Abstract
The Chinese herbal formula, Shu-Yu capsule (SYC), has been successfully used to treat depression-like disorders in clinical settings. To rapidly identify the chemical constituents of SYC and its metabolites in rat serum, a simple and sensitive liquid chromatography-tandem mass spectrometry method was established in the present study. By comparing the retention times, MS and MSn spectra data in the literature and reference standards, a total of 73 compounds were identified from SYC. In rat serum, 62 components, including 13 prototype compounds and 49 metabolites were identified. Of these components, 14 metabolites were confirmed as novel metabolites of SYC. The results of the present study indicated that certain flavonoid glycosides and monoterpene glycosides were absorbed directly. Glucuronidation and sulfation were identified as the predominant metabolic pathways of the components in SYC. In addition, certain phase I reactions, including hydrolysis, demethylation and hydroxylation occurred in the rats. These results provide scientific evidence, which support further investigations of the pharmacology and mechanism of SYC.
Keywords: Shu-Yu capsule, chemical constituents, absorption, high performance liquid chromatography-electrospray ionization tandem mass spectrometry, metabolites
Introduction
The Shu-Yu capsule (SYC) formulation is a four-herb traditional Chinese medicine (TCM) used for the treatment of clinical depression-like disorders, including premenstrual syndrome (1). It is composed of four herbal ingredients: Radix Bupleuri (Bupleurum chinense DC.), Radix Paeoniae Alba (Paeonia lactiflora Pall.), Rhizoma Cyperi (Cyperus rotundus Linn.) and Radix Glycyrrhizae (Glycyrrhiza uralensis Fisch.). According to the TCM formulation theory (2), Radix Bupleuri and Radix Paeoniae Alba are mutually complementary as the monarch herbs of the formula, which represent the components with the major therapeutic roles.
It has been demonstrated that the total glycosides of peony exert significant antidepressant-like effects by increasing the expression levels of brain derived neurotrophic factor and nerve growth factor in selective brain tissues (3). Radix Bupleuri is another monarch ingredient in SYC, which may affect the quality of prescriptions significantly. Bupleuri Radix, or prescriptions containing Bupleuri Radix as the major component, for example Xiaochaihutang, exert antidepressant-like effects by modulating serotonergic and noradrenergic systems in brain regions of rat models of depression (4,5). Our previous studies also revealed antidepressant-like effects of ethanol extracts from Paeonia lactiflora Radix and Bupleuri Radix (6). These findings confirmed that Paeoniae Radix and Bupleuri Radix have effects on the central nervous system. According to the theory of serum pharmacochemistry, only constituents absorbed into blood have the potential to exert pharmacological bioactivities (7). However, the metabolites absorbed in the blood following oral administration of SYC remain to be elucidated.
The present study aimed to confirm the absorbed components and the relative metabolites of SYC, which may be the potential bioactive components in the blood following intragastric administration of SYC. Thus, a reliable high performance liquid chromatography-electrospray ionization tandem mass spectrometry (HPLC-ESI-MSn) system was established for the detection of prototype compounds and metabolites in the rat serum following oral administration of SYC. Screening for potential bioactive components in SYC may assist in future investigations into its mechanism of action at the molecular level.
Materials and methods
Reagents
Acetonitrile, methanol and glacial acetic acid were of LC/MS reagent grade and purchased from Merck Millipore (Darmstadt, Germany), Ultra-pure water was prepared using a Milli-Q water purification system (EMD Millipore, Bedford, MA, USA). The other solvents were of analytical grade.
Radix Bupleuri, Radix Paeoniae, Radix Glycyrrhizae and Rhizoma Cyperi were collected in Shandong Province (Shandong, China) and were authenticated by Professor Hui Yun Zhang (Shandong University of Traditional Chinese Medicine, Jinan, China). SYCs were manufactured by Qingdao Haichuan Innovative Biological & Natural Drug Research Institute (Qingdao, China).
The composition and preparation of the SYC were as previously described (1).
Instrument and analysis conditions
The HPLC analysis was performed on an Agilent-1100 series liquid chromatograph system (Agilent Technologies, Inc., Santa Clara, CA, USA), equipped with a binary pump, an auto sampler, a photo-diode array detector and a column temperature controller. The analytical column was a Kromasil C18 100R (5 µm, 25×4.6 mm i.d.; AkzoNobel, Bohus, Sweden) and the oven temperature was maintained at 25°C. A mobile phase, composed of eluent A (acetonitrile) and B (0.2% acetic acid in water, v/v) with a gradient was used for the separation. The elution conditions were applied with the following linear gradient: 0–5 min, 2–5% A; 5–12 min, 5–12% A; 12–28 min, 12–16% A; 28–42 min, 16–24% A; 42–56 min, 24–36% A; 56–72 min, 36–39% A; 72–81 min, 39–64% A; 81–90 min and 64–100% A. The flow rate was 1.0 ml/min and peaks were detected at 254 nm.
In the subsequent ESI-MS/MS experiment, an MSD Trap XCT Plus mass spectrometer (Agilent Technologies) was connected to the same HPLC instrument via an electrospray ionization (ESI) interface (Agilent Technologies, Inc.). The HPLC effluent was introduced into the ESI source in a post-column splitting ratio of 1:4. The ESI-MS operating conditions (negative ion) were optimized using the SYC, as follows: Nebulizer gas pressure, 40.00 psi; dry gas flow rate, 11.00 l/min; capillary temperature, 350°C; electrospray voltage of the ion source, 3,500 V; skimmer of 40.0 V; capillary exit, 121.0 V; compound stability, 50%; trap drive level, 100%; target mass, 500 m/z; scan range, 100–14,00 m/z; collision energy, 1 V. A data-dependent program was used for the HPLC-ESI-MSn analysis, to enable the protonated or deprotonated ions to be selected for further MSn analysis. An Agilent 6300 Series Trap Control workstation (version 6.1; Agilent Technologies, Inc.) was used for data processing.
Preparation of samples for the extract
Sample preparation of the SYC was as follows: A single capsule from each of five batches of SYC were used. The powder-like contents of the capsules were mixed evenly, and 0.3 g of the mixed powder was weighed. The Radix Paeoniae, Radix Bupleuri, Rhizoma Cyperi and Radix Glycyrrhizae samples were crushed into a homogeneous size separately and sieved through a No. 40 mesh sieve (Jin Yuan Screen Factory, Yangquan, China) for further assessment.
The weighed powder was suspended in 25.0 ml of 70% (v/v) methanol and extracted in an ultrasonic water bath for 30 min at room temperature. Each resulting mixture was filtered through 0.22-µm membranes prior to use, and a 10-µl aliquot was injected into the HPLC-MS system for analysis.
In-vivo experiments
A total of 24 male Wistar rats (weight, 180–220 g; age, 8 weeks) were supplied by the Laboratory Animal Center of Shandong University of Traditional Chinese Medicine. The present study was approved by the Institutional Committee for Animal Care and Use of Shandong University of Traditional Chinese Medicine. The rats were housed in poly cages with free access to food and water, at a temperature of 22–26°C and relative humidity of 50–70%. The rats were acclimatized to the environment for 1 week prior to initiating the experiment. All rats were fasted, with free access to water, for 12 h prior to the experiment. Subsequently, 12 rats were administered with SYC at a dose of 4.1 g/kg body weight orally for 3 days, once per day. Another group of 12 rats served as a blank control group, which received physiological saline. On day 4, blood samples were collected from the inferior venae cava, 90 min after intragastric administration of SYC,and centrifuged at 1,500 × g for 15 min at 4°C to obtain serum samples, which were then frozen and stored at −80°C until analysis. Subsequently, the rats were sacrificed by cervical dislocation. The time points were selected based on our previous pharmacokinetic studies (8). All procedures were in agreement with the National Institute of Health's Guidelines on the Principles of Animal Care (9).
Sample preparation
The dried powders from the SYCs and the four herbs were weighed accurately (0.3 g), suspended in 25.0 ml of 70% (v/v) methanol, and extracted in an ultrasonic water bath (KQ-250E; Kunshan Ultrasonic Instruments Co., Ltd.) for 30 min at room temperature. Each resulting mixture was filtered through 0.22 µm polyvinylidene fluoride membranes (Merck Millipore) prior to use, and a 10 µl aliquot was injected into the HPLC-MS system for analysis.
Each collected serum sample was thawed and centrifuged at 1,500 × g for 30 min at 4°C. The supernatant (100 µl) was then mixed with 300 µl methanol and vortexed for 2 min. The denatured protein precipitate was separated by centrifugation at 16,000 × g for 30 min at 4°C, and the supernatant was separated and evaporated to dryness under a gentle nitrogen stream at 37°C. The residue was reconstituted in 100 µl methanol and centrifuged at 16,000 × g for 30 min at 4°C. An aliquot of 20 µl of the supernatant was analyzed on the HPLC-MS system.
Results
HPLC-MSn analysis of constituents in SYC extract in negative ion mode
Utilizing the optimized LC-MSn method, the components of the extracts of SYC and its single herbal extracts (Radix Paeoniae Alba, Radix Bupleuri, Radix Glycyrrhizae and Rhizoma Cyperi) were identified.
By comparing the MSn spectra data with the reference standards and literature data, 73 components, including flavonoids, terpenes and phenolic acids, were identified in the SYC extract. Among these 73 components, three predominant compounds were confirmed using reference standards. The HPLC-MSn data of the 73 tentatively identified components are summarized in Table I.
Table I.
Identification of the chemical constituents of SYC formula by liquid chromatography-tandem mass spectrometry in negative ion mode.
| No. | tRa | [M-H]− (m/z) | MSn (m/z) | Identification | Source | Molecular formula |
|---|---|---|---|---|---|---|
| 1 | 3.1 | 341 | 179, 161, 143, 119, 113 | Caffeic acid-4-O-β-D-glucopyranoside | RG | C15H18O9 |
| 2b | 4.8 | 191 | 111, 173, 129 | Quinic acid | RG | C7H12O6 |
| 3b | 6.9 | 375 | 345, 165 | Desbenzoylpaeoniflorin | RP | C16H24O10 |
| 4 | 9.3 | 169 | 125 | Gallic acid | RP | C7H6O5 |
| 5 | 11.1 | 493 | 313, 331, 283, 169 | 1′-O-galloyl-sucrose | RP | C19H26O15 |
| 6 | 11.6 | 493 | 313, 331, 169 | 6′-O-galloyl-sucrose | RP | C19H26O15 |
| 7b | 15.0 | 527 | 497, 479, 271 | 6′-O-galloyldesbenzoylpaeoniflorin | RP | C23H28O14 |
| 8 | 15.8 | 705 | 543, 421, 375 | Isomaltopaeoniflorin sulfonate | RP | C29H38O18S |
| 9b | 16.5 | 543 | 421, 375, 259, 497 | Paeoniflorin sulfonate | RP | C23H28O13S |
| 10 | 16.8 | 495 | 465, 311, 137, | Oxypaeoniflorin | RP | C23H28O12 |
| 11 | 17.9 | 495 | 465, 333, 137 | Oxypaeoniflorin or isomer | RP | C23H28O12 |
| 12 | 19.4 | 525 | 495, 167, 465, 509 | Mudanpioside E | RP | C24H30O13 |
| 13 | 21.4 | 701e | 641, 519, 611, 489 | Isomaltopaeoniflorin | RP | C29H38O16 |
| 14 | 23.2 | 641 | 519, 489, 475, 611 | 6′-O-d-glucopyranosylalbiflorin | RP | C29H18O16 |
| 15 | 24.3 | 701e | 611, 593, 641, 489, 471 | Isomaltopaeoniflorin | RP | C29H38O16 |
| 16b,c | 24.7 | 539e | 479, 357, 327, 283 | Albiflorin | RP | C23H28O11 |
| 17 | 27.6 | 495 | 465, 311, 137 | Ortho-Oxypaeoniflorin | RP | C23H28O12 |
| 18b,c,d | 27.8 | 539e | 479, 449, 327, 165 | Paeoniflorin | RP | C23H28O11 |
| 19 | 28.7 | 539e | 449, 479, 327, 165 | Paeoniflorin isomer | RP | C23H28O11 |
| 20 | 33.9 | 787 | 635, 483, 465, 617, 313 | Tetragalloylglucose | RP | C34H28O22 |
| 21 | 34.5 | 787 | 635, 465, 483, 617, 313 | Tetragalloylglucose or isomer | RP | C34H28O22 |
| 22 | 34.7 | 577 | 457, 503, 383 | Isoviolanthin | RG | C27H30O14 |
| 23b | 35.2 | 549 | 255, 417, 135, 429 | Liquiritin apioside | RG | C26H30O13 |
| 24 | 35.3 | 417 | 255, 135, 153, 119 | Neoliquiritin | RG | C21H22O9 |
| 25b | 35.7 | 453 | 417, 135, 255 | Hydrated Liquiritin | RG | C21H26O11 |
| 26b | 35.9 | 417 | 255, 135, 153, 119 | Liquiritin | RG | C21H22O9 |
| 27 | 36.5 | 539e | 479, 357, 327, 449 | Albiflorin or isomer | RP | C23H28O11 |
| 28b | 39.0 | 631 | 613, 491, 479, 465, 313 | Galloyl paeoniflorin/Galloylalbiflorin | RP | C30H32O15 |
| 29 | 39.5 | 939 | 769, 617, 447, 601, 599 | Pentagalloylglucose | RP | C41H32O26 |
| 30b | 42.0 | 631 | 313, 509, 465, 169 | Galloylpaeoniflorin/Galloylalbiflorin | RP | C30H32O15 |
| 31 | 44.0 | 433 | 271, 151 | Naringenin-O-glucose | RG | C21H22O10 |
| 32 | 45.3 | 479 | 357, 327 | Isopaeoniflorin/Albiflorin R1 | RP | C23H28O11 |
| 33 | 45.7 | 479 | 357, 397, 327, 283, 337 | Isopaeoniflorin/Albiflorin R1 | RP | C23H28O11 |
| 34 | 46.1 | 647 | 525, 479 | Benzoylpaeoniflorin sulfonate | RP | C30H32O14S |
| 35 | 46.6 | 647 | 525, 479 | Benzoylpaeoniflorin Sulfonate or isomer | RP | C30H32O14S |
| 36 | 47.1 | 549 | 255, 417, 429, 135 | Isoliquiritin apioside | RG | C26H30O13 |
| 37b | 47.6 | 445 | 269, 251 | Apigenin-7-O-β-D-glucuronide | RB | C21H18O11 |
| 38 | 48.0 | 445 | 269, 175 | Baicalin | RB | C21H18O11 |
| 39 | 48.2 | 631 | 465, 509, 313, 613, 169 | Galloylpaeoniflorin or isomer | RP | C30H32O15 |
| 40 | 48.5 | 417 | 255, 135, 153, 119 | Isoliquiritin | RG | C21H22O9 |
| 41 | 48.9 | 417 | 255, 135, 119, 153 | Neoisoliquiritin | RG | C21H22O9 |
| 42 | 49.6 | 647 | 617, 525 | Benzoylpaeoniflorin Sulfonate/isomer | RP | C30H32O14S |
| 43 | 49.5 | 417 | 255, 135, 119, 153 | Neoisoliquiritin/isomer | RG | C21H22O9 |
| 44 | 49.7 | 255 | 135, 153, 119 | Liquiritigenin | RG | C15H12O4 |
| 45 | 50.0 | 695 | 549, 531, 255 | Licorice-glycoside B | RG | C35H36O15 |
| 46 | 50.2 | 725 | 549, 531, 255 | Licorice-glycoside A | RG | C36H38O16 |
| 47 | 50.6 | 285 | 269 | Kaempferol | RB | C15H10O6 |
| 48 | 52.1 | 255 | 135, 153, 119 | Liquiritigenin/isomer | RG | C15H12O4 |
| 49 | 52.3 | 459 | 283, 268, 175 | Wogonoside | RB | C22H20O11 |
| 50b | 54.4 | 459 | 283, 268 | Wogonoside/isomer | RB | C22H20O11 |
| 51 | 54.8 | 521e | 461, 163, 265 | Lactiflorin | RP | C23H26O10 |
| 52 | 55.3 | 521e | 461, 163 | Lactiflorin/isomer | RP | C23H26O10 |
| 53 | 57.5 | 643e | 583, 553, 431, 165 | Benzoylpaeoniflorin | RP | C30H32O12 |
| 54 | 57.6 | 553 | 431, 165 | Dehydroxylate Demethylene Benzoylpaeoniflorin | RP | C29H30O11 |
| 55 | 58.4 | 643e | 583, 461, 553 | Isobenzoylpaeoniflorin | RP | C30H32O12 |
| 56 | 59.2 | 271 | 151, 119 | Naringenin | RG | C15H12O5 |
| 57 | 61.5 | 837 | 351, 193, 661, 819 | Licorice-saponin G2 | RG | C42H62O17 |
| 58 | 62.1 | 819 | 351, 193, 643, 801 | Licorice-saponin E2 | RG | C42H60O16 |
| 59 | 65.1 | 255 | 135, 153, 119 | Isoliquiritigenin | RG | C15H12O4 |
| 60 | 65.4 | 811 | 649, 471, 439 | SSb3 or SSb4 | RB | C43H72O14 |
| 61 | 65.5 | 821 | 645, 469, 351, 193 | Glycyrrhizin | RG | C42H62O16 |
| 62 | 66.3 | 821 | 645, 351, 193 | Uralsaponin A | RG | C42H62O16 |
| 63 | 66.8 | 267 | 252 | Formononetin | RG | C16H12O4 |
| 64d | 71.9 | 779 | 617, 471, 541, 439, 423 | SSa | RB | C42H68O13 |
| 65d | 72.4 | 779 | 617, 471, 541, 439 | SSd | RB | C42H68O13 |
| 66 | 72.7 | 367 | 309, 297, 352 | Glycycoumarin | RG | C21H20O6 |
| 67 | 76.5 | 283 | 268 | Wogonin | RB | C16H12O5 |
| 68 | 77.3 | 677e | 617, 471, 541, 439 | Prosaikogenin G/F | RB | C36H58O8 |
| 69 | 77.7 | 677e | 617, 471, 541, 407 | Prosaikogenin A/D | RB | C36H58O8 |
| 70 | 80.4 | 367 | 309, 281, 297, 265 | Glycycoumarin/isomer | RG | C21H20O6 |
| 71 | 82.0 | 353 | 297 | Licoisoflavone A | RG | C20H18O6 |
| 72 | 83.0 | 353 | 297 | Licoisoflavone A/isomer | RG | C20H18O6 |
| 73 | 83.3 | 381 | 366 | licoricone | RG | C22H22O6 |
tR, retention time;
absorbed compounds detected in dosed rat serum;
absorbed compounds detected in dosed rat brain;
identified by comparison with standards;
solvent adduct ion [M-H+CH3COOH]−; RP, radix paeoniae alba; RG, radix glycyrrhizae; RB, radix bupleuri. SS, saikosaponin; MS, mass spectrometry.
By comparing the chromatograms and MSn data between the extracts of SYC and the single herb extracts, the 73 identified components originated predominantly from Radix Bupleuri, Radix Paeoniae Alba and Radix Glycyrrhizae. In the SYC extracts, 34 compounds belonging to monoterpene glycosides, galloylglucoses and phenolic compounds were identified as being derived from Radix Paeoniae. In addition, 11 compounds belonging to flavonoids and triterpene glycosides were identified as ingredients of Radix Bupleuri, and 28 compounds were derived from Radix Glycyrrhizae. The identity of each component was confirmed by matching the empirical molecular formula with that of previously published information, and was further elucidated using multistage mass spectrometry, particularly for the unmatchable components in the in-house library. In addition, certain data, including retention time (tR), were also included as complementary data for identification. Acetic acid was added to the mobile phase as a modifier, and its adduct ions [M-H+CH3COOH]− were be observed in the mass spectra of certain components.
Identification of the components of SYC from Radix Paeoniae
In the present study, a total of 34 compounds were identified in the extracts of SYC from Radix Paeoniae. For compound 16 (tR=24.7 min), the solvent adduct ion [M-H+CH3COOH]− at 539 m/z was observed, and the featured fragmentation ions at 283, 327, 357 and 479 m/z were produced, consistent with the data reported in the literature (10). Compound 18 (tR=27.8 min) was identified as paeoniflorin by comparison with reference compounds, and produced the featured fragmentation ions at 165, 327 and 449 m/z, consistent with the data reported in the literature (10). Compounds 19 (tR=28.7 min) and 27 (tR=36.5 min) were induced as an isomer of albiflorin and paeoniflorin, respectively, due to a series of common characteristic ions (Table I). Compounds 32 (tR=45.3 min) and 33 (tR=45.7 min) were identified as isopaeoniflorin and albiflorin R1, sharing the molecular formula C23H28O11, according to literature references (11). Compounds 10 (tR=16.8 min), 11 (tR=17.9 min) and 17 (tR=27.6 min) were deduced as oxypaeoniflorin/oxypaeoniflorin isomer and ortho-oxypaeoniflorin (10), respectively, considering that oxypaeoniflorin exhibits a higher polarity than ortho-oxypaeoniflorin; and their ions in common at 495 m/z, 465 m/z ([M-H-HCHO]−) and 137 m/z ([C7H5O3]−). Compound 12 (tR=19.4 min) gave [M-H]− ions at m/z 525 (C24H30O13) and further produced an [M-H-OCH3]− ion at 495 m/z and an [M-H-OCH3-CH2OH]− ion at 465 m/z. Therefore, this species was tentatively identified as mudanpioside E, according to its structure reported in the literature (12).
The two isomer compounds, 51 (tR=54.8 min) and 52 (tR=55.3 min) (13), revealed the solvent adduct ion [M-H+CH3COOH]− at 521 m/z, the protonated molecular ion [M-H]− at 461 m/z and the ion at 265 m/z, which was produced via the neutral losses of CO2 (44 Da), HCHO (30 Da) and benzoyl acid (BA, 122 Da) from the precursor ion. Therefore, compound 51,52 was tentatively identified as lactiflorin or an isomer.
Compounds 53 (tR=57.5 min) and 55 (tR=58.4 min) were tentatively assigned as benzoylpaeoniflorin and isobenzoylpaeoniflorin (11) by the solvent adduct ion [M-H+CH3COOH]− at 643 m/z and the protonated molecular ion [M-H]− at 583 m/z, which further loses a benzoyl group (122 Da) to produce the ion [M-H-122]− at 461 m/z and at 553 m/z, and further loses a benzoyl group (122 Da) to produce the ion [M-H-HCHO-122]− at 431 m/z. Compound 54 may be the secondary product of benzoylpaeoniflorin in the plant due to the same [M-H]− at 553, 431 and 165 m/z as compound 55. Therefore, compound 54 was identified as dehydroxylate demethylene benzoylpaeoniflorin.
Compound 3 (tR=6.9 min) exhibited the [M-H]− ion at 375 m/z, and then produced the ion [M-H-HCHO]− at 345 m/z in the negative full scan mode. A further fragment at 165 m/z suggested the existence of a glucosyl group. Therefore, compound 3 was tentatively identified as desbenzoylpaeoniflorin, according to the literature and its fragmentation pathway (10). Compound 7 (tR=15.0 min) produced the ion [M-H]− at 527 m/z in the MS spectrum and product ion [M-H-HCHO]− at 497 m/z in the MS/MS spectrum. The MS3 spectral ions at 479 m/z ([M-H-HCHO-H2O]−) and 271 m/z were observed. And the ion at 271 m/z may have been produced by the losses of galloyl radicals (152 Da), 2HCHO (30 Da) and CO2 (44 Da). Therefore, compound 7 was confirmed as 6′-O-galloyldesbenzoylpae oniflorin. Compounds 28 (tR=39.0 min), 30 (tR=42.0 min) and 39 (tR=48.2 min) exhibited the [M-H]− ion at 631 m/z. Considering the common fragmentation ions at 613, 465, 313 and 169 m/z, these three components were assigned as galloylpaeoniflorin (14,15), galloylablbiflorin or their isomers; and this was consistent with the literature (10). Compounds 13 (tR=21.4 min) and 15 (tR=24.3 min) were two isomers with the solvent adduct ion [M-H+CH3COOH]− at 701 m/z. Compound 14 (tR=23.2 min) produced [M-H]− at 641 m/z. In addition, these compounds yielded a series of common ions, including [M-H-HCHO]− at 611 m/z, [M-H-H2O-HCHO]- at 593 m/z, [M-H-BA]− at 519 m/z and [M-H-HCHO-BA]− at 489 m/z. By referring to the literature, compounds 13, 15 and 14 were identified as isomaltopaeoniflorin or isomers, and 6′-O-d-glucopyranosylalbiflorin (16).
Compounds 5 (tR=11.07 min) and 6 (tR=11.64 min) produced fragmentation ions at 313 m/z [M-H-170]−, 331 m/z [M-H-152]− and 169 m/z [gallic acid]−. Their fragment ions indicated the loss of the gallic acid moiety (170 Da) and/or galloyl radicals (152 Da) from the precursors of [M-H]−; thus these two compounds were assigned as 1′-O-galloylsucrose, 6′-O-galloylsucrose. For compounds 19 (tR=33.9 min) and 20 (tR=34.5 min), fragmentation ions at 787 m/z [M-H]−, 617 m/z [M-H-170]−, 635 m/z [M-H-152]−, 483 m/z [M-H-2galloyl]− and 465 m/z [M-H-2galloyl-H2O]− were produced in negative ion mode. Compounds 19 and 20 were tentatively identified as tetragalloylglucose or an isomer. Similarly, compound 29 was tentatively deduced as pentagalloylglucose, according to the fragmentation pathway and data in the literature (17). However, their structures were not elucidated from the MS data due to limited information regarding the linkage position of the galloyl groups relative to the glucose unit.
Compounds 8 (tR=15.8 min), 9 (tR=16.5 min), 34 (tR=46.1 min), 35 (tR=46.6 min) and 42 (tR=49.6 min) were detected and matched with the data in the literature (10), revealing that they were identical to those of isomaltopaeoniflorin sulfonate, paeoniflorin sulfonate, benzoylpaeoniflorin sulfonate or their isomers.
Identification of the components of SYC derived from Radix Bupleuri
A total of six flavonoids, including baicalin, wogonoside, wogonin, apigenin-7-O-β-D-glucuronide and kaempferol or their isomers, were detected in the SYC extracts derived from Radix Bupleuri. Compounds 37 (tR=47.6 min) and 38 (tR=48.0 min) provided common [M-H]− ions at 445 m/z and the MS/MS ion [M-H-gluconic acid]− at 269 m/z. However, they exhibited different typical major ions at 251 m/z ([M-H-gluconic acid-H2O]−) and 175 m/z, respectively. Therefore, compound 38 was deduced as baicalin (16), and compound 37 was deduced as apigenin-7-O-β-D-glucuronide. Similarly, compounds 49 (tR=52.3 min) and 50 (tR=54.4 min) produced [M-H]− ions at 459 m/z, the MS2 ion at 283 m/z, corresponding to [M-H-gluconic acid]−, and the MS3 ion at 268 m/z, corresponding to [M-H-gluconic-acid-CH3]−. Accordingly, compounds 49,50 was tentatively identified as wogonoside or an isomer (16). Also, compound 67 (tR=76.5 min) was identified as wogonin, showing the [M-H]− ion at 283 m/z and the [M-H-CH3]− ion at 268 m/z (18). Another flavonoid had the characteristic fragment ion at 285 m/z ([M-H]−) and 269 m/z ([M-H-OH]−), therefore, compound 47 was identified as kaempferol.
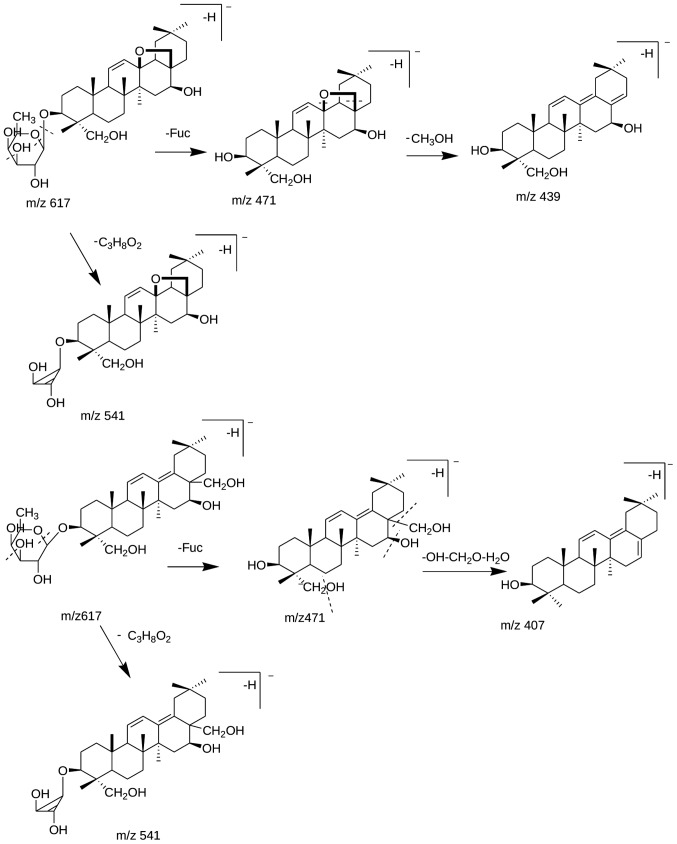
A total of five saikosaponins (SSs) from the SYC extracts were identified or tentatively identified in the negative-ion ESI mode, among which two SSs, including SSa (compound 64; tR=71.9 min) and SSd (compound 65; tR=72.4 min) were identified through comparison of their tR, and MSn data with those of the reference substances. Compound 60 (tR=65.4 min) produced the [M-H]− species at 811 m/z, and further produced the [M-H-Glc]− ion at 649 m/z and the [M-H-FucGlc-CH3OH]− ion at 471 m/z, and was tentatively identified as SS3 or SS4 (16). Compound 68 (tR=77.3 min) and compound 69 (tR=77.7 min) shared the solvent adduct ion [M-H+CH3COOH]− at 677 m/z; and the common fragmentation ions were determined at 617 m/z ([M-H]−), 541 m/z, corresponding to [M-H-C3H8O2]− and 471 m/z, corresponding to [M-H-Fuc]−. In addition, for compound 68, 439 m/z (corresponding to [M-H-Fuc-CH3OH]−), was its additional fragmentation ion; and for compound 69, 407 m/z ([M-H-Fuc-OH-CH2O-H2O]−) was its additional fragmentation ion. According to the common and additional fragmentation ions, compound 68 was tentatively deduced as prosaikogenin G or prosaikogenin F, and compound 69 was identified as prosaikogenin A or prosaikogenin D (19). The structure of compound 68/69 and the possible fragmentation patterns are summarized in Fig. 1. In addition, compounds 68 and 69 were possible secondary metabolites of the SSs in the plants during processing, storage or long-distance transport (20).
Figure 1.
Suggested fragmentation pathways for compounds 68 and 69.
Identification of the components of SYC from Radix Glycyrrhizae
A total of 28 compounds, including flavanones, isoflavanones, coumarins and saponins were detected in the SYC extracts derived from Radix Glycyrrhizae. Compounds 24 (tR=35.3 min), 26 (tR=35.9 min), 40 (tR=48.5 min), 41 (tR=48.9 min) and 43 (tR=49.5 min) exhibited the same [M-H]− ion at 417 m/z. The fragment ion at 255 m/z was observed by loss of a glucose residue (162 Da). The alycone ion at 255 m/z was then further fragmented to the product ion at 153, 135 or 119 m/z through Retro-Diels-Alder (RDA) cleavage. Thus, compounds 24, 26, 40, 41 and 43 were tentatively identified as neoliquiritin, liquiritin, isoliquiritin, neoisoliquiritin and neoisoliquiritin isomer (21).
The two isomeric compounds 23 (tR=35.2 min) and 36 (tR=47.1 min) exhibited identical MS data in the negative ion mode. With the exception of the same [M-H]− ion at 549 m/z, their MS/MS spectra exhibited [M-H-xyl]− at 417 m/z [M-H-xyl-glc]− at 255 m/z, and the product ion at 135 or 119 m/z. Thus, compound 23 was deduced as liquiritin apioside and compound 36 was tentatively identified as isoliquiritin apioside. Compound 25 (tR=35.7 min) shared a series of characteristic ions at 417, 255 and 135 m/z with liquiritin, and its molecular weight was 36 D higher than that of liquiritin, indicating that compound 25 was hydrated liquiritin.
The three isomeric compounds 44 (tR=49.7 min), 48 (tR=52.1 min) and 59 (tR=65.1 min) produced the same ion [M-H]− at 255 m/z, which was further fragmented to the product ion at 135, 153 or 119 m/z via the RDA reaction (22). Thus, compounds 44, 48 and 59 were identified as liquiritigenin, liquiritigenin isomer and isoliquiritigenin. Compounds 45 (tR=50.0 min) and 46 (tR=50.2 min) showed [M-H]− ions at 695 and 725 m/z. The two compounds exhibited the common product ion at 549, 531 and 255 m/z. In the MS/MS spectrum, compounds 45 and 46 produced [M-H-rhamnosyl]− and [M-H-176]− at 549 m/z, respectively. They subsequently produced the MS3 ion at 255 m/z by losing a glucose residue (162 Da) and an apiose residue (132 Da). Therefore, compounds 45 and 46 were identified as licorice-glycoside B and licorice-glycoside A (22).
Compound 22 (tR=34.70 min) exhibited [M-H]− at 577 m/z. In the MS/MS spectrum, the [M-H-120]− ion at 457 m/z, the [M-H-74]− ion at 503 m/z and the [M-H-120-74]− ion at 383 m/z suggested flavone C-glucosides, and these produced typical losses of 120 Da from the precursor ions; and the flavone C-rhamnosides generated the product ions by losing 74 Da. Compound 22 was deduced to be isoviolanthin (22). Compound 63 (tR=66.8 min) was identified as formononetin, according to the [M-H]− ion at 267 m/z and the [M-H-CH3]− ion at 252 m/z.
The two isomeric compounds 71 (tR=82.0 min) and 72 (tR=83.0 min) exhibited the [M-H]− ion at 353 m/z, corresponding to the product ion at 297 m/z; therefore, they were identified as licoisoflavone A or an isomer, according to the literature (23). In addition, compound 73 (tR=83.3 min) exhibited the [M-H]− ion at 381 m/z, corresponding to the product ion at 366 m/z in the MS/MS spectra. Thus, compound 73 was deduced as licoricone.
Compounds 57 (tR=61.5 min) and 58 (tR=62.1 min) exhibited the deprotonated molecule [M-H]− ion at 837 and 819 m/z, respectively. The [M-H]− ion of the two compounds fragmented into two products at 351 m/z ([glucuroglucuronic acid-H]−) and 193 m/z ([glucuronic acid-H]−). Furthermore the [M-H-gluA]− ion and the [M-H-H2O]− ion were also detected in the MS/MS spectra of the two compounds. Based upon the above fragmentation pattern and previous literature (22), compounds 57 and 58 were identified as licorice-saponin G2 and licorice-saponin E2. Compounds 61 (tR=65.5 min) and 62 (tR=66.3 min) generated [M-H]− at 821 m/z in the negative ion mode. The [M-H]− ion fragmented into two characteristic ions at 645 and 469 m/z, which corresponded to [M-H-glucuronic acid]− and [M-H-di-glucuronic acid]−, respectively. Therefore, compounds 61 and 62 were identified as glycyrrhizin and uralsaponin A, according to the literature (22).
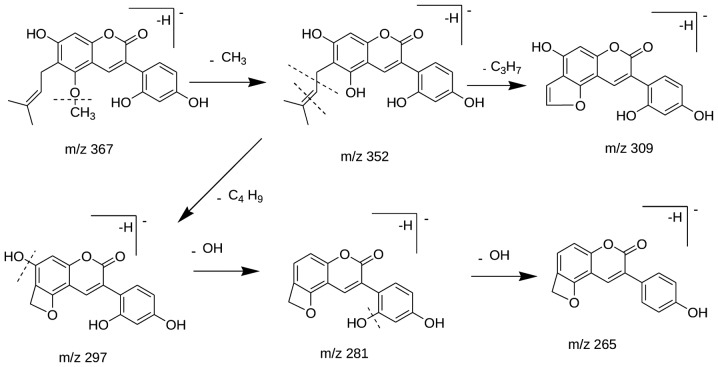
Only two coumarins were present in SYC, and these were identified as glycycoumarin. Compounds 66 (tR=72.7 min) and 70 (tR=80.4 min) exhibited the [M-H]− ion at 367 m/z, and the MS/MS spectrum produced the [M-H-CH3]− ion at 352 m/z, [M-H-CH3-C3H7]− ion at 309 m/z and [M-H-CH3-C4H0]− ion at 297 m/z. The C3H7 and C4H9 fragment ions corresponded to an isopentenyl residue. The further loss of an oxhydryl group generated an ion at 281 m/z. Compounds 66 and 70 were then identified as glycycoumarin (24). The fragmentation pathway of glycycoumarin is shown in Fig. 2.
Figure 2.
Suggested fragmentation pathway for compounds 66 and 70.
Compound 56 (tR=59.2 min) exhibited the [M-H]− ion at 271 m/z, and produced two major product ions at 151 m/z ([C7H3O4]−) and 119 m/z ([C8H7O]−), in agreement with the classic 1,3A- and 1,3B- fragments. This indicated the presence of two and one hydroxyl groups substituted on rings A and B of a flavanone, respectively, further confirming the identity of compound 56 as naringenin, a known compound in Glycyrrhizae Radix et Rhizoma (25). Compound 31 (tR=44.0 min) exhibited the deprotonated molecule [M-H]− at 433 m/z and produced predominant fragment ions at 271 m/z [M-H-glc]− in the MS2 spectra and MS3 ions at 151 and 119 m/z, which was in accordance with the fragmentation pattern of naringenin. Therefore, compound 31 was deduced as naringenin-O-glucose (26).
Identification of the absorbed components in rat serum following oral administration of SYC extracts
The absorbed components and metabolites were difficult to elucidate due to their low concentrations. In order to improve the detection sensitivity, extracted ion chromatograms (EICs) were used. First, the protonated molecular ions in Table I were used one by one to obtain EICs from the dosed rat serum, blank rat serum and SYC extract simultaneously. Subsequently, by comparing these obtained EICs, components that appeared in the dosed rat serum and SYC extract, but not in the blank rat serum, were considered to be the components absorbed into serum in the prototype. Once these components were determined as the absorbed components, they were further confirmed by carefully comparing their MS, MSn data and retention times with those in Table I. As a result, 13 prototype compounds were absorbed into the rat serum in the prototype and identified as quinic acid, desbenzoylpaeoniflorin, 6′-O-galloyldesbenzoylpaeoniflorin paeoniflorin sulfonate, albiflorin, paeoniflorin, liquiritin apioside, liquiritin, hydrated-liquiritin, galloylpaeoniflorin, galloylalbiflorin, apigenin-7-O-β-D-glucuronide and wogonoside.
Identification of the metabolites from rat serum following oral administration of SYC
Drug metabolism involves two types of enzyme-catalyzed reactions, phase I and phase II. Phase I metabolism includes oxidation, reduction, hydroxylation and desaturation; phase II metabolism includes glucuronidation, sulfation and glutathione conjugation, which may occur directly on the parent compounds, which contain appropriate structural motifs, or, as is usually the case, on functional groups added or exposed by phase I oxidation. These results increase the solubility of the drug metabolites in water, causing them to be more easily excreted from the body (27). In the present study, sulfation, glucuronidation and methylation were detected as the predominant phase II reactions, and the oxidation reaction and demethylation were observed as the predominant phase I reactions. In general, 49 compounds were considered as metabolites of SYC (Table II), in which 14 metabolites were confirmed as novel compounds.
Table II.
MS and MSn data of the identified metabolites absorbed in rat serum following oral administration of SYC in negative mode.
| No. | tRa | Formula | [M-H]−(m/z) | MSn(m/z) | Possible original compound | Identification |
|---|---|---|---|---|---|---|
| M1n | 5.6 | C22H26O11 | 465 | 345, 165, 327 | Paeoniflorin | Dehydroxylate Demethylene oxypaeoniflorin |
| M2 | 7.2 | C9H10O7S | 261 | 215, 171 | Catechin-associated | 3, 4-Dihydroxy phenylpropionic acid sulfate |
| M3n | 14.0 | C22H30O16 | 549 | 373, 197, 175 | Paeoniflorin | PaeonimetabolinIdi-glucuronide |
| M4 | 14.1 | C8H8O8S | 263 | 183, 167 | Gallic acid-associated | 4-O-Methyl gallic acid sulfate or 3-O-Methyl gallic acid sulfate |
| M5 | 16.5 | C23H28O13S | 543 | 421, 375, 259, 497 | Paeoniflorin | Paeoniflorin sulfonate |
| M6 | 18.1 | C7H6O7S | 233 | 189 | Gallic acid-associated | Protocatechuic acid-4-O-sulfate |
| M7 | 19.8 | C27H30O15 | 593 | 417, 255 | Flavonoid-associated | liquiritin glucuronide |
| M8n | 20.3 | C22H30O16 | 549 | 373, 197, 175 | Paeoniflorin | PaeonimetabolinIdi-glucuronide |
| M9 | 22.0 | C15H18O11 | 373 | 197, 175 | Paeoniflorin | PaeonimetabolinIglucuronide |
| M10 | 24.0 | C9H8O6S | 243 | 163 | Catechin-associated | m-Coumaric acid sulfate |
| M11 | 24.5 | C7H6O7S | 233 | 189 | Gallic acid-associated | Protocatechuic acid-3-O-sulfate |
| M12n | 26.8 | C17H24O10 | 387 | 211, 197, 175 | Gallic acid-associated | Trimethyl gallic acid glucuronide |
| M13n | 27.8 | C22H20O11 | 459 | 283, 267 | Flavonoid-associated | Formononetin hydroxylate glucuronide or isomer |
| M14n | 29.1 | C22H26O10 | 569Δ | 449, 327, 539, 509 | Paeoniflorin | methyl hydroxylate paeoniflorin |
| M15n | 32.1 | C21H28O13 | 487 | 311, 267 | Flavonoid-associated | Di-methyl-formononetin hydroxylate glucuronide |
| M16 | 34.3 | C26H26O12 | 529 | 353, 175 | Flavonoid-associated | Licoisoflavone A glucuronide |
| M17 | 34.7 | C21H20O10 | 431 | 255, 175, 135, 119 | Flavonoid-associated | Liquiritigenin-4, -O-glucuronide |
| M18 | 35.3 | C21H20O10 | 431 | 255, 175, 135 | Flavonoid-associated | Liquiritigenin-7-O-glucuronide |
| M19 | 35.7 | C10H12O8S | 291 | 211, 197 | Gallic acid-associated | Di-methyl C8H8O8S |
| M20 | 36.1 | C21H20O10 | 431 | 255, 175, 135 | Flavonoid-associated | Isoliquiritigenin-4, O-glucuronide |
| M21 | 36.7 | C21H20O10 | 431 | 255, 175 | Flavonoid-associated | Isoliquiritigenin-7-O-glucuronide |
| M22 | 37.3 | C21H20O11 | 447 | 271, 151 | Flavonoid-associated | Naringenin glucuronide |
| M23 | 39.9 | C7H6O7S | 233 | 189 | Gallic acid-associated | Protocatechuic acid-3-O-sulfate |
| M24 | 41.6 | C27H32O14 | 579 | 271, 151 | Flavonoid-associated | Naringenin-O-rutinoside |
| M25 | 42.4 | C13H14O8 | 297 | 175, 113 | Gallic acid-associated | Benzoyl glucuronide |
| M26 | 42.8 | C14H16O14S | 439 | 263, 121 | Gallic acid-associated | C8H8O8S glucuronide |
| M27 | 43.1 | C26H26O12 | 529 | 353, 175 | Flavonoid-associated | Na Licoisoflavone A glucuronide |
| M28 | 43.3 | C9H10O8S | 277 | 197, 169 | Gallic acid-associated | 3,4-Di-O-methyl gallic acid sulfate |
| M29 | 43.9 | C21H20O11 | 447 | 271, 151 | Flavonoid-associated | Naringenin glucuronide |
| M30 | 44.5 | C27H30O16 | 609 | 301, 286, 242 | Flavonoid-associated | Hesperidin |
| M31 | 45.7 | C15H12O7S | 335 | 255, 135, 119 | Flavonoid-associated | Liquiritigenin-4, -O-sulfate |
| M32 | 45.9 | C15H12O7S | 335 | 255, 135, 119 | Flavonoid-associated | Liquiritigenin-7-O-sulfate |
| M33 | 46.7 | C17H22O11 | 401 | 225 | Uncertain | Methyl propyl gallate glucuronide |
| M34n | 49.0 | C23H22O11 | 473 | 297, 253 | Flavonoid-associated | Methyl formononetin hydroxylate glucuronide |
| M35 | 50.3 | C21H20O10 | 431 | 255, 135, 119, 175 | Flavonoid-associated | Isoliquiritigenin glucuronide isomer |
| M36n | 50.4 | C15H18O11 | 453 | 277, 233 | Gallic acid-associated | C9H10O5 glucuronide |
| M37 | 51.1 | C17H12O8 | 343 | 229, 165, 149 | Ellagic acid | 3,7,8-Trimethyl ellagic acid |
| M38 | 51.3 | C21H22O13 | 433 | 257, 175 | Flavonoid-associated | Davidigenin glucuronide |
| M39 | 51.5 | C17H12O8 | 343 | 229, 149, 165 | Ellagic acid | 3,7,8-Trimethyl ellagic acid |
| M40n | 52.1 | C30H48O3 | 455 | 437 | Saponin-associated | Saikogenin B |
| M41 | 54.8 | C22H24O15S | 559 | 515, 383, 339 | Coumarin-associated | Glycycoumarin hydroxylate glucuronide |
| M42n | 54.9 | C22H20O11 | 459 | 283, 267 | Flavonoid-associated | Formononetin hydroxylate glucuronide or isomer |
| M43 | 56.6 | C16H12O7S | 347 | 267, 252 | Flavonoid-associated | Formononetin sulfate |
| M44 | 57.0 | C16H12O7S | 347 | 267, 252 | Flavonoid-associated | Formononetin sulfate |
| M45n | 58.3 | C26H28O11 | 515 | 353 | Flavonoid-associated | Licoisoflavone A glucose |
| M46n | 61.3 | C15H22O9 | 405b | 345, 327 | Paeoniflorin | Dehydroxylate Demethylene desbenzoylpaeoniflorin |
| M47n | 61.4 | C15H22O9 | 405b | 345, 327 | Paeoniflorin | Dehydroxylate Demethylene desbenzoylpaeoniflorin |
| M48 | 67.0 | C16H14O9 | 349 | 303, 287 | Flavonoid-associated | Dihydroxyl methyl quercetin-chalcone |
| M49 | 71.6 | C21H20O9S | 447 | 367 | Coumarin-associated | Glycycoumarin sulfate |
tR, retention time;
solvent adduct ion [M-H+CH3COOH]−; n, novel metabolites of certain constituents of SYC; SYC, Shu-Yu capsule; MS, mass spectrometry.
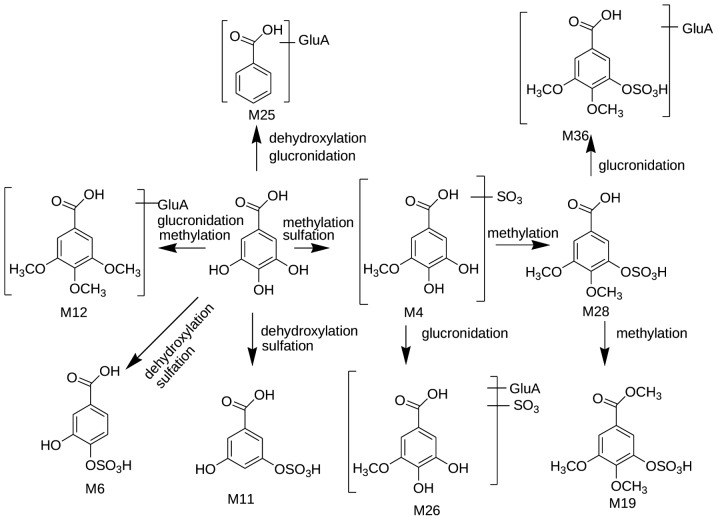
Identification of gallic acid-associated metabolites
In the present study, a total of 10 compounds were identified as metabolites of gallic acid or polyphenols, which can be degraded into gallic acid in the body. The possible metabolic pathways of gallic acid and the associated compounds are presented in Fig. 3.
Figure 3.
Suggested metabolic pathways of gallic acid-associated metabolites following oral administration with Shu-Yu capsule.
M4 exhibited the [M-H]− ion at 263 m/z, which yielded the MS/MS ion at 183 m/z with a loss of 80 Da (SO3), and the MS3 ion at 167 m/z, suggesting that gallic acid underwent the sulfation and methylation reactions. Therefore, M4 was tentatively identified as 4-O-methyl gallic acid sulfate or 3-O-methyl gallic acid sulfate. M26 exhibited the [M-H]− ion at 439 m/z and yielded the product ions at 263 m/z, corresponding to a loss of 176 Da. Thus, M26 was tentatively identified as C8H8O8S glucuronide. M28 showed an [M-H]− ion at 277 m/z, the product ion at 197 m/z and the MS3 ion at 169 m/z with a loss of 28 Da via the MS2 spectrum, suggesting that M28 was a sulfate conjugate of dimethyl gallic acid. In addition, M19 exhibited the [M-H]− ion at 291 m/z, which is 28 Da higher than M4, and the product ion at 211 m/z with a loss of 80 Da (SO3), suggesting that they were sulfate conjugates, indicating that M19 was the dimethyl conjugate of M4. M36 exhibited the [M-H]− ion at 453 m/z and the product ion at 277 m/z with a loss of 176 Da, indicating that M36 was the glucuronide conjugate of M28.
M6 and M11 exhibited an [M-H]− ion at 233 m/z, which produced product ions at 189 and 153 m/z, suggesting that they were sulfate conjugates. Thus, M6 was determined as protocatechuic acid-4-sulfate, whereas M11 was identified as protocatechuic acid-3-sulfate, with reference to previous reports (15). M12 exhibited the [M-H]− ion at 387 m/z and yielded product ions at 211, 197 and 175 m/z, indicating that M12 was a glucuronide conjugate of the three methylation products of gallic acid. M25 exhibited the [M-H]− ion at 297 m/z and the product ion at 121 m/z with a loss of 176 Da, indicating that M25 was the glucuronide conjugate of benzoic acid.
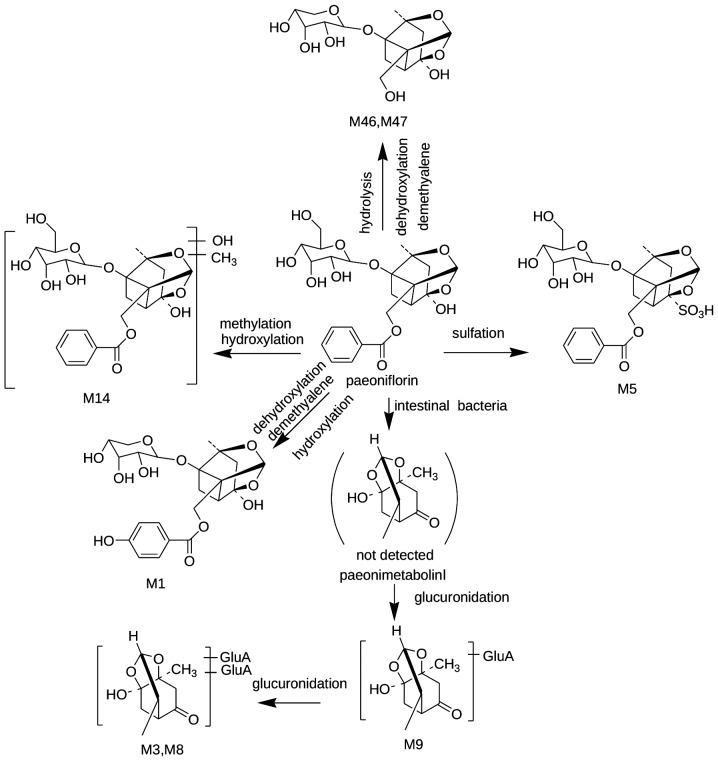
Identification of paeoniflorin-associated metabolites
In the present study, a total of eight compounds detected in the rat serum were tentatively assigned as metabolites originating from paeoniflorin, which may also be the metabolites of other paeoniflorin-associated compounds, including desbenzoylpaeoniflorin or oxypaeoniflorin. With the exception of M5 (paeoniflorin sulfonate) and M9 (paeonimetabolinIglucuronide), the other six metabolites were identified for the first time as metabolites of paeoniflorin. The suggested metabolic pathways for paeoniflorin are shown in Fig. 4.
Figure 4.
Suggested metabolic pathways of paeoniflorin-associated metabolites following oral administration with Shu-Yu capsule.
M9 exhibited the MS2 spectra [aglycon-H]− and [glucuronyl-H]− at 197 and 175 m/z, and we tentatively assigned as paeonimetabolin I glucuronides. M3 and M8 shared with M9 a series of characteristic ions at 175, 197 and 373 m/z. In addition, M3 and M8 were 176 Da higher than M9. Thus, M3 and M8 were assigned as glucuronide conjugates of M9.
Paeoniflorin sulfate absorbed M5 into the serum, and may be the metabolite of paeoniflorin through sulfation in vivo. A proportion of the paeoniflorin sulfate was most likely absorbed into the rat serum in the prototype from the SYC extract.
M14 exhibited the solvent adduct ion [M-H+CH3COOH]− at 569 m/z and shared the same characteristic ions at 165, 327, 449 and 479 m/z with paeoniflorin. In addition, compared with the molecular weight of paeoniflorin, these other species were 30 Da higher via methylation and hydroxylation reactions. Thus M14 was tentatively identified as methyl hydroxylate paeoniflorin. M1, M46 and M47 shared the same MS/MS fragmentation ion at 345 m/z and a further fragmentation ion at 327 m/z with a loss of 18 Da. In addition, M46 and M47 exhibited the solvent adduct ion [M-H +CH3COOH]− at 405 m/z, and M1 exhibited the deprotonated ion [M-H]− at 465 m/z. Based on the information presented above, M1 was tentatively identified as dehydroxylate demethylene oxypaeoniflorin. M46/M47 was tentatively identified as dehydroxylate demethylene desbenzoylpaeoniflorin or its isomer.
Identification of flavonoid-associated metabolites
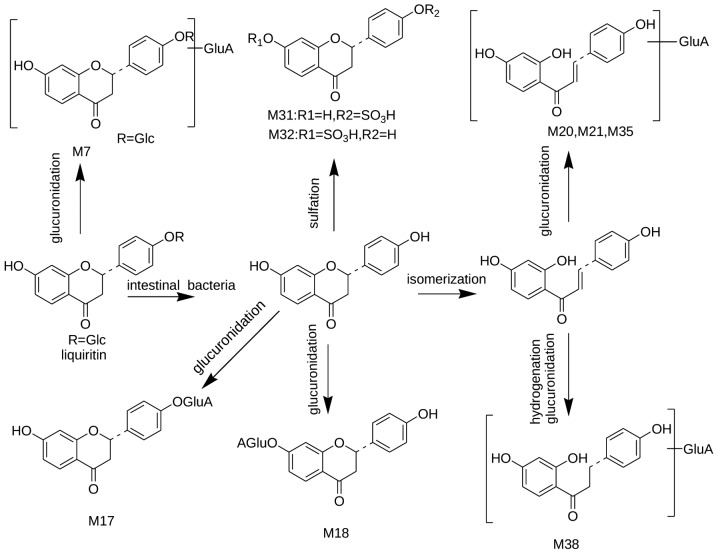
A total of 19 compounds were identified as metabolites originating from dihydroflavone, flavanones and isoflavonoids, including liquiritin, kaempferol, licoisoflavone A and formononetin. Their LC-MSn data are summarized in Table II, and the possible metabolic pathways of liguiritin are presented in Figs. 5 and 6.
Figure 5.
Suggested metabolic pathway of liquiritin following oral administration with Shu-Yu capsule.
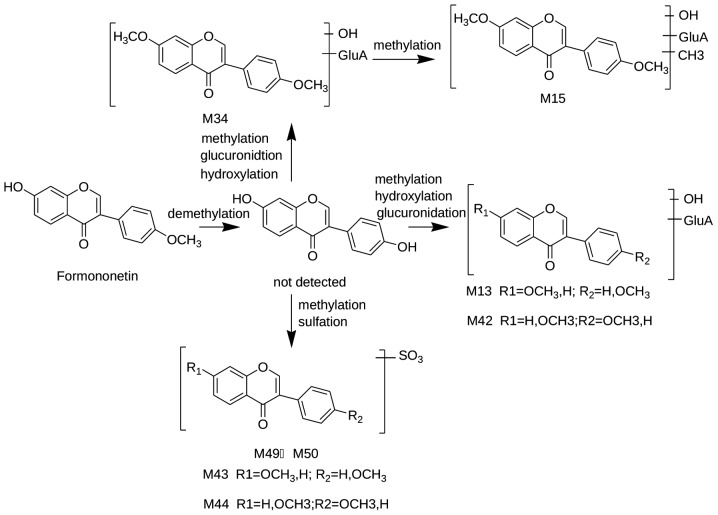
Figure 6.
Suggested metabolic pathways of formononetin following oral administration with Shu-Yu capsule.
Liquiritin was first metabolized into its aglycone liquiritigenin, and liquititigenin was then conjugated with glucuronide and sulfate. Isomerization into chalcones was also common for liquiritin. The present study revealed that M7, M17, M18, M31 and M32 were detected as major metabolites of liquiritin; and M20, M21, M38 and M35 were assigned as major metabolites of isoliquiritin. M17, M18, M20, M21 and M35 exhibited identical protonated molecules at 431 m/z, which produced identical product ions at 255, 135 and 119 m/z with a loss of 176 Da, indicating that they may be the glucuronide conjugates of liquiritigenin and isoliquiritigenin. However, the corresponding retention times differed markedly. According to the literature (23), the present study tentatively identified M17 and M18 as liquiritigenin-4′-O-glucuronide and liquiritigenin-7-O-glucuronide, respectively; and another three metabolites were identified as the glucuronide conjugates of isoliquiritigenin. Further investigation is required to determine the detailed structure of the metabolites of isoliquiritigenin. Based on the characteristic neutral loss of 80 Da and previous information (28), M31 and M32 were tentatively identified as liquiritigenin-4-O-sulfate and liquiritigenin-7-O-sulfate, respectively.
M7 exhibited the [M-H]− ion at 593 m/z, which further lost a glucuronic acid moiety (176 Da) to produce the ion at 417 m/z [M-H-176]− and 255 m/z [M-H-176-162]−, indicating that M8 was the glucuronide conjugate of liquiritin. M38 exhibited the [M-H]− ion at 433 m/z and the further product ion at 257 m/z [M-H-176]− and 175 m/z, suggesting it to be the glucuronide conjugate of davidigenin.
Formononetin has been well documented to be metabolized to daidzein (29), which is 14 Da (CH2) less than the protonated ion of formononetin. In the present study, daidzein was then conjugated with methylate, hydroxylate, sulfate and glucuroni-date. M13 and M42 exhibited the [M-H]− ion at 459 m/z, the MS2 ion at 283 m/z and the MS3 ion at 267 m/z, with a loss of 16 Da, compared with the MS2 ion. The MS2 ion was 176 Da less than the protonated ion of M13 and M42. Therefore, M13/M42 was assigned as formononetin hydroxylate glucuronide or its isomer. M43 and M48 had the same neutral loss of 80 Da in the negative ion mass spectrum, indicating that they were the sulfated conjugates of formononetin or isomers. M15 and M34 were identified as glucuronide conjugates due to the characteristic neutral loss of 176 Da (glucuronic acid). The MS2 ion at 311 m/z of M15 and the MS2 ion at 297 m/z of M34 were 44 and 30 Da higher, compared with the weight of formononetin, respectively, indicating that M15 and M34 may be formononetin or daidzein following methylation and hydroxylation. Therefore, M15 and M34 were identified as di-methyl-formononetin hydroxylate glucuronide and methyl formononetin hydroxylate glucuronide, respectively. The possible metabolic pathways of the formononetin-associated compounds are presented in Fig. 6.
M16 and M27 produced fragment ions at 529 and 353 m/z, corresponding to a loss of 176 Da. Therefore, they were tentatively identified as glucuronide conjugates of licoisoflavone A. M45 exhibited the [M-H]− ion at 515 m/z and the product ion at 353 m/z with a loss of 162 Da (glucose-H2O). Thus, M45 was identified as licoisoflavone A glucose.
M24 exhibited the [M-H]− ion at 579 m/z, and M22 and M29 exhibited the [M-H]− ion at 447 m/z; and they exhibited the same fragment ion, at 271 and 151 m/z, as naringenin. Their MS/MS spectra produced the [M-H-308]− ion at 271 m/z of M24, and the [M-H-176]− ion at 271 m/z of M22 and M29. Therefore, M24 was tentatively identified as naringenin-7-rutinoside, and M22, M29 was tentatively identified as naringenin-7-O-glucuronide or naringenin-4′-O-glucuronide. M30 exhibited the [M-H]− ion at 609 m/z, the MS2 ion [M-H-308]− ion at 301 m/z and the MS3 ion [M-H-308-CH3]− ion at 286 m/z, which were consistent with previous reports (30). Therefore, M30 was tentatively identified as hesperidin. M48 exhibited the [M-H]− ion at 349 m/z, the [M-H-CH2O2]− ion at 303 m/z in the MS2 spectrum and the [M-H-CH2O2-OH]− ion at 287 m/z in the MS3 spectrum, indicating that M48 may be the metabolite of the parent compound, quercetin. As a result, M48 was identified as dihydroxyl methyl quercetin-chalcone.
Identification of saponin-associated metabolites
M40 exhibited an [M-H]− ion at 455 m/z, which produced a product ion at 437 m/z with a loss of 18 Da. Consistent with the literature (20), the heteroannular and the homoannular saikogenins showed different characteristic fragment ions of the deprotonated molecules, including the ions [M-H-H2O]−, [M-H-CH3OH]− or [M-H-CH2O-H2O]−. In addition, the fragmentations observed at [M-H-H2O]− were most abundant for SGH (471 m/z) and SGB (455 m/z), [M-H-CH3OH]− for SGD (471 m/z) and [M-H-CH2O-H2O] for SGA (471 m/z) and SGC (455 m/z) in the negative ion MS/MS spectra. Therefore, the M40 metabolite was characterized as SGB.
Identification of coumarin-associated metabolites
M41 produced a [M-H]− ion at 559 m/z, which produced a product ion at 383 m/z with a loss of 176 Da, indicating that M41 was a glucuronide conjugate. In addition, the product ion at 383 m/z was 16 Da higher, compared with that of the molecular weight of glycycoumarin, which was tentatively assigned to hydroxylation reactions of glycycoumarin. Thus, M41 was identified as glycycoumarin hydroxylate glucuronide, which corroborates results from the literature (31).
M49 yielded a [M-H]− ion at 447 m/z, which produced a product ion at 367 m/z with a loss of 80 Da (SO3), indicating that M49 was a sulfate conjugate of glycycoumarin, according to the literature (31). Thus, M49 was tentatively identified as glycycoumarin sulfate.
Discussion
In the present study, an HPLC-ESI-MSn method was developed and applied to analyze the herbal components of SYC extracts, and the absorbed compounds and metabolites in rat serum following oral administration of SYC extracts. As a result, a total of 73 herbal components, including 28 monoterpenes, 26 flavonoids, 9 triterpenoids, 2 coumarins, and other phenolic compounds and galloyl glucoses were observed and tentatively identified. All the MSn data from these compounds in SYC were consistent with previous literature for every herb (10,18). The compounds identified from the formulation provided information to support further investigation on the absorbed components and metabolites of SYC in rat serum.
The absorbed components and metabolites identified in the present study provide an overall understanding of the absorption and metabolism of SYC in the rat body. Flavonoids were the most abundant metabolites in the drug-containing serum, and a total of 28 flavonoids, including five parent compounds and 23 metabolites, were identified in the drug-containing serum. Monoterpene glycoside compounds were found to be another primary absorbed component, including seven parent compounds and eight metabolites in the dosed serum were found. A total of eight metabolites were produced from gallic acid-related compounds and two metabolites were produced from the coumarin derivates. These results revealed that certain flavonoid glycosides and monoterpene glycosides were absorbed directly. Glucuronidation and sulfation were the predominant metabolic pathways of the components in SYC. In addition, it appeared that certain phase I reactions, including hydrolysis, demethylation and hydroxylation, also occurred.
Saponins are important in SYC, particularly the SSs, which were characterized with poor oral bioavailability. The majority of the SSs contained the unstable XIII, 28-oxide linkage, which may be hydrolyzed during extraction by organic acid or upon heating (32). Therefore, SSs exert curative effects by sequential deglycosylation metabolism in the intestine to form secondary glycoside and aglycones with improved pharmacological effects.
As is already known, TCM contains complex chemical constituents, which are directly absorbed into the blood, or indirectly absorbed via digestive intake or liver metabolism, and are then transported to target tissue through the circulation and exert effects on target tissue. The complexity and diversity of parent compounds and metabolites in the serum following the administration of TCM conforms to the theory of integrity and the synergistic effect of TCM, which is the material basis of the pharmacological actions. However, further investigations are required to clarify such pharmacological actions.
A simple and economical HPLC-ESI-MSn method was established in the present study, and led to the first report, to the best of our knowledge, on the comprehensive determination of chemical constituents in SYC, as well as its metabolites in rat serum. The results of the present study provide a basis and theoretical foundation for clarification of the chemical composition and potential bioactive compounds of SYC. The results of the present study provided useful information for the further investigation of the pharmacology and mechanism of action of SYC.
Acknowledgments
This study was supported by a grant from the 973 Program of China (grant no. 2011CB-505100).
Abbreviations
- SYC
Shu-Yu capsule
- TCM
traditional Chinese medicine
- HPLC-ESI-MSn
high performance liquid chromatography-electrospray ionization tandem mass spectrometry
References
- 1.Gao X, Sun P, Qiao M, Wei S, Xue L, Zhang H. Shu-Yu capsule, a traditional chinese medicine formulation, attenuates premenstrual syndrome depression induced by chronic stress constraint. Mol Med Rep. 2014;10:2942–2948. doi: 10.3892/mmr.2014.2599. [DOI] [PubMed] [Google Scholar]
- 2.Chen SL, Chen DX. Clinical application of combined use of Radix Bupleuri and Radix Paeoniae Alba. Shanghai Journal of TCM. 2007;41:71–74. In Chinese. [Google Scholar]
- 3.Mao QQ, Xian YF, Ip SP, Tsai SH, Che CT. Long-term treatment with peony glycosides reverses chronic unpredictable mild stress-induced depressive-like behavior via increasing expression of neurotrophins in rat brain. Behav Brain Res. 2010;210:171–177. doi: 10.1016/j.bbr.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Kwon S, Lee B, Kim M, Lee H, Park HJ, Hahm DH. Antidepressant-like effect of the methanolic extract from Bupleurum falcatum in the tail suspension test. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:265–270. doi: 10.1016/j.pnpbp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Su GY, Yang JY, Wang F, Ma J, Zhang K, Dong YX, Song SJ, Lu XM, Wu CF. Antidepressant-like effects of xiaochaihutang in a rat model of chronic unpredictable mild stress. J Ethnopharmacol. 2014;152:217–226. doi: 10.1016/j.jep.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Song CH, Li F, Guo YH, Zhang HY. Effects of Shuyu capsule and its main composition on protein expression and function of CACNA1C in hippocampus of rat with PMS liver-qi stagnation. Chinese Pharmacological Bulletin. 2014;30:1476–1477. In Chinese. [Google Scholar]
- 7.Wang XJ, Sun WJ, Sun H, Lv H, Wu Z, Wang P, Liu L, Cao H. Analysis of the constituents in the rat plasma after oral administration of Yin Chen Hao Tang by UPLC/Q-TOF-MS/MS. J Pharm Biomed Anal. 2008;46:477–490. doi: 10.1016/j.jpba.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Ge QF. Master's degree thesis. Shandong University of Traditional Chinese Medicine; 2012. Effects of ShuYu capsule on the expression of 5-HT3 receptor and its signal pathway of 5-HTR3-Ca2+-CaM in rat hippocampus neurons; pp. 32–33. In Chinese. [Google Scholar]
- 9.National Research Council . Guide for the Care and Use of Laboratory Animals. Washington, DC: The National Academies Press; 1996. [DOI] [Google Scholar]
- 10.Li SL, Song JZ, Choi FF, Qiao CF, Zhou Y, Han QB, Xu HX. Chemical profiling of Radix Paeoniae evaluated by ultra-performance liquid chromatography/photo-diode-array/quadrupole time-of-flight mass spectrometry. J Pharm Biomed Anal. 2009;49:253–266. doi: 10.1016/j.jpba.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Braca A, Kiem PV, Yen PH, Nhiem NX, Quang TH, Cuong NX, Minh CV. New monoterpene glycosides from Paeonia lactiflora. Fitoterapia. 2008;79:117–120. doi: 10.1016/j.fitote.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Lin HC, Ding HY, TW TS, Wu PL. Monoterpene glycosides from Paeonia suffruticosa. Phytochemistry. 1996;41:237–242. doi: 10.1016/0031-9422(95)00526-9. [DOI] [Google Scholar]
- 13.Su J, Zhang P, Zhang JJ, Qi XM, Wu YG, Shen JJ. Effects of total glucosides of paeony on oxidative stress in the kidney from diabetic rats. Phytomedicine. 2010;17:254–260. doi: 10.1016/j.phymed.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Liang J, Xu F, Zhang YZ, Huang S, Zang XY, Zhao X, Zhang L, Shang MY, Yang DH, Wang X, Cai SQ. The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MS(n) technique: A novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J Pharm Biomed Anal. 2013;83:108–121. doi: 10.1016/j.jpba.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 15.Xiao S, Luo K, Wen X, Fan X, Cheng Y. A pre-classification strategy for identification of compounds in traditional Chinese medicine analogous formulas by high-performance liquid chromatography-Mass spectrometry. J Pharm Biomed Anal. 2014;92:82–89. doi: 10.1016/j.jpba.2013.12.042. [DOI] [PubMed] [Google Scholar]
- 16.Yang YY, Tang YZ, Fan CL, Luo HT, Guo PR, Chen JX. Identification and determination of the saikosaponins in Radix bupleuri by accelerated solvent extraction combined with rapid-resolution LC-MS. J Sep Sci. 2010;33:1933–1945. doi: 10.1002/jssc.201000100. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Qi J, Chang YX, Zhu D, Yu B. Identification and determination of the major constituents in Traditional Chinese Medicinal formula Danggui-Shaoyao-San by HPLC-DAD-ESI-MS/MS. J Pharm Biomed Anal. 2009;50:127–137. doi: 10.1016/j.jpba.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 18.Wang DD, Liang J, Yang WZ, Hou JJ, Yang M, Da J, Wang Y, Jiang BH, Liu X, Wu WY, Guo DA. HPLC/q TOF-MS-oriented characteristic components data set and chemometric analysis for the holistic quality control of complex TCM preparations: Niuhuang Shangqing pill as an example. J Pharm Biomed Anal. 2014;89:130–141. doi: 10.1016/j.jpba.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 19.Bao Y, Li C, Shen H, Nan F. Determination of saikosaponin derivatives in Radix bupleuri and in pharmaceuticals of the chinese multiherb remedy xiaochaihutang using liquid chromatographic tandem mass spectrometry. Anal Chem. 2004;76:4208–4216. doi: 10.1021/ac0499423. [DOI] [PubMed] [Google Scholar]
- 20.Liang Z, Oh K, Wang Y, Yi T, Chen H, Zhao Z. Cell type-specific qualitative and quantitative analysis of saikosaponins in three Bupleurum species using laser microdissection and liquid chromatography-quadrupole/time of flight-mass spectrometry. J Pharm Biomed Anal. 2014;97:157–165. doi: 10.1016/j.jpba.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Yan Y, Chai CZ, Wang DW, Yue XY, Zhu DN, Yu BY. HPLC-DAD-Q-TOF-MS/MS analysis and HPLC quantitation of chemical constituents in traditional Chinese medicinal formula Ge-Gen decoction. J Pharm Biomed Anal. 2013;80:192–202. doi: 10.1016/j.jpba.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Chen L, Leng J, Chen P, Fan X, Cheng Y. Fragment ion diagnostic strategies for the comprehensive identification of chemical profile of Gui-Zhi-Tang by integrating high-resolution MS, multiple-stage MS and UV information. J Pharm Biomed Anal. 2014;98:22–35. doi: 10.1016/j.jpba.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Yan Z, Chen Y, Li T, Zhang J, Yang X. Identification of metabolites of Si-Ni-San, a traditional Chinese medicine formula, in rat plasma and urine using liquid chromatography/diode array detection/triple-quadrupole spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;885–886:73–82. doi: 10.1016/j.jchromb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, He S, Cheng X, Lu Y, Zou Y, Zhang Q. UPLC-Q-TOF-MS/MS fingerprinting of traditional Chinese formula SiJunZiTang. J Pharm Biomed Anal. 2013;80:24–33. doi: 10.1016/j.jpba.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Xiang C, Qiao X, Ye M, Guo DA. Classification and distribution analysis of compounds in Glycyrrhiza using licorice compounds database. Yao Xue Xue Bao. 2012;47:1023–1030. In Chinese. [PubMed] [Google Scholar]
- 26.Zhang W, Saif MW, Dutschman GE, Li X, Lam W, Bussom S, Jiang Z, Ye M, Chu E, Cheng YC. Identification of chemicals and their metabolites from PHY906, a Chinese medicine formulation, in the plasma of a patient treated with irinotecan and PHY906 using liquid chromatography/tandem mass spectrometry (LC/MS/MS) J Chromatogr A. 2010;1217:5785–5793. doi: 10.1016/j.chroma.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holcapek M, Kolárová L, Nobilis M. High-performance liquid chromatography-tandem mass spectrometry in the identification and determination of phase I and phase II drug metabolites. Anal Bioanal Chem. 2008;391:59–78. doi: 10.1007/s00216-008-1962-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu LL, Shu Y, Qian DW, Su SL, Duan JA, Qian YF, Xue CF. Identification of the metabolites of Sinisan extract in rat plasma, urine, feces and bile after intragastric administration. Acta Pharm Sin. 2011;46:1374–1379. In Chinese. [PubMed] [Google Scholar]
- 29.Tolleson WH, Doerge DR, Churchwell MI, Marques MM, Roberts DW. Metabolism of biochanin A and formononetin by human liver microsomes in vitro. J Agric Food Chem. 2002;50:4783–4790. doi: 10.1021/jf025549r. [DOI] [PubMed] [Google Scholar]
- 30.Su ZH, Zou GA, Preiss A, Zhang HW, Zou ZM. Online identification of the antioxidant constituents of traditional Chinese medicine formula Chaihu-Shu-Gan-San by LC-LTQ-Orbitrap mass spectrometry and microplate spectrophotometer. J Pharm Biomed Anal. 2010;53:454–461. doi: 10.1016/j.jpba.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Qiao X, Ye M, Xiang C, Wang Q, Liu CF, Miao WJ, Guo DA. Analytical strategy to reveal the in vivo process of multi-component herbal medicine: A pharmacokinetic study of licorice using liquid chromatography coupled with triple quadrupole mass spectrometry. J Chromatogr A. 2012;1258:84–93. doi: 10.1016/j.chroma.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 32.Huang HQ, Zhang X, Xu ZX, Su J, Yan SK, Zhang WD. Fast determination of saikosaponins in Bupleurum by rapid resolution liquid chromatography with evaporative light scattering detection. J Pharm Biomed Anal. 2009;49:1048–1055. doi: 10.1016/j.jpba.2009.01.011. [DOI] [PubMed] [Google Scholar]