Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is generally responsible for the cAMP/PKA regulated anion conductance at the apical membranes of secretory epithelial cells. Mutations in CFTR underlie cystic fibrosis (CF), in which the most common variant, F508del, causes protein misfolding and its proteasome-mediated degradation. A new pathway that contributes to mutant CFTR degradation is mediated by the small heat shock protein, Hsp27, which cooperates with Ubc9, the E2 enzyme for SUMOylation, to selectively conjugate mutant CFTR with SUMO-2/3. This SUMO paralog can form polychains, which are recognized by the ubiquitin E3 enzyme, RNF4, leading to CFTR ubiquitylation and recognition by the proteasome. We found also that F508del CFTR could be modified by SUMO-1, a paralog that does not support SUMO polychain formation. The use of different SUMO paralogs to modify and target a single substrate for divergent purposes is not uncommon. In this short review we discuss the possibility that conjugation with SUMO-1 could protect mutant CFTR from disposal by RNF4 and similar ubiquitin ligases. We hypothesize that such a pathway could contribute to therapeutic efforts to stabilize immature mutant CFTR and thereby enhance the action of therapeutics that correct CFTR trafficking to the apical membranes.
Keywords: SIM (SUMO-interacting motif); SUMO-specific peptidase; SUMO ligase; Ubc9 (SUMO-conjugating enzyme), multi-SUMOylation
sumo (Small Ubiquitin-like Modifier) conjugation is a posttranslational modification that regulates a vast array of nuclear as well as extranuclear functions including transcription, DNA repair, chromatin structure, apoptosis, protein transport and localization, protein degradation, channel and enzyme activity, receptor function, and more (1, 17, 18, 49). SUMO conjugation occurs through an enzymatic cascade resembling that for ubiquitin. In an ATP-dependent manner, it involves the coordinated action of an E1 ligase (SAE1/SAE2 dimer), and an E2-conjugating enzyme (Ubc9). SUMO E3 ligases may not be required for substrate modification, as the SUMO E2 can form the necessary isopeptide bond with the target protein. Rather, SUMO E3s ensure substrate selection and catalytically enhance transfer of the SUMO moiety from Ubc9 to the target lysine. Most but not all acceptor lysines lie within a consensus SUMOylation motif, ψKxE/D, where ψ represents a bulky hydrophobic amino acid (21, 24). Mammals express four SUMO paralogs, and a COOH-terminal diglycine motif in SUMO-1, -2, and -3 enables their covalent attachment to substrates. SUMO-2 and SUMO-3 differ by only four amino acids, and they share only 50% homology with SUMO-1. A consensus SUMOylation site unique to SUMO-2/3 enables them to form polychains (43). Like ubiquitylation, SUMOylation is a dynamic process that is reversed by deconjugation of the modifier via specific cysteine proteases SENP1-3, SENP5-7, USPL1, and DeSI1 and 2 (22).
Introduction to CFTR
A recent study identified CFTR (cystic fibrosis transmembrane conductance regulator) as a substrate for SUMOylation. CFTR is a member of the ATP-binding cassette (ABC) family and is composed of 1,480 amino acids. Like other ABC family members, CFTR is composed of several domains, including two membrane-spanning domains (MSD1 and MSD2), each with six transmembrane helices, two nucleotide-binding domains (NBD1 and NBD2), and a regulatory (R) domain. At the apical membrane of various epithelial cells, CFTR functions as a cAMP-regulated secretory anion channel (39). One of the most prevalent genetic disorders, cystic fibrosis (CF), is caused by mutations within the gene encoding CFTR. The most common disease causing mutation is the deletion of a phenylalanine residue at position 508 in NBD1 (40) that produces a protein folding defect (8). CFTR folding is an intricate process, so that ∼50% of the wild-type (WT) protein is degraded in most cells (23, 27, 30, 45, 48). Our goal in this brief review is to explore examples of proteins in which the SUMOylation pathway uses different SUMO isoforms (paralogs) to direct their client proteins to different functional outcomes and thereby to raise the possibility that this aspect of SUMO regulation may also apply to CFTR.
SUMO modification occurs for both F508del and WT CFTR, although the WT protein is modified to a lesser extent. CFTR contains three SUMOylation consensus sites (NBD1: FKIE aa446-449; NBD2: VKKD aa1198-1201; COOH terminus: LKEE aa1467-1470), as shown in Fig. 1; however, there may be additional nonpredicted acceptor lysines. As is evident from Fig. 2 (published as Figure 4B in Ref. 1), F508del CFTR is modified with both SUMO-1 as well as SUMO-2/3. While conjugation to both isoforms was detected, our prior studies focused on the ability of the small heat shock protein, Hsp27, to enhance the selective modification of F508del CFTR specifically with SUMO-2, which was less significant for the WT protein. Purified Hsp27 enhanced SUMOylation of F508del NBD1 in vitro, but it was not efficient at modifying WT NBD1. SUMO-2 conjugation to mutant CFTR led to its ubiquitylation by the SUMO-targeted ubiquitin ligase (STUbL), RNF4, and ultimately its degradation by the proteasome (1).
Fig. 1.
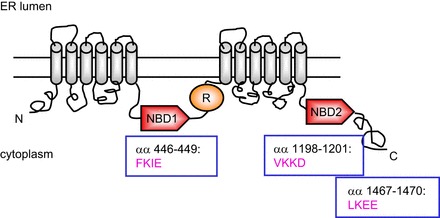
CFTR has three consensus sites for SUMOylation, ψKXE/D, followed by an acidic cluster. A schematic representation of their positions is shown.
Fig. 2.
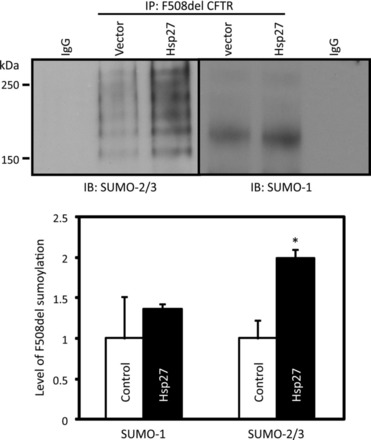
F508del CFTR can be conjugated to both SUMO-1 as well as SUMO-2/3, whereas Hsp27 promotes selective SUMOylation of F508del CFTR by SUMO-2/3. HEK 293 cells were transfected with F508del CFTR and empty vector or Hsp27 DNAs as previoulsy described (1). Modification of F508del CFTR by endogenous SUMO paralogs was assessed by CFTR immunoprecipitation (IP) followed by SUMO-1 or SUMO-2/3 immunoblotting. An irrelevant IgG served as a control. SUMO density values were normalized to CFTR levels in the IPs and mean values from six experiments are provided in the bottom panel. [From Ahner et al. (1). Reprinted with permission.]
Mechanisms of Paralog Selection
As mentioned above, modification of NBD1 was detected with SUMO-1 in addition to SUMO-2, and we hypothesize that the conjugation of different SUMO paralogs may have different functional consequences for CFTR. Furthermore, there might be a different isoform preference between WT and mutant ion channels. While this idea is relatively unconventional, it is not without precedent in the literature. Several lines of evidence suggest that SUMO-1 and SUMO-2/3 may serve different functions. Proteins conjugated exclusively to one SUMO paralog were isolated in proteomics studies (41, 46), and proteins modified with distinct SUMO isoforms localize to different subcellular compartments during the cell cycle (3). In the developing lens, SUMO paralogs exhibit very distinct expression patterns, temporally and spatially (20), and a SUMO-1 haploinsufficiency is associated with cleft lip and palate (2).
Different functional implications for various SUMO isoforms are also evident from the paralog specificity exhibited by many SUMO-interacting motifs (SIMs) that mediate protein interactions. For example, while the SIMs of CoRest1, FIP1L1, RBBP4, Usp25, MCAF1, and K-bZIP display a preference for SUMO-2/3 binding (7, 34, 38, 42), phosphorylation of the SIMs in Daxx and PML markedly increases their inclination to conjugate with SUMO-1 (5, 6).
Another selection mechanism ensures SUMO paralog specific modification through selective protection of the adduct from protease activity. In vivo, GTPase-activating protein for Ran (RanGAP1) is preferentially conjugated to SUMO-1 (32, 33). Yet in vitro, SUMO-1 and SUMO-2 modification of RanGap1 proceeds with equal efficiency (50). SUMO-1-modified RanGAP1 bound with higher affinity to RanBP2/Nup358 (SUMO E3 ligase) than SUMO-2-bound RanGAP1. In isopeptidase protection assays, both SUMO isoforms were efficiently cleaved off of RanGAP1. Yet, in the presence of Ubc9 and RanBP2/Nup358, SUMO-1-modified RanGAP1 was protected to a much higher degree from deconjugation by SENP1, 2, and 5 than SUMO2-RanGAP1. To corroborate these findings in vivo, the authors performed knockdown of either RanBP2/Nup358 or different SENPs and showed that preferential modification of RanGAP1 with SUMO-1 is dependent on the presence of both (50). While Gareau et al. (16) were not able to identify distinct amino acid side chains responsible for paralog preference, their research suggested that the RanGAP1-SUMO/Ubc9/RanBP2 complex exhibits delicate structural differences and a changing number in RanBP2-SIM-SUMO interactions depending on the paralog conjugated to the GTPase.
Since the majority of SUMO-1 is found conjugated to substrates while there exists a free pool of SUMO-2/3, and cells express 10 times more SUMO-2/3 than SUMO-1 (reviewed in Ref. 14), we can easily imagine that WT and F508del CFTR are both modified with SUMO-2/3. Yet, SUMO proteases might deconjugate the WT protein efficiently and thereby shift its paralog preference to SUMO-1, which could perhaps promote WT CFTR biogenesis. In the case of F508del, an incorrect domain assembly could interfere with protease access and prevent SUMO-2/3 cleavage, and thus destine the mutant channel for degradation.
While several groups have demonstrated that hypoxia-inducible factor-1α (Hif-1α) is SUMOylated by either SUMO-1 or SUMO-2/3, there is still controversy about the effect that the modification exerts on this transcription factor (increase in stability, decrease in stability, decrease in transactivation activity, increase in transcriptional activity).
Several SUMO E3s were able to promote Hif-1α conjugation to SUMO in vivo or in vitro, including PIASγ, RanBP2, and Cbx4/Pc2 (reviewed in Ref. 37; see also Refs. 4, 25, 29). Interestingly, K391 and K477 were identified as SUMO acceptor sites for RanBP2 and Cbx4/Pc2 in a Hif-1α fragment, yet their mutation did not abrogate Piasγ-facilitated SUMOylation of the same fragment in vitro. Furthermore, addition of Piasγ to the in vitro SUMOylation reaction induced a modified Hif-1α fragment of a different molecular weight than Cbx4/Pc2.
Taken together, these results suggest that Piasγ and Cbx4/Pc2, as well as RanBP2, can target different acceptor sites within a common substrate and might promote mono- versus poly- or multi-SUMOylation. Similarly, it might be postulated that different SUMO acceptor sites within CFTR grant access to different SUMO E3s and consequently, might be modified with different SUMO paralogs possibly promoting divergent outcomes, as discussed.
Different SUMO Paralogs Induce Divergent Fates for the Same Substrate
Tau and α-synuclein.
Tau and α-synuclein are two inherently unfolded proteins that are found at high levels in the brain, and they are associated with several neurodegenerative diseases such as Alzheimer's and Parkinson's disease (19). Both have been shown to be SUMOylated by SUMO-1 and less efficiently by SUMO-2/3 in a HEK 293 cell system overexpressing the various SUMO paralogs (12). Modification of tau and α-synuclein with SUMO-1 in a HEK 293 overexpression system diminished its ubiquitylation and degradation, as well as their solubility (31). Similarly, enhanced conjugation of α-synuclein with SUMO-1, induced via proteasome inhibition, increased α-synuclein aggregation in a Chinese hamster ovary (CHO) cell overexpression system (26).
In contrast, Krumova et al. (28) demonstrated efficient modification of α-synuclein with SUMO-2 in vitro in HEK 293 cells and in His6-SUMO-2 transgenic mice. In vitro experiments demonstrated that SUMO-2 modification of α-synuclein prevented fibril formation. Furthermore, mutation of the major SUMO acceptor sites increased aggregation and toxicity in HEK 293 cells as well as in a rat model of Parkinson's disease. The later study is supported by experiments performed in the budding yeast, Saccharomyces cerevisiae. α-Synuclein was expressed in a yeast strain with a temperature-sensitive mutation in the gene encoding a SUMO isopeptidase (ULP1). At the nonpermissive temperature, Smt3 (yeast SUMO)-modified α-synuclein accumulated and the modification was conjugated to the same lysines as in human cells. Modification with Smt3, however, prevented the formation of α-synuclein inclusions through stimulation of its autophagic clearance (42). Yeast express only one isoform of SUMO, called Smt3, and like the human SUMO-2/3, it contains a SUMOylation consensus site and forms poly-SUMO chains, though in vivo, mostly mono-SUMOylation is observed (24, 35). Thus, it is not surprising that SMT3 supports a cellular function similar to that of SUMO2/3.
Superoxide dismutase 1.
Mutations in superoxide dismutase 1 (SOD1) that disrupt its folding promote SOD1 aggregation and the formation of intracellular inclusions. This process underlies familial amyotrophic lateral sclerosis (FALS), a progressive neurodegenerative disorder. Fei et al. (13) observed that SOD1 WT and FALS-associated mutants G93A and G85R are conjugated with SUMO-1 but not SUMO-2/3 in vitro or in HEK 293 cells overexpressing SUMO-2/3. They also reported K75 as the sole SUMOylation site in WT SOD1. SUMO-1 cotransfection enhanced SOD1 stability and promoted its aggregation. Since a control was missing (i.e., use of the K75R mutant that abolishes SOD1 SUMOylation), this increase might be due to indirect effects; however, Niikura et al. (36) followed up on these findings using a system more relevant to the disorder. They found SOD1 WT and FALS mutants to be modified with SUMO-1, the latter to a much greater extent when overexpressing SUMO-1 in NSC34 cells (a motor neuron cell line). They determined that, in addition to K75, K9 is a major conjugation site for SUMO-1 in SOD1 mutants in NSC34 cells as well as HEK 293 cells. Furthermore, in NSC34 cells, FALS-associated mutants but not WT SOD1 were modified with SUMO-2/3. A K9R mutation did not significantly disturb SUMOylation; instead, SUMO-2/3 was preferentially conjugated to K75. Overexpression of SUMO-3 rather than SUMO-1 increased mutant but not SOD1 WT aggregation (in CHO cells) and its stability (in NSC34 cells). A K75R mutation abolishing SUMO-2/3 modification of SOD mutants averted both aggregation and stabilization, indicating a direct effect mediated by the SUMO site. The different observations between both studies are likely due to the use of different cell systems, the use of WT versus mutant protein, and procedural differences (like the use of N-ethylmaleimide to inhibit SUMO-specific proteases widely employed to facilitate the detection of protein SUMOylation). Yet, the finding that SOD1 mutants are not only modified by SUMO-1 as well as SUMO-2/3 but that the different paralogs, although overexpressed, exhibit a distinct preference for target lysines is very intriguing. Furthermore, while SUMO-3 promotes mutant SOD1 stability and aggregation, SUMO-1 does not. Although, the function of the latter needs to be determined, it strongly suggests distinct outcomes for the conjugation of SUMO-1 or -2/3 to SOD1. Again, these findings open the door to the concept that different SUMO acceptor sites within CFTR might be preferentially modified with different SUMO paralogs that promote independent functions.
Transcription factor specificity protein 1.
Another example of paralog specificity is provided by the transcription factor specificity protein 1 (SP1). Gong et al. (20) observed that SUMO isoforms differentially regulated the expression of the lens-specific β-crystallins during lens development through modification of SP1. In addition to a previously identified SUMOylation site at K16 they identified a SUMO acceptor site in the COOH-terminal DNA-binding domain (DBD) at K683. In vitro SUMOylation assays showed that K683 was preferentially modified by SUMO-2 over SUMO-1. Moreover, in electrophoresis mobility shift assays (EMSA) the authors demonstrated that SUMO-2-conjugated SP1 DBD exhibited a diminished binding efficacy to the βB1-crystallin gene promoter compared with its SUMO-1-modified complement. Gong et al. also reexamined the relevance of the NH2-terminal SUMOylation site K16 in human lens epithelial (HLE) cells. Cycloheximide (CHX) chase experiments demonstrated a K16-dependent acceleration of SP1 degradation upon SUMO-2 coexpression. SUMO-1 on the other hand increased SP1 protein expression, yet this effect proved to be independent of K16. Therefore, this effect might be due to enhanced transcription, indirect effects, or alternate SUMOylation sites within SP1. Curiously, the SUMO-1 effect on SP1 expression exhibits opposing effects to those of a previous report (45) stating enhanced degradation due to a relocalization to the cytoplasm and enhanced interaction with a proteasomal subunit in HeLa cells.
One possible explanation might be the use of different cell lines. However, SUMO-1 and SUMO-2/3 appear to differentially regulate SP1 protein interactions also. Quantitative chromatin immunoprecipitation (qChIP), in vitro binding assays, and coimmunoprecipitation experiments all indicated that SUMO-2 modification of SP1 disrupts its interaction with its coactivator p300. In sharp contrast, SUMO-1 conjugation enhances the association between SP1 and p300. Thus, SUMO-1 enhances SP1 transcriptional activity whereas SUMO-2 suppresses it. In this study, the authors provided compelling evidence that SUMOylation regulated three completely independent functions (DNA binding, protein interactions, and protein expression level/stability) of a single substrate. For each of these functions, SUMO-1 and SUMO-2/3 promoted opposite outcomes. The effect of the modification was mediated through two different SUMO acceptor sites, and one of these sites was shown to exhibit distinct affinities for the different isoforms. This very intricate regulatory mechanism seemed to be enabled through a tightly governed, temporally and spatially distinct expression pattern of the SUMO paralogs in the developing lens.
Implications for CFTR.
Similarly, one can imagine that CFTR, which harbors three SUMOylation consensus sites, as well as potential nonconsensus acceptor sites, might be SUMOylated at different lysines for different purposes. Beyond promoting the selective degradation of mutant CFTR, SUMO conjugation might regulate CFTR biogenesis, channel gating, CFTR trafficking, and half-life at the plasma membrane, recycling and more. For one or more of these putative functions, different paralogs might cause quite divergent consequences by promoting or inhibiting the association of CFTR with regulatory factors. Different acceptor lysines might be modified by different SUMO E3 ligases with subcellular expression patterns that correspond to the function they regulate [i.e., an E3 that promotes SUMO conjugation to regulate CFTR half-life at the plasma membrane (PM) should be localized to the PM domain]. Another regulatory mechanism might be the accessibility of the SUMOylation site (i.e., if SUMO supports folding or the degradation of the misfolded protein the correctly folded protein might obscure a previously exposed acceptor site).
Histone deacetylase 1.
With regard to a differential modification of WT and mutant CFTR, it is of particular interest that histone deacetylase 1 (HDAC1) is conjugated to different SUMO isoforms dependent on the health/disease state of the cell background, e.g., epithelial breast cancer cell line (MCF7) versus nontumorigenic mammary cell line (MCF10A). Citro et al. (10) found that HDAC1 exhibited a shorter half-life in MCF10A cells than in a breast cancer cell line. The discrepancy correlated with a preferential modification of HDAC1 with SUMO-1 in MCF10A cells but with SUMO-2 in MCF7 cells. Overexpression of SUMO-1 but not SUMO-2 increased HDAC1 ubiquitylation and facilitated its degradation in HeLa and Phoenix cells. Mutation of the two SUMO acceptor sites (K444 and K476) negated this effect. shRNA mediated knockdown of Ubc9 in MCF10A cells stabilized HDAC1. Moreover, exogenously expressed HDAC1 K444/476R was stabilized compared with exogenously expressed WT protein. No difference was observed in MCF7 cells where HDAC1 is preferentially modified with SUMO-2. To elucidate the source of this differential conjugation, the authors assessed the expression patterns of different SUMO E3 enzymes in both cell lines and found elevated PIASγ levels in the cancer cell line. Indeed, PIASγ promoted preferential conjugation of HDAC1 with SUMO-2.
Implications for CFTR.
Microarray as well as proteomic studies of cell lines/tissue samples displayed differing expression profiles not only between WT and CF cells/tissue but also between different CF mutants (9, 11, 15). While these studies are preliminary and there is some discrepancy between the candidates identified, this might be due to a difference in methodology and in the cell lines and tissue used. Thus one can imagine that though the published differentially expressed genes were not components of the SUMO pathway, they might indirectly regulate expression patterns, enzymatic activity, localization (and thus accessibility to CFTR), or target preference of SUMO pathway components, such as SUMO E3s or proteases.
Taken together, the highlighted intricacy, potency, and consequences of SUMO paralog-specific modification appear to be a legitimate regulatory mechanism for many SUMO client proteins. These differential regulatory processes appear to influence the fate and functions of other SUMO substrates, and this possibility should be explored for CFTR.
GRANTS
This work was supported in part by grants from the National Institutes of Health (RO1-DK-068196) and the Cystic Fibrosis Foundation (FRIZZE13XXO to R. A. Frizzell).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.A. and R.A.F. drafted manuscript; A.A. and R.A.F. edited and revised manuscript; X.G. analyzed data; X.G. prepared figures.
REFERENCES
- 1.Ahner A, Gong X, Schmidt BZ, Peters KW, Rabeh WM, Thibodeau PH, Lukacs GL, Frizzell RA. Small heat shock proteins target mutant cystic fibrosis transmembrane conductance regulator for degradation via a small ubiquitin-like modifier-dependent pathway. Mol Biol Cell 24: 74–84, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science 313: 1751, 2006. [DOI] [PubMed] [Google Scholar]
- 3.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell 15: 5208–5218, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berta MA, Mazure N, Hattab M, Pouyssegur J, Brahimi-Horn MC. SUMOylation of hypoxia-inducible factor-1alpha reduces its transcriptional activity. Biochem Biophys Res Commun 360: 646–652, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Cappadocia L, Mascle XH, Bourdeau V, Tremblay-Belzile S, Chaker-Margot M, Lussier-Price M, Wada J, Sakaguchi K, Aubry M, Ferbeyre G, Omichinski JG. Structural and functional characterization of the phosphorylation-dependent interaction between PML and SUMO1. Structure 23: 126–138, 2015. [DOI] [PubMed] [Google Scholar]
- 6.Chang CC, Naik MT, Huang YS, Jeng JC, Liao PH, Kuo HY, Ho CC, Hsieh YL, Lin CH, Huang NJ, Naik NM, Kung CC, Lin SY, Chen RH, Chang KS, Huang TH, Shih HM. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell 42: 62–74, 2011. [DOI] [PubMed] [Google Scholar]
- 7.Chang PC, Izumiya Y, Wu CY, Fitzgerald LD, Campbell M, Ellison TJ, Lam KS, Luciw PA, Kung HJ. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J Biol Chem 285: 5266–5273, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63: 827–834, 1990. [DOI] [PubMed] [Google Scholar]
- 9.Ciavardelli D, D'Orazio M, Pieroni L, Consalvo A, Rossi C, Sacchetta P, Di Ilio C, Battistoni A, Urbani A. Proteomic and ionomic profiling reveals significant alterations of protein expression and calcium homeostasis in cystic fibrosis cells. Mol Biosyst 9: 1117–1126, 2013. [DOI] [PubMed] [Google Scholar]
- 10.Citro S, Jaffray E, Hay RT, Seiser C, Chiocca S. A role for paralog-specific sumoylation in histone deacetylase 1 stability. J Mol Cell Biol 5: 416–427, 2013. [DOI] [PubMed] [Google Scholar]
- 11.Clarke LA, Sousa L, Barreto C, Amaral MD. Changes in transcriptome of native nasal epithelium expressing F508del-CFTR and intersecting data from comparable studies. Respir Res 14: 38, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorval V, Fraser PE. Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and alpha-synuclein. J Biol Chem 281: 9919–9924, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Fei E, Jia N, Yan M, Ying Z, Sun Q, Wang H, Zhang T, Ma X, Ding H, Yao X, Shi Y, Wang G. SUMO-1 modification increases human SOD1 stability and aggregation. Biochem Biophys Res Commun 347: 406–412, 2006. [DOI] [PubMed] [Google Scholar]
- 14.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82: 357–385, 2013. [DOI] [PubMed] [Google Scholar]
- 15.Gambardella S, Biancolella M, D'Apice MR, Amati F, Sangiuolo F, Farcomeni A, Chillemi G, Bueno S, Desideri A, Novelli G. Gene expression profile study in CFTR mutated bronchial cell lines. Clin Exp Med 6: 157–165, 2006. [DOI] [PubMed] [Google Scholar]
- 16.Gareau JR, Reverter D, Lima CD. Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2. J Biol Chem 287: 4740–4751, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Moll Cell Biol 10: 564–568, 2009. [DOI] [PubMed] [Google Scholar]
- 19.Goedert M. The significance of tau and alpha-synuclein inclusions in neurodegenerative diseases. Curr Opin Genet Dev 11: 343–351, 2001. [DOI] [PubMed] [Google Scholar]
- 20.Gong L, Ji WK, Hu XH, Hu WF, Tang XC, Huang ZX, Li L, Liu M, Xiang SH, Wu E, Woodward Z, Liu YZ, Nguyen QD, Li DW. Sumoylation differentially regulates Sp1 to control cell differentiation. Proc Natl Acad Sci USA 111: 5574–5579, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay RT. SUMO: a history of modification. Mol Cell 18: 1–12, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13: 755–766, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83: 129–135, 1995. [DOI] [PubMed] [Google Scholar]
- 24.Johnson ES. Protein modification by SUMO. Annu Rev Biochem 73: 355–382, 2004. [DOI] [PubMed] [Google Scholar]
- 25.Kang X, Li J, Zou Y, Yi J, Zhang H, Cao M, Yeh ET, Cheng J. PIASy stimulates HIF1alpha SUMOylation and negatively regulates HIF1alpha activity in response to hypoxia. Oncogene 29: 5568–5578, 2010. [DOI] [PubMed] [Google Scholar]
- 26.Kim YM, Jang WH, Quezado MM, Oh Y, Chung KC, Junn E, Mouradian MM. Proteasome inhibition induces alpha-synuclein SUMOylation and aggregate formation. J Neurol Sci 307: 157–161, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreda SM, Mall M, Mengos A, Rochelle L, Yankaskas J, Riordan JR, Boucher RC. Characterization of wild-type and deltaf508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol Biol Cell 16: 2154–2167, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krumova P, Meulmeester E, Garrido M, Tirard M, Hsiao HH, Bossis G, Urlaub H, Zweckstetter M, Kugler S, Melchior F, Bahr M, Weishaupt JH. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol 194: 49–60, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei Z, Yin QQ, Ma LN, Zhou AW, Wang LS, Yao M, Xia Q, Chen GQ. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell 25: 118–131, 2014. [DOI] [PubMed] [Google Scholar]
- 30.Lukacs GL, Mohamed A, Kartner N, Chang XB, Riordan JR, Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J 13: 6076–6086, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo HB, Xia YY, Shu XJ, Liu ZC, Feng Y, Liu XH, Yu G, Yin G, Xiong YS, Zeng K, Jiang J, Ye K, Wang XC, Wang JZ. SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc Natl Acad Sci USA 111: 16586–16591, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88: 97–107, 1997. [DOI] [PubMed] [Google Scholar]
- 33.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135: 1457–1470, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell 30: 610–619, 2008. [DOI] [PubMed] [Google Scholar]
- 35.Mullen JR, Brill SJ. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J Biol Chem 283: 19912–19921, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niikura T, Kita Y, Abe Y. SUMO3 modification accelerates the aggregation of ALS-linked SOD1 mutants. PLos One 9: e101080, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunez-O'Mara A, Berra E. Deciphering the emerging role of SUMO conjugation in the hypoxia-signaling cascade. Biol Chem 394: 459–469, 2013. [DOI] [PubMed] [Google Scholar]
- 38.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell 34: 145–154, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilewski JM, Frizzell RA. Role of CFTR in airway disease. Physiol Rev 79: S215–255, 1999. [DOI] [PubMed] [Google Scholar]
- 40.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui LC. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245: 1066–1073, 1989. [DOI] [PubMed] [Google Scholar]
- 41.Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics 4: 56–72, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekiyama N, Ikegami T, Yamane T, Ikeguchi M, Uchimura Y, Baba D, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem 283: 35966–35975, 2008. [DOI] [PubMed] [Google Scholar]
- 43.Shahpasandzadeh H, Popova B, Kleinknecht A, Fraser PE, Outeiro TF, Braus GH. Interplay between sumoylation and phosphorylation for protection against alpha-synuclein inclusions. J Biol Chem 289: 31224–31240, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276: 35368–35374, 2001. [DOI] [PubMed] [Google Scholar]
- 45.Varga K, Jurkuvenaite A, Wakefield J, Hong JS, Guimbellot JS, Venglarik CJ, Niraj A, Mazur M, Sorscher EJ, Collawn JF, Bebok Z. Efficient intracellular processing of the endogenous cystic fibrosis transmembrane conductance regulator in epithelial cell lines. J Biol Chem 279: 22578–22584, 2004. [DOI] [PubMed] [Google Scholar]
- 46.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics 5: 2298–2310, 2006. [DOI] [PubMed] [Google Scholar]
- 47.Wang YT, Chuang JY, Shen MR, Yang WB, Chang WC, Hung JJ. Sumoylation of specificity protein 1 augments its degradation by changing the localization and increasing the specificity protein 1 proteolytic process. J Mol Biol 380: 869–885, 2008. [DOI] [PubMed] [Google Scholar]
- 48.Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem 269: 25710–25718, 1994. [PubMed] [Google Scholar]
- 49.Wasik U, Filipek A. Non-nuclear function of sumoylated proteins. Biochim Biophys Acta 1843: 2878–2885, 2014. [DOI] [PubMed] [Google Scholar]
- 50.Zhu S, Goeres J, Sixt KM, Bekes M, Zhang XD, Salvesen GS, Matunis MJ. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol Cell 33: 570–580, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]


