Abstract
It has previously been demonstrated that curcumin possesses an antitumor activity, which is associated with its ability to induce G2/M cell cycle arrest and apoptosis. However the detailed underlying mechanisms remain unclear. The present study aimed to investigate the efficacy and underlying mechanism of curcumin-induced cell cycle arrest and apoptosis in U87 human glioblastoma cells. By immunofluorescence staining, subcellular fractionation and western blotting, the present study demonstrated that curcumin was able to induce G2/M cell cycle arrest and apoptosis by increasing the expression levels of cyclin G2, cleaved caspase-3 and Fas ligand (FasL), and decreasing the expression of cyclin-dependent kinase 1 (CDK1). In addition, increased expression of forkhead box protein O1 (FoxO1) and decreased expression of phosphorylated (p)-FoxO1 were detected in the curcumin-treated U87 cells. Curcumin was also able to induce the translocation of FoxO1 from the cytoplasm to the nucleus. Furthermore, following knockdown of FoxO1 expression in curcumin-treated U87 cells using FoxO1 small interfering RNA, the expression levels of cyclin G2, cleaved caspase-3 and FasL were inhibited; however, the expression levels of CDK1 were not markedly altered. Notably, following knockdown of CDK1 expression under normal conditions, the total expression of FoxO1 was not affected; however, p-FoxO1 expression was decreased and FoxO1 nuclear expression was increased. Furthermore, curcumin-induced G2/M cell cycle arrest and apoptosis could be attenuated by FoxO1 knockdown. These results indicated that curcumin may induce G2/M cell cycle arrest and apoptosis in U87 cells by increasing FoxO1 expression. The present study identified a novel mechanism underlying the antitumor effects of curcumin, and may provide a theoretical basis for the application of curcumin in glioma treatment.
Keywords: glioma, curcumin, cell cycle, forkhead box protein O1, cyclin-dependent kinase 1, apoptosis
Introduction
Glioma is the most common and aggressive type of primary brain tumor. Despite advances in treatments combining surgery, radiotherapy and chemotherapy, the survival rate remains poor (1); therefore, to improve the prognosis for patients with glioma, novel effective treatments are required.
Curcumin [1,7-bis (4-hydroxy-3-methoxyphenyl)-1, 6-hepta-diene-3,5-dione] is a lipophilic phenolic compound that is derived from the rhizome of turmeric (Curcuma longa). Turmeric is a coloring and flavoring of curry, and is a common spice, particularly in Asia. In traditional Indian medicine, turmeric is used in the treatment of several disorders, including coughs, sinusitis, hepatic disease and anorexia. The potential effectiveness of turmeric has been associated with the anti-infectious and anti-inflammatory activities of curcumin (2–6). It has previously been reported that curcumin exerts anticancer effects in several types of cancer, which are attributed to its ability to modulate numerous targets and kinases associated with survival signaling, cell proliferation and cell cycle regulation (7–9). Previous studies have also indicated that curcumin is of promising research value as a potential anticancer agent in the treatment of glioma (10,11). However, the effects and underlying mechanism of curcumin on glioma are currently unclear.
Forkhead box O (FoxO) transcription factors are a superfamily of proteins, which include FoxO1, FoxO3, FoxO4 and FoxO6 in humans. It has been reported that FoxO proteins are able to regulate cell fate by modulating the expression of genes associated with apoptosis, cell cycle transition, DNA repair, oxidative stress and longevity, and muscle growth control, as well as cell differentiation and glucose metabolism (12). Therefore, FoxOs are considered attractive candidates as tumor suppressors. The hypothesis that FoxO family members may serve as tumor suppressors has been confirmed by evidence in human cancer tissue samples. Furthermore, in nude mice experiments, IkappaB kinase-induced cell proliferation and tumorigenicity were inhibited by the expression of active FoxO3 (13). The expression of FoxO3 in breast cancer is negatively correlated with poor patient survival, and increased FoxO3a expression is common following cytotoxic drug treatment, and is associated with apoptosis and cell cycle arrest (14). Similarly, the constitutive expression of active FoxO4 can reduce tumor onset, as well as tumor size and progression, in nude mice transplanted with cells expressing the human epidermal growth factor oncogene (15). Furthermore, the tumorigenesis of phosphatase and tensin homolog-deficient tumor cells is decreased by overexpression of a constitutively active form of FoxO1 (16). Therefore, FoxO1, FoxO3 and FoxO4 may prevent tumor progression.
The present study indicated that curcumin was able to induce G2/M phase arrest and apoptosis in U87 cells, and these effects were associated with increased FoxO1 expression and FoxO1 nuclear localization.
Materials and methods
Curcumin
Curcumin (95%purity) [(E,E)-1,7-bis (4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione] was purchased from Sigma-Aldrich (St. Louis, MO, USA), and was stored as a 100 mM stock solution in dimethyl sulfoxide (DMSO) at −20°C until use. The present study was approved by the Ethical Committee of Nanjing Medical University (Nanjing, China) (no. 2014-109).
Cell culture and treatment
The human U87 glioblastoma cell line was provided by the China Center for Typical Culture Collection (Shanghai, China). The U87 cells were cultured in low-glucose (1 g/l) Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 1 mM sodium pyruvate, 100 units/ml penicillin and 100 mg/ml streptomycin (all purchased from Gibco; Thermo Fisher Scientific, Inc.). Cells were incubated at 37°C under a humidified atmosphere containing 5% CO2. The cells were incubated for 24 h, after which the medium was removed and replaced with fresh DMEM containing 20 or 40 µM curcumin or DMSO (untreated control) for 24 or 48 h. In some experiments, proliferating cells were transfected with FoxO1 small interfering (si)RNA (cat. no. sc-35382; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or cyclin-dependent kinase 1 (CDK1) siRNA (Suzhou Ribo Life Science Co., Ltd., Suzhou, China), 6 h post-treatment with Lipofectamine® 2000 in Opti-MEM serum-free medium (both purchased from Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Then, the Opti-MEM medium was changed into DMEM and cells were cultured for 24 h.
Cell viability assay
Cells were treated with 20 or 40 µM curcumin for 24 or 48 h. The effects of curcumin on cell viability were determined using the Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assay. Briefly, following incubation with the 20 or 40 µM curcumin for 24 or 48 h, the cells were incubated with 5 g/l CCK-8 solution for 2 h. Subsequently, the cells were placed in a 96-well microplate reader (BioTek, Winooski, VT, USA) for analysis and the optical density (OD) was detected at 450 nm. Cell viability was evaluated using the following formula: Cell viability (%) = [1 − (OD of the samples/OD of the control)] × 100%.
Cell proliferation assay
Cells were treated with curcumin (20 or 40 µM) for 24 h, and the control cells were untreated. The cells (1×106) were subsequently washed in phosphate-buffered saline (PBS) and fixed in ice-cold ethanol. Cell proliferation was determined by 5-ethynyl-2-deoxyuridine (EdU) incorporation using the Cell-Light EdU Imaging Detecting kit (Guangzhou RiboBio Co., Ltd., Guangzhou, China), according to the manufacturer's protocol.
Cell cycle distribution analysis
Cells were cultured in DMEM for 24 h, and were then treated with 20 or 40 µM curcumin or DMSO for 24 h. To measure cell cycle distribution, the cells were harvested and fixed in 70% ethanol overnight at 4°C. Cells were washed twice in PBS, and re-suspended in 500 µl PBS containing 20 µg/ml propidium iodide (PI; Sigma-Aldrich) for 30 min in the dark. Cell cycle distribution was analyzed by flow cytometry using CellQuest™ software (version 3.0; BD Biosciences, Oxford, UK) and a FACScan flow cytometer (BD Biosciences).
Apoptosis assay
For flow cytometric analysis, Annexin-V-fluorescein isothiocyanate (FITC) conjugate and binding buffer (Santa Cruz Biotechnology, Inc.) were used as standard reagents. Cells remained untreated, or were treated with 20 or 40 µM curcumin for 24 h. The cells (1×106) were then digested with 0.25% trypsin for 2–3 min, digestion was terminated with FBS heat-inactivated serum (Biological Industries, Kibbutz Beit Haemek, Israel), and the cells were re-suspended in PBS. Following fixing with 70% ethanol and staining with FITC and PI using an Annexin V-FITC/PI Apoptosis Detection kit (Biobyt, Ltd., Cambridge, UK), flow cytometry analyses were performed using a FACSCalibur flow cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA) and CellQuest™ software (version 3.0). Subsequently, the cells were analyzed. Fluorescent emission of FITC was measured at 515–545 nm and that of DNA-PI complexes was measured at 564–606 nm. Cell debris was excluded from analysis using an appropriate forward light scatter threshold setting. Compensation was used wherever necessary.
Subcellular fractionation
The cytoplasmic and nuclear proteins were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Treated cells were suspended in buffer A (10 mM HEPES, pH 7.9; 10 mM KCl; 0.1 mM EDTA; 0.1 mM EGTA; 1 mM dithiothreitol; 0.15% Nonidet P40; 1% protease inhibitor cocktail), were incubated for 10 min on ice, and were centrifuged at 16,000 × g for 5 min. The supernatant fraction was collected as the cytoplasmic extract. Subsequently, the pellet was washed, re-suspended in buffer B (20 mM HEPES, pH 7.9; 400 mM NaCl; 1 mM EDTA; 1 mM EGTA; 1 mM dithiothreitol; 0.5% Nonidet P40; 1% protease inhibitor cocktail) and was agitated for 15 min at 4°C. The supernatant fraction from the centrifugation (16,000 × g at 4°C for 10 min) was collected as the nuclear extract. The nuclear and cytoplasmic expression levels of FoxO1 were determined by western blot analysis.
Western blot analysis
Using the Compartment Protein Extraction kit (EMD Millipore, Billerica, MA, USA), cytoplasmic and nuclear proteins were isolated from the U87 cells. Cellular fractionation was conducted according to the manufacturer's protocol. Cells were lysed on ice in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) for 20 min in the presence of a protease inhibitor (Roche Diagnostics, Basel, Switzerland). The protein concentration was determined using the Bio-Rad Protein Assay Reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA), according to the manufacturer's protocol. Total proteins (20 µg) extracted from the untreated and treated cells were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were electophoretically transferred to a polyvinylidene difluoride membrane (EMD Millipore), according to standard procedures. Membranes were blocked for 1 h with 5% nonfat dry milk in TBS-T buffer (20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 0.1% Tween-20), followed by an overnight incubation at 4°C with the following primary antibodies: Rabbit anti-FoxO1 (1:1,000 dilution; cat. no. 2880, Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit anti-histone h4 (1:3,000 dilution; cat. no. ab61254, Abcam, Cambridge, UK), anti-phosphorylated (p)-FoxO1 (1:1,000 dilution; cat. no. sc-16307, Santa Cruz Biotechnology, Inc.), anti-CDK1 (1:1,000 dilution; cat. no. 9116, Cell Signaling Technology, Inc.), anti-cyclin G2 (1:1,000 dilution; sc-7266, Santa Cruz Biotechnology, Inc.), anti-cleaved caspase-3 (1:1,000 dilution; cat. no. 9664, Cell Signaling Technology, Inc.), anti-Fas ligand (FasL; 1:1,000 dilution; cat. no. 4273S, Cell Signaling Technology, Inc.) and anti-glyceraldehyde 3-phosphate dehydrogenase (1:1,000 dilution; cat. no. ab9484; Abcam). Blots were subsequently washed with TBS-T and were then incubated with a horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:5,000 dilution; cat. no. sc-2004, Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Proteins were detected using enhanced chemiluminescence reagents (EMD Millipore), and were exposed to X-ray films. Protein expression was semi-quantified using Scion Image Beta 4.02 (Scion Corporation, Torrance, CA, USA).
Immunofluorescence and confocal microscopy
Treated cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100. After blocking with blocking buffer containing 5% nonfat dry milk, the cells were incubated with rabbit anti-FoxO1 (1:200) overnight at 4°C, followed by incubation with a goat anti-rabbit immunoglobulin G tagged with Alexa Fluor 488 (cat. no. 4412, Cell Signaling Technology, Inc.) for 1 h at room temperature. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei. Morphological alterations in U87 cells were observed and documented under a fluorescence microscope (Zeiss LSM 510 META; Carl Zeiss AG, Oberkochen, Germany). Fields were chosen at random from various sections, in order to ensure objectivity of sampling. Morphometric analyses were performed using the Zeiss LSM510 v.3.2 analysis software (Carl Zeiss AG).
Statistical analysis
All statistical analyses were performed using SPSS software (version 19.0; IBM SPSS, Amronk, NY, USA). Quantitative data is expressed as the mean ± standard deviation and analyzed by one-way analysis of variance followed by the Tukey's post-hoc test. Comparison between the groups was made by analyzing data using a post-hoc method. Representative results from independent experiments are presented in the present study. Student's t-test or analysis of variance was used to statistically analyze the results between the control and treatment groups. P<0.05 is considered to indicate a statistically significant difference.
Results
Effects of curcumin on cell viability and proliferation in U87 human glioblastoma cells
The present study observed the effects of curcumin on the inhibition of U87 cell viability. Curcumin markedly inhibited the viability of U87 cells in a dose- and time- dependent manner. The inhibitory effects of curcumin were most obvious when used at a concentration of 40 µM (Fig. 1A). To detect whether curcumin was able to affect the proliferation of U87 cells, U87 cell growth was analyzed using the EdU incorporation assay following treatment with various concentrations of curcumin for 24 h. Similarly, cell proliferation was markedly inhibited by curcumin when used at 20 or 40 µM. In addition, the effect of 40 µM curcumin was more obvious than that of 20 µM curcumin (Fig. 1B).
Figure 1.
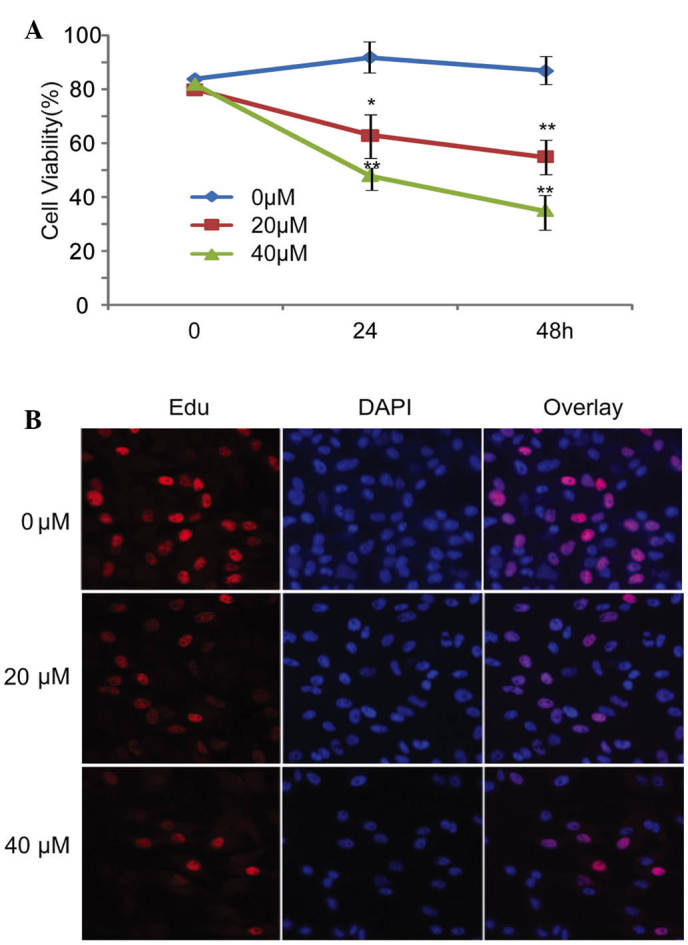
Effects of curcumin on cell viability and proliferation in U87 human glioblastoma cells. (A) U87 cells were treated with various concentrations of curcumin (20 and 40 µM). Following a 24 or 48 h incubation, Cell Counting kit-8 cytotoxicity assays were used to determine cell viability. (B) U87 cells were left untreated or were treated with curcumin (20 and 40 µM) for 24 h. The proliferative ability of U87 cells was determined using 5-ethynyl-2-deoxyuridine (Edu) incorporation assays. *P<0.05, **P<0.01 vs. 0 µm. The results are presented as the mean ± standard deviation, and are representative of three independent experiments (magnification, ×20). DAPI, 4′,6-diamidino-2-phenylindole.
Effects of curcumin on G2/M cell cycle arrest and apoptosis in U87 cells
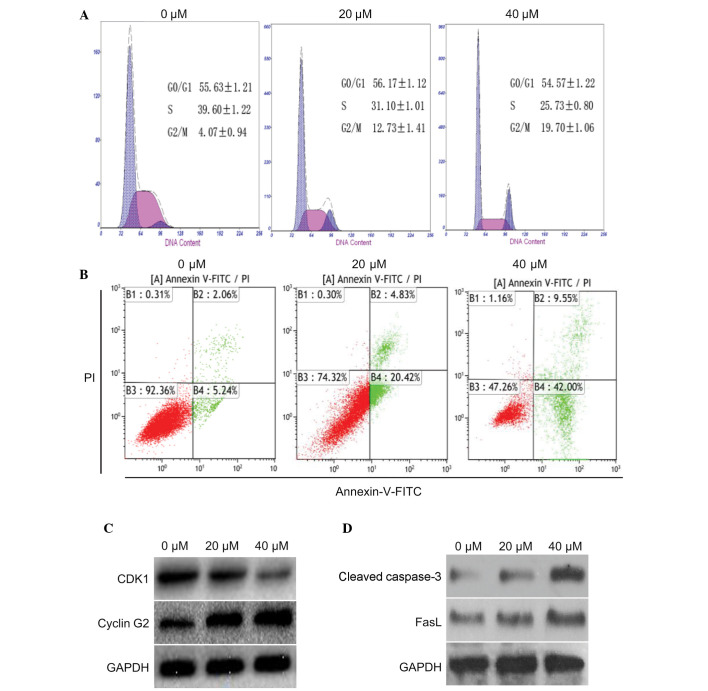
The present study determined the effects of curcumin on cell cycle progression and apoptosis in U87 cells. To determine whether curcumin had an effect on cell cycle distribution and apoptosis, U87 cells were treated with 0, 20 or 40 µM curcumin for 24 h. According to the results of a flow cytometric analysis, an increased number of cells were detected in G2/M phase, and a corresponding reduction in the number of cells was detected in S phase (Fig. 2A). In addition, the number of apoptotic cells was increased in a dose-dependent manner. Curcumin (40 µM) was able to markedly induce apoptosis of U87 cells (Fig. 2B). These results indicate that curcumin may effectively induce G2/M arrest and apoptosis in U87 cells.
Figure 2.
Effects of curcumin on G2/M cell cycle arrest and apoptosis in U87 cells. (A) U87 cells were left untreated (control) or were treated with 20 or 40 µM curcumin for 24 h. Subsequently, the cells were harvested and stained with propidium iodide (PI), in order to determine cell cycle distribution by flow cytometry. (B) Effects of curcumin on apoptosis were analyzed by flow cytometry using Annexin-V-fluorescein isothiocyanate (FITC) and PI. (C) After U87 cells treated with 20 or 40 µM curcumin for 24 h, total proteins were collected. Cell cycle-related proteins cyclin-dependent kinase 1 (CDK1) and cyclin G2 were determined by western blotting. (D) Western blotting was conducted to detect the expression levels of apoptotic proteins cleaved caspase-3 and Fas ligand (Fas)L. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
The expression levels of proteins associated with the cell cycle and apoptosis were also detected in the cells following treatment with 20 or 40 µM curcumin for 24 h. Compared with the control group, cyclin G2 was markedly increased in the U87 cells; however, the expression levels of CDK1 were markedly decreased (Fig. 2C). Furthermore, two apoptotic proteins: Cleaved caspase-3 and FasL, were markedly upregulated (Fig. 2D). These data indicate that curcumin may induce G2/M cell cycle arrest and apoptosis in U87 cells by decreasing CDK1 expression and increasing the expression of cyclin G2, cleaved caspase-3 and FasL.
Curcumin induces expression and nuclear localization of FoxO1
To investigate the mechanism underlying regulation of the previously mentioned proteins, the present study detected the expression levels of FoxO1 in the presence of 40 µM curcumin. The expression levels of FoxO1 in the nucleus and cytoplasm were increased in response to curcumin in a dose-dependent manner. Notably, the expression levels of p-FoxO1 were markedly decreased (Fig. 3A). In addition, the nuclear translocation of FoxO1 was detected following treatment with curcumin by immunofluorescence, which demonstrated that the nuclear expression of FoxO1 was markedly increased (Fig. 3B). These findings were in accordance with the results presented in Fig. 3A. These results indicate that FoxO1 may have an important role in curcumin-induced regulation of the cell cycle and apoptosis.
Figure 3.
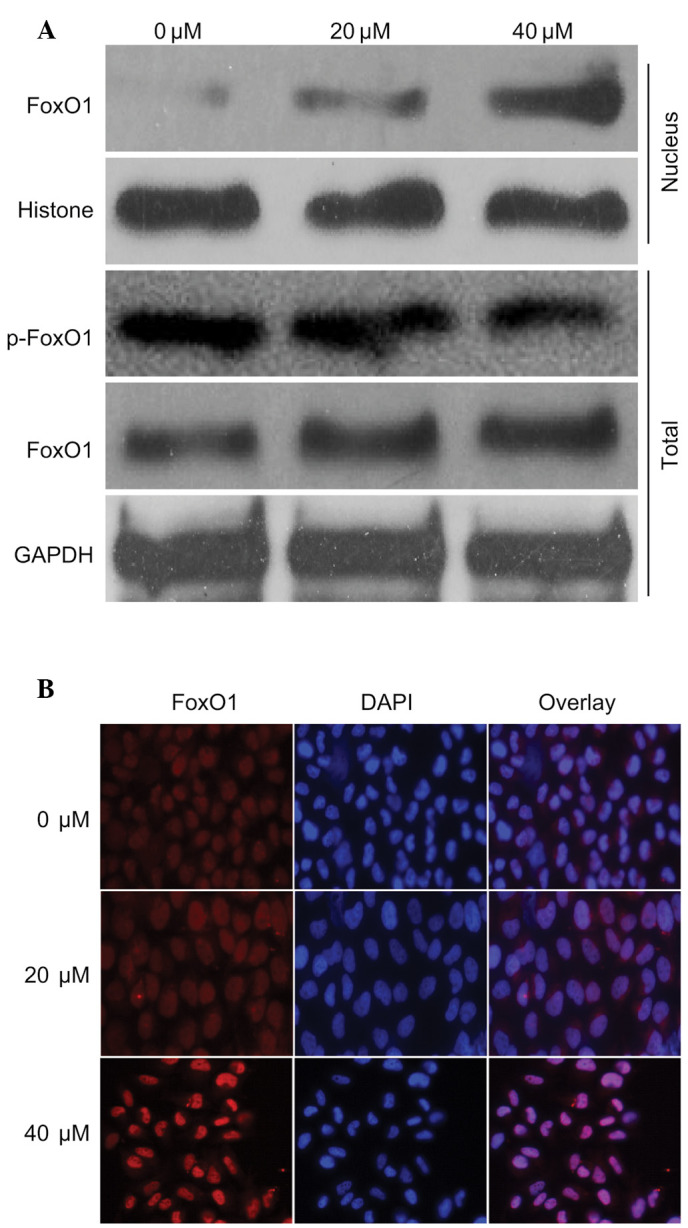
Curcumin induced expression and localization of forkhead box O1 (FoxO1). (A) U87 cells were left untreated or were treated with 20 or 40 µM curcumin for 24 h. Nuclear and cytoplasmic proteins were extracted according to the nuclear extraction kit protocol. Subsequently, FoxO1 and phosphorylated (p)-FoxO1 expression in the nucleus and cytoplasm were detected by western blotting. (B) After the U87 cells were treated with 20 or 40 µM curcumin, immunofluorescence was conducted to determine the effects of curcumin on nuclear translocation of FoxO1 (magnification, ×20). GAPDH, glyceraldehyde 3-phosphate dehydrogenase; DAPI, 4′,6-diamidino-2-phenylindole.
Effects of FoxO1 on the expression of target proteins associated with the cell cycle and apoptosis
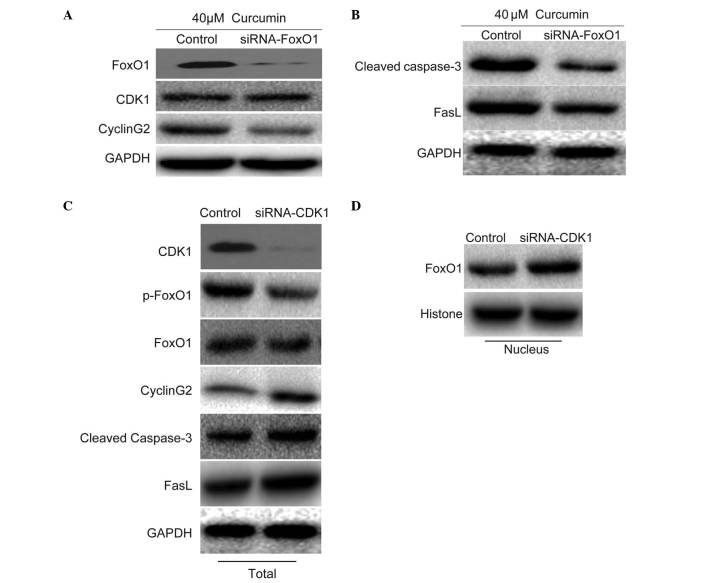
To investigate the role of FoxO1, the present study determined the effects of FoxO1 on the expression of CDK1, cyclin G2, cleaved caspase-3 and FasL in the presence of 40 µM curcumin. Following knockdown of FoxO1 with FoxO1 siRNA, the expression levels of CDK1 were not affected. Following the knockout of CDK1 by CDK1-siRNA, the expression of p-FoxO1 decreased; although the total expression of FoxO1 was not affected, the expression of FoxO1 in the nucleus decreased. However, the expression levels of cyclin G2 were inhibited (Fig. 4A), thus indicating that cyclin G2 may be regulated by FoxO1. In addition, the expression levels of cleaved caspase-3 and FasL were markedly inhibited (Fig. 4B), thus indicating that cleaved caspase-3 and FasL may be downstream of FoxO1. To determine whether FoxO1 was regulated by CDK1, CDK1 expression was knocked down using CDK1 siRNA in untreated cells. In cells transfected with CDK1 siRNA, the expression levels of total FoxO1 were not affected; however, the expression levels of p-FoxO1 were decreased. Furthermore, the expression levels of cyclin G2, cleaved caspase-3 and FasL were increased (Fig. 4C), and the nuclear expression of FoxO1 was increased (Fig. 4D). These results indicate that the nuclear translocation of FoxO1 may be dependent on the effects of CDK1 on FoxO1 phosphorylation.
Figure 4.
Effects of forkhead box O1 (FoxO1) on the expression of target proteins associated with cell cycle progression and apoptosis. (A) U87 cells were treated with 40 µM curcumin and were transfected with FoxO1 small interfering (si)RNA for 24 h. Western blotting was conducted to determine the effect of FoxO1 knockdown on cyclin-dependent kinase 1 (CDK1) and cyclin G2 expression. (B) Expression levels of cleaved caspase-3 and Fas ligand (FasL) were determined by western blotting following FoxO1 knockdown and curcumin treatment. (C) U87 cells were transfected with CDK1 siRNA under normal conditions for 24 h. Western blotting was conducted to determine the effects of CDK1 knockdown on phosphorylated (p)-FoxO1, FoxO1, cyclin G2, cleaved caspase-3 and FasL expression. (D) Nuclear expression of FoxO1 was detected by western blotting following CDK1 knockdown. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
FoxO1 is required for curcumin-induced G2/M cell cycle arrest and apoptosis in U87 cells
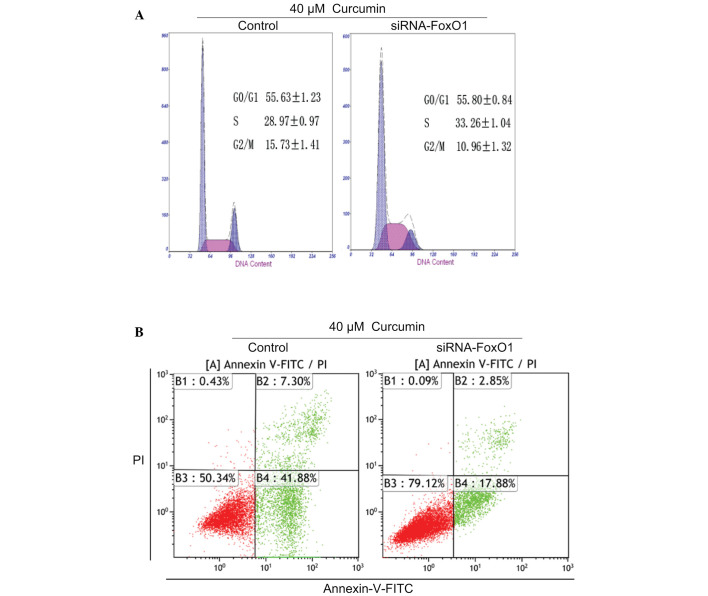
The present study also tested the function of FoxO1 in U87 cells. Following FoxO1 knockdown with FoxO1 siRNA in the presence of 40 µM curcumin, cell cycle distribution and apoptosis were detected in U87 cells. Curcumin-induced G2/M arrest and apoptosis were partly inhibited by FoxO1 knockdown (Fig. 5A and B). These data suggest that FoxO1 may have an important role in curcumin-induced G2/M cell cycle arrest and apoptosis in U87 cells.
Figure 5.
Forkhead box O1 (FoxO1) is required for curcumin-induced G2/M cell cycle arrest and apoptosis in U87 cells. (A) Following treatment with 40 µM curcumin, U87 cells were transfected with FoxO1 small interfering (si)RNA for 24 h. Subsequently, the cells were harvested and stained with propidium iodide (PI) to determine cell cycle distribution by flow cytometry. (B) Apoptosis was analyzed by flow cytometry using Annexin-V-fluorescein isothiocyanate (FITC) and PI in cells transfected with FoxO1 siRNA and treated with 40 µM curcumin.
Discussion
Curcumin is a botanical polyphenol that is extracted from the rhizomes of Curcuma longa. Curcumin has garnered increasing attention due to its low toxic side effects and promising antitumor effects. Numerous studies have demonstrated that curcumin has strong inhibitory effects on three phases of tumor formation (initiation, promotion and progression) (17,18). In pre-clinical and clinical trials, researchers have reported that curcumin exerts anticancer effects, particularly in glioma (19). Although the underlying mechanism remains to be elucidated, research has focused on several factors that may affect its anticancer effects, including oxygen effect, glutathione content, ability to repair radiation damage, cell cycle phase and cell apoptosis (20).
In the present study, following treatment with curcumin, compared with the control group, the proliferation of U87 cells was inhibited, and the ratio of cells in G2/M phase and the rate of apoptosis were significantly increased in a dose-dependent manner. These findings were concordant with the results of other studies. Although curcumin has been shown to induce G2/M arrest and apoptosis (21–23), the detailed mechanism underlying the effects of curcumin on the cell cycle and apoptosis have yet to be investigated. Therefore, the present study also detected the expression levels of proteins associated with G2/M phase and apoptosis, and demonstrated that curcumin was able to induce the expression of cyclin G2 and FasL. Cyclin G2 accumulates in the G2 phase, at the G2/M border, and hinders cell cycle transition from G2 to M phase (24). In addition, the proapoptotic gene FasL encodes a protein that activates the death receptor Fas/CD95/apoptosis antigen 1 and promotes mitochondria-independent apoptosis (25). These findings may explain why curcumin induces G2/M arrest and apoptosis.
FoxO members contain a conserved DNA binding domain and bind to a consensus DNA binding sequence (TTGTTTAC) in target genes (26,27). FoxO proteins can be phosphorylated at their highly conserved sites by the survival kinase Akt (a downstream target of phosphoinositide 3-kinase signaling) within and nearby their forkhead domains (28,29). In addition, FoxO1 can be phosphorylated by CDK1 and CDK2 at serine 249 in vitro and in vivo (30). The exportation of FoxO proteins from the nucleus to the cytoplasm is controlled by phosphorylation. Once phosphorylated, FOXO proteins are exported from the nucleus to the cytoplasm and become inactive (12). The present study demonstrated that curcumin was able to inhibit the expression of CDK1 and induce the nuclear expression of FoxO1. These findings may be the result of decreased FOXO1 phosphorylation, due to the inhibition of CDK1 by curcumin.
The activity of FoxO transcription factors is important for cell cycle transition and cell fate. FoxOs are able to increase the expression of p27, p21 and p130, and inhibit the expression of cyclin D1 and cyclin D2, which inhibit transition of the cell cycle from G1 phase to S phase. Growth arrest and DNA-damage-inducible, alpha and cyclin G2 can also be increased by active FoxOs, which may result in G2/M arrest (24). In addition, active FoxO proteins regulate cell survival by modulating the expression of death receptor ligands, which function in autocrine and paracrine pathways (25,31), and are also associated with transactivation of B-cell lymphoma 1 (Bcl-2) interacting mediator of cell death, a gene that encodes a member of the proapoptotic BH3-only subgroup of Bcl-2 family proteins, which functions in the 'intrinsic', mitochondrial apoptotic pathway (32,33). These results suggested that FoxO transcription factors can induce cell death through mitochondria-dependent and -independent mechanisms. However, the role of FoxO1 in cell cycle progression and apoptosis remains unclear. The results of the present study demonstrated that following knockdown of FoxO1 by siRNA, curcumin-induced G2/M arrest and apoptosis were inhibited, and the expression levels of cyclin G2 and FasL were decreased; however, the expression of CDK1 was not effected by FoxO1 knockdown. The subsequent results supported that FoxO1 was a substrate of CDK1. Once CDK1 was inhibited by curcumin, the phosphorylation of FoxO1 was decreased; therefore, active FoxO1 was translocated from the cytoplasm to the nucleus where it triggered the expression of target genes. However, the present study did not investigate the mechanism underlying curcumin-induced CDK1 inhibition and FoxO1 upregulation; therefore, further studies are required.
In conclusion, the present study demonstrated that curcumin leads to G2/M phase arrest and apoptosis, which may be associated with the expression of active FoxO1 in the nucleus, and may mediate the proliferation and apoptosis of U87 cells. The results indicated that this process was associated with the curcumin-induced inhibition of CDK1. In addition, cyclin G2 and FasL were identified as potential target genes of FoxO1. The present study provides evidence for a novel mechanism to explain the antitumor effects of curcumin, and provides a theoretical basis for the application of curcumin in glioma.
Acknowledgments
The present study was supported by grants from the National Natural Science Foundation China (grant nos. 81272791 and 81502159) and the Social Programs of Wuxi Technology Bureau (grant no. CSE01N1107).
References
- 1.Grossman SA, Ye X, Piantadosi S, Desideri S, Nabors LB, Rosenfeld M, Fisher J, NABTT CNS Consortium Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin Cancer Res. 2010;16:2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garodia P, Ichikawa H, Malani N, Sethi G, Aggarwal BB. From ancient medicine to modern medicine: Ayurvedic concepts of health and their role in inflammation and cancer. J Soc Integr Oncol. 2007;5:25–37. doi: 10.2310/7200.2006.029. [DOI] [PubMed] [Google Scholar]
- 3.Cole GM, Teter B, Frautschy SA. Neuroprotective effects of curcumin. Adv Exp Med Biol. 2007;595:197–212. doi: 10.1007/978-0-387-46401-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang SE, Kuo ML, Hsu CH, Chen CR, Lin JK, Lai GM, Hsieh CY, Cheng AL. Curcumin-containing diet inhibits diethylnitrosamine-induced murine hepatocarcinogenesis. Carcinogenesis. 2000;21:331–335. doi: 10.1093/carcin/21.2.331. [DOI] [PubMed] [Google Scholar]
- 5.Lim GP, Chu T, Yang F, Beech W, Frautschy SA, Cole GM. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J Neurosci. 2001;21:8370–8377. doi: 10.1523/JNEUROSCI.21-21-08370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiyagarajan M, Sharma SS. Neuroprotective effect of curcumin in middle cerebral artery occlusion induced focal cerebral ischemia in rats. Life Sci. 2004;74:969–985. doi: 10.1016/j.lfs.2003.06.042. [DOI] [PubMed] [Google Scholar]
- 7.LoTempio MM, Veena MS, Steele HL, Ramamurthy B, Ramalingam TS, Cohen AN, Chakrabarti R, Srivatsan ES, Wang MB. Curcumin suppresses growth of head and neck squamous cell carcinoma. Clin Cancer Res. 2005;11:6994–7002. doi: 10.1158/1078-0432.CCR-05-0301. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Veena MS, Stevenson K, Tang C, Ho B, Suh JD, Duarte VM, Faull KF, Mehta K, Srivatsan ES, Wang MB. Liposome-encapsulated curcumin suppresses growth of head and neck squamous cell carcinoma in vitro and in xenografts through the inhibition of nuclear factor kappaB by an AKT-independent pathway. Clin Cancer Res. 2008;14:6228–6236. doi: 10.1158/1078-0432.CCR-07-5177. [DOI] [PubMed] [Google Scholar]
- 9.Lin YG, Kunnumakkara AB, Nair A, Merritt WM, Han LY, Armaiz-Pena GN, Kamat AA, Spannuth WA, Gershenson DM, Lutgendorf SK, et al. Curcumin inhibits tumor growth and angiogenesis in ovarian carcinoma by targeting the nuclear factor-kappaB pathway. Clin Cancer Res. 2007;13:3423–3430. doi: 10.1158/1078-0432.CCR-06-3072. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang W, Long L, Zheng B, Ji W, Yang N, Zhang Q, Liang Z. Curcumin promotes differentiation of glioma-initiating cells by inducing autophagy. Cancer Sci. 2012;103:684–690. doi: 10.1111/j.1349-7006.2011.02198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A, Ahuja A, Ali J, Baboota S. Curcumin-loaded lipid nanocarrier for improving bioavailability, stability and cytotoxicity against malignant glioma cells. Drug Deliv. 2016;23:214–229. doi: 10.3109/10717544.2014.909906. [DOI] [PubMed] [Google Scholar]
- 12.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 13.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/S0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 14.Taylor S, Lam M, Pararasa C, Brown JE, Carmichael AR, Griffiths HR. Evaluating the evidence for targeting FOXO3a in breast cancer: A systematic review. Cancer Cell Int. 2015;15:1. doi: 10.1186/s12935-015-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Zhao R, Yang HY, Lee MH. Constitutively active FOXO4 inhibits Akt activity, regulates p27 Kip1 stability, and suppresses HER2-mediated tumorigenicity. Oncogene. 2005;24:1924–1935. doi: 10.1038/sj.onc.1208352. [DOI] [PubMed] [Google Scholar]
- 16.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/S1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 17.Prakobwong S, Gupta SC, Kim JH, Sung B, Pinlaor P, Hiraku Y, Wongkham S, Sripa B, Pinlaor S, Aggarwal BB. Curcumin suppresses proliferation and induces apoptosis in human biliary cancer cells through modulation of multiple cell signaling pathways. Carcinogenesis. 2011;32:1372–1380. doi: 10.1093/carcin/bgr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moragoda L, Jaszewski R, Majumdar AP. Curcumin induced modulation of cell cycle and apoptosis in gastric and colon cancer cells. Anticancer Res. 2001;21:873–878. [PubMed] [Google Scholar]
- 19.Shao J, Zheng D, Jiang Z, Xu H, Hu Y, Li X, Lu X. Curcumin delivery by methoxy polyethylene glycol-poly (caprolactone) nanoparticles inhibits the growth of C6 glioma cells. Acta Biochim Biophys Sin (Shanghai) 2011;43:267–274. doi: 10.1093/abbs/gmr011. [DOI] [PubMed] [Google Scholar]
- 20.Vallianou NG, Evangelopoulos A, Schizas N, Kazazis C. Potential anticancer properties and mechanisms of action of curcumin. Anticancer Res. 2015;35:645–651. [PubMed] [Google Scholar]
- 21.Weir NM, Selvendiran K, Kutala VK, Tong L, Vishwanath S, Rajaram M, Tridandapani S, Anant S, Kuppusamy P. Curcumin induces G2/M arrest and apoptosis in cisplatin-resistant human ovarian cancer cells by modulating Akt and p38 MAPK. Cancer Biol Ther. 2007;6:178–184. doi: 10.4161/cbt.6.2.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luthra PM, Kumar R, Prakash A. Demethoxycurcumin induces Bcl-2 mediated G2/M arrest and apoptosis in human glioma U87 cells. Biochem Biophys Res Commun. 2009;384:420–425. doi: 10.1016/j.bbrc.2009.04.149. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Xu YM, Ye ZQ, Yu JH, Hu XY. Curcumin induces cell cycle arrest and apoptosis of prostate cancer cells by regulating the expression of IkappaBalpha, c-Jun and androgen receptor. Pharmazie. 2013;68:431–434. [PubMed] [Google Scholar]
- 24.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 25.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 26.Biggs WH, III, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome. 2001;12:416–425. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- 27.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/bj3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 29.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 31.Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–47937. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- 32.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/S0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 33.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]