Abstract
Annexin A1 (ANXA1) is a member of the annexin superfamily. Previous studies have reported that ANXA1 is highly expressed in various types of malignant tumor; however, its role in the progression of non-small cell lung cancer (NSCLC) remains to be fully clarified. The present study aimed to investigate the oncogenic role of ANXA1 in NSCLC cells in vitro. RNA interference was used to down-regulate ANXA1 expression in A549 and H1299 cells using a small interfering RNA lentiviral vector. Subsequently, cell proliferation and migration were detected using Cell Counting kit-8, clone formation, wound healing and Transwell chamber assays. Successful transfection was confirmed using fluorescence microscopy, which demonstrated that ANXA1 had been efficiently inhibited. ANXA1 knockdown suppressed the proliferation, migration and invasion of NSCLC cells. In conclusion, the present study provided evidence suggesting that ANXA1 may contribute to the growth and invasion of NSCLC cell lines, and ANXA1 may be exploited as an in vitro therapeutic target for the treatment of NSCLC.
Keywords: annexin A1, non-small cell lung cancer, small interfering RNA
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide. Non-small cell lung cancer (NSCLC) accounts for 80% of lung cancer cases, and the reported overall 5-year survival rate is <5% (1). The main factors associated with NSCLC are tumor invasion and metastasis; therefore, a better understanding regarding the cellular and molecular mechanisms underlying metastatic dissemination of NSCLC cells is required.
Annexin A1 (ANXA1) is a member of the annexin superfamily of calcium and phospholipid-binding proteins, which has been detected in various organisms, including vertebrates, invertebrates and plants (2). ANXA1 is an endogenous mediator of the anti-inflammatory effects of glucocorticoids, which acts via the inhibition of phospholipase A2 (3). Functionally, ANXA1 has been reported to be involved in intracellular signaling, cell growth and cell differentiation (4). Growing evidence has suggested that ANXA1 contributes to the pathological consequence and sequelae of a number of severe human diseases, including cancer (5). As a potential marker for malignant progression, ANXA1 expression levels have been demonstrated to be upregulated in breast cancer (6), and knockdown of ANXA1 by specific small interfering (si)RNA resulted in a significant reduction in the invasiveness of breast cancer cells (7). The expression of ANXA1 protein is also upregulated in human hepatocellular carcinoma (5). Furthermore, a previous study demonstrated that ANXA1 was overexpressed in melanoma, and may promote metastasis via formyl peptide receptor stimulation and matrix metalloproteinase 2 expression (8,9). ANXA1 expression is also dysregulated in esophageal squamous cell carcinoma (10). As a novel mechanism of post-transcriptional regulation, microRNA (miR)-196a targets ANXA1 expression, and miR-196a has been identified as a marker of esophageal cancer (11). Upregulation of ANXA1 expression is correlated with increased recurrence rate and decreased overall survival in esophageal cancer (12). Furthermore, the expression of ANXA1 has been reported to be dysregulated in prostate carcinogenesis, resulting in enhanced tumor aggressiveness via the upregulation of interleukin-6 expression and activity (13). These previous studies have reported diverse roles of ANXA1 in various types of human cancer; however, the relevant biological function of ANXA1 in NSCLC remains to be elucidated.
The present study aimed to investigate the effects of ANXA1 on NSCLC, and determine the effects of ANXA1 knockdown on cell proliferation and metastatic ability. The aim of this study was to evaluate the relationship between ANXA1 expression and the biological function of NSCLC.
Materials and methods
Ethics statement
The present study was approved by the institutional review board (CWO) of Guangzhou Medical University (Guangzhou, China). All patients provided written informed consent.
Tissue collection
Lung tumor tissue samples were harvested from 10 patients (aged, 64.9±6.8 years; male, n=8; female, n=2) at the Cancer Center of Guangzhou Medical University on July 15, 2014 during surgery. Matched healthy paracarcinoma tissue samples were also harvested from normal lung tissue.
Cell culture
The human NSCLC cell lines (BEAS-2B, A549, H460, H1299, 95D, H520 and PAa) and 293T cells were purchased from American Type Culture Collection (Manassas, VA, USA). All cells were cultured as monolayers in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) in a humidified atmosphere containing 5% CO2 at 37°C.
Preparation of ANXA1 siRNA lentiviral vectors
The siRNA duplexes targeting ANXA1 (accession number, NM_000700) were designed online (http://rnaidesigner.invitrogen.com/rnaiexpress/rnaiexpress.jsp). The sequences are shown in Table I. Hairpin DNA oligomers were synthesized and annealed. According to the manufacturers protocol (Takara LA Taq; Takara Bio, Inc., Otsu, Japan), each PCR reaction mixture (10 µl) contained the following reagents: 5 µl 10X LA PCR Buffer II, 1.6 µl dNTP mixture, 0.2 µM of each primer (forward: 5′-AGC GTC AAC AGA TCA AAG CAG CAT-3′ and reverse: 5′-AGA CCC TGT TAA TGT CTC TGA TTT-3′), 1.0 µl LA Taq and 171 ng genomic DNA. The PCR cycling conditions were as follows: Initial denaturation at 94°C for 5 min followed by 30 cycles with denaturation at 98°C for 10 sec, annealing at 55°C for 30 sec, extension at 72°C for 60 sec; then a final extension at 72°C for 10 min. All PCR products were purified and sequenced by Sangon Biotech Co., Ltd. (Shanghai, China) using the ABI 3730XL DNA Sequencer (Applied Biosystems, Foster City, CA, USA). The annealed double-stranded siRNA oligonucleotides were then cloned into a GV115 (Shanghai Genechem Co., Ltd., Shanghai, China) lentiviral vector that was driven by the U6 promoter and carried the transgene for green fluorescent protein. A control siRNA unrelated to human gene sequences was used as a negative control (5′-TTC TCC GAA CGT GTC ACGT-3′) (NC). The accuracy of the inserted vector sequences was verified by sequencing. The most efficient recombinant vector was selected following transfection of A549 cells with LV-ANXA1-RNAi-A, LV-ANXA1-RNAi-B and LV-ANXA1-RNAi-C. After around 24 h, single cells were transfected using an established protocol (Lipofectamine® 2000 Reagent kit; Invitrogen; Thermo Fisher Scientific, Inc.), Briefly, cells were exposed to 30 nM concentration of either scrambled siRNAs (Shanghai Genechem Co., Ltd.) and diluted in Opti-MEM (Life Technologies; Thermo Fisher Scientific, Inc.) along with DharmaFECT (GE Dharmacon, Lafayette, CO, USA) in transfection medium (complete RPMI medium without antibiotics and BSA). Cells were incubated with these transfection complexes for 24 h and then the medium was changed back to complete RPMI. After 4 days, the protein was harvested and extracted using lysis buffer and inhibition levels were detected using western blotting. The selected vector was subsequently denoted as ANXA1 siRNA. The 293T cells were infected with ANXA1 siRNA and NC lentiviral vectors using virion-packaging elements (pHelper 1.0 and pHelper 2.0; Shanghai Genechem Co., Ltd.) (14). ANXA1 siRNA and the NC vector were separately cotransfected into 293T cells with packing plasmids by calcium phosphate precipitation. Following a 24 h culture, the viral supernatant of each clone was collected and the viral titer was measured according to standard protocols.
Table I.
Sequences of ANXA1 siRNA.
| siRNA | Accession no. | Target sequence | CDS | GC % |
|---|---|---|---|---|
| LV-ANXA1-RNAi-A | NM_000700 | ATTCTATCAGAAGATGTAT | 75..1115 | 26.32 |
| LV-ANXA1-RNAi-B | NM_000700 | CTTGTATGAAGCAGGAGAA | 75..1115 | 42.11 |
| LV-ANXA1-RNAi-C | NM_000700 | AGCGCAATTTGATGCTGAT | 75..1115 | 42.11 |
| LV-ANXA1-RNAi-NC | TTCTCCGAACGTGTCACGT |
siRNA, small interfering RNA; ANXA1, annexin A1; CDS, coding region.
Selection of cells in which lentiviral vectors exhibit stable expression
The 293T cells were washed and resuspended in complete medium following transfection. Stable cell lines containing ANXA1 siRNA and NC lentiviral vectors were selected and 25 µg/ml puromycin was added to the medium. Following 6 weeks of culturing in the presence of puromycin, the remaining cells were isolated and transferred into 24-well dishes. An aliquot of the selected clones was removed for subsequent experimentation, and the remaining clones were frozen for future use.
Transfection of A549 and H1299 cells with ANXA1 siRNA
A549 and H1299 cells were subcultured into 6-well plates, at a density of 8×104/cm2 in a volume of 1 ml. Subsequently, 3.2×109 TU/ml lentiviral vector and NC-infected cells, together with 10 µl polybrene. The cells were allowed to adhere overnight in serum-containing antimicrobial-free RPMI-1640 medium in an atmosphere containing 5% CO2 at 37°C. After 24 h, the transduction medium was replaced with serum-containing medium.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Approximately 1 µg total RNA was used to generate cDNA using the Primescript RT-PCR kit (Takara Bio, Inc.). According to the manufacturers protocol, each RT reaction mixture (10 µl) contained the following reagents: 2 µl 5X primescript buffer, 0.5 µl primescript RT enzyme mix I, 0.5 µl oligo-dT primer (50 µM), 0.5 µl random 6 mers (100 µM) and 500 ng genomic DNA. The conditions were as follows: 37°C for 15 min, 85°C for 5 sec and 4°C for 10 min. All PCR products were synthesized by Biometra (Göttingen, Germany). The following primers (Takara Bio, Inc.) were used in the present study: ANXA1, upstream 5′-AGC GTC AAC AGA TCA AAG CAG CAT-3′, downstream 5′-AGA CCC TGT TAA TGT CTC TGA TTT-3′; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), upstream 5′-CCA TCA CCA TCT TCC AGGAG-3′ and downstream 5′-CCT GCT TCA CCA CGT TCTTG-3′. According to the manufacturers protocol (SYBR® Premix Ex Taq™ II; Tli RNaseH Plus; Takara Bio Inc., Japan), each PCR reaction mixture (25 µl) contained the following reagents: 12.5 µl 2X SYBR Premix Ex Taq II, 1.0 µl PCR forward primer (10 µM), 1.0 µl PCR reverse primer (10 µM), 0.5 µl 50X Reference Dye II and 2 µl genomic DNA. PCR was performed using the Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). PCR was performed using Platinum Taq polymerase (Invitrogen; Thermo Fisher Scientific, Inc.) under the following conditions: 37°C for 15 min, 85°C for 5 sec followed by 40 cycles at 95°C for 30 sec, 95°C for 5 sec, 60°C for 34 sec, 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. GAPDH was amplified as an internal control. Data were analyzed using the comparative quantification cycle method (2−ΔΔCq) (15). Three separate experiments were performed.
Western blot analysis
Cells from each group were harvested and proteins were extracted using lysis buffer [1 ml 1 mol/l Tris-HCl, 4 ml 10% sodium dodecyl sulphate (SDS), 40 µl 0.5 mol/l EDTA, 10 µl protease inhibitor and 14.96 ml ddH2O]. The protein content was quantified using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, a working reagent was prepared by mixing 50 parts of BCA Reagent A and 1 part BCA Reagent B. The PRP pellet was resus-pended into 25 µl of mammalian protein extraction reagent, and 200 µl of working reagent was added to the solution. After 30 min of incubation, the absorbance was measured at 562 nm on a BioTek Synergy 2 96-well plate reader (BioTek, Winooski, VT, USA) and converted to a concentration using a calibration curve. Cell extracts were boiled for 5 min in loading buffer, and an equal amount of protein (40 µg) was separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE). Separated protein bands were transferred onto nitrocellulose membranes (8-µm pores; Millipore, Billerica, MA, USA) and the membranes were blocked in 5% skimmed milk powder. Standard western blotting was performed using a rabbit polyclonal anti-ANXA1 antibody (cat. no. ab137745; 1:1,000 dilution; 4°C for 16 h; Abcam, Cambridge, UK) and a horseradish peroxidase-conjugated rabbit anti-rat IgG H&L polyclonal antibody (cat. no. ab6734; 1:5,000 dilution; at room temperature for 1 h; Abcam). Equal protein sample loading was monitored by probing the same membrane filter with mouse monoclonal anti-β-actin antibody (cat. no. ab6276; 1:5,000 dilution; at 4°C for 16 h; Abcam), rabbit polyclonal anti-GAPDH antibody (cat. no. ab70699; 1:2,000 dilution; 4°C for 16 h; Abcam) and rabbit polyclonal anti-tubulin antibody (cat. no. ab150729; 1:1,000 dilution; 4°C for 16 h; Abcam), which was used as an internal control. Blots were visualized using enhanced chemiluminescence (Millipore) and were exposed to chemiluminescent film (Pierce; Thermo Fisher Scientific, Inc.). Data were measured using ImageJ 1.48u software (National Institutes of Health, Bethesda, MD, USA).
Wound healing assay
Transduced cells were incubated until they had reached 90–100% confluence. The cells were scratched using a P-10 pipette tip, and were then incubated for various durations. Phase contrast images were captured at 0 and 24 h using a Nikon microscopy system (Nikon Eclipse Ti-s; Nikon Corporation, Tokyo, Japan). The wound healing distance was measured using ImageJ software (1.48u; National Institutes of Health). All assays were conducted in triplicate, and the mean values were calculated.
Migration and invasion assays
The migratory ability of human A549 and H1299 NSCLC cells transduced with ANXA1 siRNA and NC siRNA vectors was determined using Corning Transwell insert chambers (Corning, Inc., Corning, NY, USA). Briefly, during the logarithmic growth phase, cells were trypsinized with 1X trypsin, and were resuspended in 200 µl (2×105 cell/ml) serum-free RPMI-1640 medium. The cells were placed in the upper chamber of the insert without Matrigel. Medium containing 5% FBS was added to the lower chamber as a chemoattractant. Following a 24 h incubation, the cells on the upper membrane were carefully removed, and cells that had migrated through the membrane were manually counted at 200x magnification from 10 fields per filter using a Nikon microscope (Nikon Eclipse Ti-s; Nikon Corporation). All experiments were independently repeated at least three times.
The invasive ability of human A549 and H1299 NSCLC cells transduced with ANXA1 siRNA and NC siRNA vectors was determined using Matrigel-coated cell culture chambers (8 µm pore size; EMD Millipore, Billerica, MA, USA). Briefly, the cells were transduced and cultured to ~90% confluence in 24-well dishes. Subsequently, the cells were resuspended in 200 µl (1×106 cell/ml) serum-free RPMI-1640 medium and were placed in the upper chamber of the insert with Matrigel. Medium containing 5% FBS was added to the lower chamber as a chemoattractant. Following a 24 h incubation, the cells that remained on the upper membrane were carefully removed. Cells that had invaded through the membrane were manually counted at 200× magnification from 10 fields per filter using a Nikon microscope (Nikon Eclipse Ti-s; Nikon Corporation). All experiments were independently repeated at least three times.
Cell proliferation assay
Cells were seeded into 96-well plates at a density of 2×103 cells/well. Cell viability was assessed using the Cell Counting kit (CCK)-8 assay (Beyotime Institute of Biotechnology, Shanghai, China). Briefly, cells were seeded into 96-well plates (2.0×103 cells per well) and incubated in α-MEM supplemented with 10% FBS for 4 days. CCK-8 reagent (10 µl, 1 mg/ml) was added and incubated for 3 h at 37°C. The absorbance of each well was measured using a spectrophotometer (51119200; Thermo Fisher Scientific, Inc.) at 450 nm. Three independent experiments were performed.
Clone formation assay
For the clone formation assay, 125 cells/4 ml were plated onto 6-well plates and were incubated at 37°C. Once the cells grew to visible colonies, the colonies were washed once with phosphate-buffered saline and were fixed with 4% paraformaldehyde for 20 min. Subsequently, the cells were stained with crystal violet (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), and the number of clones per well was counted. All assays were conducted in triplicate, and the mean values were calculated.
Statistical analysis
All assays were conducted in triplicate, and the mean values were calculated. Data are presented as the mean ± standard deviation. All statistical analyses were performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Unpaired sets of data were compared using unpaired Student's t-test (two-tailed). P<0.05 was considered to indicate a statistically significant difference.
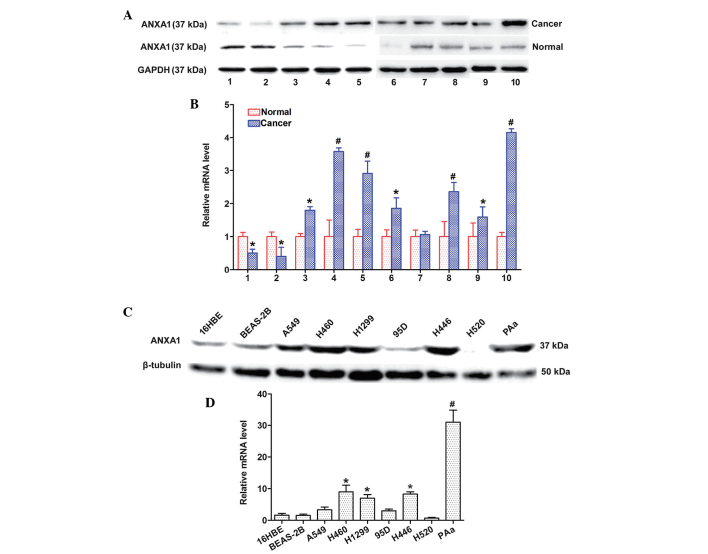
ANXA1 expression is upregulated in NSCLC tissues and cell lines
The expression levels of ANXA1 were detected in 10 matched clinical tissue samples and seven NSCLC cell lines by western blotting and qPCR. The protein expression levels of ANXA1 were increased in the 8 matched cancer tissues, as compared with in the normal tissues (Fig. 1A). In addition, the mRNA expression levels of ANXA1 were markedly upregulated in the eight matched cancer tissues, as compared with in the normal tissues (Fig. 1B). The protein expression levels of ANXA1 were also upregulated in the five NSCLC cell lines (Fig. 1C). Similarly, ANXA1 mRNA expression was increased in the five NSCLC cell lines (Fig. 1D) as compared with in the normal cell line. The cancer and normal tissues were obtained from the same patients. These results indicate that ANXA1 expression may be upregulated at both the mRNA and protein level in NSCLC.
Figure 1.
Relative expression levels of annexin A1 (ANXA1) in non-small cell lung cancer (NSCLC) tissues and cell lines. (A) Western blotting indicated that ANXA1 expression was markedly increased in NSCLC tissues. *P<0.05, #P<0.01 vs. normal lung tissues. (B) Quantitative polymerase chain reaction analysis indicated that the mRNA expression levels of ANXA1 were upregulated in NSCLC tissues. (C) ANXA1 protein expression levels were increased in NSCLC cell lines. (D) ANXA1 mRNA expression levels were downregulated in the normal cell line. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-tubulin were used as endogenous controls. Data are presented as the mean ± standard deviation for triplicate determinations. *P<0.05, #P<0.01 vs. 16HBE cells.
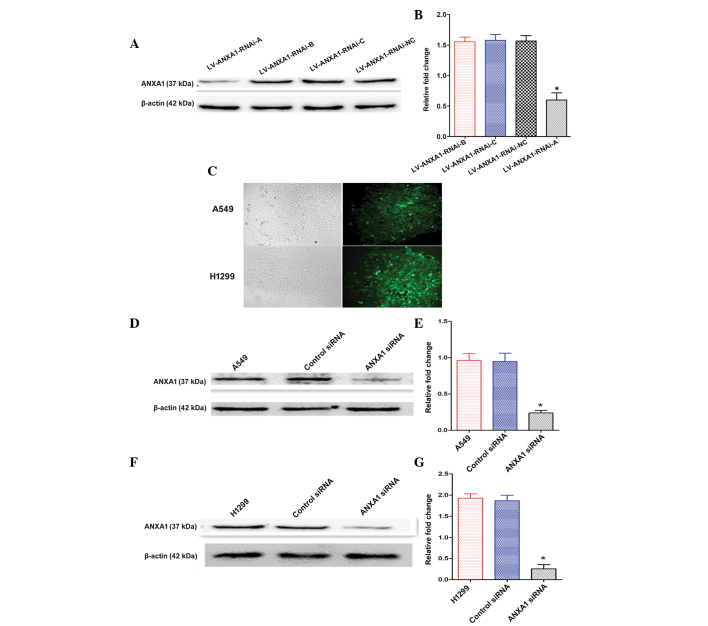
Construction and selection of the most effective ANXA1 siRNA expression vector
The expression levels of ANXA1 were detected by western blotting 72 h post-transfection of A549 cells with the three siRNAs. The ANXA1 siRNA sequences are presented in Table I. The transfection of LV-ANXA1-RNAi-A inhibited the expression of ANXA1 in A549 cells (Fig. 2A). In particular, the most obvious gene-silencing effect was observed following transfection with LV-ANXA1-RNAi-A, which reduced ANXA1 expression by 90.1% (Fig. 2B). Therefore, LV-ANXA1-RNAi-A was selected for further experimentation; its viral titer was 3.2×109 TU/ml.
Figure 2.
Establishment and functional analyses of annexin A1 (ANXA1) knockdown cells. (A and B) Total cellular proteins were extracted following transduction with three small interfering (si)RNA vectors. The most efficient recombinant vector was selected for further experimentation: LV-ANXA1-RNAi-A. (C) After stable transduction the cells were observed under a fluorescence microscope (magnification, 100x). (D) ANXA1 protein expression was markedly inhibited in the A549 ANXA1 siRNA group, as determined by western blotting. (E) Quantification of ANXA1 protein expression in A549 cells. (F) Western blotting indicated that ANXA1 expression was markedly decreased in the H1299 ANXA1 siRNA group. (G) Quantification of ANXA1 protein expression in H1299 cells. β-actin was used as an endogenous control. Data are presented as the mean ± standard deviation for triplicate determinations. *P<0.05, compared with the control siRNA and untreated groups.
Knockdown of ANXA1 expression with ANXA1 siRNA
The stably transduced cells were selected and observed under a fluorescence microscope, in order to detect green fluorescence. No alterations in cellular morphology were detected post-transduction (Fig. 2C). The A549 and H1299 cells were shown to exhibit a higher ANXA1 gene expression in preliminary experiments (Fig. 1C and D). The present study aimed to examine the effects of ANXA1 siRNA on A549 and H1299 cells. Western blot analysis demonstrated that the protein expression levels of ANXA1 in A549 cells of the ANXA1 siRNA group were significantly decreased, as compared with in the cells of the siRNA NC and untreated groups (Fig. 2D and E). Similar results were obtained from the H1299 cells (Fig. 2F and G). Statistical analysis revealed that the protein expression levels of ANXA1 were markedly downregulated by ANXA1 siRNA, as compared with in the NC siRNA group (P<0.05). In addition, no significant difference was detected between the NC siRNA and untreated A549 cell groups (Fig. 2E). Similar results were obtained from the H1299 cells (Fig. 2G). These results indicate that ANXA1 siRNA was successfully constructed.
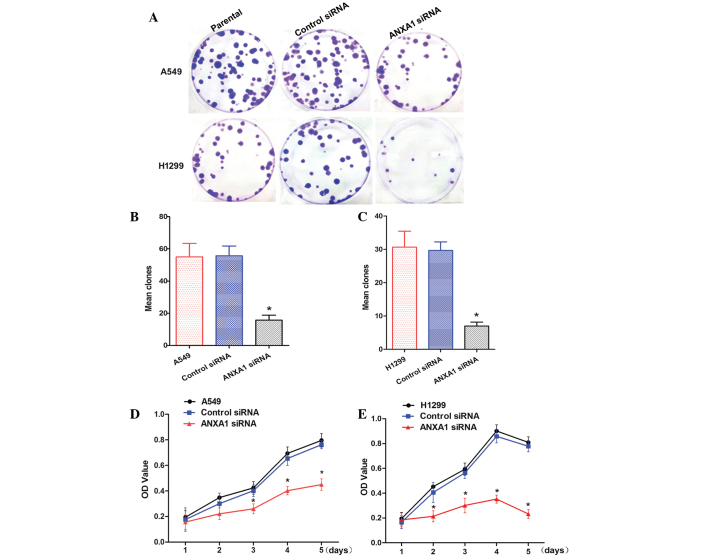
ANXA1 siRNA inhibits clone formation and cell proliferation
To determine whether knockdown of ANXA1 expression affected biological function of NSCLC cells, cellular activities were analyzed using clone formation and CCK-8 assays. ANXA1 knockdown inhibited the clone forming ability of A549 and H1299 cells (Fig. 3A). Statistical analysis demonstrated that clone formation was inhibited by ANXA1 siRNA, as compared with in the NC siRNA group (P<0.05). In addition, there was no significant difference between the NC siRNA and untreated cell groups (Fig. 3B and C). Cells from the ANXA1 siRNA, NC siRNA and untreated groups were cultured for 5 days. The CCK-8 assay indicated that A549 cell growth in the ANXA1 siRNA group was significantly decreased (P<0.05). Statistical analysis revealed that the proliferation rate was significantly decreased, and no significant differences were detected between the NC siRNA and untreated groups (Fig. 3D). Similarly, ANXA1 knockdown inhibited H1299 cell proliferation (Fig. 3E; P<0.05). These data suggest that ANXA1 knockdown inhibits NSCLC cell proliferation, and may have a critical role in NSCLC.
Figure 3.
Annexin A1 (ANXA1) small interfering (si)RNA inhibits cell proliferation and clone formation in A549 and H1299 cells. (A) Clone formation assay of each group. Quantification of clone forming ability of (B) A549 cells and (C) H1299 cells. Proliferation of (D) A549 cells and (E) H1299 cells, as detected by Cell Counting kit-8 assay. Data are presented as the mean ± standard deviation for triplicate determinations. *P<0.05, compared with the control siRNA and untreated groups. OD, optical density.
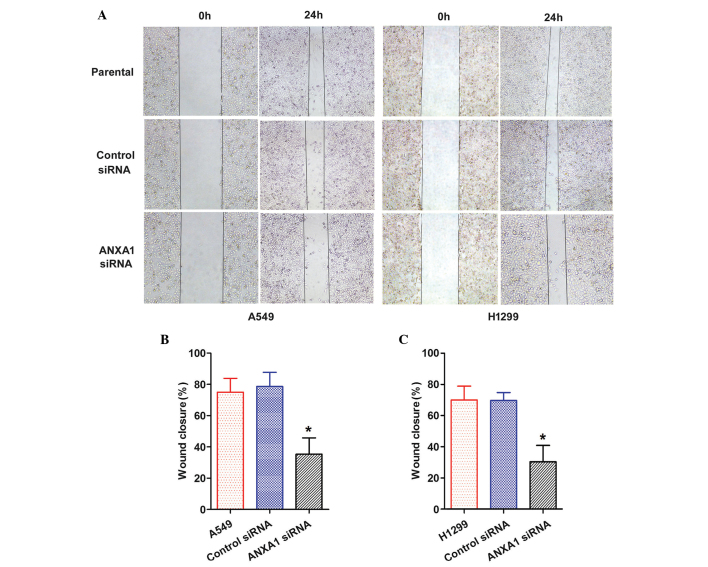
ANXA1 knockdown suppresses the migration and invasion of NSCLC cells
The role of ANXA1 in the migration of NSCLC cells was evaluated by wound healing and Transwell chamber assays. A scratch was made in the cell layer, and wound closure was monitored over the course of 24 h. A gradual decrease in wound closure was recorded in both the ANXA1 siRNA groups (Fig. 4A). Statistical analysis revealed that ANXA1 knockdown significantly suppressed migration of the cells, as compared with in the NC siRNA group (P<0.05), and there was no significant difference between the NC siRNA and untreated groups (Fig. 4B and C).
Figure 4.
Annexin A1 (ANXA1) small interfering (si)RNA inhibits wound healing in A549 and H1299 cells. (A) Cell migration was determined using a wound healing assay (magnification, ×4). Quantification of wound area changes 0 and 24 h after scratch damage in (B) A549 and (C) H1299 cells. Data are presented as the mean ± standard deviation for triplicate determinations. *P<0.05, compared with the control siRNA and untreated groups.
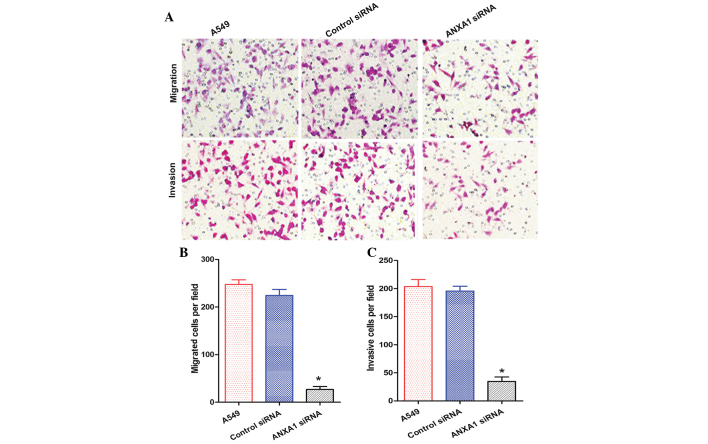
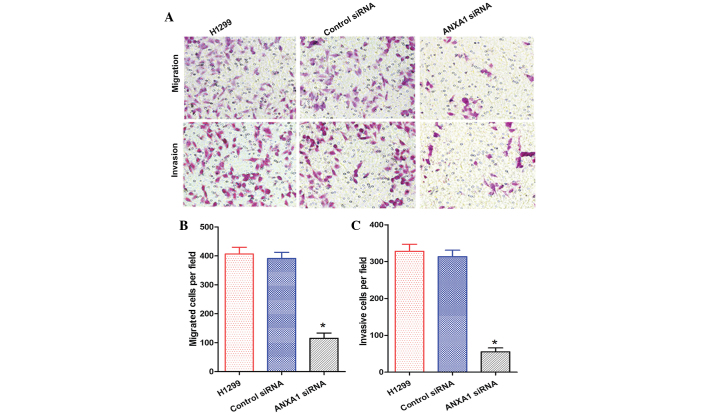
The migration and invasion of A549 cells in the ANXA1 siRNA group was decreased, as compared with that of the NC siRNA and untreated groups (Fig. 5A). Statistical analysis revealed that ANXA1 siRNA significantly suppressed the metastasis and invasion of A549 cells (P<0.05), and there was no significant difference between the NC siRNA and untreated groups (Fig. 5B and C). In addition, the migration and invasion of H1299 cells in the ANXA1 siRNA group was decreased, as compared with in the NC siRNA and untreated groups (Fig. 6A). Statistical analysis revealed that ANXA1 siRNA significantly inhibited the metastasis and invasion of H1299 cells (Fig. 6B and C; P<0.05). Cell migration is a critical step in metastasis, and these results suggest that ANXA1 may have a critical role in the metastatic behavior of cancer cells.
Figure 5.
(A) Annexin A1 (ANXA1) small interfering (si)RNA suppressed the migration and invasion of A549 cells (original magnification, ×200). Quantification of the (B) migration and (C) invasion of A549 cells. Data are presented as the mean ± standard deviation for triplicate determinations. *P<0.05, compared with the control siRNA and untreated groups.
Figure 6.
(A) Annexin A1 (ANXA1) small interfering (si)RNA suppressed the migration and invasion of H1299 cells (original magnification, ×200). Quantification of the (B) migration and (C) invasion of H1299 cells. Data are presented as the mean ± standard deviation for triplicate determinations. *P<0.05, compared with the control siRNA and untreated groups.
Discussion
Human lung cancer is a major cause of cancer-associated mortality worldwide, and is associated with a low 5-year survival rate. Lung cancer-associated mortality is often caused by extensive metastasis (16,17). Tumor metastasis is a complex multi-step and multistage process (18,19); therefore, the identification of novel diagnostic methods and treatment biomarkers is required. A previous study demonstrated that ANXA1 expression was significantly higher in patients with NSCLC, as compared with in control subjects (20). Increased ANXA1 expression has also been detected in lung squamous carcinoma cell (21). Previous 2D-PAGE results indicated that ANXA1 protein is upregulated in lung cancer and lung cancer/chronic obstructive pulmonary disorder groups, as compared with in the control group (22). In addition, ANXA1 expression is significantly upregulated in tumor tissues, as compared with in the normal tissues of patients with lung cancer (23,24). The present study demonstrated that ANXA1 was upregulated in 10 matched cancer tissues, thus indicating that ANXA1 may exhibit increased expression in NSCLC. However, the possible biological function of ANXA1 in NSCLC remains to be elucidated.
The present study revealed that ANXA1 was functionally involved in NSCLC progression and metastatic formation, and ANXA1 was shown to be upregulated in NSCLC tissues and cell lines. In addition, knockdown of ANXA1 suppressed the proliferation of NSCLC cell lines, and inhibited invasion and migration. These results indicated that silencing ANXA1 expression in A549 and H1299 cells may inhibit metastasis in vitro.
The present study was designed to estimate the effects of ANXA1 on the migration of cancer cells, and to investigate the underlying mechanism. It has previously been suggested that ANXA1 may regulate miR-26b and miR-562 via down-regulation of nuclear factor (NF)-κB activity, which may lead to higher endothelial cell tube formation and inhibit wound healing capacity (25). Furthermore, ANXA1 has been shown to regulate tumor necrosis factor-β-induced proliferation and inflammation in lung fibroblasts, via effects on the extracellular signal-regulated kinases and NF-κB pathways (26). ANXA1 has also been reported to promote metastasis via activation of the transforming growth factor-β/Smad signaling pathway (27).
In conclusion, the present study aimed to investigate the role of ANXA1 in NSCLC, and to evaluate the potential of using ANXA1 as a marker for NSCLC diagnosis and treatment. However, further investigations are required regarding the mechanism by which ANXA1 is associated with NSCLC metastasis.
Acknowledgments
The present study was funded by the National Nature Science Foundation of China (grant no. 81401391); the Ph.D. Programs Foundation of Ministry of Education of China (grant no. 20134423110001); the National Nature Science Foundation of Guangdong Province (grant no. S2012010010181); the Science and Technology Project of Guangzhou City (grant no. 2014Y2-00171); and the Education System Innovative Academic Team of Guangzhou City (grant no. 13C06).
References
- 1.Ni L, Zhu X, Gong C, Luo Y, Wang L, Zhou W, Zhu S, Li Y. Trichosanthes kirilowii fruits inhibit non-small cell lung cancer cell growth through mitotic cell-cycle arrest. Am J Chin Med. 2015;43:349–364. doi: 10.1142/S0192415X15500238. [DOI] [PubMed] [Google Scholar]
- 2.Gerke V, Moss SE. Annexins: From structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 3.Guo C, Liu S, Sun MZ. Potential role of Anxa1 in cancer. Future Oncol. 2013;9:1773–1793. doi: 10.2217/fon.13.114. [DOI] [PubMed] [Google Scholar]
- 4.Bist P, Shu S, Lee H, Arora S, Nair S, Lim JY, Dayalan J, Gasser S, Biswas SK, Fairhurst AM, Lim LH. Annexin-A1 regulates TLR-mediated IFN-β production through an interaction with TANK-binding kinase 1. J Immunol. 2013;191:4375–4382. doi: 10.4049/jimmunol.1301504. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y, Lin G, Fang W, Zhu H, Chu K. Increased expression of annexin A1 predicts poor prognosis in human hepatocellular carcinoma and enhances cell malignant phenotype. Med Oncol. 2014;31(327) doi: 10.1007/s12032-014-0327-7. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Zhang C, Chen C, Sun S, Zheng H, Wan S, Meng Q, Chen Y, Wei J. Investigation of circulating antibodies to ANXA1 in breast cancer. Tumour Biol. 2015;36:1233–1236. doi: 10.1007/s13277-014-2751-x. [DOI] [PubMed] [Google Scholar]
- 7.Okano M, Kumamoto K, Saito M, Onozawa H, Saito K, Abe N, Ohtake T, Takenoshita S. Upregulated Annexin A1 promotes cellular invasion in triple-negative breast cancer. Oncol Rep. 2015;33:1064–1070. doi: 10.3892/or.2015.3720. [DOI] [PubMed] [Google Scholar]
- 8.Boudhraa Z, Merle C, Mazzocut D, Chezal JM, Chambon C, Miot-Noirault E, Theisen M, Bouchon B, Degoul F. Characterization of pro-invasive mechanisms and N-terminal cleavage of ANXA1 in melanoma. Arch Dermatol Res. 2014;306:903–914. doi: 10.1007/s00403-014-1517-z. [DOI] [PubMed] [Google Scholar]
- 9.Boudhraa Z, Rondepierre F, Ouchchane L, Kintossou R, Trzeciakiewicz A, Franck F, Kanitakis J, Labeille B, Joubert-Zakeyh J, Bouchon B, et al. Annexin A1 in primary tumors promotes melanoma dissemination. Clin Exp Metastasis. 2014;31:749–760. doi: 10.1007/s10585-014-9665-2. [DOI] [PubMed] [Google Scholar]
- 10.Han G, Tian Y, Duan B, Sheng H, Gao H, Huang J. Association of nuclear annexin A1 with prognosis of patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:751–759. [PMC free article] [PubMed] [Google Scholar]
- 11.Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, et al. MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 12.Wang KL, Wu TT, Resetkova E, Wang H, Correa AM, Hofstetter WL, Swisher SG, Ajani JA, Rashid A, Hamilton SR, Albarracin CT. Expression of annexin A1 in esophageal and esophagogastric junction adenocarcinomas: Association with poor outcome. Clin Cancer Res. 2006;12:4598–4604. doi: 10.1158/1078-0432.CCR-06-0483. [DOI] [PubMed] [Google Scholar]
- 13.Inokuchi J, Lau A, Tyson DR, Ornstein DK. Loss of annexin A1 disrupts normal prostate glandular structure by inducing autocrine IL-6 signaling. Carcinogenesis. 2009;30:1082–1088. doi: 10.1093/carcin/bgp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z, Zhu JS, Xu ZP, Zhang Q. Lentiviral vector-mediated siRNA knockdown of the YAP gene inhibits growth and induces apoptosis in the SGC7901 gastric cancer cell line. Mol Med Rep. 2011;4:1075–1082. doi: 10.3892/mmr.2011.543. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Bonomi M, Pilotto S, Milella M, Massari F, Cingarlini S, Brunelli M, Chilosi M, Tortora G, Bria E. Adjuvant chemotherapy for resected non-small-cell lung cancer: Future perspectives for clinical research. J Exp Clin Cancer Res. 2011;30(115) doi: 10.1186/1756-9966-30-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z. Effect of daily aspirin on risk of cancer metastasis: A study of incident cancers during randomised controlled trials. Lancet. 2012;379:1591–1601. doi: 10.1016/S0140-6736(12)60209-8. [DOI] [PubMed] [Google Scholar]
- 18.Soda K. The mechanisms by which polyamines accelerate tumor spread. J Exp Clin Cancer Res. 2011;30:95. doi: 10.1186/1756-9966-30-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JD, Nakamura I, Roberts LR. The tumor microenvironment in hepatocellular carcinoma: Current status and therapeutic targets. Semin Cancer Biol. 2011;21:35–43. doi: 10.1016/j.semcancer.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Guan S, Sun S, Jin Y, Lee KH, Chen Y, Wei J. Detection of circulating antibodies to linear peptide antigens derived from ANXA1 and DDX53 in lung cancer. Tumour Biol. 2014;35:4901–4905. doi: 10.1007/s13277-014-1643-4. [DOI] [PubMed] [Google Scholar]
- 21.Nan Y, Yang S, Tian Y, Zhang W, Zhou B, Bu L, Huo S. Analysis of the expression protein profiles of lung squamous carcinoma cell using shot-gun proteomics strategy. Med Oncol. 2009;26:215–221. doi: 10.1007/s12032-008-9109-4. [DOI] [PubMed] [Google Scholar]
- 22.Pastor MD, Nogal A, Molina-Pinelo S, Meléndez R, Salinas A, González De la Peña M, Martín-Juan J, Corral J, García-Carbonero R, Carnero A, Paz-Ares L. Identification of proteomic signatures associated with lung cancer and COPD. J Proteomics. 2013;89:227–237. doi: 10.1016/j.jprot.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Rho JH, Roehrl MH, Wang JY. Glycoproteomic analysis of human lung adenocarcinomas using glycoarrays and tandem mass spectrometry: Differential expression and glycosylation patterns of vimentin and fetuin A isoforms. Protein J. 2009;28:148–160. doi: 10.1007/s10930-009-9177-0. [DOI] [PubMed] [Google Scholar]
- 24.Xu QY, Gao Y, Liu Y, Yang WZ, Xu XY. Identification of differential gene expression profiles of radioresistant lung cancer cell line established by fractionated ionizing radiation in vitro. Chin Med J (Engl) 2008;121:1830–1837. [PubMed] [Google Scholar]
- 25.Anbalagan D, Yap G, Yuan Y, Pandey VK, Lau WH, Arora S, Bist P, Wong JS, Sethi G, Nissom PM, et al. Annexin-A1 regulates microRNA-26b* and microRNA-562 to directly target NF-κB and angiogenesis in breast cancer cells. PloS One. 2014;9:e114507. doi: 10.1371/journal.pone.0114507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia Y, Morand EF, Song W, Cheng Q, Stewart A, Yang YH. Regulation of lung fibroblast activation by annexin A1. J Cell Physiol. 2013;228:476–484. doi: 10.1002/jcp.24156. [DOI] [PubMed] [Google Scholar]
- 27.de Graauw M, van Miltenburg MH, Schmidt MK, Pont C, Lalai R, Kartopawiro J, Pardali E, Le Dévédec SE, Smit VT, van der Wal A, et al. Annexin A1 regulates TGF-beta signaling and promotes metastasis formation of basal-like breast cancer cells. Proc Natl Acad Sci USA. 2010;107:6340–6345. doi: 10.1073/pnas.0913360107. [DOI] [PMC free article] [PubMed] [Google Scholar]