Abstract
Background:
Antipsychotic-induced weight gain (AIWG) is a serious concern in therapy with antipsychotic medications. To identify single nucleotide polymorphisms (SNPs) associated with AIWG, we conducted a genome-wide association study (GWAS) for antipsychotic treatment.
Methods:
The discovery cohort consisted of 534 patients with schizophrenia, who underwent 8-week treatment with antipsychotics and were genotyped using the Illumina Human 610-Quad BeadChip. The independent replication cohort consisted of 547 patients with schizophrenia, treated with similar antipsychotics, and genotyped using the Sequenom MassARRAY platform. Two hundred and thirty-six drug-naive patients treated with risperidone or quetiapine were analyzed independently. Additionally, we conducted pathway and expression analyses using several public bioinformatics databases.
Results:
After correction for age and gender, the top 2 genome-wide significant SNPs with AIWG were located in the PTPRD gene (protein tyrosine phosphatase, receptor type D, 9p24-p23; rs10977144, P GWAS = 9.26E-09; rs10977154, P GWAS = 4.53E-08). The third most significant SNP was in the GFPT2 gene (glutamine-fructose-6-phosphate amidotransferase 2, 5q35.3; rs12386481, P GWAS = 1.98E-07). These results were validated in the replication cohort (rs10977144, P Replication = 4.30E-03; rs10977154, P Replication = 6.33E-03; rs12386481, P Replication =7.65E-03). These results were also verified in those patients initially exposed to risperidone and quetiapine (rs10977144, P = 1.97E-05; rs10977154, P = 2.04E-05; rs12386481, P = 1.97E-04). Pathway analyses showed that AIWG may involve in multiple pathways related to metabolic processes. Moreover, PTPRD mRNA might be highly expressed in brain regions, and the SNPs (rs10977144, rs1097154) also showed significant expression quantitative trait locus effects.
Conclusions:
Our findings indicate that PTPRD polymorphisms might modulate AIWG.
Key words: schizophrenia, antipsychotic-induced weight gain (AIWG), genome-wide association study, protein tyrosine phosphatase, receptor type, D (PTPRD)
Introduction
Schizophrenia is a well-known severe psychiatric disorder with a lifetime risk of approximately 1% in the general population.1 Exposure to the treatment of the typical or atypical antipsychotic medications (APMs) may be a critical cause of weight gain in patients with schizophrenia, which may impede the recovery process.2,3 Numerous studies have shown that antipsychotic treatment is often associated with medical complications such as obesity, diabetes, lipid disturbances, cancers, and coronary heart disease; premature mortality is also documented.4–9 Treatment-emergent weight gain varies within the broad class of antipsychotics.4,6,8 However, the individual’s propensity to develop weight gain following antipsychotics treatment shows substantial variability, which largely depends on genetic factors.6–8
Excessive weight gain is an unfavorable side effect occurring frequently in patients who are undergoing antipsychotic drug treatments, especially atypical antipsychotics. Twin and sibling studies have demonstrated similar degrees in weight gain profiles upon receiving antipsychotic medications.9 Therefore, the interindividual variability across unrelated patients receiving the same antipsychotic medications is thought to be caused by genetic causes.9,10 For this reason, pharmacogenetics research efforts have focused on the identification of genetic variants contributing to individual variability regarding several antipsychotic-related phenotypes.
The candidate gene approach has been used to identify susceptibility genes of antipsychotic-induced weight gain (AIWG) including the following promising genes, such as the fat mass and obesity associated gene (FTO); 5-hydroxytryptamin 2C (HTR2C), leptin (LEP), dopamine D3 receptor (DRD3), tumor necrosis factor (TNF), synaptomal-associated protein 25kDa (SNAP25), cannabinoid 1 receptor (CNR1), guanine nucleotide binding protein (GNB3), insulin-induced gene 2 (INSIG2), adrenergic receptor α2a (ADRA2A), and NADH-ubiquinone oxidoreductase Fe-S protein 1 (NDUFS1).6–11
In contrast to candidate gene-based methods, the genome-wide association study (GWAS) approach allows unbiased, “hypothesis-free” detection of DNA variants associated with the phenotype of interest.12,13 The first genome-wide linkage study of AIWG found a locus with suggestive multipoint logarithm of the odds of 2.74 at chromosome 12q24, located in less than 1cM from the pro-melanin-concentrating (PMCH) gene, a neuropeptide involved in the control of food intake and energy expenditure.14 Malhotra et al have implemented a GWAS including 139 pediatric patients with mood and psychotic disorders, who were treated for the first time with 1 of 3 second-generation antipsychotics. The strongest associated single nucleotide polymorphism (SNP) rs489693 was located in the downstream of the melanocortin 4 receptor (MC4R) gene.15 Replication of these findings needs to be implemented in independent cohorts. Here, we describe a GWAS for AIWG.
Methods
Subjects in Discovery Cohort
We performed an open-labeled GWAS involving 746 patients hospitalized with schizophrenia, as we described in a previous GWAS project.16 Additionally, 534 patients (71.58%), who received less than 4-week treatment of antipsychotics before enrollment, finally completed 8 weeks of antipsychotic monotherapy and related clinical assessment (258 males and 276 females, mean age 26.4±5.3 y). These patients received one of the following atypical antipsychotics: risperidone, quetiapine, olanzapine, clozapine, aripiprazole, or ziprasidone (table 1).
Table 1.
Description of Discovery and Replication Schizophrenia Cohorts
| Discovery Cohort | Replication Cohort | |||
|---|---|---|---|---|
| Mean ± SD or n (%) | Dose (mg/d) | Mean ± SD or n (%) | Dose (mg/d) | |
| Mean ± SD | Mean ± SD | |||
| N | 534 | NA | 547 | NA |
| Age (y) | 26.4±5.3 | NA | 27.4±7.7 | NA |
| Males | 258 (48.3%) | NA | 271 (49.5%) | NA |
| Baseline BMI (kg/m2) | 22.29±7.47 | NA | 21.65±6.38 | NA |
| Aripiprozole (ARI) | 58 (10.86%) | 21.08±5.13 | 77 (14.08%) | 20.83±6.44 |
| Ziprasidone (ZIP) | 55 (10.31%) | 103.97±35.36 | 72 (13.16%) | 104.02±33.55 |
| Risperidone (RIS) | 194 (36.33%) | 5.12±1.68 | 121 (22.12%) | 5.09±1.22 |
| Quetiapine (QUE) | 71 (13.29%) | 545.38±182.88 | 82 (14.99%) | 533.98±196.22 |
| Olanzapine (OLZ) | 80 (14.98%) | 18.05±5.99 | 96 (17.55%) | 17.47±5.29 |
| Clozapine (CLZ) | 76 (14.23%) | 306.59±127.72 | 99 (18.10%) | 315.85±101.75 |
Note: BMI, body mass index; NA, not applicable.
Subjects in Replication Cohort
The replication cohort included 547 patients with schizophrenia (271 males and 276 females, mean age 27.4±7.7 y), who received less than 4-week treatment of antipsychotics before enrollment. The replication samples were enrolled from other hospitals (from Liaoning, Hebei, and Henan provinces). The diagnosis and assessment of the replication cohort was similar to the discovery cohort. They were also followed up during the 8-week treatment course with the same range of antipsychotic monotherapy. The demographic characteristics of all patients were described in table 1.
Briefly, patients were enrolled into the present study according to the following criteria: (1) Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) criteria for schizophrenia (and then the subjects were remained in follow-up for 8-week monotherapy with certain antipsychotics); (2) physically healthy with normal hematological and biochemical parameters; (3) precluding a current or a past DSM-IV-TR diagnosis of mood disorders or substance abuse. Furthermore, none of the patients received psychiatric medications other than antipsychotics, except hypnotics when medically needed, and continuing medication for physical health problems.
Exclusion criteria included a current or past diagnosis of an eating disorder, biochemical evidence of thyroid dysfunction, and any acute nonpsychiatric medical disorders. Participants who were pregnant or breastfeeding were excluded. The specific choice, dosage, and titration schedule of the specific antipsychotic medications were based on clinical indications.
All patients were clinically assessed after 8 hours or more of overnight fasting. Then, they were weighed and assessed before treatment and at 2, 4, 6, and 8 weeks posttreatment. The body mass index (BMI) was calculated as weight in kilograms divided by height in meters squared (kg/m2). The standing height was measured using the stadiometers. The objectives and procedures of the study were explained to all subjects and written informed consent was obtained. The study was approved by the research ethical committees of local hospitals. All procedures were conducted in accord with principles expressed in the Declaration of Helsinki. The study was also registered for clinical trial number as ChiCTR-RNC-09000522 (http://www.chictr.org/).
Considering the different mechanism of various antipsychotics, we selected risperidone and quetiapine, which have been reported to cause moderate effects of AIWG, in order to make a drug-specific analysis. To control the confounding factors of prior treatment histories, we selected first-episode, drug-naive patients for further analysis. From above-mentioned subjects (534 patients in discovery cohort and 547 patients in replication cohort), we further analyzed the association of top significant SNP with 236 first-episode, drug-naive patients (29.7±9.5 y old, 117 males and 119 females) treated with risperidone and quetiapine and followed up for 8 weeks.
Genotyping
Genomic DNA was extracted from whole blood using a Qiagen QIAamp DNA Mini Kit (Qiagen GmbH). All DNA samples were subjected to rigorous quality control (QC) to check for fragmentation and amplification. DNA was normalized to a concentration of 50ng/ml (diluted in 10mM Tris/1mM EDTA) with a Nanodrop Spectrophotometer (ND-1000, NanoDrop). Approximately 200ng of genomic DNA was used for genotyping analysis. Briefly, each sample was whole-genome amplified, fragmented, precipitated and resuspended in appropriate hybridization buffer.
For the discovery cohort, the genotyping of denatured samples was performed on Illumina HumanHap610-Quad BeadChips, which include 620901 SNPs and copy number variation (CNV) probes in total, as we descripted previously.16 For the replication cohort, Sequenom MassArray methods were used to genotype the top 20 associated SNPs from the discovery cohort. To further evaluate the quality of the genotype data for the validation analysis, we selected 6 SNPs (showing significant association evidence in both the GWAS and validation studies) to be re-genotyped in 100 randomly selected GWAS samples by using the DNA sequencing methods on an ABI PRISM 377–96 DNA Sequencer (Applied Biosystem). The concordance rate between DNA sequencing and Illumina or Seuquenom MassArray genotyping was 99.8%.
Statistical Analysis
Initially, 534 cases of discovery cohort were genotyped with 620901 SNPs and CNV probes. The Quanto v1.2 software was used to calculate the statistic power of the association.17 To conduct the GWAS with the BMI gain, we used PLINK software version 1.07.18 The QC procedures to filter the cases and SNPs were similar to those previously used, including the examination of potential genetic association based on pairwise identity by state, identification of a first- or second-degree relative pair, identification of low genotyping rate (MIND > 0.1), and principal-component analysis for population stratification using EIGENSTRAT.16,19 The total genotyping rate in remaining individuals was >0.98. After stringent quality control, we excluded 24889 SNPs with a call rate <90%, 101966 SNPs with minor allele frequency <1%, and 1699 SNPs with significant deviation from Hardy–Weinberg equilibrium (P < 5.0E-05) in the controls. The final postquality control data set ready for analysis consisted of 495371 SNPs and 534 schizophrenia cases of Chinese Han descent (table 1). The BMI gain was used as the phenotype for the quantitative trait locus analyses. To control the potential confounding effects of age and gender, we conducted log-linear analyses using age and gender as covariates.
According to the stringent Bonferroni multiple correction, the genome-wide significance threshold was set as traditional 5.0E-08, the standard GWAS threshold. For gene annotation, we used University of California Santa Cruz Genome Browser (http://genome.cse.ucsc.edu/) and NCBI databases (http://www.ncbi.nlm.nih.gov). To control the false positive error rate during the process of multiple tests, we performed Bonferroni corrections.
To evaluate whether susceptibility loci located in genes involved in particular biological pathways show enrichment of significant P values for association with AIWG, we performed pathway analysis using the Knowledge-based mining system for Genome-wide Genetic studies software,20 which controls for the number of SNPs and gene size. The pathway analysis was based on the results of our discovery data set. We elevated pathways from canonical databases Gene Ontology (GO). More detailed description about pathway analyses are listed in the supplementary materials.
Furthermore, to explore the expression patterns of the top significantly susceptible genes in human tissues, we used the Gene Enrichment Profiler database (http://xavierlab2.mgh.harvard.edu/EnrichmentProfiler/), as well as the Human Brain Transcriptome (http://hbatlas.org/pages/hbtd).21,22 Moreover, to detect the functional effects of the at-risk SNPs in the susceptibility gene, we analyzed their associations with gene expression levels in BRAINEAC database (http://www.braineac.org/).23
Results
Five hundred and thirty-four schizophrenia patients in the discovery cohort received 8 weeks of treatment with atypical antipsychotic drugs (risperidone, quetiapine, olanzapine, clozapine, aripiprazole, or ziprasidone). For the replication cohort, 547 patients with schizophrenia were enrolled in the 8-week antipsychotic treatment course. Demographic and medication data for this cohort are also listed in table 1.
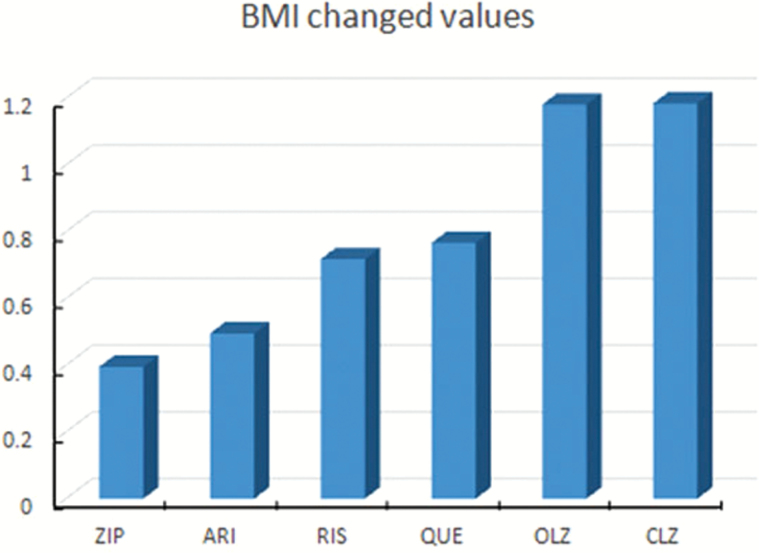
A weight gain “case” was defined as a patient who gained 7% or more of his or her baseline body weight in a short-term trial.24 The proportion of patients meeting this criterion was 17.01% (91/534). Over the 8-week course of treatment, the average weight gain ratio in 2 cohorts was 2.43% (SD = 0.64%). A chi-square test of the distributions displayed in demonstrated significant effects of antipsychotics (χ2 = 62.39, P < .001). Figure 1 shows the BMI gain profile induced by 3 different groups of antipsychotics following the 8-week treatment in 2 cohorts (F = 9.337, P < .001). Olanzapine- and clozapine-treated patients demonstrated a markedly different distribution, with the majority of subjects experiencing extreme AIWG. In contrast, aripiprazole and ziprasidone showed mild effects, and risperidone and quetiapine showed moderate effects on weight gain.
Fig. 1.
Profile of BMI gain (kg/m2) in 1,081 patients with schizophrenia following 8-week treatment with atypical antipsychotic drugs. The y-axis represents the BMI change values. Abbreviation of the antipsychotic medications: ARI, aripiprazole; ZIP, ziprasidone; RIS, risperidone; QUE, quetiapine; OLZ, olanzapine; CLZ, clozapine.
To calculate the statistic power, the following assumptions were made: the additive model was chosen, marker allele frequency was set from 0.05 to 0.8, the main effect of genotype on weight gain (βG) was 1, and minor allele frequency was set as 0.05. Using these assumptions, with 1081 patients (534 patients in discovery cohort and 547 ones in replication cohort), we obtained the power of ~82%.
For the 534 patients with schizophrenia in the discovery cohort, we implemented the GWAS analyses, using the BMI gain as the dependent variable after controlling for age and gender as confounding factors. The Manhattan plot and the quantile-quantile (Q-Q) plot of GWAS results were shown in supplementary figures S1 and S2. The Lambda genomic control inflation factor (λGC) was about 1.06, which showed no significant population stratification among the subjects in discovery cohort.
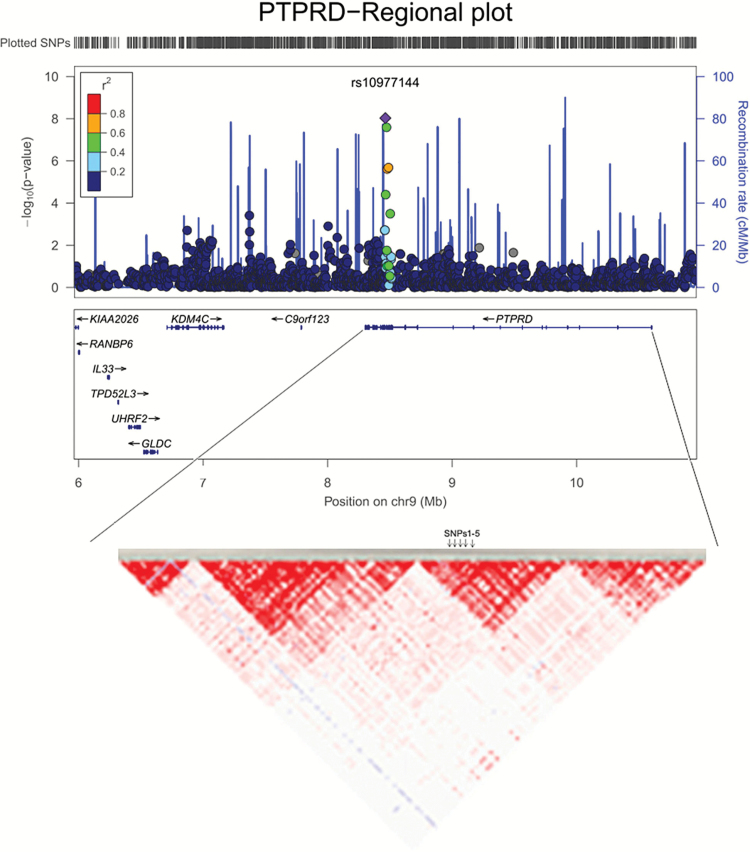
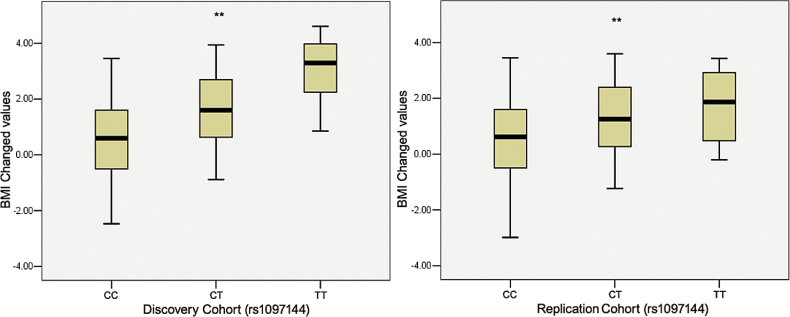
Moreover, table 2 showed the results of top 20 significant SNPs associated with AIWG in discovery cohort, as well as the results of replication cohorts, using the log-linear analyses with BMI gain as dependent variable after correcting by age and gender. The top 2 significantly associated SNPs were located in the protein tyrosine phosphatase, receptor type D gene (PTPRD, 9p24-p23) (rs10977144, P GWAS = 9.26E-09; rs10977154, P GWAS = 4.53E-08, figure 2). These results were further validated in an independent replication cohort (n = 547; rs10977144, P Replicated = 4.31E-03; rs10977154, P Replicated = 6.33E-03; table 2, figure 3). The third significant SNP, close to the genome-wide significance level, was rs12386481 (P GWAS = 1.98E-07), which was located in the glutamine-fructose-6-phosphate amidotransferase 2 (GFPT2, 5q35.3). There were 3 other SNPs (rs10124277, rs2296102, rs13286274) in the PTPRD gene that were close to suggestive genome-wide significance (P GWAS < 5.0E-05, table 2, figure 2). In total, the 5 SNPs in PTPRD gene associated with AIWG were in strong linkage disequilibrium (LD), which further suggested the PTPRD polymorphisms might be participating in mediating AIWG (r 2 > 0.9 using our Illumina 610K healthy control data16).
Table 2.
Association Results of Top 20 Significant SNPs With AIWG in Discovery and Replication Cohorts
| Chromosome | SNP | Position | Genes | Location | Allelea | NMISS | Discovery Cohort | Replication Cohort | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BETAb | STATc | P GWAS d | BETA | STAT | P Replication | |||||||
| 9 | rs10977144 | 8464233 | PTPRD | intron | T/C | 534 | 1.421 | 5.857 | 9.26E-09 | 0.392 | 2.868 | 4.30E-03 |
| 9 | rs10977154 | 8473347 | PTPRD | intron | T/C | 534 | 0.9929 | 5.549 | 4.53E-08 | 0.3184 | 2.741 | 6.33E-03 |
| 5 | rs12386481 | 179766091 | GFPT2 | 5UTR | A/G | 533 | −0.8954 | −5.306 | 1.98E-07 | −0.4976 | −2.677 | 7.65E-03 |
| 2 | rs2046545 | 114417849 | ACTR3 | intron | G/A | 534 | −0.9236 | −4.737 | 2.79E-06 | −0.3423 | −2.722 | 6.71E-03 |
| 6 | rs1886243 | 35225377 | TCP11 | 5UTR | G/T | 534 | −0.5323 | −4.719 | 3.03E-06 | −0.4961 | −2.596 | 9.70E-03 |
| 9 | rs10124277 | 8489295 | PTPRD | intron | T/C | 534 | 0.4381 | 4.711 | 3.15E-06 | 0.2754 | 2.831 | 4.81E-03 |
| 9 | rs2296102 | 8479105 | PTPRD | intron | T/G | 534 | 0.9216 | 4.692 | 3.45E-06 | 0.4739 | 2.776 | 5.70E-03 |
| 5 | rs3853114 | 13534754 | LOC391738 e | 3UTR | C/T | 534 | −0.4231 | −4.644 | 4.32E-06 | −0.1973 | −2.125 | 0.03401 |
| 1 | rs4649328 | 231737008 | KCNK1 | 5UTR | T/C | 534 | 0.4078 | 4.434 | 1.13E-05 | 0.3708 | 2.287 | 0.0226 |
| 10 | rs7906629 | 90048617 | RNLS f | intron | A/C | 534 | −0.4843 | −4.301 | 2.02E-05 | −0.4075 | −2.666 | 7.90E-03 |
| 10 | rs1426621 | 90085709 | RNLS | intron | T/G | 534 | −0.432 | −4.287 | 2.15E-05 | −0.2575 | −2.645 | 8.40E-03 |
| 5 | rs6449807 | 65635596 | FLJ46010 | intron | T/C | 534 | 0.4527 | 4.286 | 2.17E-05 | 0.1686 | 1.693 | 0.091 |
| 4 | rs4861124 | 41337646 | LIMCH1 | intron | G/A | 534 | −0.4329 | −4.237 | 2.68E-05 | 0.1376 | 0.8643 | 0.3878 |
| 3 | rs7611808 | 132828435 | CPNE4 | intron | G/A | 534 | −1.363 | −4.199 | 3.15E-05 | −0.5232 | −3.026 | 2.59E-03 |
| 9 | rs13286274 | 8466459 | PTPRD | intron | T/C | 534 | 1.213 | 4.187 | 3.31E-05 | 0.1529 | 1.172 | 0.2418 |
| 8 | rs2929104 | 40030807 | INDOL1 | 3UTR | T/G | 534 | 1.388 | 4.18 | 3.41E-05 | 0.2708 | 2.098 | 0.03636 |
| 6 | rs6915627 | 35236308 | TCP11 | 5UTR | A/G | 534 | −0.3937 | −4.163 | 3.66E-05 | −0.1294 | −1.163 | 0.2452 |
| 6 | rs2195729 | 123631825 | TRDN | intron | C/T | 534 | −0.3874 | −4.152 | 3.83E-05 | 0.2817 | 2.201 | 0.02818 |
| 9 | rs882895 | 129854187 | C9orf90 | 3UTR | A/G | 532 | −0.3952 | −4.115 | 4.49E-05 | −0.2115 | −2.287 | 0.02256 |
| 3 | rs13076635 | 31825509 | OSBPL10 | intron | A/G | 533 | 0.564 | 4.115 | 4.49E-05 | 0.2754 | 2.831 | 4.81E-03 |
aMinor/major alleles.
bBETA means the regression coefficient of the log-linear analysis.
cSTAT refers to the coefficient t-statistic of log-linear analysis.
d P GWAS are P values corrected by using log-linear association at GWAS level, with age and gender as covariates.
eLOC391738 is also known as RPL29P13.
fRNLS is also known as C10orf59.
Fig. 2.
Regional association plot of the indicated PTPRD rs10977144 and rs1097154 with BMI gain (kg/m2). (A) The purple diamond indicated the top associated SNP located on the chromosomal 9p24-p23. (B) The linkage disequilibrium (LD) pattern of SNPs at PTPRD gene. SNPs1-5 showed five associated SNPs with AIWG, which were in stron LD (r 2 > .9), were listed as follows: SNP1 (rs10977144, chr9:8464233); SNP2 rs13286274, chr9:8466459); SNP3 (rs10977154, chr9:8473347); SNP4 (rs2296102, chr9:8479105); SNP5 (rs10124277, chr9:8489295).
Fig. 3.
Box plot of PTPRD rs10977144 T allele and antipsychotic drug-induced BMI gain (kg/m2) in two cohorts of patients. (A) Discovery cohort. (B) Replication cohort. **P < .01.
Among the above-mentioned subjects, 236 drug-naive patients treated with risperidone or quetiapine were analyzed independently, in order to control for potential confounding factors, such as previous and concomitant medicine medical history. These results also were verified in patients firstly exposed to risperidone and quetiapine (rs10977144, P = 1.97E-05; rs10977154, P = 2.04E-05; rs12386481, P = 1.97E-04). Furthermore, in the drug-naive cohorts, the rs10977144 T allele carriers showed different effects by treatment trajectory on the risk of AIWG (figure 4).
Fig. 4.
PTPRD rs10977144 genotype (TT+CT vs CC carriers) modulated BMI gain (kg/m2) over 8 weeks of risperidone and quetiapine treatment in the 236 first-episode, drug-naive schizophrenia patients. Considering number of TT carriers was only 3, we combined TT and CT carriers in one group. *P < .05; **P < .0001.
For the discovery cohort, there were 34 SNPs in total that achieved nominal significance (P < 1.01E-05). Other suggestive associations included the actin-related protein 3 (ACTR3, 2q14.1, rs2046545); t-complex 11, testis-specific (TCP11, 6p21.31, rs1886243); renalase (RNLS, 10q23.31, rs7906629 and rs1426621), copine IV (CPNE4, 3q22.1, rs7611808), and the oxysterol-binding protein-like protein 10 (OSBPL10, 3p22.3, rs13076635), with both P GWAS < 10–5 and P Replicated < 10–3.
Previously reported susceptibility genes for AIWG gain were also analyzed in our sample. The most significant SNPs in previously reported genes were listed in supplementary table 2, including MC4R, FTO, DRD3, ADRA2A, LEP, GNB3, SNAP25, and NDUFS1.6–11 Of these genes, LEPR (rs3790426, P = 3.19E-03), FTO (rs1075440, P = 1.13E-03), PAM (rs2657477, P = 6.28E-04), MC4R (rs10460146, P = 0.01397), and SNAP25 (rs481302, P = 0.01218) (supplementary table 1) achieved modest association (P < .05) in our sample.
To investigate the network effects of multiple genes on AIWG, we conducted pathway analyses using GWAS data. Of the 825 GO pathways tested, 10 pathways indicated significant enrichment of associated genes after the hybrid set-based test and hypergeometric test (P < .05) (supplementary table 3). These pathways were mainly involved in metabolic processes and regulation of phosphorylation, such as glucosamine metabolism, amino sugar metabolism, and regulation of peptidyl tyrosine phosphorylation (P < .05).
To test the biological plausibility of PTPRD in the pathogenesis of AIWG or schizophrenia, we investigated expression enrichment profiling of the PTPRD polymorphic gene in multiple human tissues. We found that PTPRD mRNAs were preferentially expressed in human brain tissues such as the whole brain, frontal cortex, and subthalamic nucleus, all of which showed higher enrichment scores (supplementary figure S3). Temporal expression analyses showed that the expression level of PTPRD was relatively high at the late fetal and infant stages of life, as well as in the later stages of life. The results of gene expression analysis further supported the putative role of PTPRD in regulating weight and brain function. The GFPT2 mRNAs are mainly expressed in smooth muscle and other peripheral tissues; brain expression level of GFPT2 was, however, relatively high during late fetal and infant stages (supplementary figure S4).
To explore the potential effect of polymorphisms on the expression of susceptible genes, we conducted an expression quantitative trait loci (eQTL) analysis using the gene expression data from the BRAINEAC database; we then performed eQTL analyses for the 2 SNPs (rs10977144, rs10977154) at PTPRD across 10 brain tissues in the brains of 130 healthy subjects. Moreover, the top 3 significant SNPs were found to be associated with the expression level of PTPRD in the hippocampus or thalamus (for exon-specific probesets of mRNA expression: PTPRD [transcript ID 3198375] in hippocampus, rs10977144, P = .0028; rs10977154, P = .0037; GFPT2 [transcript ID 2890692] in thalamus, rs2386481, P = .0002; respectively; supplementary figures S3 and S4).
Discussion
We have performed a GWAS to identify genetic susceptibility variants associated with AIWG. The two most significantly associated SNPs (rs10977144, P GWAS = 9.26E-09; rs10977154, P GWAS = 4.53E-08) were located in an intron of the PTPRD gene. PTPRD is a receptor-type protein tyrosine phosphatase expressed in particular brain regions (eg, hippocampal CA2 and CA3) in B lymphocytes, and in the thymic medulla. The eQTL analysis showed that rs10977154 may have a potential cis-eQTL effect in the human hippocampus (supplementary table 2 and figure S3), also supporting previous reports that PTPRD may be associated with cognitive function.25,26
To elucidate the physiologic roles of PTPRD, Uetani et al found that PTPdelta-deficient mice were semilethal due to insufficient food intake.26 The results also suggested that PTPRD might play an important role in the mechanisms underlying weight gain. Moreover, a recent report has identified a PTPRD polymorphism that may be associated with type 2 diabetes mellitus (T2DM).27 Thus, we hypothesized that PTPRD may participate in the underlying genetic mechanism of metabolic disturbance of fat or glucose related to antipsychotic therapy.
For the 236 first-episode, drug-naive patients on risperidone and quetiapine therapy, the PTPRD rs10977144 also showed association with AIWG (P = 1.97E-05 at 8-week treatment, figure 4). This result further suggested the association of PTPRD rs10977144 with AIWG, excluding the confounding effects of other medications, or prior treatment histories.
The third most significant locus rs12386481 (P GWAS = 1.98E-07) located in the glutamine-fructose-6-phosphate transaminase 2 (GFPT2; also known as the GFAT2) gene. This gene has also been reported to be involved in metabolic disorders. For example, Zhang et al reported that GFPT2 sequence variation contributed to susceptibility to T2DM and diabetic nephropathy in Caucasian and African American individuals,28 and furthermore suggested that GFPT2 may play a role in the metabolic abnormality. The eQTL analysis showed that GFPT2 rs12386481 genotype also showed significant cis-eQTL effects in several brain tissues (supplementary table 3 and figure S4), which suggested that rs12386481 polymorphism might mediate the expression level of GFPT2 in human brain regions. However, this finding requires replication in other populations and the mechanisms of GFPT2 in regulating obesity or AIWG need further exploration.
Other associations identified at the suggestive level of significance (P GWAS < 10–5 and P Replicated < 10–3) were listed as follows: the actin-related protein 3 (ACTR3, 2q14.1, rs2046545); t-complex 11, testis-specific (TCP11, 6p21.31, rs1886243); renalase (RNLS, 10q23.31, rs7906629 and rs1426621), copine IV (CPNE4, 3q22.1, rs7611808), and the oxysterol-binding protein-like protein 10 (OSBPL10, 3p22.3, rs13076635).
The ACTR3 is a major constituent of the ARP2/3 complex, which is necessary for nucleating actin polymerization at filament branches.29 Xu et al demonstrated that RNLS was secreted from transfected human embryonic kidney cells, and it metabolized catecholamines and regulate blood pressure, with dopamine as the preferred substrate, followed by epinephrine and norepinephrine.30 To our best knowledge, there have been no reports of the association of ACTR3 or RNLS polymorphisms with AIWG in schizophrenia or with obesity in the general population. However, OSBPL10, as a lipid receptor implicated in hyperlipidemia, has been reported to transfer phosphatidylserine and is thought to be involved in proposed mechanisms underlying cancer, dyslipidemia, and the metabolic syndrome.31–33
Although our best findings differ to those in the literature, we also identified other possible susceptibility genes for AIWG (supplementary table 2) that have been previously reported. These findings included the DRD3, ADRα2a, LEP, GNB3, SNAP25, and NDUFS1 genes. Modest associated loci in our study included PAM, FTO, MC4R, and ADRA2A (P < .05).
Malhotra et al were the first to report the association of MC4R rs489693 with AIWG.15 Moreover, Czerwensky et al reported an independent replication of the association with MC4R rs17782313, in other multiple independent cohorts.34,35 However, we were unable to validate this finding in our Chinese Han samples since our Illumina 610-Quad BeadChips did not include rs489693 or rs17782313. Other potential explanations for this lack of validation include clinical heterogeneity (different clinical characteristics of Han Chinese patients), population heterogeneity, and natural selection. Clinically, our study focused on acute effects of antipsychotic drugs on weight gain of adult inpatients, whereas Malhotra et al conducted a follow-up study of weight gain during a 12-week trial in 139 pediatric outpatients.34 Genetically, according to the HapMap database, the minor allele frequencies (MAF) of rs489693 (risk allele A) were 0.189 in Han Chinese in Beijing and 0.408 in Caucasian European residents from Utah. The allelic differences between European and Han Chinese populations might possibly be due to different population structure, evolutionary forces, etc. Moreover, rs489693 has been reported to have recessive effects,15 requiring significantly larger sample size to replicate, especially given the lower MAF in Chinese Han.
Moreover, the GWAS-based pathway analyses indicated that multiple genes in metabolic processes might jointly contribute to AIWG, such as glucosamine metabolic process, amino sugar metabolic process, and regulation of peptidyl tyrosine phosphorylation (P < .05). These results further confirmed the earlier observation that metabolic pathways were involved in AIWG in schizophrenia.8,36,37 Even though there is scant literature reporting genetic factors underlying individual differences of AIWG, our findings provide further support for particular susceptibility genes contributing to metabolic pathology. Future research should explore the genetic background of metabolic syndrome accompanied by AIWG, particularly when one intends to elevate fasting plasma glucose or type 2 diabetes as the long-term consequences of obesity and the development of the metabolic syndrome.
Expression profiling analysis suggested that the PTPRD mRNA was preferentially expressed in human brain tissues. However, GFPT2 mRNA was mainly expressed in smooth muscle and other peripheral tissues and expressed relatively lower in brain. Considering the human brain temporo-spatial expression analysis of PTPRD and the eQTL results of SNPs rs10977144 and rs10977154 in the hippocampus, we speculate that PTPRD might play an important role for the neuropsychiatric disorders. However, the specific roles of PTPRD in the pathogenesis of obesity or AIWG should also be further explored in future studies.
One limitation of our study involves the short-term follow-up of participants and the lack of a washout period for individuals before commencing the 8-week treatment. Previous studies have also reported that immediate/initial and longer term effects of AIWG are associated with different genetic factors.8,38,39 In the future, we will further explore the genetic factors underlying the long-term effects of AIWG. On the other hand, although we replicated the associated results in 236 drug-naive patients who then were treated with risperidone and quetiapine, we could not verify whether the whole effect was driven by the risperidone and quetiapine subgroups. For this reason, a better-designed randomized clinical trial with all kinds of antipsychotics should also be implemented to validate the GWAS discovery of susceptible genes associated with AIWG.
In summary, our study has identified genetic associations with AIWG, especially with the genes related to metabolic process or diabetes. These findings require replication and validation in other cohorts. A better understanding of these mechanisms and the role of specific polymorphisms could eventually help tailor individualized antipsychotic medications with minimal toxicity for patients with schizophrenia.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org
Funding
National Natural Science Foundation of China (81222017, 91232305, 81361120395, 81221002); National Key Technology R&D Program of China (2015BAI13B01, 2012BAI01B06); Program for New Century Excellent Talents in University (NCET-12-0008); the National High Technology Research and Development Program of China (2008AA02Z401, 2009AA022702).
Supplementary Material
Acknowledgments
We thank all family members that participated in this study. We extend our gratitude to all subjects who participated in this study. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. [DOI] [PubMed] [Google Scholar]
- 2. Lett TA, Wallace TJ, Chowdhury NI, Tiwari AK, Kennedy JL, Müller DJ. Pharmacogenetics of antipsychotic-induced weight gain: review and clinical implications. Mol Psychiatry. 2012;17:242–266. [DOI] [PubMed] [Google Scholar]
- 3. Mas S, Llerena A, Saiz J, et al. Strengths and weaknesses of pharmacogenetic studies of antipsychotic drugs: the potential value of the PEPs study. Pharmacogenomics. 2012;13:1773–1782. [DOI] [PubMed] [Google Scholar]
- 4. Henderson DC. Schizophrenia and comorbid metabolic disorders. J Clin Psychiatry. 2005;66:11–20. [PubMed] [Google Scholar]
- 5. Henderson DC, Cagliero E, Copeland PM, et al. Glucose metabolism in patients with schizophrenia treated with atypical antipsychotic agents: a frequently sampled intravenous glucose tolerance test and minimal model analysis. Arch Gen Psychiatry. 2005;62:19–28. [DOI] [PubMed] [Google Scholar]
- 6. Musil R, Obermeier M, Russ P, Hamerle M. Weight gain and antipsychotics: a drug safety review. Expert Opin Drug Saf. 2015;14:73–96. [DOI] [PubMed] [Google Scholar]
- 7. Hamilton SP. The promise of psychiatric pharmacogenomics. Biol Psychiatry. 2015;77:29–35. [DOI] [PubMed] [Google Scholar]
- 8. Reynolds GP. Pharmacogenetic Aspects of Antipsychotic Drug-induced Weight Gain - A Critical Review. Clin Psychopharmacol Neurosci. 2012;10:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gebhardt S, Theisen FM, Haberhausen M, et al. Body weight gain induced by atypical antipsychotics: an extension of the monozygotic twin and sib pair study. J Clin Pharm Ther. 2010;35:207–211. [DOI] [PubMed] [Google Scholar]
- 10. Choong E, Quteineh L, Cardinaux JR, et al. ; ODEX team. Influence of CRTC1 polymorphisms on body mass index and fat mass in psychiatric patients and the general adult population. JAMA Psychiatry. 2013;70:1011–1019. [DOI] [PubMed] [Google Scholar]
- 11. Gonçalves VF, Zai CC, Tiwari AK, et al. A hypothesis-driven association study of 28 nuclear-encoded mitochondrial genes with antipsychotic-induced weight gain in schizophrenia. Neuropsychopharmacology. 2014;39:1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cichon S, Craddock N, Daly M, et al. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crowley JJ, Sullivan PF, McLeod HL. Pharmacogenomic genome-wide association studies: lessons learned thus far. Pharmacogenomics. 2009;10:161–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chagnon YC, Mérette C, Bouchard RH, Emond C, Roy MA, Maziade M. A genome wide linkage study of obesity as secondary effect of antipsychotics in multigenerational families of eastern Quebec affected by psychoses. Mol Psychiatry. 2004;9:1067–1074. [DOI] [PubMed] [Google Scholar]
- 15. Malhotra AK, Correll CU, Chowdhury NI, et al. Association between common variants near the melanocortin 4 receptor gene and severe antipsychotic drug-induced weight gain. Arch Gen Psychiatry. 2012;69:904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yue WH, Wang HF, Sun LD, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011;43:1228–1231. [DOI] [PubMed] [Google Scholar]
- 17. Gauderman W, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006. http://hydra.usc.edu/gxe.
- 18. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 20. Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benita Y, Cao Z, Giallourakis C, Li C, Gardet A, Xavier RJ. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood. 2010;115:5376–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramasamy A, Trabzuni D, Guelfi S, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lieberman JA, Stroup TS, McEvoy JP, et al. ; Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) Investigators. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353:1209–1223. [DOI] [PubMed] [Google Scholar]
- 25. Yim YS, Kwon Y, Nam J, et al. Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc Natl Acad Sci USA. 2013;110:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uetani N, Kato K, Ogura H, et al. Impaired learning with enhanced hippocampal long-term potentiation in PTPdelta-deficient mice. EMBO J. 2000;19:2775–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maruthur NM, Gribble MO, Bennett WL, et al. The pharmacogenetics of type 2 diabetes: a systematic review. Diabetes Care. 2014;37:876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang H, Jia Y, Cooper JJ, Hale T, Zhang Z, Elbein SC. Common variants in glutamine:fructose-6-phosphate amidotransferase 2 (GFPT2) gene are associated with type 2 diabetes, diabetic nephropathy, and increased GFPT2 mRNA levels. J Clin Endocrinol Metab. 2004;89:748–755. [DOI] [PubMed] [Google Scholar]
- 29. Kim J, Lee JE, Heynen-Genel S, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu J, Li G, Wang P, et al. Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J Clin Invest. 2005;115:1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maeda K, Anand K, Chiapparino A, et al. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013;501:257–261. [DOI] [PubMed] [Google Scholar]
- 32. Koriyama H, Nakagami H, Katsuya T, et al. Variation in OSBPL10 is associated with dyslipidemia. Hypertens Res. 2010;33:511–514. [DOI] [PubMed] [Google Scholar]
- 33. Koriyama H, Nakagami H, Katsuya T, et al. Identification of evidence suggestive of an association with peripheral arterial disease at the OSBPL10 locus by genome-wide investigation in the Japanese population. J Atheroscler Thromb. 2010;17:1054–1062. [DOI] [PubMed] [Google Scholar]
- 34. Czerwensky F, Leucht S, Steimer W. MC4R rs489693: a clinical risk factor for second generation antipsychotic-related weight gain? Int J Neuropsychopharmacol. 2013;16:2103–2109. [DOI] [PubMed] [Google Scholar]
- 35. Yilmaz Z, Davis C, Loxton NJ, et al. Association between MC4R rs17782313 polymorphism and overeating behaviors. Int J Obes (Lond). 2015;39:114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuo PH, Kao CF, Chen PY, et al. Polymorphisms of INSIG2, MC4R, and LEP are associated with obesity- and metabolic-related traits in schizophrenic patients. J Clin Psychopharmacol. 2011;31:705–711. [DOI] [PubMed] [Google Scholar]
- 37. Newcomer JW. Second-generation (atypical) antipsychotics and metabolic effects: a comprehensive literature review. CNS Drugs. 2005;19(Suppl 1):1–93. [DOI] [PubMed] [Google Scholar]
- 38. Chintoh AF, Mann SW, Lam L, et al. Insulin resistance and decreased glucose-stimulated insulin secretion after acute olanzapine administration. J Clin Psychopharmacol. 2008;28:494–499. [DOI] [PubMed] [Google Scholar]
- 39. Reynolds GP, Yevtushenko OO, Gordon S, Arranz B, San L, Cooper SJ. The obesity risk gene FTO influences body mass in chronic schizophrenia but not initial antipsychotic drug-induced weight gain in first-episode patients. Int J Neuropsychopharmacol. 2013;16:1421–1425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.