in a pre-heated oil bath at 70 °C for 12 hours. The product polymer P1 was then purified by precipitation in cold ether (20 mL) to yield the random copolymer. Yield: 90%. GPC (THF) Mn: 20 K. Đ: 1.5. 1H-NMR (400 MHz, CDCl3) δ: 8.45, 7.66, 7.09, 4.20–4.06, 3.80–3.42, 3.01, 2.10–1.65, 1.10–0.80. The molar ratios between the two monomers was determined by the integration of the methoxy proton in the polyethylene glycol unit and the aromatic proton in the pyridine and found to be 48%:52% (PEG:PDS).
Synthesis of nanogel-caspase conjugates
NG Empty
The polymer (3 mg) was dissolved in 1mL 1X PBS buffer pH 7.4 and the solution was left stirring at 20 °C for 15 minutes. A calculated amount of DTT was added to the micellar aggregates and the solution was allowed to crosslink for 1 hour at 20 °C. The resulting nanogels were dialyzed at 20 °C using a 7,000 Da MWCO membrane.
NG-Casp-In
The polymer (3 mg) was dissolved in 1mL 1X PBS buffer pH 7.4 and the solution was left stirring at 20 °C for 15 minutes. To this micellar aggregate solution, 0.06 mg of caspase-3 was added and the mixture was left reacting for 1 hour at 20 °C. Then a calculated amount of DTT was added to the solution and was stirred for another hour at 20 °C to allow for crosslinking. The resulting nanogels were dialyzed at 20 °C using a 7,000 Da MWCO membrane and unbound caspase-3 was removed by Amicon Ultra Centrifugal Filters MWCO 100,000.
NG-Casp-Out
The polymer (3 mg) was dissolved in 1 mL PBS buffer pH 7.4 and the solution was left stirring at 20 °C for 15 minutes. A calculated amount of DTT was added to the micellar aggregates and the solution was allowed to crosslink for 1 hour at 20 °C. Then, 0.06 mg of caspase-3 was added and the mixture was left reacting for another hour at 20 °C. The resulting nanogels were dialyzed at 20 °C using a 7,000 Da MWCO membrane and unbound caspase-3 was removed by Amicon Ultra Centrifugal Filters MWCO 100,000.
NG EmptyRRR
To functionalize the surface of the nanogels, the same procedure described above for nanogels was followed. In addition, CRRR peptide (50% by weight compared to the polymer) was added and then stirred for another hour at 20 °C. The resulting nanogels were dialyzed at 20 °C using a 7,000 Da MWCO membrane.
NG-Casp-InRRR
To functionalize the surface of the nanogels, the same procedure described above for NG-Casp-In was followed. In addition, an excess of the ligand (5 mg), CRRR, was added and then stirred for another hour at 20 °C. The resulting nanogels were dialyzed at 20 °C using a 7,000 Da MWCO membrane and unbound caspase-3 was removed by Amicon Ultra Centrifugal Filters MWCO 100,000 Da.
NG-Casp-OutRRR
To functionalize the surface of the nanogels, the same procedure described above for NG-Casp-Out was followed. In addition, CRRR peptide (50% by weight compared to the polymer) was added and then stirred for another hour at 20 °C. The resulting nanogels were dialyzed at 20 °C using a 7,000 MWCO membrane and unbound caspase-3 was removed by Amicon Ultra Centrifugal Filters MWCO 100,000.
Activity assay
For measurement of caspase-3 activity, various amounts of nanogel, ranging from 13 to 230 micrograms, were used in order to release 50 nM caspase-3 in each experiment. These nanogel-caspase conjugates were incubated in 100 mM DTT for 1 hour in order to fully release the cargo caspase-3. Identical samples were subject to 0.5 mM DTT treatment in an identical fashion in order to assay for any free, or unbound, caspase-3. The caspase-3 activity was then assayed over a 7 minute time course in caspase-3 activity assay buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, and 10% PEG 400. In this experiment, caspase-3 hydrolyzes the peptide substrate, N-acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin, resulting in the release of the 7-amino-4-methylcoumarin (AMC) moiety as a fluorophore that can be quantified over time. The fluorogenic substrate N-acetyl-Asp-Glu-Val-Asp-AMC, Enzo Lifesciences (Ac-DEVD-AMC), Ex 365/Em495, was added to a final concentration of 100 µM to initiate the reaction. These 100 µ assays were performed in duplicate in a 96-well microplate at 37 °C using a spectramax M5 spectrophotometer.
RESULTS AND DISCUSSION
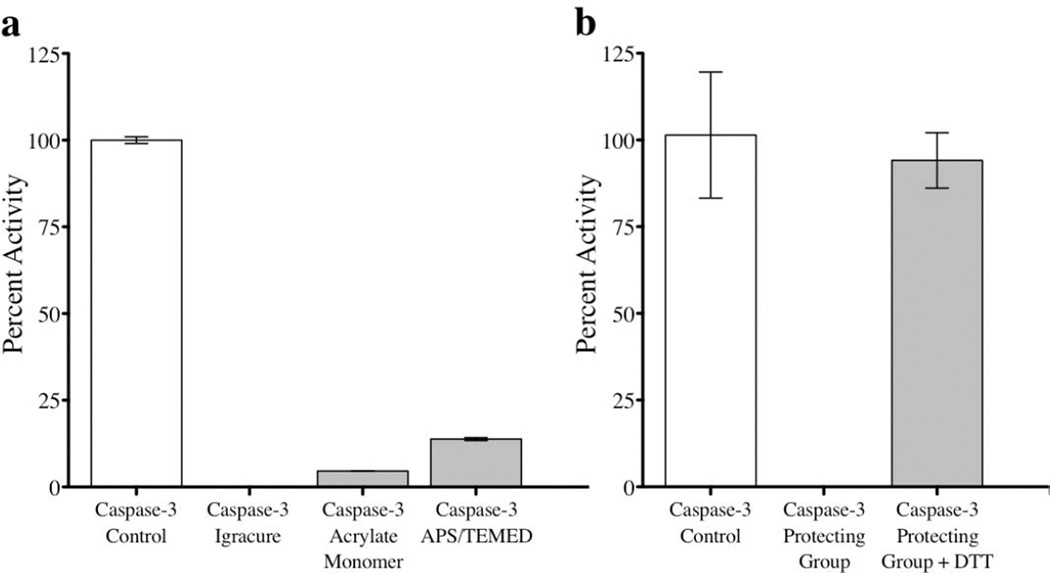
Since caspases are active in the cytosol, it is clear that these proteins are active under the reducing conditions of high glutathione (GSH) concentrations. Therefore, we were interested in developing a nanogel where the crosslinkers are based on disulfides. In this case, the protein will be stably encapsulated in the low GSH concentration (µM) of the serum, but will be released at high GSH concentration of the cytosol (mM) due to the disulfide bond cleavage in the polymeric nanogel. Accordingly, we attempted the preparation of caspase-encapsulated nanogels using inverse emulsion polymerization. In this case, free radical polymerization of a variety of PEG-functionalized acrylates was carried out in the presence of caspase-3 as the cargo and a diacrylamide derived from cystine as the crosslinker. Although we were able to achieve protein encapsulation, we also found that the process had an irreversibly detrimental effect on caspase-3 activity (data not shown). A number of experiments were then performed to assess the effect of the photoinitiator (Igracure) with light, the acrylate monomer alone, and another common initiator/catalyst combination (APS/TEMED) on caspase-3 activity. The results (Figure 1a) led us to conclude that an irreversible reaction between the surface functional groups of caspase-3 and the reactants utilized for this inverse emulsion polymerization, specifically the acrylic monomers, destroyed the protein’s activity. In fact, incubation of caspase-3 with the PEG-acrylate monomer alone indeed irreversibly damaged the activity of the protein.
Figure 1.
Recovery of caspase-3 activity after treatment with various polymerization agents. (a) Caspase-3 was incubated with a set of polymerization agents, all of which had severe effects on the activity compared to the control. These agents included: (1) a common photoinitiator, Igracure, followed by UV treatment (2) an acrylate monomer frequently used in inverse emulsion polymerization methods and (3) an ammonium persulfate (APS) initiator and catalyst tetramethylethylenediamine (TEMED). (b) Caspase-3 was silenced when reacted with cysteinyl-2-pyridyl disulfide (protecting group) in the absence of reductant. However, this silenced caspase regained 94% activity after treatment with the reductant DTT.
This observation of irreparable damage to the protein function led us to reassess our polymer linkage to this fragile enzyme. We hypothesized that the reactive cysteines at the active site of the protein were interacting with the polymerization agents and asked if we could use reversible thiol chemistry to our advantage. Since pyridyldisulfide undergoes a selective thiol-disulfide exchange with reduced thiols of cysteines, we treated caspase-3 with cysteinyl-2-pyridyl disulfide. The product disulfide acts as a protecting group to completely silence the caspase-3 activity in an oxidized state, but upon exposure to a reducing environment the protein is able to regain full activity (Figure 1b).
We were inspired by these results at the possibility of a reactive self-assembly approach to encapsulate proteins in a way that would turn-off its activity and then reversibly recover the activity under the native conditions of the enzyme. There are five reduced cysteines that are surface exposed in a caspase monomer (Figure S1). If we were to mix caspase-3 with a copolymer that contains pyridyldisulfide moieties as side chains, then we hypothesized that the cysteines in the protein would react with the polymer to afford a self-assembled protein-polymer conjugate due to the thiol-disulfide exchange reaction. Pyridyldisulfide (PDS) is a hydrophobic functional group and when copolymerized with a PEG-containing monomer, the copolymer forms amphiphilic assembly. We have previously used a 7:3 ratio of PDS and oligoethyleneglycol (OEG) based monomers to obtain robust amphiphilic assemblies to sequester hydrophobic guest molecules.34 However, for the reactive self-assembly with a protein that we envision here, it is essential that the polymer is more dynamic. Therefore, we targeted a polymer that contains a lower percentage of the hydrophobic PDS-based monomer and used a 1:1 ratio of these two monomers for the self-assembly.
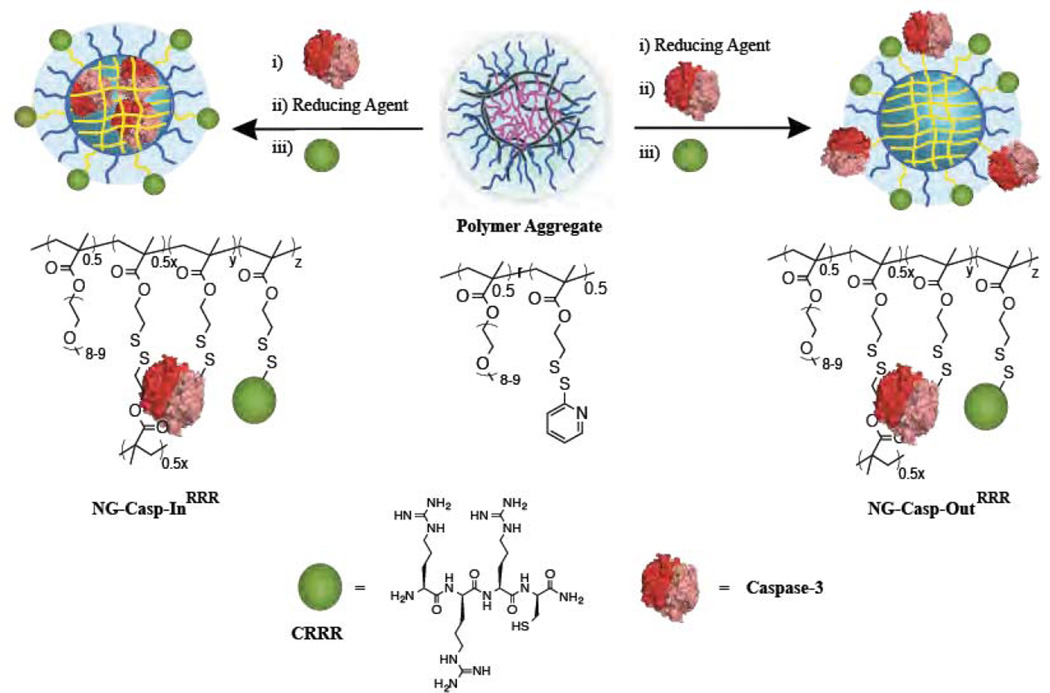
The random copolymer nanogel precursor was obtained by the reversible addition-fragmentation chain transfer (RAFT) polymerization of OEG-methacrylate and PDS-methacrylate. The feed ratio of the monomers was 50:50 and experimentally the resulting copolymer was found to contain 48% OEG units and 52% PDS groups, as discerned by NMR (Figure S2). Polydispersity and Mn was found to be 1.5 and 20K, respectively. To encapsulate the protein using the reactive self-assembly strategy, caspase-3 and the amphiphilic polymer were added simultaneously to aqueous media. The protein was presumably wrapped in the assembly by initiating thiol-disulfide exchange reactions between PDS groups; the assembly was further locked-in with the addition of a precise amount of DTT (Scheme 1, NG-Casp-In). It is likely that the exposed cysteines on the caspase play a critical role in this reactive self-assembly process.
Scheme 1.
Preparation of nanogel-caspase-3 conjugates. Covalent conjugation of caspase-3 in the interior or on the surface of polymeric redox sensitive nanogels through disulfide linkages
Concurrently, we also targeted a control nanogel, where the nanoassembly did not contain any encapsulated protein (NG-empty). In an aqueous solution, the polymer forms an aggregate, which was locked-in using a self-crosslinking process enabled by intra- and inter-chain disulfide cross-linking of the PDS groups in the presence of the reducing agent dithiothreitol (DTT). The nanogel formation process was monitored by tracing the absorption spectra of 2-pyridinethione (byproduct of the disulfide crosslinking) at 343 nm (Figure S4).34 Based on the 2-pyridinethione released, the crosslink density of these nanogels was 18% (Figure S4a). As an additional control, we also prepared a nanogel, where the caspase-3 is covalently conjugated to the surface of the nanogel. Here, we first prepared the cross-linked nanogels (similar to NG-empty); caspase-3 was then covalently attached on its surface using the cysteine residues of the protein and unreacted PDS groups (Scheme 1, NG-Casp-Out). This effectively decorated the nanogel with caspase-3 attached to the outside. Due to their hydrophobic nature, we suspected that the unreacted PDS groups might collapse into the core of the nanogels and therefore might not be available for surface functionalization. This seems to be not the case and surface functionalization can indeed be achieved with reasonable efficiency.
To ultimately evaluate these nanogels in both passive and activated cellular uptake pathways, we also prepared nanogels with cell penetrating capabilities by incorporating a cysteine-containing tri-arginine peptide35–37 on the surface of these nanogels to generate NG-EmptyRRR, NG-Casp-InRRR, NG-Casp-OutRRR (Scheme 1). Incorporation of the peptide on the surface was confirmed by further increase in the 2-pyridinethione absorption spectrum (Figure S4d, e and f). Based on the 2-pyridinethione released, we found that the presence or absence of caspase had no overall impact on the ability to conjugate a second molecular entity. The crosslinking densities of these nanogels NG-EmptyRRR, NG-Casp-InRRR, NG-Casp-OutRRR were each found to be ~18%, suggesting that peptide had been incorporated.
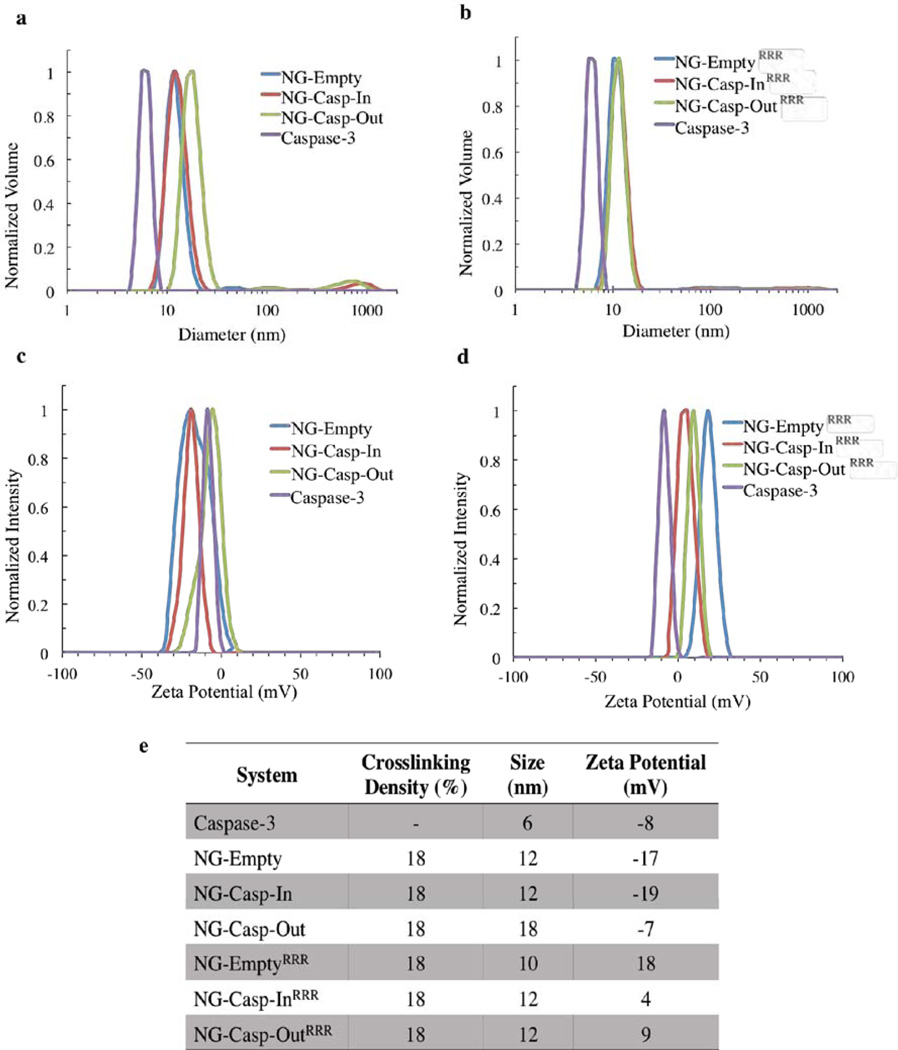
To further characterize these nanogel-protein conjugates, we evaluated their size by dynamic light scattering (DLS) and found that the hydrodynamic diameter of free caspase-3, NG-Empty and NG-EmptyRRR were 6, 12, and 10 nm respectively (Figure 2). After the conjugation of caspase to the nanogels, there was an increase in the size of the conjugates, indicative of protein conjugation (12 nm – 18 nm, Figure 2a and b). Caspase-3 is negatively charged (pI 6.1), whereas CRRR is positively charged due to the arginine residues. These differences led us to evaluate the change in the surface charge of these conjugates by zeta potential measurements. After conjugation of caspase-3, the surface charge of NG-Casp-In (−19 mV) did not change significantly from the value observed in the original NG-Empty (−17 mV, Figure 2c). In the case of NG-Casp-Out, the zeta potential value obtained shifted towards −7 mV, similar to the value observed for free caspase-3 (−8 mV). Although these are PEG-functionalized surfaces, these studies suggest that the nanogels carry a slight, apparent negative charge, which has been previously seen with PEG-functionalized surfaces.38–39 After surface decoration of nanogels with CRRR, we expected that NG-EmptyRRR would show a positive zeta potential value since these nanogels do not have caspase-3 conjugated. As expected, the surface charge for NG-EmptyRRR was found to be +18 mV. With caspase-3 conjugation, the surface charge became less positive: +4 mV for NG-Casp-InRRR and +9 mV for NG-Casp-OutRRR (Figure 2d). These results, along with the UV-vis absorption spectra recorded during the nanogel-caspase conjugates synthesis (Figure S4), suggest that unreacted PDS groups from the nanogels were sterically available to covalently react with the surface exposed cysteine residues from caspase-3. Furthermore, despite the presence of a large and bulky biomacromolecule on these polymeric nanogels, subsequent surface functionalization by CRRR addition can also be achieved, demonstrating the prevalent accessibility and reactivity of the PDS groups. The properties of these nanogel-caspase conjugates are summarized in Figure 2e.
Figure 2.
Nanogel-Caspase Conjugates Characterization. (a) DLS of nanogel-caspase conjugates (b) DLS of nanogel-caspaseRRR conjugates (c) ζ-potential of nanogel-caspase conjugates (d) ζ-potential of nanogel-caspaseRRR conjugates (e) Summary table of the properties of nanogels, caspase-3 and nanogel-caspase conjugates.
To further assess the covalent protein conjugation to the nanogels, we utilized SDS-PAGE to assess caspase dissociation from the nanogels in a reducing environment (Figure 3). When treated with sodium dodecyl sulfate (SDS) and boiled, caspase-3 dissociates into the constituent large and small subunits (17 and 12 kDa, respectively), which migrate as two characteristic bands on SDS-PAGE. We expected that caspase-3 conjugated to the inside or outside of nanogels should not be observable by SDS-PAGE, because it will remain bound to and migrate (or fail to migrate) with the nanogel. We expected to observe the two caspase-3 bands only after treatment of the nanogels with a reducing agent. As anticipated, we did not observe the bands corresponding to caspase-3 in the NG-Casp-In or in the NG-Casp-Out samples in the absence of reductant (Figure 3a). This suggests that caspase-3 is covalently conjugated through disulfide bonds and not simply associated through physical adsorption. Since disulfide bonds can be cleaved at high concentrations of the reducing agent, we hypothesized that we should be able to “unlock” the protein from the assembly by exposing the conjugates to DTT. The appearance of the characteristic caspase-3 bands for the large and small subunits in both samples, NG-Casp-In and NG-Casp-Out, indicated that the protein was released after the reduction of disulfide bonds. This confirmed the conjugation of caspase-3 in both the “in” and “out” configurations (Figure 3b) and demonstrated that these nanogels respond to a specific redox stimulus, namely DTT. We estimated the concentration of caspase-3 released from each conjugate by comparing the intensity of the bands from nanogel-released caspase to known concentrations loaded into neighboring wells. Both nanogel-caspase conjugates (50 µg) released about 2 µg of caspase-3. Similar results were observed for the nanogels functionalized with the cell-penetrating peptide, RRR. Under non-reducing conditions, no caspase bands were observed for the NG-Casp-InRRR and NG-Casp-OutRRR samples (Figure 3c). When the samples were exposed to reducing conditions, we observed the appearance of the bands indicative of free caspase-3 released from NG-Casp-InRRR and NG-Casp-OutRRR (Figure 3d). The concentration of released caspase-3 from 50 µg NG-Casp-InRRR and NG-Casp-OutRRR was found to be 1 µg and 0.5 µg, respectively.
Figure 3.
SDS-PAGE gel validating the nanogel-caspase conjugation through reducible disulfide linkages. (a) Nanogel-caspase conjugates under non-reducing conditions (b) Nanogel-caspase conjugates under reducing conditions (c) Nanogel-caspaseRRR conjugates under non-reducing conditions (d) Nanogel-caspaseRRR under reducing conditions.
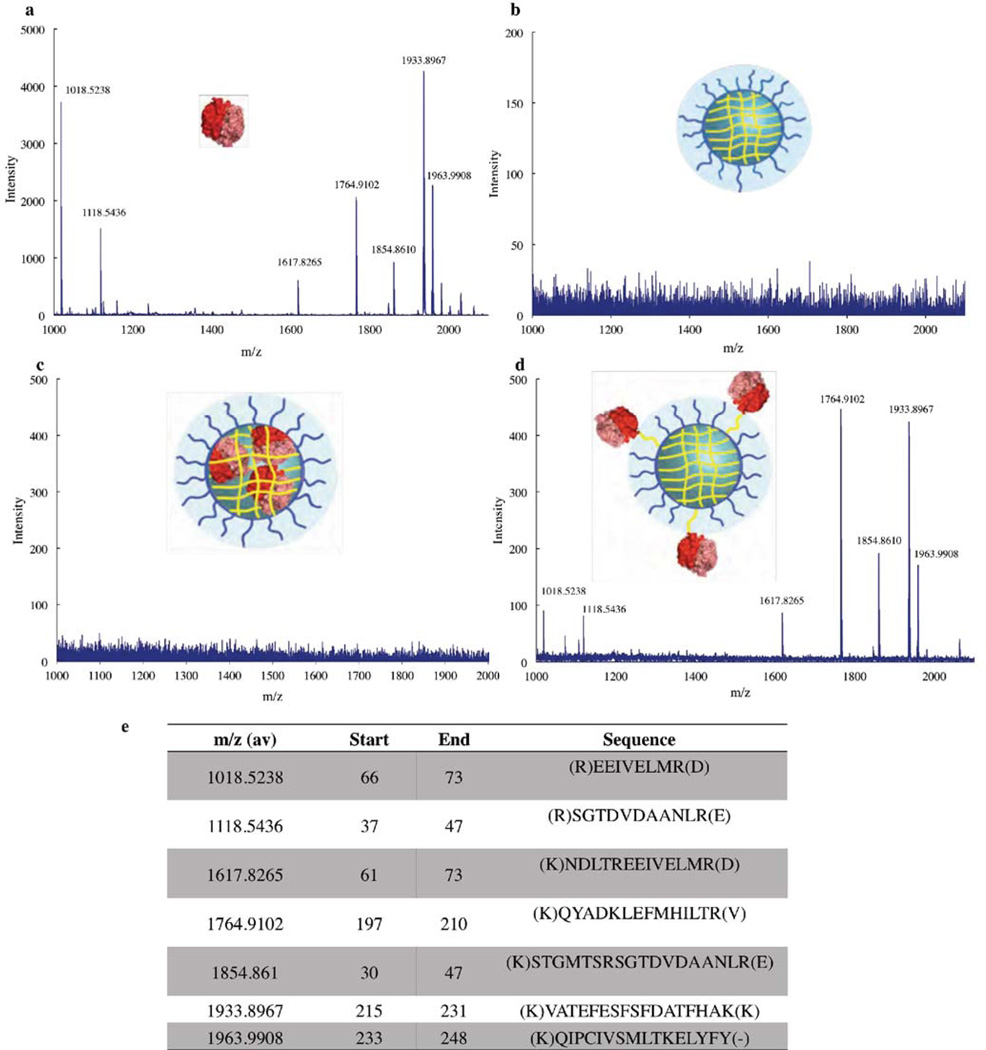
We had hypothesized that the encapsulation of proteins would protect these cargo from protease degradation. We had also hypothesized that the proteins could be encapsulated within the interior of these nanogels or conjugated to the surface of the nanogels by simply altering the order of protein conjugation and crosslinking steps. To test both of these hypotheses, we performed an enzymatic degradation study. The conjugates were exposed to acetonitrile (20% of the total volume) to denature the protein, then added trypsin, a serine protease that hydrolyzes peptide bonds strictly after the basic residues arginine and lysine. These fragments were then analyzed by mass spectrometry (MS). First, caspase-3 itself was subjected to trypsin digest and the MS analysis displayed seven major peaks (Figure 4a). The caspase-3 fragments that each of these peaks represent are shown in Figure 4e.
Figure 4.
Mass Spectrometry of Nanogels-Caspase Conjugates. Mass spectra of (a) caspase-3 (b) NG-Empty (c) NG-Caps-In (d) NG-Caps-Out.
We also analyzed empty nanogels by the same method; as expected, no peaks were observed in the MS (Figure 4b). In the case of NG-Casp-In, we expected that if the caspase-3 was in fact encapsulated within the crosslinked core of the assembly, trypsin would not be able to reach the caspase-3 and therefore no signal would be observed. Indeed, none of the peaks observed previously for caspase-3 were present, suggesting that the protein is “wrapped” and protected from proteolytic digestion within the nanogels (Figure 4c). On the other hand, in the case of NG-Casp-Out, peaks with m/z matching those observed for caspase-3 were detected, demonstrating that these caspase molecules are indeed on the outside of the nanogel assemblies and are not protected from proteolysis like those in the NG-Casp-In state (Figure 4d). These results demonstrate the versatility of the reactive self-assembly to covalently bind proteins and “cage” the protein cargo within the nanogel, protecting it from enzymatic degradation.
A unique aspect of this system is the versatility of these polymeric nanogels to silence enzymatic activity, when conjugated. This capability is only useful if protein cargos retain enzymatic activity upon release from the nanogels. The enzymatic activities of caspase-3 conjugated to the nanogels or released after redox stimulus were assessed using a fluorogenic substrate cleavage assay. Based on quantification of caspase release (Figure 3), the activity from the precise quantity of nanogels encapsulating 50 nM caspase-3 was measured under non-reducing conditions (Figure 5). The catalytic cysteine of caspase-3 requires a reducing environment to be active, so a small amount of DTT (0.5 mM) was added for the “non-reducing conditions.” This was enough reductant to assess activity of any unbound caspase, but not enough to promote disassembly of the nanogels. Under these conditions, none of the conjugates exhibited caspase-3 activity, suggesting that there is no unbound active caspase and the enzyme conjugated to the nanogel is effectively catalytically silenced. This silencing could be due to the structural constraints imposed by the nanogels and/or the lack of accessibility to the peptide substrate. The fact that no caspase-substrate cleavage was observed, even in the NG-Casp-Out nanogels, suggests that even non-encapsulated caspase have been catalytically silenced. Caspase-3 contains a free and highly reactive surface cysteine as part of its catalytic diad (Cys-285). Due to its reactivity, it is likely that this cysteine is involved in the conjugation to the nanogel. Thus, the surface incorporated caspase could be catalytically silent due to covalent conjugation of the active site cysteine-285.
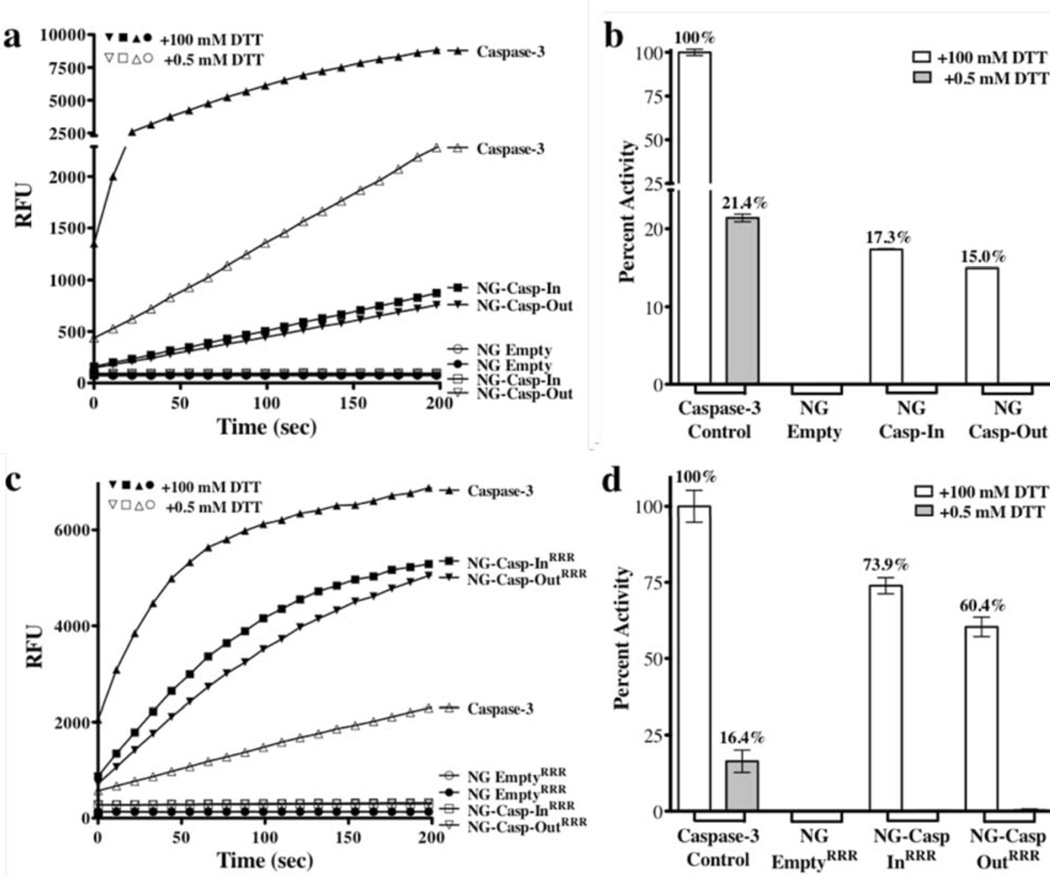
Figure 5.
Protein Enzymatic Activity. (a) Enzymatic activity of caspase-3 (b) Caspase-3 percent activity recovered from nanogel-caspase conjugates was measured under essentially non-reducing (0.5 mM DTT) or fully-reducing (100 mM DTT) conditions. At 0.5 mM DTT no disassembly of nanogels was observed, whereas at 100 mM DTT, full disassembly was observed (c) enzymatic activity of caspase-3 (d) percent activity recovered from nanogel-caspase RRR conjugates. This experiment was performed in duplicate on each of two days.
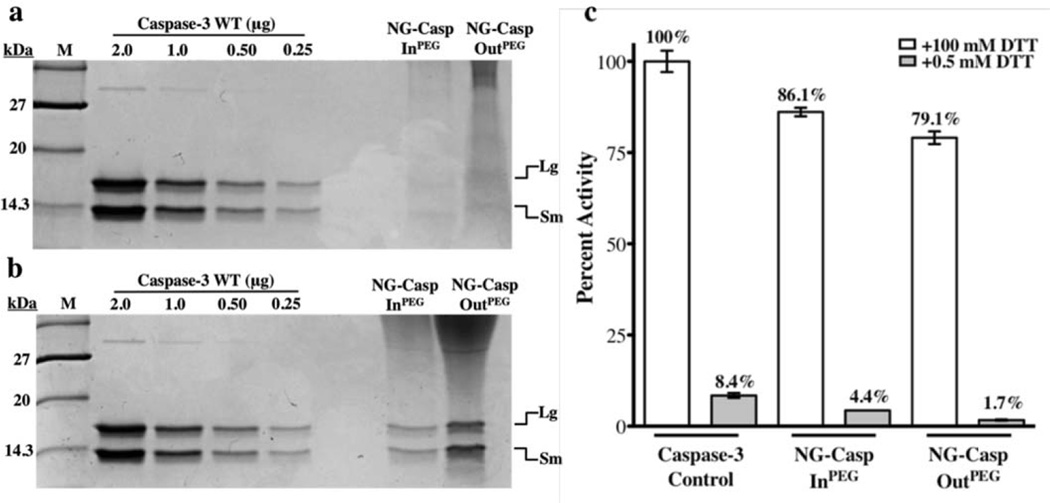
The nanogel-caspase conjugates were then incubated in the presence of 100 mM DTT to release the caspase-3 cargo. Recovery of enzymatic activity under these conditions confirmed that the protein is active upon release from the nanogel (Figure 5a and c). Released caspase-3 activity reached 17% (Figure 5b) upon release from NG-Casp-In and 15% for NG-Casp-Out and 74% for NG-Casp-InRRR and 60% for NG-Casp-OutRRR (Figure 5d). An earlier protocol for the construction of caspase-containing nanogels utilized lyophilization. Caspase-3 is an obligate heterotetramer, which does not refold spontaneously with high yields, so it is not surprising that dramatically lower yields of functional protein (0.24–3%) were released from lyophilized nanogels using the older protocol (Figure S5). Given the 15–74% recovery rates observed currently, it is safe to conclude that the conjugation process itself is mild and that the nanogels protect the caspase, helping it to avoid denaturation and thus retain enzymatic activity. We were surprised to note the higher fraction of recovered caspase-3 activity upon release from the triarginine-containing nanogels, NG-Casp-InRRR and NG-Casp-OutRRR (Figure 5d), compared to those from NG-Casp-In and NG-Casp-Out (Figure 5b). This result was puzzling, since both these types of nanogels had very similar abilities to induce cellular apoptosis (vide infra). A key difference between these nanogels is that the non-functionalized nanogels (NG-Casp-In, NG-Casp-Out) contain unreacted PDS moieties, while these functional groups have been consumed during conjugation of the RRR peptide in the functionalized nanogels (NG-Casp-InRRR and NG-Casp-OutRRR). We hypothesized that, during the release from the non-functionalized nanogels, the added DTT is initially increasing the percent crosslinking (toward 100%) leaving only a fraction of DTT to liberate the caspase from what is then a much more extensively crosslinked nanogel. To test this hypothesis, we prepared the non-functionalized nanogel-conjugates as before, with an 18% crosslinking density and then reacted away the remaining PDS groups using thiol-terminated PEG (MW 1k) to generate NG-Casp-InPEG and NG-Casp-OutPEG. Next, the enzymatic activity of capase-3 was assessed after exposing the conjugates to 100 mM DTT. SDS-PAGE of the nanogel-caspase PEG-thiol conjugates validated the caspase conjugation (Figure 6a and b). Once the non-functionalized nanogel-conjugates were protected from additional crosslinking by the addition of PEG groups, it was possible to release and recover a high percentage of caspase activity (79% and 86%, Figure 6c) comparable to those observed for the nanogel-caspaseRRR conjugates. The extent of recovered caspase-3 activity is quite remarkable considering the lability of the caspase heterotetramer and its dependence on proper formation of the active-site loop bundle for activity. This high yield indicates that these nanogels incorporate their cargo without damaging it, while at the same time remaining robust and responding specifically to a redox stimulus.
Figure 6.
SDS-PAGE gel validating the nanogel-caspaseconjugation through reducible disulfide linkages. (a) Nanogel-caspasePEG conjugates under non-reducing conditions (b) nanogel-caspasePEG conjugates under reducing conditions (c) percent activity recovered from nanogel-caspasePEG conjugates.
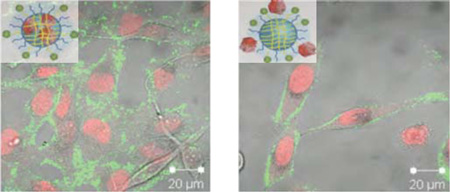
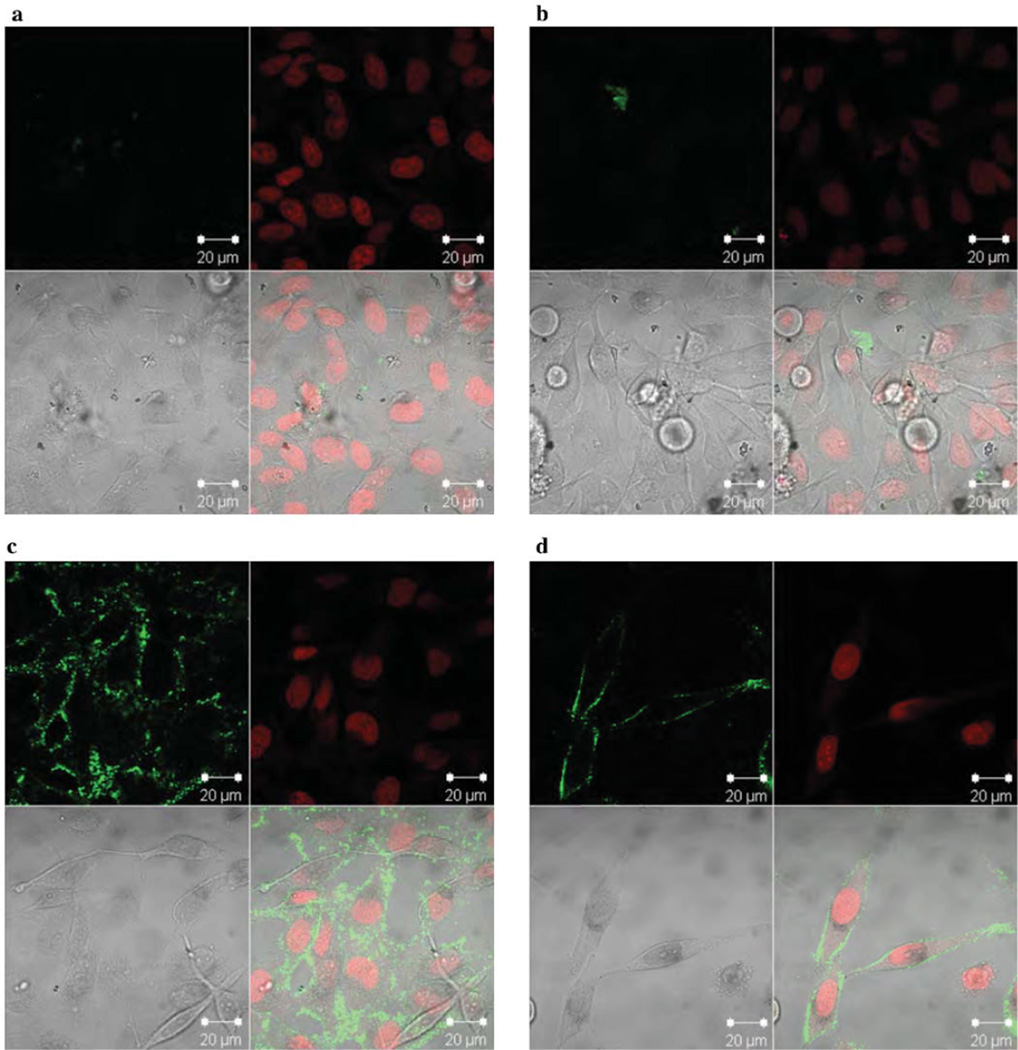
The ultimate goal for the use of these nanogel-protein conjugates is to deliver enzymes in their inactive form and activate them using the innate intracellular environment in mammalian cells. In this case, such caspase delivery is expected to result in cell killing. To determine whether caspase-conjugated nanogels are capable of internalization in living cells, we monitored the cellular uptake of nanogels upon incubation of the conjugates with HeLa cells. Caspase-3 was labeled with fluorescein isothiocyanate (FITC) to enable intracellular visualization; the cell nucleus was stained with DRAQ5. The fluorescence distribution of FITC and DRAQ5 was observed by confocal fluorescence microscopy (Figure 7, Figure S6). FITC-caspase-3 was observed for nanogels caspase on the surface or inside of nanogels (Figure 7a, b), whereas no fluorescence was visible in cells treated with FITC-labeled caspase-3 (Figure S6), suggesting that the protein is not capable of penetrating the cells by itself and requires nanogel conjugation for efficient internalization. Cellular uptake of nanogel conjugates is slow unless peptides for uptake are attached. This trend mirrors previous reports showing no significant internalization of polymeric nanogels at doses of 0.1 mg/mL after 6 hours in HeLa cells.40 As expected, incorporation of triarginine containing peptides onto caspase-containing nanogels (NG-Casp-OutRRR and NG-Casp-InRRR; Scheme 1) displayed higher accumulation of caspase-3 both on the membrane and in the cell within this short time frame. This is likely due to the increased local concentration of the positively charged RRR and the negatively charged cellular membrane.
Figure 7.
Cellular internalization. (a) NG-FITC-Casp-In (b) NG-FITC-Casp-Out (c) NG-FITC-Casp-InRRR (d) NG-FITC-Casp-OutRRR at 0.5 mg/mL on HeLa cells. Within each image set, the top left panel is the FITC channel (green; caspase-3), top right is the DRAQ5 channel (red; nucleus), bottom left is the Diffrential interference contrast (DIC) image and bottom right is the overlap of all three. This experiment was performed with triplicate visualization on one day. One representative field is shown for each condition.
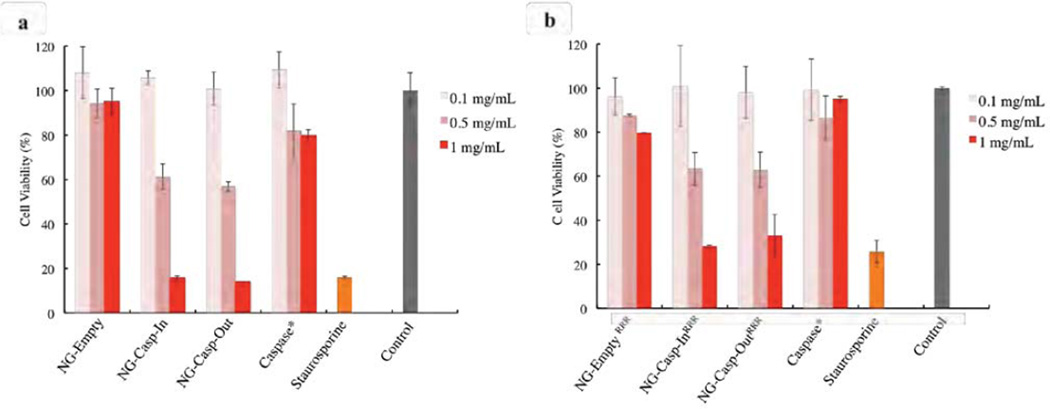
Caspase-3 plays a critical role during the apoptotic process, so we anticipated strong cell death-inducing potential of these nanogel-caspase conjugates, which release up to 75% of the caspase-3 cargo in an active form. The extent of cell death was measured in HeLa cells treated with increasing doses of the nanogel conjugates (Figure 8). Staurosporine, a protein kinase inhibitor known to induce apoptosis41 was used as a positive control. After 24 hours, cell viability was measured using an Alamar Blue assay. Apoptosis is characterized by the marked changes in cell morphology such as cell shrinkage and membrane blebbing.42 HeLa cells treated with nanogel caspase conjugates appear to be rounded and shrinking similar to those undergoing apoptosis induced by staurosporine (Figure S8), suggesting that killing was via an apoptotic route.
Figure 8.
Cell viability after 24 hours exposure of HeLa cells with the conjugates. (a) nanogel-caspase conjugates (b) nanogel-caspaseRRR. The concentration in the caspase-3 samples is the feed amount of caspase-3 used when preparing 0.1 mg/mL, 0.5 mg/mL and 1 mg/mL solutions. Nanogel:Caspase-3 (50:1). Data in (a) were collected from a single experiment performed in triplicate on one day. Data in (b) were collected from experiments were performed in triplicate on two separate days. Data from one day is shown. The second day data is shown in Figure S7.
We anticipated that bare nanogels would be relatively non-toxic and that those conjugated to caspase-3 should exhibit significantly higher rates of cell death induction. NG-Empty exhibited low cellular toxicity at concentrations up to 1 mg/mL (Figure 8a), whereas nanogel-caspase conjugates displayed a strong dose response for cell death. At a concentration of 1 mg/mL, the cell viability for both NG-Casp-In and NG-Casp-Out was reduced to nearly 20%. To confirm that the cell death observed in the nanogel-caspase conjugates was induced by the intracellular release of active caspase-3 aided by the nanogels and not by the action of caspase-3 in solution, cells were exposed to free caspase-3 utilizing the amount of protein fed during the synthesis of 0.1, 0.5 and 1.0 mg/mL nanogels (50:1 weight ratio, nanogel:caspase-3). Since caspase-3 alone is not expected to effectively penetrate the cell membrane, we anticipated that the protein itself should not induce cell death. Indeed, the cell viability observed for caspase-3 was approximately 80% for a concentration up to 1 mg/mL, indicating that the vast majority of the cell death observed corresponded to apoptosis induced by the intracellular release of active caspase-3 from the polymeric nanogels. Similar results were observed for the case of nanogels decorated with RRR peptide (Figure 8b). At a concentration of 1 mg/mL, the cell viability for NG-EmptyRRR was about 80% (Figure 8b); this may be because positively charged RRR peptides directly penetrate the cell membrane, causing rupture or damage, thus introducing slightly higher toxicity. Similar to the nonfunctionalized nanogel caspase conjugates, the RRR caspase nanogels induced cell death in a dose responsive manner. Cell viability for NG-Casp-InRRR and NG-Casp-OutRRR was reduced to about 30–35% at a concentration up to 1 mg/mL. Although nanogels lacking any targeting peptides (Figure 7a, b) show much less cell internalization than the RRR nanogels, they are more capable of inducing cell death. At first this result was perplexing, with cell internalization studies seemingly uncorrelated with increases in cell death. However, upon further investigation we observed that the RRR-decorated nanogels appear to be accumulating on cell membranes, rather than being fully incorporated in the cytoplasm where caspases can be released and activated. Note that caspases work optimally under the reducing and the neutral pH conditions of the cytosol. The activity of these enzymes are substantially reduced under non-native conditions. For examples, although caspase-3 is optimally active at pH 7.4, its activity significant decreases at lower pH values,43–44 dropping to just 10% remaining activity at pH 6.0.33 Thus, although far fewer NG-Casp exist in cells and NG-CaspRRR are more abundant overall, the NG-Casp have greater cell-killing potential, presumably due to their more favorable intracellular localization. This points to an important finding. Optimal delivery of caspases ultimately require localization in the cytosol. The fact that the nanogels (without the cell penetrating peptides) exhibit excellent apoptotic efficiency suggests that these nanocarriers are already doing well in this capacity. Future generations of caspase-containing nanogels that utilize alternate targeting ligands that are capable of cytosolic delivery, rather than the triarginine peptides, should yield even more productive delivery systems, capable of selective cell killing via apoptosis at even lower doses.
Conclusions
We report a novel reactive self-assembly strategy to conjugate active enzymes to polymeric nanogels with responsive characteristics, where caspase-3 has been used as the active protein cargo. In this study, we show that: (i) the proteins can be effectively encapsulated inside a polymeric nanogel by simply mixing the polymer and the protein in this reactive self-assembly strategy. (ii) Conjugation of the protein after the nanogel formation affords a control conjugate, where the protein is attached to the surface of the nanogel. (iii) The activity of the protein is completely turned off in both these approaches, a feature that is critical when delivering cargos that could have deleterious consequences in off-target locations. (iv) The redox sensitive unlocking event causes the protein to be reactivated, allowing recovery of ~80% of the activity. In addition to showing the versatility of these approaches, the fact that these have been demonstrated with a protein that is very prone to irreversible unfolding suggests that this approach is broadly applicable to other proteins. (v) The recovery of activity under reducing conditions can be utilized to recover the enzymatic activity of caspases inside cells, where the reducing environment of the cytosol due to high glutathione concentrations is targeted as the triggering mechanism. This was discerned by the fact that both conjugation approaches provide robust cellular entry, protein release, and apoptotic activity. (vi) The nanogel-protein conjugate was able to gain cellular entry, while the protein by itself did not enter the cells. These results, combined with the fact that the nanogel by itself is not cytotoxic, suggest that these conjugates are promising protein carriers. (vii) The protein conjugated to the surface of the nanogel is prone to proteolysis, while the encapsulated protein is not accessible to proteolytic enzymes. These results suggest that while both approaches are versatile for in vitro protein delivery, the encapsulation approach is more promising for future in vivo applications. (viii) The nanogel-protein conjugates, decorated with cell-penetrating peptides, gain cellular entry much more rapidly compared to the unfunctionalized nanogels. However, the overall apoptotic efficiency of the unfunctionalized nanogels is comparable to (or even better than) those functionalized with RRR. These results show that the unfunctionalized nanogels end up in the cytosol more effectively, as these are not hampered by electrostatic association with cellular membranes. The fact that the unfunctionalized nanogels are slow in cellular uptake, but are more effective in releasing the cargo, bodes well for utilizing these vehicles for targeted delivery of a broad range of protein cargos. These efforts are currently underway in our laboratories.
Supplementary Material
Acknowledgments
Support from the NCI of the National Insitutes of Health (CA169140) is acknowledged. Support from National Science Foundation (CHE-1307118 to ST) and a fellowship for JV and DGT (DGR-0654128) are gratefully acknowledged.
Footnotes
Supporting Information Available: Experimental details, synthetic procedures, NMR, absorption spectra of nanogels-caspase conjugates synthesis, crosslinking density and caspase-3 concentration calculations. This material is available free of charge via the internet at http://pubs.acs.org.
Contributor Information
Jeanne A. Hardy, Email: hardy@chem.umass.edu.
S. Thayumanavan, Email: thai@chem.umass.edu.
References
- 1.Gu Z, Biswas A, Zhao M, Tang Y. Chem. Soc. Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 2.Leader B, Baca QJ, Golan DE. Nat. Rev. Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 3.Joralemon MJ, McRae S, Emrick T. Chem. Commun. 2010;46:1377–1393. doi: 10.1039/b920570p. [DOI] [PubMed] [Google Scholar]
- 4.Abuchowski A, Vanes T, Palczuk NC, Davis FF. J. Biol. Chem. 1977;252:3578–3581. [PubMed] [Google Scholar]
- 5.Gauthier MA, Klok H-A. Polym. Chem. 2010;1:1352–1373. [Google Scholar]
- 6.Al-Tahami K, Singh J. Recent Pat. Drug Deliv. Formul. 2007;1:65–71. doi: 10.2174/187221107779814113. [DOI] [PubMed] [Google Scholar]
- 7.Cobo I, Li M, Sumerlin BS, Perrier S. Nat. Mater. 2015;14:143–159. doi: 10.1038/nmat4106. [DOI] [PubMed] [Google Scholar]
- 8.Pisal DS, Kosloski MP, Balu-lyer SV. J. Pharm. Sci. 2010;99:2557–2575. doi: 10.1002/jps.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murthy N, Xu MC, Schuck S, Kunisawa J, Shastri N, Fréchet JMJ. Proc. Natl. Acad. Sci. 2003;100:4995–5000. doi: 10.1073/pnas.0930644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen JA, Beaudette TT, Tseng WW, Bachelder EM, Mende I, Engleman EG, Fréchet JMJ. Bioconjugate Chem. 2009;20:111–119. doi: 10.1021/bc800338n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Toro DC, Thayumanavan S. Eur. Polym. J. 2013;49:2906–2918. doi: 10.1016/j.eurpolymj.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalia J, Raines RT. Curr. Org. Chem. 2010;14:138–147. doi: 10.2174/138527210790069839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Droumaguet B, Nicolas J. Polym. Chem. 2010;1:563–598. [Google Scholar]
- 14.Broyer RM, Grover GN, Maynard HD. Chem. Commun. 2011;47:2212–2226. doi: 10.1039/c0cc04062b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fierer JO, Veggiani G, Howarth M. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1176–1181. doi: 10.1073/pnas.1315776111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noren CJ, Anthonycahill SJ, Griffith MC, Schultz PG. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 18.Joshi NS, Whitaker LR, Francis MB. J. Am. Chem. Soc. 2004;126:15942–15943. doi: 10.1021/ja0439017. [DOI] [PubMed] [Google Scholar]
- 19.Boutureira O, Bernardes GJL. Chem. Rev. 2015;115:2174–2195. doi: 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- 20.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. Angew. Chemie - Int. Ed. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 21.Gong Y, Leroux J-C, Gauthier MA. Bioconjugate Chem. 2015 doi: 10.1021/bc500611k. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 22.Ayame H, Morimoto N, Akiyoshi K. Bioconjugate Chem. 2008;19:882–890. doi: 10.1021/bc700422s. [DOI] [PubMed] [Google Scholar]
- 23.Lee AL, Wang Y, Ye WH, Yoon HS, Chan SY, Yang YY. Biomaterials. 2008;29:1224–1232. doi: 10.1016/j.biomaterials.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh P, Yang X, Arvizo R, Zhu Z-J, Agasti S, Mo Z, Rotello VM. J. Am. Chem. Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.González-Toro DC, Ryu J-H, Chacko RT, Zhuang J, Thayumanavan S. J. Am. Chem. Soc. 2012;134:6964–6967. doi: 10.1021/ja3019143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachelder EM, Beaudette TT, Broaders KE, Dashe J, Fréchet JMJ. J. Am. Chem. Soc. 2008;130:10494–10495. doi: 10.1021/ja803947s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azagarsamy MA, Alge DL, Radhakrishnan SJ, Tibbitt MW, Anseth KS. Biomacromolecules. 2012;13:2219–2224. doi: 10.1021/bm300646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molla MR, Marcinko T, Prasad P, Deming D, Garman SC, Thayumanavan S. Biomacromolecules. 2014;15:4046–4053. doi: 10.1021/bm501091p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence MJ, Rees GD. Adv. Drug Delivery Rev. 2000;45:89–121. doi: 10.1016/s0169-409x(00)00103-4. [DOI] [PubMed] [Google Scholar]
- 30.Pop C, Salvesen GS. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuñez G, Benedict MA, Hu Y, Inohara N. Oncogene. 1998;17:3237–3245. doi: 10.1038/sj.onc.1202581. [DOI] [PubMed] [Google Scholar]
- 32.McStay GP, Salvesen GS, Green DR. Cell Death Differ. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Calvo M, Peterson EP, Rasper DM, Vaillancourt JP, Zamboni R, Nicholson DW, Thornberry NA. Cell Death Differ. 1999;6:362–369. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- 34.Ryu J-H, Jiwpanich S, Chacko R, Bickerton S, Thayumanavan S. J. Am. Chem. Soc. 2010;132:8246–8247. doi: 10.1021/ja102316a. [DOI] [PubMed] [Google Scholar]
- 35.Fuchs SM, Raines RT. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y, Takehashi M, Tanaka S, Ueda K, Simpson JC, Jones AT, Sugiura Y, Futaki S. Mol. Ther. 2004;10:1011–1022. doi: 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Nakase I, Tadokoro A, Kawabata N, Takeuchi T, Katoh H, Hiramoto K, Negishi M, Nomizu M, Sugiura Y, Futaki S. Biochemistry. 2007;46:492–501. doi: 10.1021/bi0612824. [DOI] [PubMed] [Google Scholar]
- 38.Avgoustakis K, Beletsi A, Panagi Z, Klepetsanis P, Karydas AG, Ithakissios DS. J. Controlled Release. 2002;79:123–135. doi: 10.1016/s0168-3659(01)00530-2. [DOI] [PubMed] [Google Scholar]
- 39.Sezgin Z, Yüksel N, Baykara T. Eur. J. Pharm. Biopharm. 2006;64:261–268. doi: 10.1016/j.ejpb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Ryu J-H, Bickerton S, Zhuang J, Thayumanavan S. Biomacromolecules. 2012;13:1515–1522. doi: 10.1021/bm300201x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang XD, Gillespie SK, Hersey P. Mol. Cancer Ther. 2004;3:187–197. [PubMed] [Google Scholar]
- 42.Ziegler U, Groscurth P. News Physiol. Sci. 2004;19:124–128. doi: 10.1152/nips.01519.2004. [DOI] [PubMed] [Google Scholar]
- 43.Paroutis P, Touret N, Grinstein S. Physiology. 2004;19:207–215. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- 44.Geisow MJ, Evans WH. Exp. Cell. Res. 1984;150:36–46. doi: 10.1016/0014-4827(84)90699-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.