Summary
Background
Active pulmonary tuberculosis is difficult to diagnose and treatment response is difficult to effectively monitor. A WHO consensus statement has called for new non-sputum diagnostics. The aim of this study was to use an integrated multicohort analysis of samples from publically available datasets to derive a diagnostic gene set in the peripheral blood of patients with active tuberculosis.
Methods
We searched two public gene expression microarray repositories and retained datasets that examined clinical cohorts of active pulmonary tuberculosis infection in whole blood. We compared gene expression in patients with either latent tuberculosis or other diseases versus patients with active tuberculosis using our validated multicohort analysis framework. Three datasets were used as discovery datasets and meta-analytical methods were used to assess gene effects in these cohorts. We then validated the diagnostic capacity of the three gene set in the remaining 11 datasets.
Findings
A total of 14 datasets containing 2572 samples from 10 countries from both adult and paediatric patients were included in the analysis. Of these, three datasets (N=1023) were used to discover a set of three genes (GBP5, DUSP3, and KLF2) that are highly diagnostic for active tuberculosis. We validated the diagnostic power of the three gene set to separate active tuberculosis from healthy controls (global area under the ROC curve (AUC) 0·90 [95% CI 0·85–0·95]), latent tuberculosis (0·88 [0·84–0·92]), and other diseases (0·84 [0·80–0·95]) in eight independent datasets composed of both children and adults from ten countries. Expression of the three-gene set was not confounded by HIV infection status, bacterial drug resistance, or BCG vaccination. Furthermore, in four additional cohorts, we showed that the tuberculosis score declined during treatment of patients with active tuberculosis.
Interpretation
Overall, our integrated multicohort analysis yielded a three-gene set in whole blood that is robustly diagnostic for active tuberculosis, that was validated in multiple independent cohorts, and that has potential clinical application for diagnosis and monitoring treatment response. Prospective laboratory validation will be required before it can be used in a clinical setting.
Funding
National Institute of Allergy and Infectious Diseases, National Library of Medicine, the Stanford Child Health Research Institute, the Society for University Surgeons, and the Bill and Melinda Gates Foundation.
Introduction
Tuberculosis is a worldwide public health issue, with 9·6 million new infections and 1·5 million deaths in 2014.1 Despite advances in diagnosis and treatment, there is still a large burden of disease, and accurate diagnosis of tuberculosis remains difficult. Traditional methods such as tuberculin skin testing and interferon γ release assays are unable to distinguish between latent tuberculosis and active tuberculosis, and have reduced sensitivity in HIV-positive patients.2 The more recently developed Xpert MTB/RIF assay has greatly improved diagnostic power, but is associated with reduced accuracy in HIV-positive patients and is not useful for monitoring treatment response.3,4 Further, Xpert MTB/RIF relies on induced sputum, which is difficult to obtain from adults after symptomatic improvement and from paediatric patients at any time.
In addressing the need for better diagnostics, WHO recently released a consensus target product profile defining the ideal features that a new diagnostic should have.5 The WHO target product profile highlighted a strong need for a new diagnostic with excellent sensitivity that (1) uses non-sputum samples (such as blood), (2) maintains an overall sensitivity of greater than 80% in patients with HIV co-infection, (3) attains a sensitivity of greater than 66% in children with culture-positive tuberculosis, and (4) is relatively simple to run.
Several studies have investigated the host response to pulmonary tuberculosis infection with microarray-based whole genome expression profiles in peripheral blood. However, the results from these studies have not translated into clinical practice so far, because of poor generalisability. For instance, seven different studies have proposed ten different gene signatures (with small amounts of overlap) to distinguish active tuberculosis from other diseases or latent tuberculosis in children and adults.6–12 Many of these studies have now been deposited in publically accessible databases such as the National Institutes of Health Gene Expression Omnibus (NIH GEO), allowing their re-use for further analysis.
We hypothesised that integration of gene expression data from heterogeneous patient populations with active tuberculosis across a wide variety of ages, countries, and inclusion criteria would yield a set of conserved genes that are indicative of active tuberculosis and could be generalised across cohorts. Therefore the aim of this study was to use an integrated multicohort analysis of samples from publically available datasets to derive a diagnostic gene set in the peripheral blood of patients with active tuberculosis.
Methods
Systematic search and multicohort analysis
We searched two public gene expression microarray repositories (NIH GEO and ArrayExpress) for all human gene expression datasets that matched any of the following search terms: tuberculosis, TB, and mycobact[wildcard]. We retained datasets that examined clinical cohorts of active pulmonary tuberculosis infection in whole blood for further study, and excluded datasets that examined only vaccine response, were done only in cell culture, used on-chip two-sample arrays, or were done in tissues or were obtained from sample types other than whole blood. The remaining 14 datasets contained 2572 samples from 10 countries from both adult and paediatric patients (table). All latent tuberculosis and other diseases samples were Mycobacterium tuberculosis smear-negative and culture-negative; culture status of patients with active tuberculosis are defined per dataset.
Table.
Summary table of all datasets that matched inclusion criteria (whole blood, clinically active pulmonary tuberculosis)
| Year | Reference | Platform | Use | Country | Age | HIV Status |
Active tuberculosis culture or smear |
Healthy controls |
Latent tuberculosis |
Other disease |
Active tuberculosis |
Treatment | Total | Miscellaneous | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSE19491 | 2010 | Berry8 | GPL6947 | Discovery | South Africa, UK, USA |
Adults | Negative | Positive | 86 | 69 | 193 | 31 | .. | 409 | Other disease breakdown: 28 ASLE, 82 PSLE, 31 Still’s, 52 Streptococcus and/or Staphylococcus infection; post-treatment samples not used. |
| GSE25534 | 2010 | Maertzdorf30 | GPL1708 | Validation | South Africa |
Adults | Negative | Positive | 6 | 19 | .. | 19 | .. | 44 | Two-colour array (on-chip comparisons between healthy controls, latent tuberculosis, and active tuberculosis) |
| GSE28623 | 2011 | Maertzdorf22 |
GPL4133/ GPL6480 |
Validation | The Gambia |
Adults | Negative | Positive | 37 | 25 | .. | 46 | .. | 108 | .. |
| Cliff Combined Dataset |
2013 | Cliff13 | GPL570 | Validation | South Africa |
Adults | Negative | Positive | .. | .. | .. | 36 | 117 | 153 | Treatment measured at 1, 2,4, and 26 weeks |
| GSE34608 | 2012 | Maertzdorf24 |
GPL4133/ GPL6480 |
Validation | Germany | Adults | Negative | Positive | 18 | .. | 18 | 8 | .. | 44 | Other diseases all sarcoid |
| GSE37250 | 2014 | Kaforou7 | GPL10558 | Discovery | Malawi, South Africa |
Adults | Positive and negative |
Positive | .. | 167 | 175 | 195 | .. | 537 | See reference for other disease distributions; 194 patients with other diseases reported but only 175 available with microarrays. |
| GSE39939 | 2014 | Anderson6 | GPL10558 | Validation | Kenya | Child- ren |
Positive and negative |
Positive and negative |
.. | 14 | 64 | 44 negative, 35 positive |
.. | 157 | Other diseases breakdown: 33 pneumonia, 5 sepsis, 7 malnutrition, 19 other |
| GSE39940 | Anderson6 | Validation | Malawi, South Africa |
Child- ren |
Positive and negative |
Positive | .. | 54 | 169 | 111 | .. | 334 | Other diseases breakdown: 86 pneumonia, 8 CLD, 11 URI, 34 other infections, 12 malignancy, 18 other |
||
| GSE40553 | 2012 | Bloom9 | GPL10558 | Validation | South Africa, UK |
Adults | Negative | Positive | .. | .. | .. | 36 | 130 | 166 | Treatment measured at 0·5, 2, 4, 6, and 12 months. Two cohorts followed. Latent tuberculosis not used; overlaps with GSE19491 |
| GSE41055 | 2013 | Verhagen10 | GPL5175 | Validation | Venezuela | Child- ren |
Negative | Positive and negative |
9 | 9 | 7 negative; 2 positive |
.. | 27 | .. | |
| GSE42834 | 2014 | Bloom9 | GPL10558 | Discovery | UK, France | Adults | Negative | Positive | 118 | .. | 123 | 40 | .. | 281 | Other diseases breakdown: 83 sarcoidosis, 24 pneumonia, 16 cancer |
| GSE56153 | 2012 | Ottenhoff23 | GPL6883 | Validation | Indonesia | Adults | Negative | Positive | 18 | .. | 18 | 35 | 71 | Treatment measured at 8 and 28 weeks |
|
| GSE62147 | 2015 | Tientcheu29 | GPL6480 | Validation | The Gambia |
Adults | Negative | Positive | .. | .. | .. | 26 | 26 | 52 | M africanum and M tuberculosis |
| GSE74092 | 2015 | Maertzdorf12 | RT-PCR array GPL21040 |
Validation | India | Adults | Negative | Positive | 76 | .. | .. | 113 | .. | 189 | KLF2 not present in these data |
ASLE=adult systemic lupus erythematosus. PSLE=paediatric systemic lupus erythematosus. CLD=chronic lung disease. URI=upper respiratory infection.
Two gene expression datasets in the GEO (GSE19491 and GSE42834) contained multiple subcohorts. For these datasets, we removed the non-whole-blood samples, normalised the remaining samples, and then treated them as single cohorts. One pair of datasets (GSE31348 and GSE36238) is a single clinical cohort from Cliff and colleagues.13 For this cohort, we downloaded the raw Affymetrix files and co-normalised them using gcRMA14 (R package ‘affy’) to make a single cohort, which we refer to as the Cliff Combined in this report. When comparing between datasets, it is important to ensure similar normalisation methods. Thus, all Affymetrix datasets were gcRMA renormalised from raw data. For all non-Affymetrix arrays, we downloaded data in non-normalised form, background corrected using the normal-exponential method, and then quantile normalised (R package ‘limma’).15 We log2 transformed all data before use. We downloaded all probe-to-gene mappings from the GEO from the most current SOFT fi les on Jan 9, 2015.
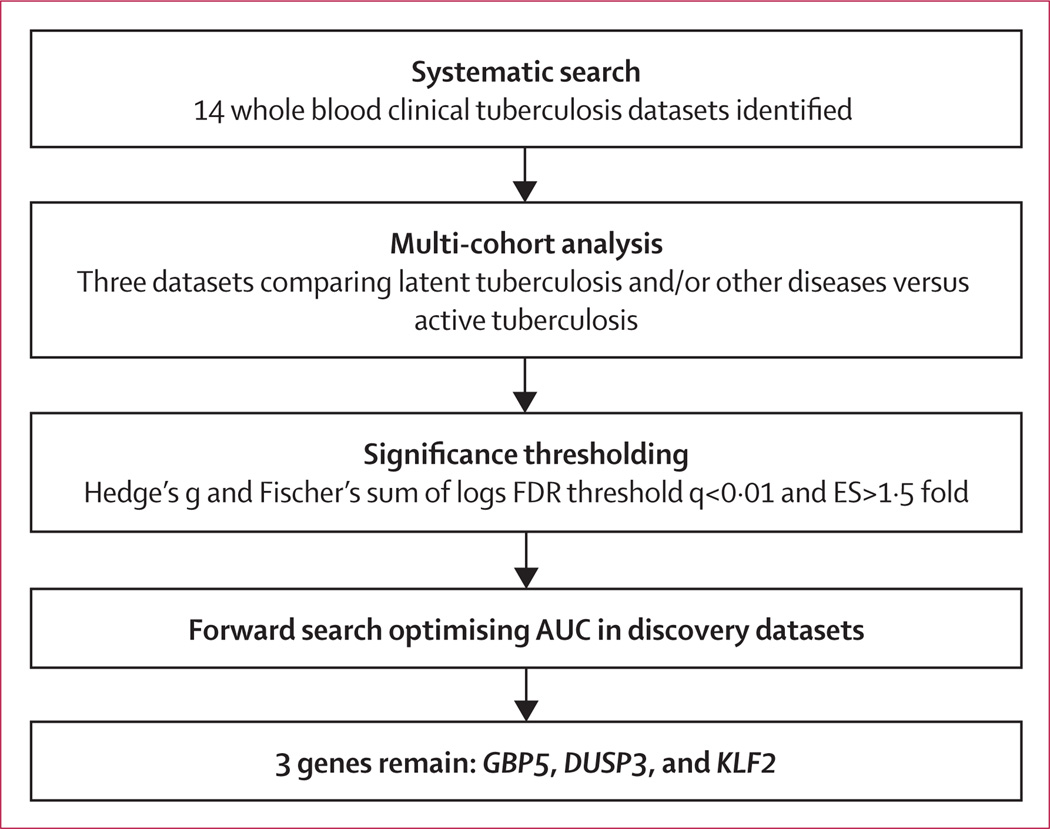
We compared gene expression in patients with either latent tuberculosis or other diseases versus patients with active tuberculosis using our validated multicohort analysis framework, as previously described.16–19 We used three datasets (GSE19491, GSE37250, and GSE42834) as the discovery datasets, and removed genes not present in all three datasets. These datasets were chosen because they were the largest datasets comparing the groups of interest; the remaining datasets were left out specifically to allow for independent validation of results. We applied two meta-analytical methods: (1) combining gene expression effect sizes (Hedges’ g) using a DerSimonian-Laird random-effects model (using R package ‘rmeta’) and (2) combining p values with Fisher’s sum of logs method (figure 1); both were then corrected to false discovery rate (FDR) via Benjamini-Hochberg method. We set significance thresholds for differential expression at FDR less than 1% and an effect size greater than 1·5 fold (in non-log space).
Figure 1. Multicohort analysis.
Schematic of the multicohort analysis workflow
TB score
We did a forward search as previously described,17 with the slight modification to the way the tuberculosis score is calculated. Briefly, the algorithm starts with the single gene with the best discriminatory power, and then at each subsequent step adds the gene with the best possible increase in weighted AUC (area under the curve; the sum of the AUC for each dataset times the number of samples in that dataset) to the set of genes, until no further additions can increase the weighted AUC more than some threshold amount (here 0·005 × the total number of samples). At each iteration of the greedy forward search, when adding a new gene, we defined a tuberculosis score as follows: for each sample, the mean expression of the down-regulated genes is subtracted from the mean expression of the up-regulated genes to yield a tuberculosis score. The forward search always optimises only the discovery datasets, so that the validation datasets are truly independent tests. The final tuberculosis score is thus calculated as: (GBP5 + DUSP3) / 2 – KLF2. This tuberculosis score was then directly tested for diagnostic power using receiver operating characteristic (ROC) curves.
For validation, violin plots show the tuberculosis score for a given dataset across all subsets of patient samples. Violin plot error bars show IQR because they cannot be assumed to have normal distributions within subsets. All ROC curves show comparison to active tuberculosis patients within a given dataset.
Validation of TB score
We validated the three-gene set in 11 independent clinical tuberculosis gene expression datasets, comparing its ability to differentiate between four types of comparisons: (1) healthy controls versus active tuberculosis, (2) latent tuberculosis versus active tuberculosis, (3) other diseases versus active tuberculosis, and (4) response to treatment in longitudinal cohorts of active tuberculosis patients. Seven of the validation datasets include multiple patient classes (ie, healthy controls, latent tuberculosis, and active tuberculosis); in such cases, we compared each patient class separately against the active tuberculosis group in the same dataset, wherein active tuberculosis was always defined as culture-positive or smear-positive cases.
To assess the performance of the tuberculosis score in a so-called real-world manner, we constructed global expression matrices, in which all datasets for each type of comparison were merged into a single matrix, and then x tested the tuberculosis score for a single global cutoff across all datasets. However, because cohorts were profiled on a variety of microarray technologies with different processing methods, the background levels of gene expression between the cohorts varied substantially. Thus, we re-scaled the datasets to make baseline gene expression levels match. To do this, we computed the mean for each gene across all samples (the global mean), and then subtracted from the mean within each dataset, such that each gene within each dataset had the same mean as all other datasets. This method preserves the relative differences of a gene between samples within a dataset. We calculated two sets of global cutoffs: one used the Youden method20 to jointly maximise sensitivity and specificity, the other was set at a sensitivity of 95% to test resulting specificity. To show violin plots and gene levels on the same background, the expression values of the target genes were mean centered. This was only for visualisation and does not affect the tuberculosis score, as the subtraction of a common value falls out of the equation.
Comparison with previous gene sets
In testing diagnostic gene or transcript sets of other groups, transcripts were always summarised to genes. In every case, the gene set was tested according to its original described model, reconstructed by us using the entire discovery dataset. In the case of the three-gene set of Laux da Costa and colleagues11 (who used targeted PCR instead of microarrays), they validated their model by building a random forest classifier in GSE42834 (Bloom and colleagues9); we thus re-built their model in GSE42834 and tested in the remaining datasets. In reports that provided multiple gene sets, the gene sets with the best original diagnostic power were tested (for instance, Kaforou and colleagues7 provide five gene sets for testing active tuberculosis against other diseases, latent tuberculosis, or both; we only used the best signatures for other diseases and latent tuberculosis). Summary ROCs for the previous gene sets were calculated including both discovery and validation datasets.
Summary ROC curves
We constructed summary ROC curves according to the method of Kester and Buntinx,21 which incorporates information from the entirety of an ROC curve, rather than relying on a single summary point (Q*). Briefly, each ROC curve is modelled as a logistic function of its sensitivity and specificity at each cutoff point; the parameters for the ROC curve (α and β) are estimated with weighted linear regression, with errors estimated with a bootstrap of 10 000 repetitions with replacement. The summary α and β parameters are combined using a random-effects model, with errors carried through from the bootstrap. The summary α and β are then re-transformed to construct a summary ROC curve (appendix p 1). We constructed upper and lower summary ROC CIs with the upper and lower bounds on α and β, respectively, reflecting uncertainty for curve skewness. AUCs of the summary curves are calculated with the trapezoidal method with 1000 points.
Briefly, to test gene signatures in gene expression patterns from known sorted cells, we aggregated public gene expression data from several immune cell types and then calculated the relevant tuberculosis score in each cell type genome, as described previously.17
Between-groups tuberculosis score comparisons were done with the Wilcoxon rank sum test. Significance levels were set at two-tailed p<0·05, unless specified otherwise. All computation and calculations were done in the R language for statistical computing (version 3.0.2).
Role of the funding source
The funders had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Results
We identified 14 publically available datasets composed of 2572 patient samples that matched the inclusion criteria (table).6–10,12,13,22–29 We applied our multicohort analysis framework16–18 to three of these datasets (GSE19491, adults;8 GSE37250, adults,7 and GSE42834, adults9), composed of 1023 whole blood samples (236 patients with latent tuberculosis, 491 patients with other diseases, and 296 patients with active tuberculosis) to compare patients with latent tuberculosis or other diseases to patients with active tuberculosis (figure 1). ‘Other diseases’ in the samples included patients with sarcoidosis, pulmonary and non-pulmonary infections, autoimmune diseases, and lung cancer.
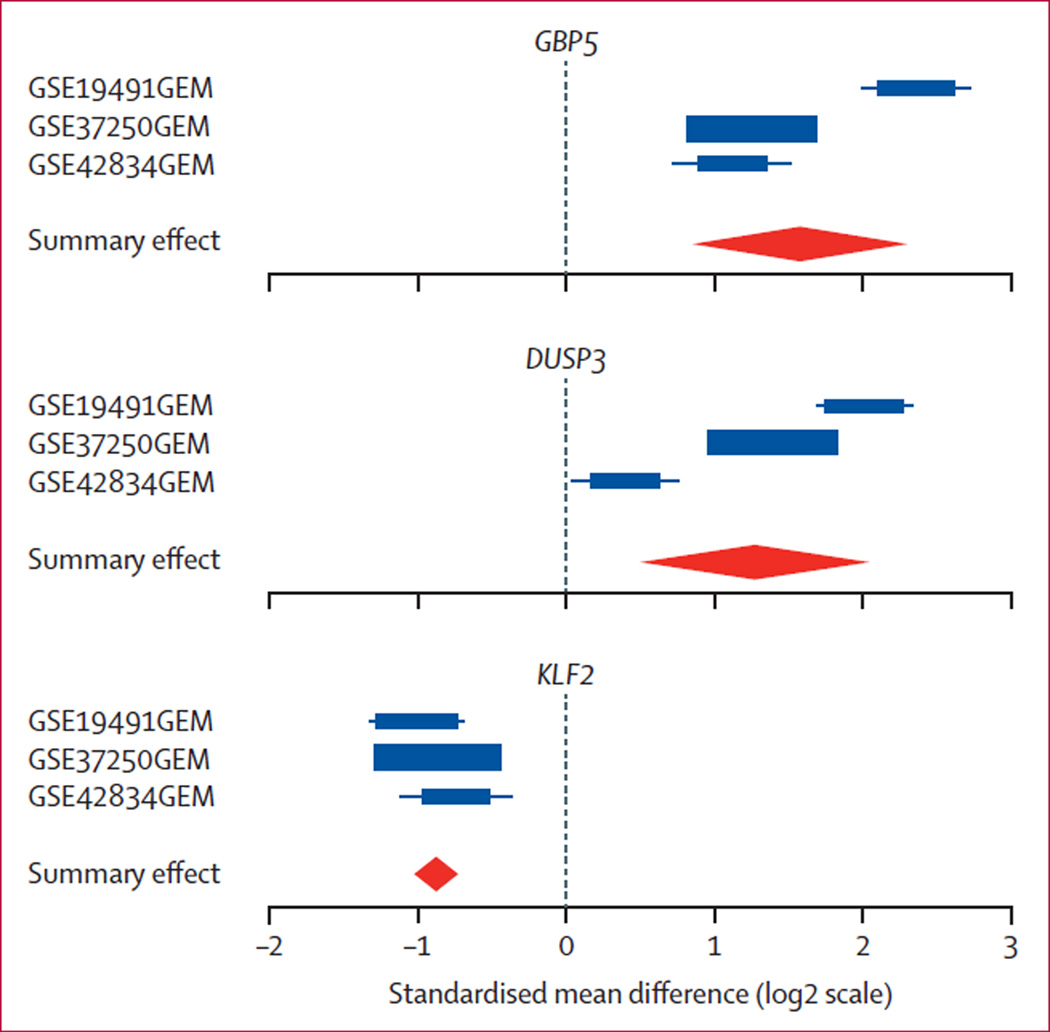
We identified 266 genes that were significantly differentially expressed (158 over-expressed and 108 under-expressed) in active tuberculosis compared with latent tuberculosis and other diseases at FDR of 1% or less and effect size greater than 1·5 fold (appendix p 2). We applied a greedy forward search17 to obtain a set of genes optimised for diagnostic power, resulting in a three-gene set (GBP5, DUSP3, and KLF2; figure 2).
Figure 2. Forest plots for each of the three genes derived in the forward search.
Forest plots for each of the three genes derived in the forward search. The x axis represent standardised mean difference between latent tuberculosis and other diseases versus active tuberculosis. The size of the blue rectangles is inversely proportional to the SE of mean in the study. Whiskers represent the 95% CI. The orange diamonds represent overall, combined mean difference for a given gene. Width of the diamonds represents the 95% CI of overall combined mean difference. FDR=false discovery rate.
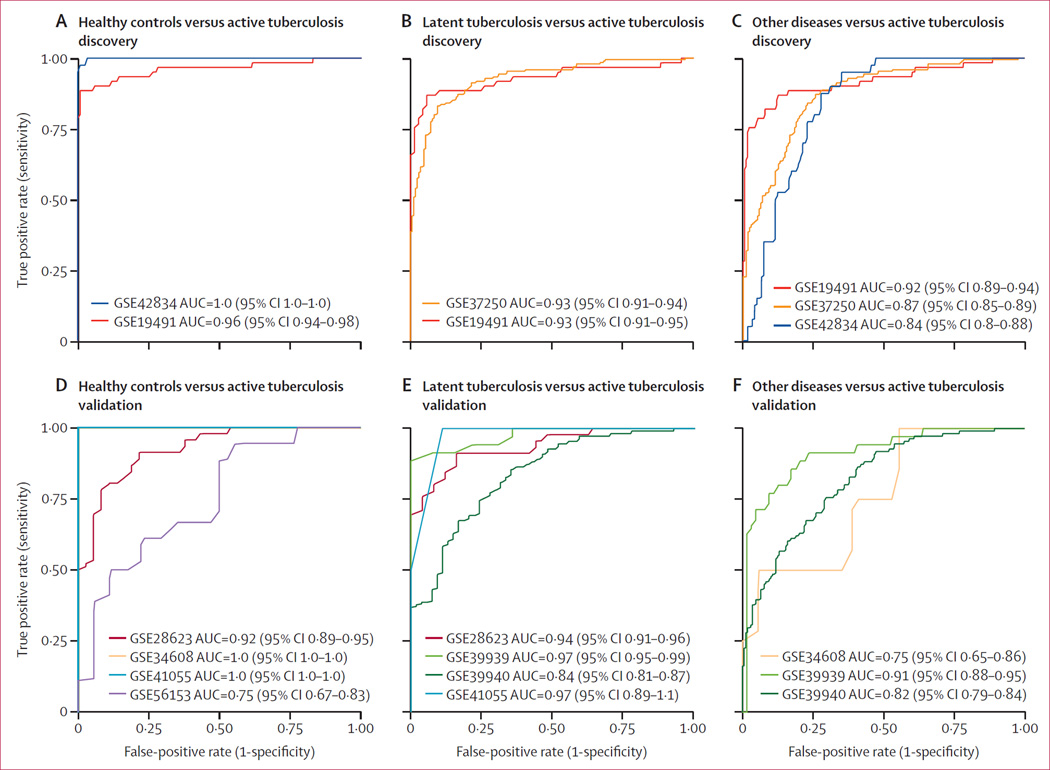
As expected, in the discovery datasets, the three-gene set distinguished active tuberculosis from healthy controls (AUCs of 0·96 [95% CI 0·94–0·98] and 1·0 [95% CI 1–1], mean sensitivity 0·93, mean specificity 0·97), latent tuberculosis (AUCs of 0·93 [95% CI 0·91–0·95] and 0·93 [95% CI 0·91–0·94], mean sensitivity 0·88, mean specificity 0·85), and other diseases (mean AUC of 0·88 [range 0·84–0·92]; mean sensitivity 0·82, mean specificity 0·79; figure 3A–C; appendix p 3). Individual dataset test characteristics (sensitivity, specificity, negative predictive value, positive predictive value, and accuracy) are shown in the appendix (p 4). A breakdown of the other disease category by disease class is shown in the appendix (p 5). The tuberculosis score did well across all classes of other diseases (AUC≥0·85) except sarcoidosis (AUC=0·79), which might be the result of the interferon response common to these two diseases.24
Figure 3. Performance of the three-gene set in the discovery datasets.
ROC curves in discovery cohorts showing healthy controls (A), patients with latent tuberculosis (B), and patients with other diseases (C) versus patients with active tuberculosis. Healthy patients were not included in the multicohort analysis but are shown here. ROC curves in four validation cohorts comparing healthy controls with active tuberculosis (D), patients with latent tuberculosis with patients with active tuberculosis, and ROC curves in three validation cohorts comparing patients with other diseases with active tuberculosis (E). Violin plots with patient-level data are shown in figure 6 and appendix pp 3, 5, 6. ROC=receiver operating characteristic. AUC=area under the curve.
There were four independent datasets comparing healthy controls with active tuberculosis patients: GSE28623 (Maertzdorf and colleagues, adults),22 GSE34608 (Maertzdorf and colleagues, adults),24 GSE41055 (Verhagen and colleagues, children),10 and GSE56153 (Ottenhoff and colleagues, adults);23 these datasets contained a total of 82 healthy controls and 74 patients with active tuberculosis (table). Despite substantial clinical heterogeneity in these datasets, including age, country of origin, and inclusion criteria, patients with active tuberculosis had a significantly higher score than healthy controls (Wilcoxon p values: GSE28623, p=2·41 e-13; GSE34608: p=1·28 e-6; GSE41055, p=0·036; GSE56153, p=0·012) in all datasets, with a mean AUC of 0·92 (range 0·75–1·0, mean sensitivity 0·86, mean specificity 0·81; figure 3D; appendix p 6; individual dataset test characteristics in appendix p 7). The cause of the relatively low AUC in GSE56153 could be either clinical or technical factors. A fifth dataset, GSE74092 (Maertzdorf and colleagues, adults)12, a targeted RT-PCR study of 189 adults from India, included two of the three genes (GBP5 and DUSP3); these two genes alone had an AUC of 0·94, but were left out of calculation of mean AUC because of the missing third gene (appendix p 8). A sixth dataset, GSE25534 (Maertzdorf and colleagues, adults),30 which used a two-channel array design, the three-gene set perfectly classified healthy versus active tuberculosis samples, although no ROC curve could be constructed because each sample has a control on-chip, rather than a separate group of healthy controls (N=25; appendix p 9).
There were four independent datasets comparing patients with latent tuberculosis and those with active tuberculosis patients (GSE28623,22 GSE39939 [Anderson and colleagues, children],6 GSE39940 [Anderson and colleagues, children]),6 and GSE41055;10 102 patients with latent tuberculosis, 194 patients with active tuberculosis; table). Patients with active tuberculosis had significantly higher tuberculosis scores than those with latent tuberculosis (Wilcoxon p<0·05) in all datasets. The four cohorts had a mean AUC of 0·93 (range 0·84–0·97; mean sensitivity 0·87, mean specificity 0·85; figure 3E; appendix p 6; individual dataset test characteristics in the appendix (p 7). Furthermore, in GSE25534 (which had a two-channel array), the three-gene set classified latent tuberculosis versus active tuberculosis samples with 97% accuracy (N=38 samples, appendix p 9). These results provide strong evidence that the three-gene set separates active tuberculosis from latent tuberculosis.
There were three independent datasets comparing patients with other diseases versus those with active tuberculosis (GSE34608,24 GSE39939,6 GSE39940;6 251 patients with other diseases, 154 patients with active tuberculosis; table). In these cohorts, the other disease category included mainly patients with pneumonia, but also patients with chronic lung diseases (such as sarcoidosis), non-pulmonary infections, or malignancies. Patients with active tuberculosis had higher tuberculosis scores than those with other diseases (Wilcoxon p<0·05) in all datasets. The three cohorts had a mean AUC of 0·83 (range 0·75–0·91; mean sensitivity 0·65, mean specificity 0·74; figure 3F; appendix p 6; individual dataset test characteristics in appendix p 7). Even in the difficult case of separating active tuberculosis from other diseases, the three-gene set performs well.
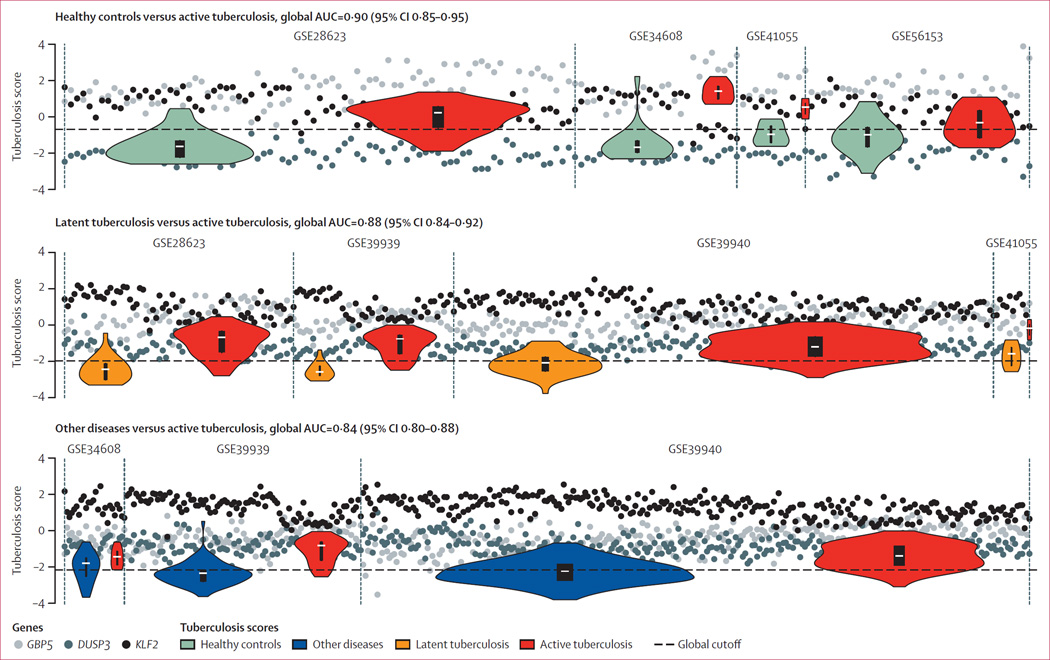
The test characteristics reported for each of the comparisons above used different tuberculosis score thresholds for each dataset to maximise joint specificity and sensitivity within a given dataset. However, a real-world clinical application would require a single threshold that can be applied universally across all patients (instead of using different thresholds for different cohorts). We thus combined and re-scaled the available validation datasets into single so-called global matrices for each comparison type, from which we were able to evaluate a single global ROC AUC for each comparison, and estimate test characteristics from optimal cutoffs. The AUCs using a global cutoff across all datasets are 0·90 (95% CI 0·85–0·95, sensitivity 0·85, specificity 0·93) for healthy controls versus active tuberculosis, 0·88 (95% CI 0·84–0·92 sensitivity 0·80, specificity 0·86) for latent tuberculosis versus active tuberculosis, and 0·84 (95% CI 0·80–0·88, sensitivity 0·81, specificity 0·74) for other diseases versus active tuberculosis, across all validation datasets (figure 4). To more closely match the desired sensitivity of the WHO consensus target product profile, we also tested thresholds set at a minimum sensitivity of 0·95 (rather than maximising the sum of sensitivity and specificity as above). Maximised for sensitivity, the global threshold test characteristics across all validation datasets were: sensitivity 0·96 and specificity 0·56 for healthy controls versus active tuberculosis; sensitivity 0·95 and specificity 0·51 for latent tuberculosis versus active tuberculosis; and sensitivity 0·95 and specificity 0·47 for other diseases versus active tuberculosis. We note that an NPV calculated at 95% sensitivity and 50% specificity for a disease with a 10% prevalence is 99%. The effects of the re-scaling, and the effects of including the discovery datasets in the global expression matrices, are shown in the appendix (pp 10–12). Notably, no major prescaling differences occured between datasets run on the same microarray type (eg, GSE37250, GSE39939, GSE39940, and GSE42834, all run on GPL10558 [Illumina HumanHT-12 V4; Illumina Inc, San Diego, CA, USA]). These results show that even when we enforce a global threshold, our gene signature was able to maintain accurate partitioning of patients with active tuberculosis from the healthy controls, latent tuberculosis, and other diseases cohorts.
Figure 4. Establishment of a single global test cutoff in the validation datasets.
Sample-level normalised gene scores and group tuberculosis score distributions. Cohorts are shown. Bars within violin plots show IQR; white dashes show medians. By centering the genes within each dataset to their global mean, a single cutoff across multiple datasets can be established.
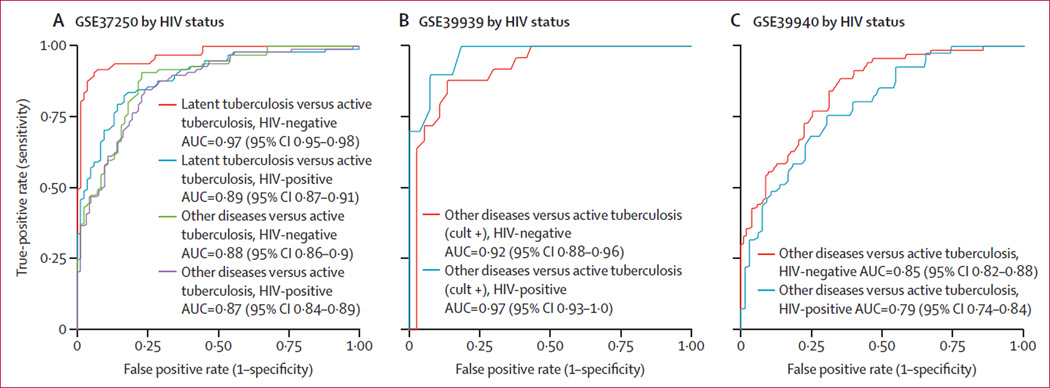
Next we investigated the effect of several confounding factors (HIV co-infection, tuberculosis drug resistance, and culture status, disease severity, and BCG vaccination) on the three-gene set. In three datasets (GSE37250, GSE39939, and GSE39940) that included patients with active tuberculosis with or without HIV co-infection, no difference was noted in the tuberculosis score AUCs for other diseases versus active tuberculosis with or without HIV co-infection (figure 5A – C). In GSE37250, we noted a decrease in tuberculosis score AUC for latent tuberculosis versus active tuberculosis with HIV co-infection, although the AUC remained high for both groups (HIV-negative AUC 0·97 (95% CI 0·95–0·98); HIV-positive AUC 0·89 (0·87–0·91). Additionally, one dataset, GSE50834, examined peripheral blood mononuclear cells from HIV-positive patients with and without tuberculosis co-infection; here, the tuberculosis score had an AUC of 0·85 (95% CI 0·82–0·88), although no non-HIV infected cohorts were included (appendix p 13).
Figure 5. Effect of HIV co-infection on the diagnostic power of the tuberculosis score.
In GSE37250, GSE39939, and GSE39940, no significant difference was noted in the diagnostic power for other diseases versus active tuberculosis based on HIV status. In GSE37250, there was a decrease in ROC AUC from 0·96 to 0·89 in latent tuberculosis versus active tuberculosis in HIV-positive patients. ROC=receiver operating characteristic. AUC=area under the curve.
We then examined confounders other than HIV co-infection. In GSE19491 the tuberculosis score did not differ due to BCG vaccination status or M tuberculosis drug resistance. Additionally, the tuberculosis score was positively correlated with disease severity (Jonckheere– Terpstra test; p<0·001) as defined by chest radiography (appendix p 14). The effects of culture status were pronounced in children. Two paediatric datasets, GSE39939 and GSE41055 (of which GSE41055 is underpowered), included cohorts of patients with culture-negative active tuberculosis. In these datasets, the tuberculosis scores in such patients were significantly lower than those in culture-positive active tuberculosis (p<0·05; appendix p 6). However, in GSE19491, in adults with culture-positive active tuberculosis the degree of smear positivity or a negative culture from either sputa or bronchoalveolar lavage when the other is positive did not affect tuberculosis score (appendix p 15). These results suggest that a positive active tuberculosis classification via tuberculosis score in children would be highly specific for active tuberculosis, although might not be sensitive to children with culture-negative active tuberculosis.
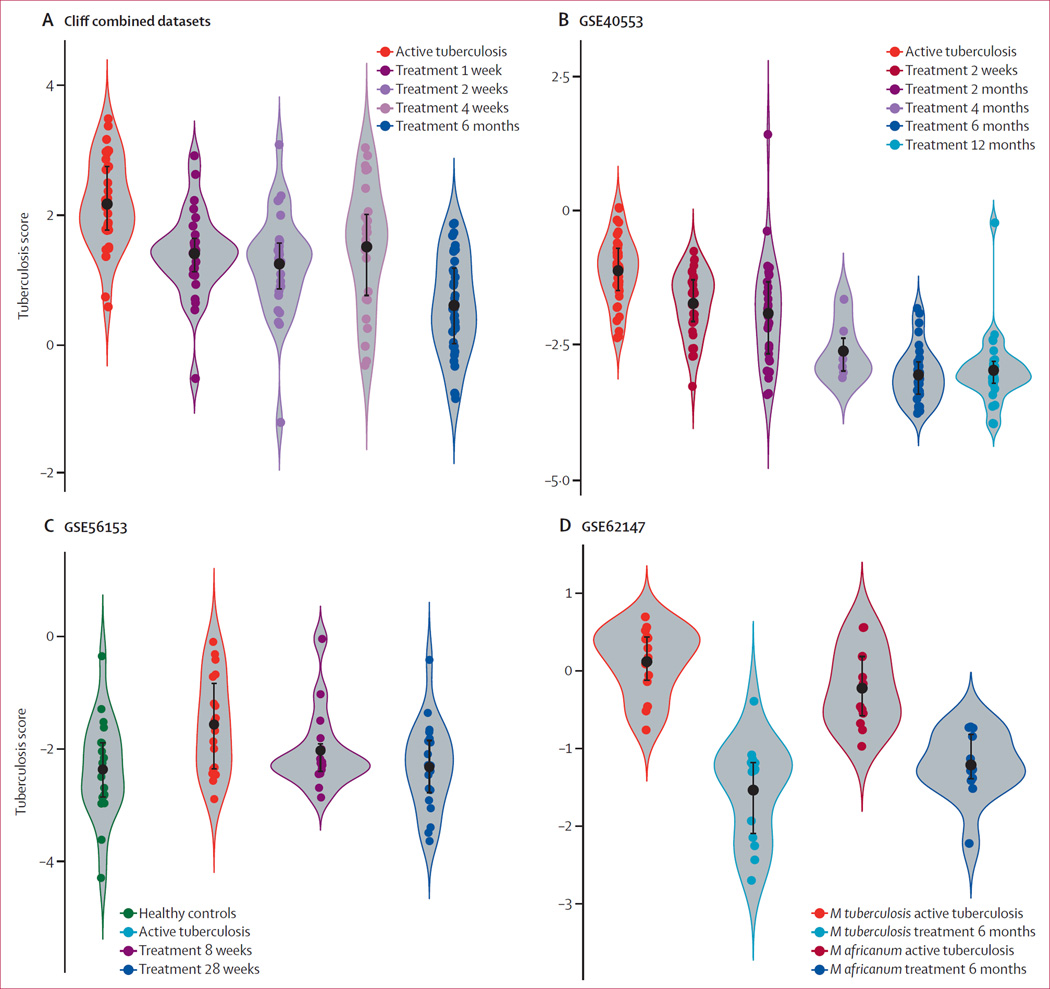
Next, we examined the four datasets that profiled patients with active tuberculosis longitudinally during treatment (the Cliff Combined dataset,13 GSE40553,25 GSE56153,23 and GSE62147;29 table). Each of the four datasets followed patients with active tuberculosis for up to 6 months or 12 months. In each dataset, the tuberculosis score showed a significant decreasing trend as treatment progressed (figure 6; regression models in the appendix p 16). Furthermore, most patients showed individual trends of a decreasing score over time (appendix p 17). In GSE56153, the tuberculosis scores of patients at recovery (treatment week 28) were not different from those of healthy controls (Wilcoxon p>0·05). In GSE62147, patients with active tuberculosis due to Mycobacterium africanum were also examined; here, too, the tuberculosis score fell with treatment. These results suggest that the tuberculosis score could be a useful biomarker for clinical response to treatment, and could potentially identify treatment non-responders, although no non-responders were available for study here.
Figure 6. Violin plots showing the performance of the three-gene set in longitudinal validation datasets.
Four validation datasets examined active tuberculosis patients during treatment and recovery. All four datasets took samples before and during treatment. The tuberculosis score falls over time of treatment. GSE56153 also included healthy controls; the tuberculosis score returned to normal after treatment (Wilcoxon p=not significant between cured cases and healthy controls; C). GSE62147 also examined active Mycobacterium africanum infections (D).
All datasets examined so far in this report examined pulmonary active tuberculosis; one question is whether the three-gene set might also be useful for diagnosis of extrapulmonary tuberculosis. One dataset, GSE63548, compared tuberculosis-infected lymph node tissue to lymph nodes from healthy controls;31 here, the tuberculosis score had an ROC AUC of 0·98 (appendix p 18). However, because this study was conducted in actual lymph node tissue, not peripheral blood, further work will be needed to assess the use of the tuberculosis score in extrapulmonary tuberculosis.
Several of the studies used in our analysis have previously identified transcripts or gene sets for diagnosing patients with active tuberculosis.6–12 However, these gene sets either contain large number of genes or are not generalisable, or both. We tested nine previously published diagnostic gene sets from five studies for their ability to discriminate other diseases and latent tuberculosis from active tuberculosis in all datasets examined here (appendix pp 19–25). Each gene set was tested across all datasets using the method described in its original paper; for methods that require models, such as k-nearest neighbours (Berry and colleagues8), support vector machines (Bloom and colleagues9), or random forest (Verhagen and colleagues10, Laux da Costa and colleagues11), the model was constructed using the entire original discovery cohort, and then tested in the other independent cohorts. The four-gene set by Maertzdorf and colleagues12 was not included here because its original parameterisation in PCR data (given as –Ct) does not allow its reapplication in microarray data without re-estimating parameters in each new dataset. All but one gene set (Kaforou and colleagues7) show a significant drop in discriminatory power in independent validation datasets. The three-gene random forest model of Laux da Costa and colleagues (GBP5, CD64 [also known as FCGR1A]) and GZMA) did not generalise well. Only the two gene sets from Kaforou and colleagues did as well as our three-gene set when comparing basic diagnostic power. While the two gene sets from Kaforou and colleagues did as well as our three-gene set when comparing basic diagnostic power, they contain 71 genes (including only one of our genes DUSP3), precluding their clinical application in resource-limited environments.
Finally, we investigated the expression of both the entire set of 266 significant genes and the diagnostic three-gene set in publicly available whole genome expression profiles from 25 different types of immune cells. Both gene sets showed a statistically significant enrichment in M1 macrophages (p<0·05; appendix p 26), the workhorses of the interferon γ-mediated host response to active tuberculosis.
Discussion
A crucial requirement to reduce the global burden of tuberculosis disease is better instruments for diagnosis and for monitoring treatment response.32 Here, we used a multicohort analysis of three public tuberculosis gene expression datasets composed of 1023 whole blood patient samples across a range of ages, countries and inclusion criteria to find genes that are statistically differentially expressed in active tuberculosis compared with latent tuberculosis and other diseases. We identified a three-gene set, and validated it in 11 additional independent whole blood datasets composed of 1345 samples. The results showed that it is robustly diagnostic for active tuberculosis versus healthy controls, latent tuberculosis, and other diseases, that its diagnostic capability is not affected by HIV status and BCG vaccination status, and is significantly correlated with severity of active tuberculosis.
Our results provide strong evidence that the three-gene set could be used to address some of the major challenges in the diagnosis of active tuberculosis as defined by the recent WHO consensus target product profile.5 First, the three-gene set is based on peripheral blood rather than sputum samples. Second, our three-gene set performed well in the diagnosis of latent tuberculosis versus culture-positive active tuberculosis in children, with a mean sensitivity of 0·86 and mean specificity of 0·86 (appendix p 7); substantially higher than the target sensitivity of 0·66 in the consensus target product profile. Third, HIV status did not change the diagnostic power of the three-gene set for comparisons of other diseases versus active tuberculosis, and although HIV-positive patients had a lower AUC for latent tuberculosis versus active tuberculosis, it was still high (0·97 HIV-negative, 0·89 HIV-positive). Finally, the high parsimony and internal normalisation of the three-gene set could allow for a point-of-care application, because it might be possible to assay three genes with solar-powered PCR instruments.33 Although the three-gene set does not meet all needs of the consensus target product profile (eg, overall sensitivity >0·95 can only be achieved with specificity in the range of 0·50), it is able to fulfill several of the requirements of the consensus target product profile.
Several tuberculosis diagnostic gene sets have been proposed by others; five of the datasets used here were used to derive diagnostic gene sets in their original manuscripts. These gene sets were all derived using statistical techniques that are designed to optimise a set of genes within the discovery cohorts; because different discovery cohorts were used, naturally different gene sets were identified. However, single-study discovery analyses that rely on machine learning models are prone to overfitting, and thus suffer from an absence of generalisability (appendix pp 19–25). Furthermore, when comparing these published gene signatures, there is surprisingly little overlap between them. Here, we tested each gene set using its original described model against all available validation data to reasonably estimate their performance in real-world application settings, in which biological and technological confounding factors can have a substantial impact. Notably, although the three-gene set of Laux da Costa and colleagues11 is also parsimonious, their model does not generalise well, with poor AUCs in independent test sets. Their selection process was essentially a vote-counting method of previous gene sets; thus, their three genes are not specifically selected to act together, as our forward search does. Similarly, Maertzdorf and colleagues12 published a four-gene diagnostic set (all four of which were noted to be significant in the meta-analysis here). However, the microarray validations done by this group required re-estimation of model parameters in each dataset, meaning their validation AUCs carry a high positive bias. Finally, the gene set of Kaforou and colleagues7 requires measurement of 71 genes in adults, which is possible with high-powered expensive laboratory equipment (eg, Fluidigm Biomark [South San Francisco, CA, USA], Nanostring Technologies nCounter [Seattle, WA, USA]), but not in remote and resource-poor settings where tuberculosis is prevalent. By contrast, our three-gene set could be optimised to a low-cost PCR-based assay, which is a standard used for many other infectious disease applications. Overall, compared with all other published gene sets for tuberculosis, ours is parsimonious (only three genes), can distinguish both other diseases and latent tuberculosis from active tuberculosis with one test in multiple clinical groups, and performs well in independent external datasets. The multiple gene expression diagnostics that have been derived all suggest that host-response gene expression assays are likely to eventually play an important role in tuberculosis diagnostics.
Another crucial and unmet need is the ability to undertake quantitative monitoring of tuberculosis treatment response. The current standard in clinical trials for new drugs for tuberculosis treatment requires waiting for 2 years after treatment to observe relapse rates. Improved monitoring techniques might allow non-responders to be identified earlier. The three-gene set increases with disease severity and decreases with time of treatment (returning to the same level as healthy controls at the end of treatment) with remarkably similar coefficients across datasets (the tuberculosis score fell by 0·02 to 0·05 per week). This consistency across multiple datasets suggests the potential to detect deviations from the standard treatment response using our three-gene set, and identify treatment non-responders substantially earlier. The correlation of the tuberculosis score with disease severity also suggests that it might be possible to leverage the test for a predictive enrichment strategy for new drug trials.34 Using the three-gene set to improve tuberculosis drug trials is thus an interesting possibility that requires further study.
The small size of the three-gene set will be important in its ultimate clinical application, reducing costs and complexity relative to larger gene sets. A small number of PCR targets can be run in parallel using low-power, low-cost equipment. For instance, Cepheid’s GeneXpert MTB/RIF assay measures the expression of five loci, and costs between US$10–$20 per cartridge.35 Using this assay as an approximate benchmark, an assay to measure the three-gene set could probably be provided at similar cost after commercial optimisation.
Finally, the importance of the innate immune response and lung-resident macrophages in the establishment of tuberculosis infection is well known.36 However, understanding of the specific cellular mechanisms enlisted during a host response to mycobacteria is lacking. We have identified host response genes to active tuberculosis that are strongly associated with innate immune cells, in particular M1 macrophages. The relation of interferon γ to active tuberculosis has been previously shown by Berry and colleagues8 and Cliff and colleagues;13 however, interferon γ activation causes several downstream events. Thus, the three-gene set might give insight into which interferon γ related pathways are crucial and specific to the host response to active pulmonary tuberculosis. The three genes are known to have roles in immune regulation and infection response. GBP5 promotes assembly of both the AIM2 and NLRP3 inflammasome assembly in response to pathogenic bacteria.37,38 DUSP3 is a known regulator of both JNK and ERK signalling.39,40 KLF2 has been shown to be down-regulated in macrophages in response to bacterial stimulation; further, knockdown and knockout studies have shown that decreased KLF2 leads to a pro-inflammatory phenotype.41–43 Further hypothesis-driven studies of these three genes could provide better insight into both the global and the local immune response during tuberculosis infection and might help to design more effective therapeutics and vaccines.
One limitation of our study is that the global ROCs required a re-centering of means to accommodate for changes in baseline gene expression measurement by different technologies. However, such a centering is justified because in a real-world application of the three-gene set, the same technology with a global mean will be used across all cohorts. Furthermore, when the three-gene set is reduced to a targeted assay, the available public data can be mapped to the background gene expression levels of the final clinical platform to use the publically available data to set optimal cutoffs for future diagnosis. Thus, although the optimal cutoffs could change in the final commercial form, our results show that the three-gene set could be developed as a clinical test with a single cut-off for diagnosis of active tuberculosis. A second limitation of our study is that, because of the enormous possible search space for potential gene sets, we used a greedy algorithm to identify the three-gene set. It thus remains possible that a different gene set with similar diagnostic power could still be identified. A third weakness is that the measure of AUC is a relatively blunt measure of diagnostic power, because we are interested here mainly in the portion of the curve corresponding to high sensitivity; however, sensitivity and specificity have been provided for all ROC curves. Additionally, the datasets used here do not publicly provide enough standard diagnostic criteria to allow the calculation of a net reclassification index. Thus, the question of diagnostic net benefit of a molecular diagnostic test will have to wait for a large prospective trial.
Overall, these data show that our three-gene set is robustly diagnostic for active tuberculosis across 14 datasets containing 2572 clinical samples. The three-gene set could improve clinical diagnosis and treatment response monitoring, although this needs to be confirmed by prospective validation with a targeted assay. The gene set is based on whole blood and is robust to multiple clinical confounders. The parsimony of the three-gene set should ease translation to clinical practice and might prove cost effective in the resource-poor environments in which tuberculosis is prevalent.
Supplementary Material
Research in context.
Evidence before this study
Several studies have been published so far describing gene signatures for the diagnosis of active pulmonary tuberculosis. However, almost all of these studies have been based on single dataset analysis. In a few cases in which the studies analysed samples from multiple countries, they exclusively focused on a certain age group (eg, children or adults). The derived gene sets often contain dozens of transcripts, and show diminished diagnostic power in external independent cohorts. A 2014 WHO target product profile for new tuberculosis diagnostics described a strong need for a new diagnostic with excellent sensitivity that uses non-sputum samples (such as blood), with good diagnostic performance in both HIV-positive patients and in children, and that is relatively simple to run.
Added value of this study
We have derived a 3-gene signature (GBP5, DUSP3, and KLF2) in whole blood that is robustly diagnostic for active tuberculosis across multiple validation datasets irrespective of age.
Implications of all the available evidence
Multiple diagnostic host gene signatures for tuberculosis have been published. We show here that our signature is either smaller, or has better test characteristics, or both, when compared to prior diagnostics. The increasing interest in using host gene expression signatures to diagnose tuberculosis suggests that these methods may become part of the clinical toolkit in the near future.
Acknowledgments
TES reports grants from National Library of Medicine (2T15LM007033), the Stanford Child Health Research Institute Young Investigator Award (through the Institute for Immunity, Transplantation and Infection), the Society for University Surgeons, and has been a scientific advisor for Multerra Biosciences outside of the submitted work. In addition, TES and PK have has a patent pending “Methods for diagnosis of tuberculosis” (provisional application number 62/241,506), filed by Stanford OTL 10/14/2015, inventors PK and TES. PK reports grants from the Bill and Melinda Gates Foundation and National Institute of Allergy and Infectious Disease (1U19AI109662, U19AI057229, U54I117925, and U01AI089859) outside of the submitted work.
We thank the authors of the datasets that were re-analysed in this report. There are too many to name individually, but without their valuable prior insights and data, this work would not have been possible. We are enormously grateful for their continued support for open data sharing.
Footnotes
Contributors
TES and PK conceived and designed the study. TES and LB collected the data. TES did all experiments. TES, CMT, and PK interpreted the data. TES, CMT, and PK critically revised the report.
Declaration of interests
All other authors declare no competing interests.
References
- 1.Global Tuberculosis Programme, WHO. Global tuberculosis report. Geneva: World Health Organisation; 2015. [Google Scholar]
- 2.Wallis RS, Pai M, Menzies D, et al. Biomarkers and diagnostics for tuberculosis: progress, needs, and translation into practice. Lancet. 2010;375:1920–1937. doi: 10.1016/S0140-6736(10)60359-5. [DOI] [PubMed] [Google Scholar]
- 3.Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;1:CD009593. doi: 10.1002/14651858.CD009593.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedrich SO, Rachow A, Saathoff E, et al. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir Med. 2013;1:462–470. doi: 10.1016/S2213-2600(13)70119-X. [DOI] [PubMed] [Google Scholar]
- 5.WHO. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. Geneva: World Health Organization; 2014. [Google Scholar]
- 6.Anderson ST, Kaforou M, Brent AJ, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaforou M, Wright VJ, Oni T, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 2013;10:e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloom CI, Graham CM, Berry MP, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhagen LM, Zomer A, Maes M, et al. A predictive signature gene set for discriminating active from latent tuberculosis in Warao Amerindian children. BMC Genomics. 2013;14:74. doi: 10.1186/1471-2164-14-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laux da Costa L, Delcroix M, Dalla Costa ER, et al. A real-time PCR signature to discriminate between tuberculosis and other pulmonary diseases. Tuberculosis (Edinb) 2015;95:421–425. doi: 10.1016/j.tube.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maertzdorf J, McEwen G, Weiner J, et al. Concise gene signature for point-of-care classification of tuberculosis. EMBO Mol Med. 2015 doi: 10.15252/emmm.201505790. published online Dec 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cliff JM, Lee JS, Constantinou N, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis. 2013;207:18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Sta Assoc. 2004;99:909–917. [Google Scholar]
- 15.Smyth G. Limma: linear models for microarray data. In: Gentleman RCV, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 16.Khatri P, Roedder S, Kimura N, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7:287ra71. doi: 10.1126/scitranslmed.aaa5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li MD, Burns TC, Morgan AA, Khatri P. Integrated multi-cohort transcriptional meta-analysis of neurodegenerative diseases. Acta Neuropathol Commun. 2014;2:93. doi: 10.1186/s40478-014-0093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andres-Terre M, McGuire H, Pouliot Y, Sweeney T, Tato C, Khatri P. Transcriptional signatures of viral infection across multiple respiratory viruses derived from integrated, multi-cohort analysis. Immunity. 2015;43:1199–1211. doi: 10.1016/j.immuni.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Kester AD, Buntinx F. Meta-analysis of ROC curves. Med Decis Making. 2000;20:430–439. doi: 10.1177/0272989X0002000407. [DOI] [PubMed] [Google Scholar]
- 22.Maertzdorf J, Ota M, Repsilber D, et al. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS One. 2011;6:e26938. doi: 10.1371/journal.pone.0026938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ottenhoff TH, Dass RH, Yang N, et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One. 2012;7:e45839. doi: 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maertzdorf J, Weiner J, Mollenkopf HJ, et al. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A. 2012;109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloom CI, Graham CM, Berry MP, et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One. 2012;7:e46191. doi: 10.1371/journal.pone.0046191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu LS, Lee SW, Huang KY, Lee TY, Hsu PW, Weng JT. Systematic expression profiling analysis identifies specific microRNA-gene interactions that may differentiate between active and latent tuberculosis infection. Biomed Res Int. 2014;2014:895179. doi: 10.1155/2014/895179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Y, Yang Q, Tang Y, et al. Increased complement C1q level marks active disease in human tuberculosis. PLoS One. 2014;9:e92340. doi: 10.1371/journal.pone.0092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawany N, Showe LC, Kossenkov AV, et al. Identification of a 251 gene expression signature that can accurately detect M. tuberculosis in patients with and without HIV co-infection. PLoS One. 2014;9:e89925. doi: 10.1371/journal.pone.0089925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tientcheu LD, Maertzdorf J, Weiner J, et al. Differential transcriptomic and metabolic profiles of M africanum- and M tuberculosis-infected patients after, but not before, drug treatment. Genes Immun. 2015;16:347–355. doi: 10.1038/gene.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maertzdorf J, Repsilber D, Parida SK, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12:15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 31.Maji A, Misra R, Kumar Mondal A, et al. Expression profiling of lymph nodes in tuberculosis patients reveal inflammatory milieu at site of infection. Sci Rep. 2015;5:15214. doi: 10.1038/srep15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denkinger CM, Kik SV, Cirillo DM, et al. Defining the needs for next generation assays for tuberculosis. J Infect Dis. 2015;211:S29–S38. doi: 10.1093/infdis/jiu821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang L, Mancuso M, Lu Z, Akar G, Cesarman E, Erickson D. Solar thermal polymerase chain reaction for smartphone-assisted molecular diagnostics. Sci Rep. 2014;4:4137. doi: 10.1038/srep04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Temple R. Enrichment of clinical study populations. Clin Pharmacol Ther. 2010;88:774–778. doi: 10.1038/clpt.2010.233. [DOI] [PubMed] [Google Scholar]
- 35.WHO. Roadmap for Rolling Out Xpert MTB/RIF for Rapid Diagnosis of TB and MDR-TB. Geneva: World Health Organization; 2010. [Google Scholar]
- 36.Dorhoi A, Kaufmann SH. Perspectives on host adaptation in response to Mycobacterium tuberculosis: modulation of inflammation. Semin Immunol. 2014;26:533–542. doi: 10.1016/j.smim.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Shenoy AR, Wellington DA, Kumar P, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 38.Meunier E, Wallet P, Dreier RF, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishibashi T, Bottaro DP, Chan A, Miki T, Aaronson SA. Expression cloning of a human dual-specificity phosphatase. Proc Natl Acad Sci USA. 1992;89:12170–12174. doi: 10.1073/pnas.89.24.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso A, Saxena M, Williams S, Mustelin T. Inhibitory role for dual specificity phosphatase VHR in T cell antigen receptor and CD28-induced Erk and Jnk activation. J Biol Chem. 2001;276:4766–4771. doi: 10.1074/jbc.M006497200. [DOI] [PubMed] [Google Scholar]
- 41.Mahabeleshwar GH, Qureshi MA, Takami Y, Sharma N, Lingrel JB, Jain MK. A myeloid hypoxia-inducible factor 1α-Krüppel-like factor 2 pathway regulates gram-positive endotoxin-mediated sepsis. J Biol Chem. 2012;287:1448–1457. doi: 10.1074/jbc.M111.312702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das M, Lu J, Joseph M, et al. Kruppel-like factor 2 (KLF2) regulates monocyte differentiation and functions in mBSA and IL-1β-induced arthritis. Curr Mol Med. 2012;12:113–125. doi: 10.2174/156652412798889090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lingrel JB, Pilcher-Roberts R, Basford JE, et al. Myeloid-specific Krüppel-like factor 2 inactivation increases macrophage and neutrophil adhesion and promotes atherosclerosis. Circ Res. 2012;110:1294–1302. doi: 10.1161/CIRCRESAHA.112.267310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.