Abstract
Neutrophil elastase (NE) is a neutrophil-derived serine proteinase with specificity for a broad range of substrates. NE has been reported to be associated with the pathogenesis of several conditions, particularly that of pulmonary diseases. Previous studies have shown that NE can cleave the pro-myelocyte - retinoic acid receptor-alpha chimeric protein and is important for the development of acute pro-myelocytic leukemia. To further elucidate the role of NE in acute pro-myelocytic leukemia, the present study successfully constructed a lentiviral vector containing the NE gene (LV5-NE), which was transfected into NB4 acute pro-myelocytic leukemia cells. The effects of NE overexpression in NB4 cells were detected using a Cell-Counting Kit-8 assay, flow cytometry and western blot analysis. The results showed that NE significantly promoted the proliferation of NB4 cells, inhibited cell apoptosis and apoptotic signaling, and led the activation of Akt. In an additional experiment, a vector expressing small hairpin RNA targeting NE was constructed to assess the effects of NE knockdown in U937 cells. Western blot analysis revealed that apoptotic signaling was increased, while Akt activation was decreased following silencing of NE. The results of the present study may indicate that NE activates the phosphoinositide-3 kinase/Akt signaling pathway in leukemia cells to inhibit apoptosis and enhance cell proliferation, and may therefore represent a molecular target for the treatment of pro-myelocytic leukemia.
Keywords: neutrophil elastase, LV5-NE, proliferation, apoptosis, phosphoinositide-3 kinase/Akt, NB4 cells, leukemia cells
Introduction
Acute pro-myelocytic leukemia (APL), which accounts for ~10% of all acute myeloid leukemia (AML) cases, is classified as the M3 subtype of AML within the French-American-British morphological classification system (1), A reciprocal chromosomal translocation affects the long arm of chromosomes 17 and 15 at the site of the retinoic acid receptor alpha (RARα) gene and the pro-myelocyte (PML) gene, respectively, resulting in a PML-RARA fusion gene, which leads to formation of the PML-RARα chimeric protein in 95% of APL cases (2–7). The PML and RARα oncoproteins, whose roles in leukemogenesis have been elucidated, are known to be targeted by specific biomolecules that are active in the disease (8).
Neutrophil elastase (NE), also known as human leukocyte elastase, is a neutrophil-derived serine proteinase with specificity for a broad range of substrates and is encoded by Elane, formerly known as Ela2 (9). The major physiological role of NE is to kill engulfed microorganisms within neutrophil phagolysosomes (10–13). NE has been reported to be associated with the pathogenesis of several diseases, including acute lung injury, cystic fibrosis, emphysema and leukemia (14–17). NE has also been characterized by its ability to cleave extracellular matrix proteins (18). NE is able to cleave the fusion protein PML-RARα into two different protein variants (16,18,19). This type of cleavage action has a role in the development of APL (20,21). A previous study by our group showed that NE has a role in the development of leukemia (22). However, the mechanism underlying the effect of NE in APL remain to be fully elucidated.
In order to explore the biological functions of NE in APL, NE was overexpressed using a constructed lentiviral vector. The effects of NE on NB4 cell proliferation and apoptosis were assessed and the possible mechanistic functions of NE were explored in APL cell lines. In addition, NE was silenced using small hairpin (sh)RNA to clarify its effect on the PI3K/Akt pathway in U937 cells.
Materials and methods
Cell lines and culture
Human 293T cells (Chinese Academy Cell Bank; Shanghai, China) were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco). Human NB4 cells (Chinese Academy Cell Bank) were maintained in RPMI 1640 medium (Gibco) with 10% FBS. Human U937 cells (Chinese Academy Cell Bank) were maintained in RPMI-1640 medium (Gibco) with 10% FBS. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2, and the culture medium was replaced at two-day intervals.
Construction of lentivirus
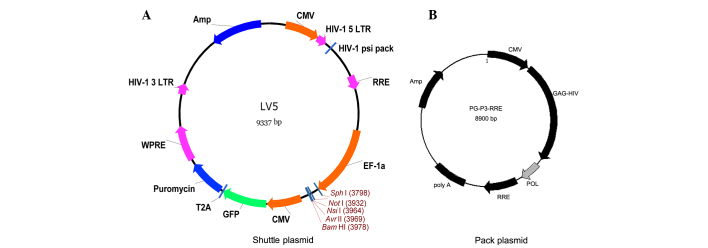
Shuttle plasmid and packing plasmid vector maps (Genepharma, Shanghai, China) are shown in Fig. 1A and B, respectively. A recombinant plasmid containing a lentiviral vector and three original auxiliary packaging vectors was constructed. NE was inserted into the LV5 plasmid but was not inserted for the negative control group. The three different plasmids were extracted with endo-free and high purity kits (Takara Bio, Inc., Otsu, Japan). The restriction enzymes used were NotI and BamHI (Takara Bio, Inc.). The recombinant plasmid and vectors were co-transfected into 293T cells using RNAi-Mate transfection reagent (Genepharma). The medium was replaced with complete medium after transfection for 6 h. After culture for 72 h, the cell supernatant was collected, which contained the lentivirus. Following concentration of the supernatant, the viral titer was determined.
Figure 1.
Vector maps. (A) Shuttle plasmid and (B) package vector. CMV, cytomegalovirus; GFP, green fluorescence protein; HIV, human immunodeficiency virus; LV, lentivirus; LTR, long terminal repeat; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element; RRE, Rev response element; Amp, ampicillin resistance gene; poly A, polyadenylation site.
Titer detection
293T cells were cultured in 96-well plates at a density of 0.5–10×103 cells/100 µl per well for 24 h. Then 90 µl complete medium (DMEM+10% FBS) was added to 7–10 EP tube. Original lentivirus (10 µl) was added to the first tube. Then 10 µl of the mixed suspension from the first tube was added to the second tube. This was repeated until each tube had been filled. Then, 90 µl of culture medium was discarded and the mixed suspension by concentration gradient (lowest to highest) was added to the cells. When cells had been cultured at 37°C in a humidified atmosphere containing 5% CO2 for 48 h, 100 µl of culture medium was added to each well. The cells were cultured for a further 72 h and subsequently counted based on the expression of green fluorescent protein (GFP) under an Eclipse 80i, Nikon fluorescence microscope (Nikon, Tokyo, Japan). The titer was calculated using the following formula: TU/µl = (PxN/100xV)/DF, where P is the percentage of GFP-positive cells, N is the number of cells, V is the virus dilution volume and DF is the dilution factor.
Lentiviral transfection
NB4 cells in the logarithmic growth phase (1×105 cells/well) were seeded in a 24-well plate. These cells were transfected with the GFP-expressing lentiviral vector LV5-NC and the NE-expressing recombinant lentivirus LV5-NE, and 1 µg/ml polybrene (Genepharma) was added. After culture for 24 h, the medium was refreshed. Fluorescence was detected following 48–72 h of incubation using the fluorescence microscope. The LV5-NE- and LV5-NC-transfected NB4 cells were screened with puromycin (Sigma-Aldrich, St. Louis, MO, USA) and successful transfectants were used for subsequent experiments. Experiments were performed on untransfected NB4 cells as well as on LV5-NE- and LV5-NC-transfected NB4 cells.
Transient transfection
U937 cells (2.5×105/ml) in the logarithmic growth phase were seeded in six-well plates. For transfection, 5 µl siRNA and 5 µl Lipofectamine 2000 (Invitrogen; Thermo Fisher Scentific, Inc.) were diluted in 100 μl Opti-MEM (Invitrogen) separately. The siRNA and Lipofectamine 2000 were then gently mixed and incubated for 25 min at room temperature. The siRNA-Lipofectamine 2000 complexes were subsequently added to each well and mixed by gentle agitation. The siRNA was purchased from RiboBio (Guangzhou, China) and the sequences of the NE-specific siRNAs were as follows: 5′-GAUCGACUCUAUCAUCCAAdTdT-3′ and 3′-dTdTCUAGCUGAGAUAGUAGGUU-5′. The resulting solution was added to each well, followed by incubation for 6 h. Subsequently, 1 ml fresh complete medium was added. The transfected cells were named as U937/NE-shRNA. In the negative control group, U937/shRNA-NC cells were processed similarly. Following 48 h of transfection, the successfully transfected cells were verified by fluorescent microscopy. The transfection efficiency was expressed as the percentage of red fluorescence protein-positive cells. In subsequent experiments, native U937 cells as well as U937/shRNA-NC and U937/NE-shRNA cells were used.
RNA isolation and reverse-transcription quantitative polymerase chain reaction (RT-qPCR) analysis
In each group of cells, TRIzol (Invitrogen, Thermo Fisher Scientific, Inc.) was used to extract total RNA, which was reverse transcribed into complementary DNA. In brief, 10 µl RNA solution (Takara Bio, Inc.) was subjected to RT. PCR amplification was then performed in a ThermoScript™ RT-PCR system (Invitrogen) using a PreTaq kit (Takara Bio, Inc.). All the primers were synthesized from Sangon Biotech, Co., Ltd. (Shanghai, China). The specific primers for NE of RT-PCR were 5′-CGG AAT TCA TGA CCC TCG GCC GCC GA-3′ (forward) and 5′-CCGCTCGAGTCAGTGGGTCCTGCTGGC-3′ (reverse). The reaction mixture contained 1 µl PrimeScriptRT Enzyme mix I (Takara Bio, Inc.), 1 µl (10 µM of each primer and 500 ng RNA in a final volume of 20 µl). Thermocycling conditions were as follows: Reverse transcription at 37°C for 15 min, then reverse transcriptase inactivation reaction at 85°C for 5 sec. The RT-PCR products were verified by agarose gel (Invitrogen; Thermo Fisher Scentific, Inc.). The specific primers for NE used for qPCR were 5′-ACTGCGTGGCGAATGTAA-3′ (forward) and 5′-CGATGTCGTTGAGCAAGTT-3′ (reverse). Primers for GAPDH were 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′ (reverse). The reaction mixture contained 5 µl PreTaq Enzyme, 0.5 µl (10 µM) of each primer and 100 ng cDNA in a final volume of 10 µl. Thermocycling conditions were as follows: Pre-denaturation at 95°C for 5 min, 39 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec, and a final extension at 95°C for 5 min. GAPDH was used as a reference gene and the RT-PCR products were verified by agarose gel with the DL 2,000 DNA maker (Takara Bio, Inc.). Quantity One Software (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for quantification of mRNA levels. Expression of NE, relative to GAPDH was determined using the 2−ΔΔCq method (23).
Western blot assay
For extraction of total protein, cells in each group were washed with ice-cold phosphate-buffered saline and lysed in radioimmunoprecipitation solu tion containing a protease inhibitor cocktail (Roche, Los Angeles, CA, USA). Following centrifugation at 13,000 × g per minute for 30 min, protein was stored at -80°C following determination of the concentration was via the bicinchoninic acid method (Beyotime Institute of Biotechnology, Shanghai, China). A total of 100 µg protein per group was subjected to 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). Membranes were then blocked for 2 h at room temperature in 5% skimmed milk, followed by then incubation with primary antibody overnight at 4°C. The following primary antibodies were used: Mouse anti-β-actin monoclonal antibody (1:1,000 dilution; cat. no. 3700s; Cell Signaling Technology, Inc., Danvers, MA, USA); rabbit anti-NE polyclonal antibody (pAb; cat. no. sc-9520), rabbit anti-human polyclonal B-cell lymphoma 2 (Bcl-2; cat. no. sc-492) and rabbit anti-human polyclonal Bcl-2-associated X protein (Bax; cat. no. sc-6236) antibodies (all 1:1,000 dilution; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); and rabbit anti-Akt pAb (pAb; cat.no. ab8805) and rabbit anti-p-Akt (308; cat. no. ab38449) (both from 1:1,000 dilution; Abcam, Cambridge, MA, USA). Membranes were then incubated with secondary antibodies [goat anti-mouse IgG antibody (cat. no. ZM-0491) or goat anti-rabbit IgG antibody (cat. no. ZA-0448); 1:1,000 dilution; Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China] for 1 h at 37°C. After washing three times in Tris-buffered-saline with Tween 20 (TBST), immunoreactive complexes were visualized using an enhanced chemiluminescence system (Bio-Rad Laboratories, Inc.). β-actin served as an internal positive control. Protein bands were quantitatively analyzed using Quantity One Software 4.5.2 (Bio-Rad Laboratories, Inc.).
Cell viability assay
The Cell Counting Kit-8 (CCK-8) assay (7Sea Biotech, Shanghai, China) was used to assess cell viability. Cells in each group (1.0×104/well) were seeded in 96-well plates and incubated for 1–4 days. Subsequently, 10 µl CCK-8 solution (5 mg/ml) was added to each well, and cells were incubated for 2 h at 37°C. The absorbance at 450 nm was measured using an Eon spectrophotometer (Bio-Rad Laboratories, Inc.) and plotted to obtain cell growth curves. The experiment was repeated three times in triplicate wells for each condition.
Flow cytometric analysis
The cells were routinely collected and centrifuged at 500 × g for 5 min at room temperature. After the cells had been washed twice with phosphate-buffered saline (PBS), a staining mixture was prepared containing 5 µl Annexin-V-fluorescein isothiocyanate fluorescent dye (Sigma-Aldrich) and 5 µl propidium iodide (Sigma-Aldrich). The rate of cell apoptosis was analyzed using a FACsorter (BD Biosciences, San Jose, CA, USA) following a 15 min incubation at room temperature. Furthermore, 5×105 cells were collected, centrifuged at 500 × g for 5 min and washed with pre-cooled PBS twice. The supernatant was discarded, precooled 70% ethanol (1 ml) was added and the samples were fixed overnight at 4°C. Following fixation, the samples were centrifuged at 500 × g for 5 min and washed with 1 ml PBS twice. The cells were resuspended in 100 µl of 1 mg/ml RNase solution (Takara Bio, Inc.). in a 37°C water bath for 30 min. Following incubation, 150 µl of 50 µg/ml propidium iodide staining solution was added and incubated for 30 min in dark at room temperature. The cell cycle distribution was detected using a FACsorter. Experiments were repeated three times.
Statistical analysis
Values are expressed as the mean ± standard deviation. Statistical analysis was performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). An independent samples t-test was employed for comparing the means between two groups. P<0.05 was considered to indicate a statistically significant difference. Each experiment was repeated at least three times.
Results
Vector-mediated expression of NE in NB4 cells
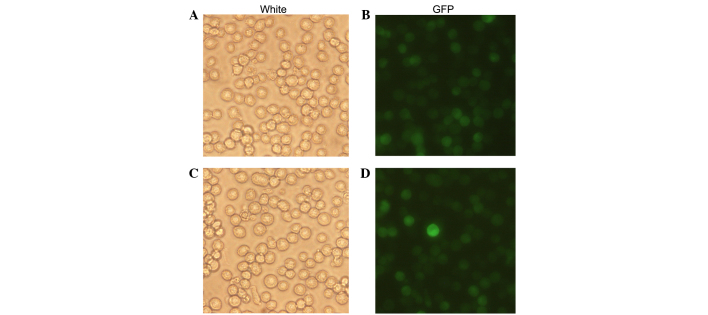
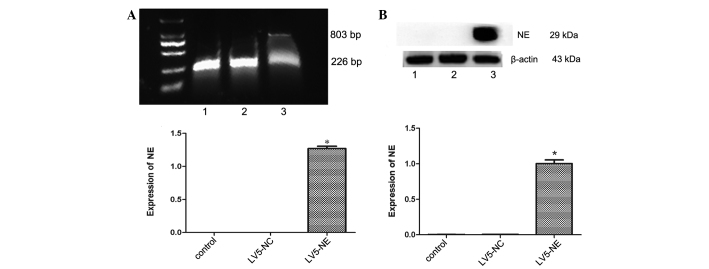
The recombinant lentivirus LV5-NE was transfected into NB4 cells to induce NE overexpression. The percentage of GFP-positive cells was ~100%, demonstrating that after screening with puromycin, all cells were positive transfectants (Fig. 2). RT-PCR and qPCR assays showed that NE mRNA was expressed in the LV5-NE-transfected cells (P<0.05 vs. control), whereas native NB4 cells or LV5-NC-transfected cells showed no expression of NE (Fig. 3A). Furthermore, western blot analysis also showed that NE protein was expressed in the LV5-NE-transfected cells (P<0.05 vs. control), while NE protein expression was low in native NB4 cells and LV5-NC-transfected cells (Fig. 3B).
Figure 2.
Fluorescence microscopic assessment of NB4 cell transfection. (A) Light microscopy and (B) fluorescent microscopy images of NB4 cells were all transfected with LV5-NC. (C) Light microscopy and (D) fluorescent microscopy images of NB4 cells were transfected with LV5-NE. Green fluorescent protein expression in infected cells indicated successful expression of lentivirus containing NE. GFP, green fluorescence protein; NE, neutrophil elastase. Magnification, ×200.
Figure 3.
Effects of transfection with LV5-NC or LV5-NE on NE expression in NB4 cells. (A) The mRNA expression of NE was assessed by reverse-transcription semi-quantitative PCR analysis. The upper panel shows an electrophoretic gel containing the PCR products, which were quantified by densitometry (lower panel). The mRNA of NE was expressed in LV5-NE-transfected NB4 cells, but not in untreated or LV5-NC-transfected cells. The 803 bp band was mRNA of NE and 226 bp was mRNA of glyceraldehyde 3-phosphate dehydrogenase. (B) The protein expression of NE was assessed by western blot analysis. The upper panel shows a representative blot, and quantitative analysis of protein levels was performed by densitometric analysis. The NE protein was highly expressed in LV5-NE-transfected NB4 cells, while expression was low in untreated or LV5-NC-transfected cells. Lanes: M, marker; 1, NB4 cells; 2, NB4 cells transfected with LV5-NCs; 3, NB4 cells transfected with LV5-NE. Values are expressed as the mean ± standard deviation. *P<0.05 vs. control/LV5-NC groups. LV5-NC, negative control vector; LV5-NE, lentiviral vector overexpressing neutrophil elastase; PCR, polymerase chain reaction.
NE enhances the proliferation of NB4 cells
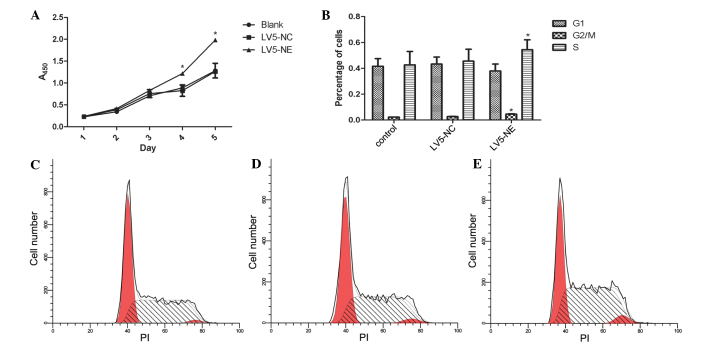
The CCK-8 assay showed that overexpression of NE promoted the proliferation of NB4 cells when compared with that of the control cells (P<0.05) (Fig. 4A). Furthermore, flow cytometric analysis showed that the percentage of LV5-NE-transfected NB4 cells in S phase was significantly higher than that of native or LV5-NC-transfected NB4 cells (P<0.05) (Fig. 4B–D).
Figure 4.
Effects of NE on the proliferation and the cell cycle of NB4 cells. (A) The Cell Counting Kit-8 assay was used to assess cell proliferation on days 1, 2, 3, 4 and 5 post-transfection. Absorbance was measured at 450 nm. At days 3 and 4, the proliferation of NB4 cells was significantly increased following transfection with LV5-NE, while not being affected by transfection with LV5-NC. (B) Flow cytometric analysis showed that transfection with LV5-NE significantly increased the S-phase population of NB4 cells, while the cell cycle was not significantly affected by transfection with LV5-NC. Values are expressed as the mean ± standard deviation. *P<0.05 vs. control. Cell cycle distribution profiles of (C) NB4 cells, (D) NB4 cells transfected with LV5-NC and (E) NB4 cells transfected with LV5-NE. A, absorbance; PI, propidium iodide; LV5-NC, negative control vector; LV5-NE, lentiviral vector overexpressing neutrophil elastase.
NE inhibits apoptosis in NB4 cells
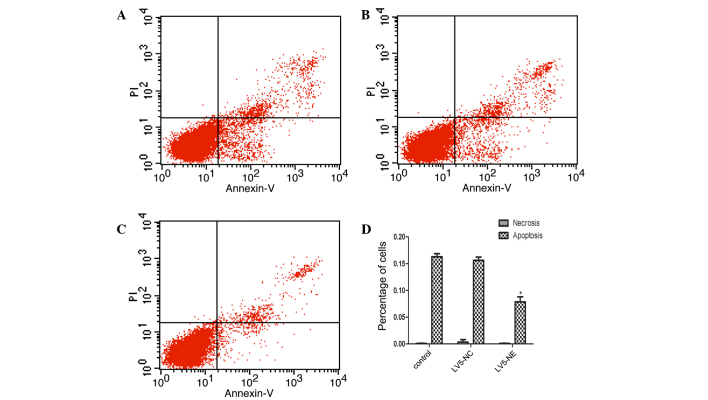
Flow cytometric analysis showed that the apoptotic rate of NB4 cells was significantly decreased following LV5-NE transfection, while LV5-NC had no significant effect (Fig. 5). Furthermore, western blot analysis demonstrated that the expression of anti-apoptotic protein B-cell lymphoma 2 (Bcl-2) was upregulated in NB4 cells following LV5-NE transfection (P<0.05), while LV5-NC had no effect on Bcl-2 levels (Fig. 6). The apoptotic protein Bcl-2-associated X (Bax) was not affected by any of the vectors. These results indicated that NE inhibits apoptosis of NB4 cells by upregulating the expression of the cell survival protein Bcl-2.
Figure 5.
Effect of NE on apoptosis of NB4 cells. Flow cytometric dot plots of Annexin V/PI stained (A) untreated NB4 cells, (B) NB4 cells transfected with LV5-NC and (C) NB4 cells transfected with LV5-NC. (D) Quantification of Annexin V-positive cells provided the apoptotic rate, while Annexin V− and PI-positive cells were regarded as necrotic. Transfection with LV5-NE significantly reduced the apoptotic rate of NB4 cells. Values are expressed as the mean ± standard deviation. *P<0.05 vs. control. LV5-NC, negative control vector; LV5-NE, lentiviral vector overexpressing neutrophil elastase; PI, propidium iodide.
Figure 6.
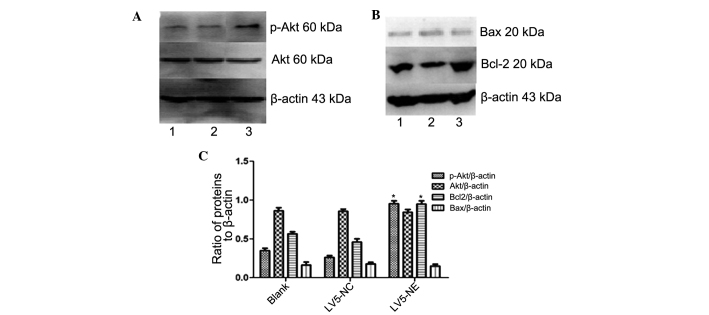
Effects of NE on Akt and apoptotic signaling in NB4 cells. (A and B) The protein expression of Akt, p-Akt, Bax and Bcl-2 in NB4 cells following transfection with LV5-NC or LV5-NE was assessed by western blot analysis. Lanes: 1, NB4 cells; 2, NB4 cells transfected with LV5-NCs; 3, NB4 cells transfected with LV5-NE. (C) Densitometric quantification of protein expression. Overexpression of NE led to a significant increase in Akt phosphorylation and Bcl-2 expression, while total Akt and Bax were not significantly affected. Values are expressed as the mean ± standard deviation. *P<0.05 vs. control. LV5-NC, negative control vector; LV5-NE, lentiviral vector overexpressing neutrophil elastase; p-Akt, phosphorylated Akt; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein.
NE activates the phosphoinositide-3 kinase (PI3K)/Akt pathway in NB4 cells
Western blot analysis showed increased expression of phosphorylated (activated) Akt in NB4 cells following transfection with LV5-NE (P<0.05), while LV5-NC had no obvious effect on Akt phosphorylation and levels of total Akt were not affected by any of the vectors (Fig. 6A). This result indicated that NE may promote the proliferation of NB4 cells by activation of the PI3K/Akt pathway.
Knockdown of NE in U937 cells
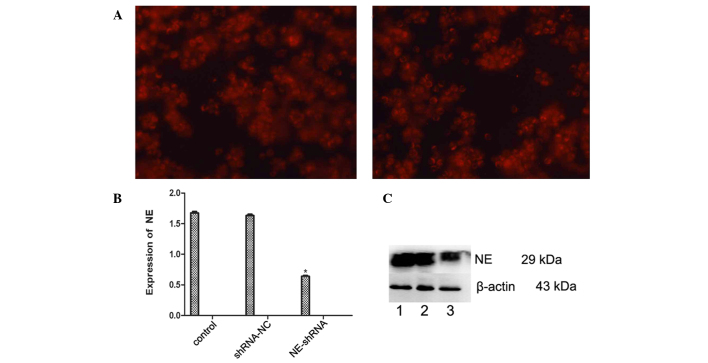
NE-shRNA was used to silence NE in U937 cells. Transfection with NE-shRNA and shRNA-NC vectors was confirmed by fluorescence microscopy (Fig. 7A), and RT-qPCR demonstrated that the mRNA expression of NE was efficiently reduced in the U937/NE-shRNA cells (P<0.05), while shRNA-NC did not affect the mRNA levels of NE (Fig. 7B). Western blot analysis showed that the protein expression of NE in U937 cells was also decreased by NE-shRNA (P<0.05), while not being affected by shRNA-NC (Fig 7C).
Figure 7.
Knockdown of NE in U937 cells with NE-shRNA. (A) Microscopic images of U937 cells transfected with shRNA-NC (left) and NE-shRNA, (right). Magnification, ×200. (B) The mRNA expression of NE was detected by reverse-transcription quantitative polymerase chain reaction. NE-shRNA efficiently decreased the expression of NE in U937 cells. Values are expressed as the mean ± standard deviation. *P<0.05 vs. control. (C) The protein expression of NE was assessed by western blot analysis. The expression of NE was obviously decreased following transfection with NE-shRNA. Lanes: 1, NB4 cells; 2, NB4 cells transfected with LV5-NCs; 3, NB4 cells transfected with LV5-NE. shRNA-NC, negative control small hairpin RNA; shRNA-NE, small hairpin RNA targeting neutrophil elastase.
NE knockdown deactivates the PI3K/Akt pathway and induces apoptotic signaling in U937 cells
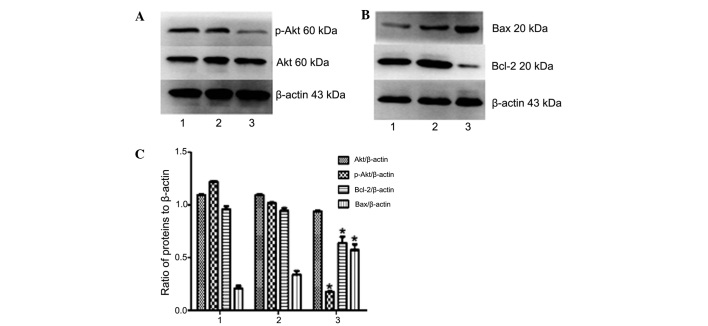
Western blot analysis indicated that the expression of phosphorylated (activated) Akt and Bcl-2 in U937/NE-shRNA cells were decreased compared with those in U937 or U937/shRNA-NC cells (P<0.05) (Fig. 8). However, the protein expression levels of total Akt and Bax were in U937 cells were not significantly affected by shRNA-NE (P>0.05) (Fig 8). Collectively, these results further confirmed that NE has an oncogenic role in leukemia cells, and that its knockdown leads to deactivation of the PI3K/Akt pathway and induction of apoptosis.
Figure 8.
Effects of NE knockdown on Akt and apoptotic signaling in U937 cells. (A and B) The protein expression of Akt, p-Akt, Bax and Bcl-2 in U937 cells following transfection with shRNA-NC or shRNA-NE was assessed by western blot analysis. Lanes: 1, NB4 cells; 2, NB4 cells transfected with shRNA-NC; 3, NB4 cells transfected with shRNA-NE. (C) Densitometric quantification of protein expression. Following knockdown of NE, the protein expression of Bax in U937 cells was significantly increased, while the expression of Bcl-2 and levels of p-Akt were significantly increased, and total Akt was not affected. Values are expressed as the mean ± standard deviation. *P<0.05 vs. control. shRNA-NC, negative control small hairpin RNA; shRNA-NE, small hairpin RNA targeting neutrophil elastase; p-Akt, phosphorylated Akt; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein.
Discussion
NE belongs to the chymotrypsin family of serine proteases and has been reported to be associated with numerous pathological conditions. Previous studies have shown that NE can promote the proliferation, diffusion and metastasis of tumor cells, and is closely associated with tumor occurrence, development and prognosis. NE is also crucially involved in the defense against bacterial pathogens through various mechanisms, including cell lysis (24), degradation of virulence factors (12), or regulation of the inflammatory response by targeting chemokines, cytokines and their receptors (25,26). The underlying mechanisms of the roles of NE in diseases have been elucidated in pulmonary conditions in particular, and it is known that NE causes tissue damage to decrease the tolerance of the host towards pulmonary infection with Burkholderia species (27). NE was first reported to promote cellular proliferation in psoriasis, a benign neoplastic disorder of keratinocytes (28). NE has been reported to promote lung cancer cell proliferation in vitro and in vivo by degrading a novel target substrate, insulin receptor substrate-1, leading to hyperactivity of PI3K and ultimately resulting in enhanced tumor cell proliferation (29,30). NE can also selectively bind to the cancer cell surface and undergo classic clathrin pit-mediated endocytosis (31). The ability of NE to enter cancer cells while inducing their proliferation has been demonstrated in a number of cell types, most notably in breast cancer cells (32); however, its role in acute pro-myelocytic leukemia has not been reported.
APL is a rare form of cancer, and targeted therapy has proven to successfully eradicate leukemia stem cells in the majority of affected patients (33,34). Exploration of the possible mechanism of the roles of NE in APL may provide a foundation for further developing targeted therapies for this disease. NE inhibitors have been used for the clinical treatment of a variety of conditions. Sirtinol inhibits NE activity and was shown to attenuate lipopolysaccharide-mediated acute lung injury in mice (35). NE inhibitors were further shown to prevent liver injury induced by lipopolysaccharide and the inhibition of NE induced hepatic microvascular responses (36,37).
A previous study by our group showed that NE promoted the proliferation and inhibited apoptosis of K562 cells (38). In the present study, a lentivirus expressing the NE gene was constructed and transfected into NB4 cells, which promoted proliferation and inhibited apoptosis; furthermore, the S-phase population of the cell cycle was increased. This was paralleled by increased activation of Akt, suggesting that NE may promote NB4 cell proliferation by activation of the PI3K/Akt pathway.
Acknowledgments
The current study was supported by the National Natural Science Foundation of China (no. 81171658) and the Natural Science Foundation Project of CQ CSTC (no. 2011BA5037).
References
- 1.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukemia. Ann Intern Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 2.Lo Coco F, Diverio D, D'Adamo F, Avvisati G, Alimena G, Nanni M, Alcalay M, Pandolfi PP, Pelicci PG. PML/RAR-alpha rearrangement in acute promyelocytic leukaemias apparently lacking the t(15; 17) translocation. Eur J Haematol. 1992;48:173–176. doi: 10.1111/j.1600-0609.1992.tb00592.x. [DOI] [PubMed] [Google Scholar]
- 3.Berger R, Bernheim A, Daniel MT, Valensi F, Flandrin G. Cytological types of mitoses and chromosomal abnormalities in acute leukemia. Leuk Res. 1983;7:221–236. doi: 10.1016/0145-2126(83)90011-5. [DOI] [PubMed] [Google Scholar]
- 4.de Thé H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15; 17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 5.Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyeloeytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 6.Rowley JD, Golomb HM, Dougherty C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet. 1977;1:549–550. doi: 10.1016/S0140-6736(77)91415-5. [DOI] [PubMed] [Google Scholar]
- 7.Kakizuka A, Miller WH, Jr, Umesono K, Warrell RP, Jr, Frankel SR, Murty W, Dmitrovsky E, Evans RM. Chromosomal translocation t(15; 17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-C. [DOI] [PubMed] [Google Scholar]
- 8.de Thé H, Chen Z. Acute promyelocytic leukaemia: Novel insights into the mechanisms of cure. Nat Rev Cancer. 2010;10:775–783. doi: 10.1038/nrc2943. [DOI] [PubMed] [Google Scholar]
- 9.Baugh RJ, Travis J. Human leukocyte granule elastase: Rapid isolation and characterization. Biochemistry. 1976;15:836–841. doi: 10.1021/bi00649a017. [DOI] [PubMed] [Google Scholar]
- 10.Belaaouaj A, McCarthy R, Baumann M, Gao Z, Ley TJ, Abraham SN, Shapiro SD. Mice lacking neutrophil elastase reveal impaired host defense against gram-negative bacterial sepsis. Nat Med. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 11.Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 2000;289:1185–1188. doi: 10.1126/science.289.5482.1185. [DOI] [PubMed] [Google Scholar]
- 12.Weinrauch Y, Drujan D, Shapiro SD, Weiss J, Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417:91–94. doi: 10.1038/417091a. [DOI] [PubMed] [Google Scholar]
- 13.Gabay JE, Scott RW, Campanelli D, Griffith J, Wilde C, Marra MN, Seeger M, Nathan CF. Antibiotic proteins of human polymorphonuclear leukocytes. Proc Natl Acad Sci USA. 1989;86:5610–5614. doi: 10.1073/pnas.86.14.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaynar AM, Houghton AM, Lum EH, Pitt BR, Shapiro SD. Neutrophil elastase is needed for neutrophil emigration into lungs in ventilator-induced lung injury. Am J Respir Cell Mol Biol. 2008;39:53–60. doi: 10.1165/rcmb.2007-0315OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro SD, Goldstein NM, Houghton AM, Kobayashi DK, Kelley D, Belaaouaj A. Neutrophil elastase contributes to cigarette smoke-induced emphysema in mice. Am J Pathol. 2003;163:2329–2335. doi: 10.1016/S0002-9440(10)63589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senior RM, Tegner H, Kuhn C, Ohlsson K, Starcher BC, Pierce JA. The induction of pulmonary emphysema with human leukocyte elastase. Am Rev Respir Dis. 1997;116:469–475. doi: 10.1164/arrd.1977.116.3.469. [DOI] [PubMed] [Google Scholar]
- 17.Lane AA, Ley TJ. Neutrophil elastase cleaves PML-RAR and is important for the development of acute promyelocytic leukemia in mice. Cell. 2003;115:305–318. doi: 10.1016/S0092-8674(03)00852-3. [DOI] [PubMed] [Google Scholar]
- 18.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 19.Lane AA, Ley TJ. Neutrophil elastase is important for PML-retinoic acid receptor activities in early myeloid cells. Mol Cell Biol. 2005;25:23–33. doi: 10.1128/MCB.25.1.23-33.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao YM, Zhong L, Zhang X, Hu XX, Liu BZ. PML (NLS(−)) inhibits cell apoptosis and promotes proliferation in HL-60 cells. Int J Med Sci. 2013;10:498–507. doi: 10.7150/ijms.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu XX, Zhong L, Zhang X, Gao YM, Liu BZ. NLS-RARα promotes proliferation and inhibits differentiation in HL-60 cells. Int J Med Sci. 2014;11:247–254. doi: 10.7150/ijms.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang KL, Ma PP, Yang XQ, Zhong L, Wang H, Zhu XY, Liu BZ. Neutrophil elastase and its therapeutic effect on leukemia cells. Mol Med Rep. 2015;12:4165–4172. doi: 10.3892/mmr.2015.3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Belaaouaj A. Neutrophil elastase-mediated killing of bacteria: Lessons from targeted mutagenesis. Microbes Infect. 2002;4:1259–1264. doi: 10.1016/S1286-4579(02)01654-4. [DOI] [PubMed] [Google Scholar]
- 25.Kessenbrock K, Dau T, Jenne DE. Tailor-made inflammation: How neutrophil serine proteases modulate the inflammatory response. J Mol Med (Berl) 2011;89:23–28. doi: 10.1007/s00109-010-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham CT. Neutrophil serine proteases: Specific regulators of inflammation. Nat Rev Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 27.Sahoo M, Del Barrio L, Miller MA, Re F. Neutrophil elastase causes tissue damage that decreases host tolerance to lung infection with burkholderia species. PLoS Pathog. 2014;10:e1004327. doi: 10.1371/journal.ppat.1004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiedow O, Wiese F, Streit V, Kalm C, Christophers E. Lesional elastase activity in psoriasis, contact dermatitis and atopic dermatitis. J Invest Dermatol. 1992;99:306–309. doi: 10.1111/1523-1747.ep12616644. [DOI] [PubMed] [Google Scholar]
- 29.Houghton AM, Rzymkiewicz DM, Ji H, Gregory AD, Egea EE, Metz HE, Stolz DB, Land SR, Marconcini LA, Kliment CR, et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med. 2010;16:219–223. doi: 10.1038/nm.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metz HE, Houghton A. Insulin receptor substrate regulation of phosphoinositide 3-kinase. Clin Cancer Res. 2011;17:206–211. doi: 10.1158/1078-0432.CCR-10-0434. [DOI] [PubMed] [Google Scholar]
- 31.Gregory AD, Hale P, Perlmutter DH, Houghton AM. Clathrin pit-mediated endocytosis of neutrophil elastase and cathepsin G by cancer cells. J Biol Chem. 2012;287:35341–35350. doi: 10.1074/jbc.M112.385617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittendorf EA, Alatrash G, Qiao N, Wu Y, Sukhumalchandra P, St John LS, Philips AV, Xiao H, Zhang M, Ruisaard K, et al. Breast cancer cell uptake of the inflammatory mediator neutrophil elastase triggers an anticancer adaptive immune response. Cancer Res. 2012;72:3153–3162. doi: 10.1158/0008-5472.CAN-11-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasr R, Guillemin MC, Ferhi O, Soilihi H, Peres L, Berthier C, Rousselot P, Robledo-Sarmiento M, Lallemand-Breitenbach V, Gourmel B, et al. Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nat Med. 2008;14:1333–1342. doi: 10.1038/nm.1891. [DOI] [PubMed] [Google Scholar]
- 34.de Thé H, Le Bras M, Lallemand-Breitenbach V. The cell biology of disease: Acute promyelocytic leukemia, arsenic and PML bodies. J Cell Biol. 2012;198:11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai YF, Yu HP, Chang WY, Liu FC, Huang ZC, Hwang TL. Sirtinol inhibits neutrophil elastase activity and attenuates lipopolysaccharide-mediated acute lung injury in mice. Sci Rep. 2015;5:8347. doi: 10.1038/srep08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon AH, Qiu Z. Neutrophil elastase inhibitor prevents endotoxin-induced liver injury following experimental partial hepatectomy. Br J Surg. 2007;94:609–619. doi: 10.1002/bjs.5625. [DOI] [PubMed] [Google Scholar]
- 37.Ishii K, Ito Y, Katagiri H, Matsumoto Y, Kakita A, Majima M. Neutrophil elastase inhibitor at tenuates lipopolysaccharide-induced hepatic micro vascular dysfunction in mice. Shock. 2002;18:163–168. doi: 10.1097/00024382-200208000-00013. [DOI] [PubMed] [Google Scholar]
- 38.Jiang K, Ma P, Yang X, Zhong L, Wang H, Zhu X, Liu B. Over-expression of neutrophil elastase promotes proliferation and inhibits apoptosis in K562 cells. Chin J Cell Mol Immunol. 2015;31:159–162. 167. In Chinese. [PubMed] [Google Scholar]