Abstract
A comprehensive survey was conducted along 10 km of geothermal rivers in southwestern Iran. A total of 40 water samples were tested for the presence of Acanthamoeba spp., and genotypes were determined by targeting the diagnostic fragment 3 region of the 18S rRNA gene. The pathogenic potential of all positive isolates was also identified using tolerance ability test. High occurrences of Acanthamoeba (50%) were detected in the sampling areas. Based on sequencing analysis, isolates belonging to T4 (93.7%) and T2 (6.25%) genotypes were reported. Thermo- and osmotolerance tests revealed that five strains are highly pathogenic. Since every collection site of this study was associated with high human activity, posting of warning signs, monitoring of recreational water sources, and awareness of high-risk people are of utmost importance. To the best of our knowledge, the present research is the first to report T2 genotype from geothermal water sources in Iran.
Keywords: Acanthamoeba, geothermal rivers, Iran
Introduction
Acanthamoeba spp. is the causative agent of blindness due to keratitis, rare and fatal encephalitis, cutaneous ulcers, and sinusitis.1–3 The ubiquity of Acanthamoeba genotypes leads to high exposure of people to this unicellular parasite. According to a serological survey carried out by Chappell et al, nearly 100% of human beings encounter Acanthamoeba spp.4 Interestingly, there are recent reports regarding amoebae isolated from the mucosal tissue of healthy and immunocompromised patients.5,6 Acanthamoeba is distributed in both man-made and natural niches, such as soil, dust, fresh water, sea water, hydrotherapy tubs, artificial lakes, rain water, industrial treatment discharges, wastewaters, and textile industrial water.1,7,8 To date, there are 20 known genotypes of Acanthamoeba (T1–T20) according to the sequencing of the diagnostic fragment 3 (DF3) region of the 18S rRNA gene.9–11 T4 is the most prevalent genotype both in clinical and environmental sources; however, not all T4 s are pathogenic.12,13 It is worth mentioning that Khan et al (2009) developed a simple plating assay in order to differentiate pathogenic isolates from the nonpathogenic ones. The set of results from their studies revealed that the ability of strains to grow at higher temperature and osmolarity is the hallmark of pathogenic amoebae.7 This may be due to the higher level secretion of heat shock protein in the pathogenic strains. According to the study by Khan et al.14, amoebae with thermotolerance and osmotolerance abilities seem to be pathogenic. In addition, thermotolerant Acanthamoeba spp. proliferated successfully in water bodies during warmer months of the year and in geothermal-heated waters.
Most diseases related to Acanthamoeba occur as the corneal infection of people in Iran and around the world who use soft contact lens.7,8 However, Acanthamoeba could be the causative agent of other diseases, including Acanthamoeba Granulomatose Encephalitis (AGE). It is worth mentioning that the general lack of awareness among physicians regarding AGE has resulted in its misdiagnosis worldwide. Furthermore, due to the unawareness of physicians regarding the signs, symptoms, and diagnosis procedure of AGE in developing countries, there exist very few reports regarding this disease. However, an increase in the incidences of cerebral infections related to Acanthamoeba spp. in the USA and worldwide has been documented. In Iran, amoebic keratitis is the only reported disease of Acanthamoeba, and it is caused by the genotypes T2, T3, T4, and T11.15,16 Most patients of amoebic keratitis reported coming in contact with contaminated water, hot springs, homemade saline solution, soil, and dust before the onset of the disease.8 Moreover, Acanthamoeba plays both direct (using its own pathogenic factors) and indirect roles as a transmission vehicle for bacteria, viruses, and fungi. It also acts as a reservoir for pathogenic bacteria, including Pseudomonas aeruginosa and Mycobacterium; thus, it also plays an indirect role in public health.17
Ilam Province, located in the southwest of Iran, has several attractions for tourists due to the presence of recreational areas, including thermal springs, natural warm pools, and mud baths; thus, people frequently come in contact with unfiltered water in the recreational geothermal springs and pools. Moreover, there are several reports regarding the occurrence of amoebic keratitis in the country, which resulted from the contact of high-risk people with such waters.18
Thus, the present research was conducted to identify Acanthamoeba genotypes in geothermal water sources and their pathogenic potential in the southwest of Iran, as there were no surveys on its occurrence. To the best of our knowledge, this is the first of such study in the southwest of Iran.
Materials and Methods
Sampling location
Dehloran is a city in Ilam Province, southwestern Iran. Its population during the 2012 census was 30,989, with 7,623 families. The most popular and attractive points in the area are geothermal rivers and pools (Dehloran River and Tar River). These thermal rivers are located 3 km from Dehloran and they are used by the local people and tourists for therapeutic and recreational purposes. Along the Dehloran River, there are seven hot springs and two warm swimming pools where water originates from geothermal springs, and these pools serve as recreational areas for tourists and endemic people. A mud bath and springs are also present in the area having a high population of sick people, including people with arthritis, lumbar pain, and muscular disorders (Fig. 1). Thus, the area is also very popular for various recreational sites of thermal rivers and therapeutic springs. Figure 1 shows the sites of the sampling area.
Figure 1.
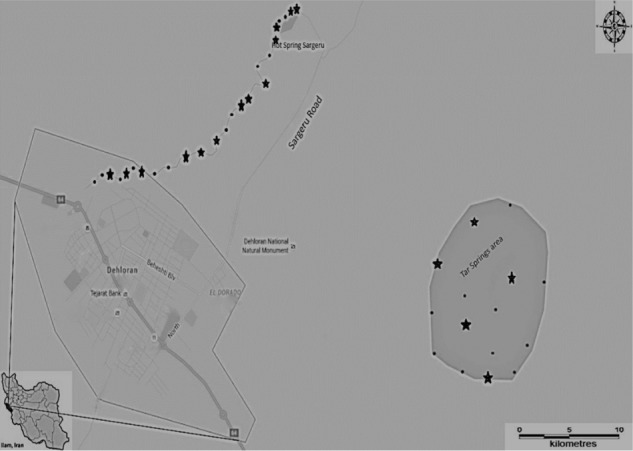
Map showing the sampling area in Ilam Province, southwestern Iran (solid circles indicate the sites of sampling and stars indicate the positive sites).
Cultivation and morphological identification
A total of 40 geothermal water samples with an average temperature of 30 °C–50 °C were collected along 10 km of thermal rivers (Dehloran River and Tar River). One sample per site was used for the study. Overall, 40 sites frequently used by people and tourists were included in the present study. Sampling was done within two days. To this end, 500 mL sterile containers were used to collect water samples at a depth range of 5–10 cm below the surface. All samples were transferred to the laboratory within 24 hours of collection. Approximately, 400 mL of each sample was filtered using nitrocellulose filters (pore size: 0.45 μm; Sigma-Aldrich) and cultivation was done using 2% non-nutrient agar (Difco) covered with Escherichia coli. Briefly, a non-nutrient agar medium was prepared using distilled water and Bacto agar (Difco). The medium was then autoclaved. The filters were placed at the surface of the agar. On the second day of the experiment, the perimeter around the sample was cut out and subcultured onto a fresh plate. This procedure was done in order to reduce fungal and bacterial contamination. The presence of Acanthamoeba spp. was observed using an inverted microscope. Acanthamoeba was detected using the characteristic of endocysts, the shape of trophozoites, and the formation of pseudopodia. After 24 hours of incubation, microscopic detection was carried out. Axenic cultures were then achieved by subsequent cultures of the sourced culture. Briefly, a single trophozoite or cyst was first observed using an inverted microscope, and with the aid of a sterile scalpel, the amoebae were transferred onto a new culture agar. A pure culture was then achieved after several passages.
DNA extraction and polymerase chain reaction analysis
DNA extraction was carried out using a modified phenol–chloroform method.8,19 Amplification of the DF3 region of 18S rRNA (rDNA) was performed using JDP1–2 primer pairs.20,21 The sequences of JDP primers were as follows: JDP1 5′-GGCCCAGATCGTTTACCGTGAA-3′ and JDP2 5′-TCTCACAAGCTGCTAGGGAGTCA-3′.20 Polymerase chain reaction (PCR) was performed in a thermal cylinder (Techne) using ready-made master mix (Taq DNA Polymerase Master Mix RED, Denmark) at a final volume of 30 μL. The master mix contained MgCl2, dNTPs, Taq DNA polymerase, red dye, and stabilizer. Briefly, 25 μL of Taq Master Mix was mixed with 5 ng template DNA, 0.1 μM of each primer, and distilled water.
The thermal cycling conditions were set at an initial denaturing step of 94 °C for 1 minute and 35 repetitions at 94 °C for 35 seconds; annealing step was 56 °C for 45 seconds, and 72 °C for 1 minute. PCR amplicons were separated using 1.5% agarose gel stained with a solution of ethidium bromide and observed under ultraviolet light. A DNA ladder was used to estimate the size of the amplicon.
Sequencing, homology analysis, and nucleotide sequence accession numbers
PCR amplicons were purified and resolved using the ABI 3130X automatic sequencer in Takapouzist Company. The amplification primers (JDP1 and JDP2) were submitted as sequencing primers targeting the diagnostic fragment of the 18S rRNA gene. Genotype identification was performed using nucleotide similarity search with the aid of the Basic Local Alignment Search Tool (BLASTn) to search for the most similar reference sequences. DNA sequences for the new strains were submitted in the genetic sequence database at the National Center for Biotechnical Information (NCBI) using the Sequin program (version 10.3) under the accession number: KU356836-51.
Tolerance assays
Two physical pathogenicity assays were performed on the positive isolates according to previous studies.3 Each positive strain was subcultured and incubated at two temperatures (37 °C and 44 °C) for 24, 48, and 72 hours. Inverted microscopy was used to monitor the outgrowths of the amoebae. For osmotolerance assay, each positive isolate was subcultured in two plates at different molarities (0.5 and 1 M), using D-Mannitol (Merck). The outgrowths of amoebae were then monitored within 24–72 hours.
Results
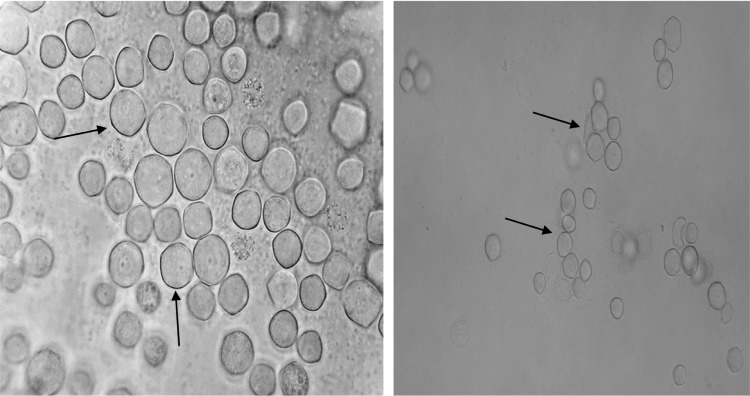
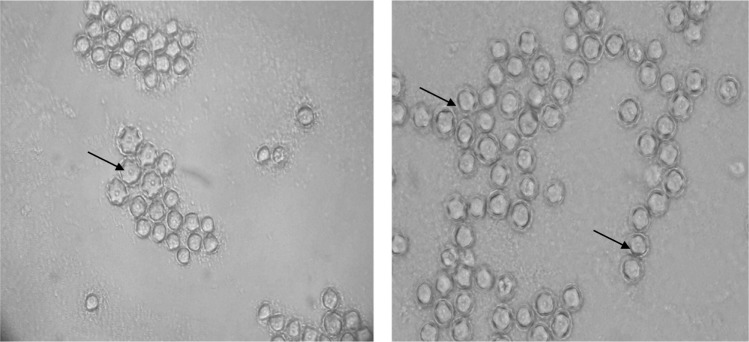
Among the 40 geothermal water samples, 20 (50%) were positive for Acanthamoeba based on the morphological characteristics of amoebae. It is also important to note that 42.8% of hot springs were contaminated with Acanthamoeba. Axenic culture was successfully carried out on 16 isolates; however, four positive plates were not successful in axenic cultivation due to high bacterial contamination. Thus, 16 strains were submitted for molecular analysis. Acanthamoeba revealed the presence of double-walled cysts measuring 20–25 μm (Figs. 2 and 3) and trophozoites (Fig. 4) having flat shape and spine-like structures. Genotyping of the DF3 region revealed 15 isolates (93.75%), which belonged to the T4 genotype, and a single strain labeled as NS11 was identified as T2 genotype (Table 1). The identity and query coverage of isolates in comparison with available genes in the gene data bank were 92%–99% and 83%–99%, respectively (Table 1). Two strains labeled NS14 and NS28 exhibited high identity with Acanthamoeba polyphaga and Acanthamoeba castellanii. Interestingly, the mud bath (isolated strain: NS22) used by the majority of people with disability was contaminated with the highly pathogenic Acanthamoeba (Table 1). It was also observed that five Acanthamoeba strains (NS1, NS5, NS10, NS14, and NS22) possessed the ability to grow at all incubation temperatures and revealed a fast growth at both 37 °C and 44 °C and high osmolarity (1 M). The aforementioned isolates were considered as highly pathogenic amoebae. It is worth mentioning that isolates with the ability to grow at higher temperature and osmolarity are considered as highly pathogenic strains. On the other hand, if the amoebae growth is detectable at 37 °C with a 0.5 M molarity, they are considered as low pathogens. Acanthamoeba belonging to the T2 genotype (NS11) exhibited a fast growth at a temperature of 37 °C and 0.5 M osmolarity and is thus classified as a strain with low pathogenicity potential (Table 1).
Figure 2.
Photomicrograph of Acanthamoeba genotype T2 with low pathogenicity isolated from a thermal pool in Ilam Province (left magnification ×1,000; right: ×400).
Figure 3.
Photomicrograph of pathogenic Acanthamoeba genotype cysts isolated from a river in Ilam Province.
Figure 4.
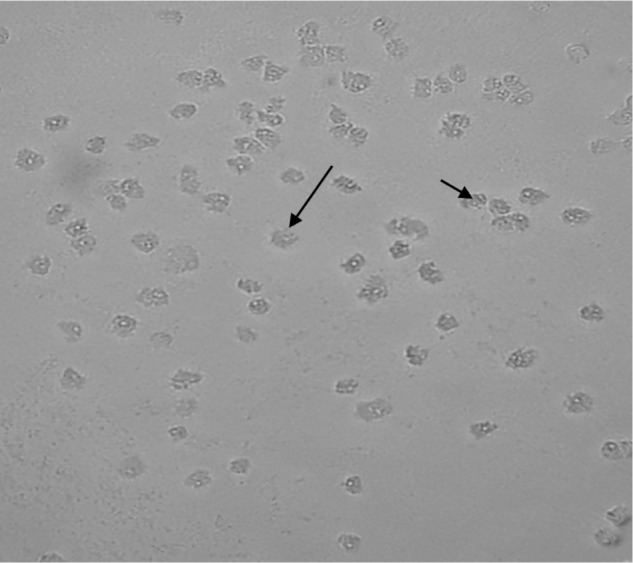
Photomicrograph of Acanthamoeba trophozoites isolated from a river in Ilam Province.
Table 1.
Data regarding the positive strains of Acanthamoeba genotypes isolated from geothermal waters in Ilam Province, Iran.
| CODE | SAMPLING SITES | NNA CULTURE/PCR | TEMPERATURE TOLERANCE (37 °C/44 °C) | OSMOTOLERANCE 0.5/1 M | GENOTYPE | MAX IDENTITY/QUERY COVERAGE | ACCESSION NUMBER |
|---|---|---|---|---|---|---|---|
| NS 1 | Hot spring | +/+ | +/+ | +/+ | T4 | 97/97 | KU356836 |
| NS 2 | River | +/+ | −/− | −/− | T4 | 96/99 | KU356837 |
| NS 5 | River | +/+ | +/+ | +/+ | T4 | 98/98 | KU356838 |
| NS 6 | River | +/+ | −/− | −/− | T4 | 99/94 | KU356839 |
| NS 9 | Hot spring | +/+ | −/+ | +/− | T4 | 92/83 | KU356840 |
| NS 10 | Swimming pool | +/+ | +/+ | +/+ | T4 | 99/97 | KU356841 |
| NS 11 | Swimming pool | +/+ | +/− | +/− | T2 | 99/98 | KU356842 |
| NS 14 | River | +/+ | +/+ | +/+ | T4 | 99/98 | KU356843 |
| NS 15 | River | +/+ | −/− | −/− | T4 | 99/96 | KU356844 |
| NS 16 | Spring | +/+ | −/− | −/− | T4 | 98/96 | KU356845 |
| NS 18 | River | +/+ | +/− | +/− | T4 | 99/95 | KU356846 |
| NS 22 | Hot spring (Tar mud bath) | +/+ | +/+ | +/+ | T4 | 99/95 | KU356847 |
| NS 28 | Swimming pool | +/+ | −/− | −/− | T4 | 99/97 | KU356848 |
| NS 31 | Tar river | +/+ | −/− | −/− | T4 | 99/96 | KU356849 |
| NS 35 | Tar spring | +/+ | −/− | −/− | T4 | 99/97 | KU356850 |
| NS 37 | Tar spring | +/+ | −/− | −/− | T4 | 98/97 | KU356851 |
| NS 20 | River | +/− | −/− | −/− | NA | NA | NA |
| NS 32 | River | +/− | −/− | −/− | NA | NA | NA |
| NS 36 | River | +/− | −/− | −/− | NA | NA | NA |
| NS 39 | River (Tar) | +/− | −/− | −/− | NA | NA | NA |
Abbreviations: NNA, non-nutrient agar; NA, not applicable.
Discussion
The contamination of geothermal springs and rivers with potentially pathogenic Acanthamoeba could be a health threat for human beings, since these springs were mainly used for therapeutic purposes. All samples from springs and rivers contain mineral compounds, and their temperatures ranged from 30 °C to 50 °C. The sanitary conditions of some of the springs have been improved, and the mud baths were specially used for therapeutic purposes in the country. The main sanitary improvement of such springs included free flow of water to pools and artificial springs. The geothermal springs of the studied area were chosen because a lot of ill people visited these springs while others use it for drinking. Several studies revealed a range of 5.9%–21% of contamination of Acanthamoeba in geothermal water sources worldwide.22–24 A study reported the contamination of hot springs by Acanthamoeba in the northwest of Iran, and this contamination was estimated to be 20%, of which isolates belonging to T4 (10 strain) and T3 (2 strain) were prevalent,24 while in southern Taiwan, the contamination of thermal springs was estimated to be 15%. The result of these researchers showed that out of a total of 60 water samples from four thermal springs, Acanthamoeba was detected in nine occasions, and the detected genotypes belonged to T2, T4, and T15.23 It is worth mentioning that hot spring recreation areas in northern Taiwan were surveyed for the occurrence of Acanthamoeba genotypes and a contamination of 21.2% was reported.22 Of a total of 51 analyzed samples, Acanthamoeba (A. polyphaga and Acanthamoeba jacobsi), Hartmannella, and Naegleria had 5.9% (3/51), 52.9% (27/51), and 5.9% (3/51) prevalences, respectively.
The present research showed a higher contamination, and half (50%) the geothermal water samples were contaminated due to the potentially pathogenic Acanthamoeba. The higher contamination may be due to the less periodical surveillance and microbial monitoring in this region. Moreover, this result was in opposition to that of Kao et al, which indicated that the T15 genotype, corresponding to A. jacobsi, was the most prevalent type in the thermal spring environment of Taiwan.23 These researches also mentioned that 15% of hot springs were contaminated with Acanthamoeba, which belonged to T15, T4, and T2 genotypes. Another study conducted in Turkey revealed the presence of Acanthamoeba T15 type in water samples of hot springs and swimming pools.25 In accordance with the aforementioned studies, the most frequently identified Acanthamoeba genotype was T15, followed by T6, and then T5 in Taiwan.26 Thus, it seems that the T15 genotype is frequently isolated from warm waters, such as hot springs. In Iran, the T15 genotype was only detected in one occasion from cold rivers of Tehran.27 Another study conducted in weak alkaline carbonate spring water in Taiwan revealed an isolation of 5.9% of Acanthamoeba, while the most isolated amoeba was Vermamoeba vermiformis (52.9%). Interestingly in this research, T15 was isolated in two samples as well as the T4 genotype.26
According to thermo- and osmotolerance abilities, five strains also demonstrated pathogenic potential. However, it is recommended that more studies should be carried out using in vivo tests. As a matter of fact, in vivo tests, including animal assays or cell culture tests, and evaluation of cytopathic effect are the best complementary tests for pathogenicity.
To the best of our knowledge, the present research is the first report of T2 genotype from hot springs in Iran and the second report of T2 from water sources in the country. Interestingly, this strain showed a pathogenic potential according to its growth at high temperature and osmolarity. T2 has been reported as a causative agent of keratitis in Iran and worldwide.15
Conclusion
Overall, as both isolated genotypes (T4 and T2) are related to Acanthamoeba keratitis due to high human activity in the studied area, it is important to monitor geothermal waters. It is worth mentioning that more studies should be conducted for genotype distribution in waters associated with human activity.
Recommendation
As most amoebic keratitis patients report a history of contact with contaminated water sources, knowledge regarding the distribution of pathogenic Acanthamoeba and implication of preventive measures are of utmost importance. It is recommended that there should be increased awareness among the high-risk people, such as contact lens users, and posting of warning signs near recreational and therapeutic water sources.
Indeed, the present research highlights the contamination of geothermal water sources by potentially pathogenic Acanthamoeba genotypes, which can result in Acanthamoeba-related disease; thus, more research on survey of recreational waters is suggested in order to provide awareness to individuals at high risk and those who come in contact with such waters.
Acknowledgments
The authors thank Dr. Niloofar Taghipour for her valuable comments and appreciate Radan English Edit Institute for improving the manuscript language. The authors thank Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Footnotes
ACADEMIC EDITOR: Timothy Kelley, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2076 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE). Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived and designed the experiments and made critical revisions and approved the final version: MN, RS. Performed the experiments: AL, ZL. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Marciano-Cabral F, Cabral G. Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev. 2003;16(2):273–307. doi: 10.1128/CMR.16.2.273-307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free- living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50(1):1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 3.Lorenzo-Morales J, Khan NA, Walochnik J. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite. 2015;22:10. doi: 10.1051/parasite/2015010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chappell CL, Wright JA, Coletta M, Newsome AL. Standardized method of measuring Acanthamoeba antibodies in sera from healthy human subjects. Clin Diagn Lab Immunol. 2001;8(4):724–30. doi: 10.1128/CDLI.8.4.724-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memari F, Niyyati M, Haghighi A, Seyyed Tabaei SJ, Lasjerdi Z. Occurrence of pathogenic Acanthamoeba genotypes in nasal swabs of cancer patients in Iran. Parasitol Res. 2015;114(5):1907–12. doi: 10.1007/s00436-015-4378-2. [DOI] [PubMed] [Google Scholar]
- 6.Cabello-Vílchez AM, Martín-Navarro CM, López-Arencibia A, et al. Genotyping of potentially pathogenic Acanthamoeba strains isolated from nasal swabs of healthy individuals in Peru. Acta Trop. 2014;130:7–10. doi: 10.1016/j.actatropica.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Khan NA. Acanthamoeba: Biology and Pathogenesis. Norfolk: Horizon Scientific Press; 2009. [Google Scholar]
- 8.Niyyati M, Rezaeian M. Current status of Acanthamoeba in Iran: a narrative review article. Iran J Parasitol. 2015;10(2):157–63. [PMC free article] [PubMed] [Google Scholar]
- 9.CAST RJ Development of an Acanthamoeba-specific reverse dot-blot and the discovery of a new ribotype. J Eukaryot Microbiol. 2001;48(6):609–15. doi: 10.1111/j.1550-7408.2001.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst PA, Booton GC, Crary M. Phylogenetic analysis and the evolution of the 18S rRNA gene typing system of Acanthamoeba. J Eukaryot Microbiol. 2015;62(1):69–84. doi: 10.1111/jeu.12186. [DOI] [PubMed] [Google Scholar]
- 11.Corsaro D, Walochnik J, Köhsler M, Rott MB. Acanthamoeba misidentification and multiple labels: redefining genotypes T16, T19, and T20 and proposal for Acanthamoeba micheli sp. nov. (genotype T19) Parasitol Res. 2015;114(7):2481–90. doi: 10.1007/s00436-015-4445-8. [DOI] [PubMed] [Google Scholar]
- 12.Walochnik J, Haller-Schober E-M, Kölli H, et al. Discrimination between clinically relevant and nonrelevant Acanthamoeba strains isolated from contact lens-wearing keratitis patients in Austria. J Clin Microbiol. 2000;38(11):3932–6. doi: 10.1128/jcm.38.11.3932-3936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirjalali H, Niyyati M, Abedkhojasteh H, Babaei Z, Sharifdini M, Rezaeian M. Pathogenic assays of Acanthamoeba belonging to the t4 genotype. Iran J Parasitol. 2013;8(4):530–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Khan NA, Jarroll EL, Paget TA. Molecular and physiological differentiation between pathogenic and nonpathogenic Acanthamoeba. Curr Microbiol. 2002;45(3):197–202. doi: 10.1007/s00284-001-0108-3. [DOI] [PubMed] [Google Scholar]
- 15.Maghsood AH, Sissons J, Rezaian M, Nolder D, Warhurst D, Khan NA. Acanthamoeba genotype T4 from the UK and Iran and isolation of the T2 genotype from clinical isolates. J Med Microbiol. 2005;54(8):755–9. doi: 10.1099/jmm.0.45970-0. [DOI] [PubMed] [Google Scholar]
- 16.Niyyati M, Lorenzo-Morales J, Rezaie S, et al. Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Exp Parasitol. 2009;121(3):242–5. doi: 10.1016/j.exppara.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Paterson GN, Rittig M, Siddiqui R, Khan NA. Is Acanthamoeba pathogenicity associated with intracellular bacteria? Exp Parasitol. 2011;129(2):207–10. doi: 10.1016/j.exppara.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Rezaeian M, Niyyati M. Pathogenic Free Living Amoeba. Iran: Tehran University Publication; 2009. [Google Scholar]
- 19.Behniafar H, Niyyati M, Lasjerdi Z. Molecular characterization of pathogenic Acanthamoeba isolated from drinking and recreational water in East Azerbaijan, Northwest Iran. Environ Health Insights. 2015;29(9):7–12. doi: 10.4137/EHI.S27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder JM, Booton GC, Hay J, et al. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of Acanthamoebae from humans with keratitis and from sewage sludge. J Clin Microbiol. 2001;39(5):1903–11. doi: 10.1128/JCM.39.5.1903-1911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Booton G, Kelly D, Chu Y-W, et al., editors. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lenses, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. J Clin Microbiol. 2002;40(5):1621–5. doi: 10.1128/JCM.40.5.1621-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang SW, Hsu BM, Chen NH, et al. Isolation and identification of Legionella and their host amoebae from weak alkaline carbonate spring water using a culture method combined with PCR. Parasitol Res. 2011;109(5):1233–41. doi: 10.1007/s00436-011-2366-8. [DOI] [PubMed] [Google Scholar]
- 23.Kao PM, Hsu BM, Chen NH, et al. Isolation and identification of Acanthamoeba species from thermal spring environments in southern Taiwan. Exp Parasitol. 2012;130(4):354–8. doi: 10.1016/j.exppara.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Solgi R, Niyyati M, Haghighi A, et al. Thermotolerant Acanthamoeba spp. isolated from therapeutic hot springs in Northwestern Iran. J Water Health. 2012;10(4):650–6. doi: 10.2166/wh.2012.032. [DOI] [PubMed] [Google Scholar]
- 25.Evyapan G, Koltas IS, Eroglu F. Genotyping of Acanthamoeba T15: the environmental strain in Turkey. Trans R Soc Trop Med Hyg. 2015;109(3):221–4. doi: 10.1093/trstmh/tru179. [DOI] [PubMed] [Google Scholar]
- 26.Huang SW, Hsu BM. Isolation and identification of Acanthamoeba from Taiwan spring recreation areas using culture enrichment combined with PCR. Acta Trop. 2010;115(3):282–7. doi: 10.1016/j.actatropica.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Niyyati M, Nazar M, Haghighi A, Nazemalhosseini Mojarad E. Screening of recreational areas of rivers for potentially pathogenic free-living amoebae in the suburbs of Tehran, Iran. J Water Health. 2012;10(1):140–6. doi: 10.2166/wh.2011.068. [DOI] [PubMed] [Google Scholar]