Abstract
Juvenile idiopathic arthritis (JIA) is the most common chronic inflammatory arthropathy of childhood. Juvenile idiopathic arthritis is believed to be a complex genetic trait influenced by both genetic and environmental factors. Twin and family studies suggest a substantial role for genetic factors in the predisposition to JIA. Describing the genetics is complicated by the heterogeneity of JIA; the International League of Associations for Rheumatology (ILAR) has defined seven categories of JIA based on distinct clinical and laboratory features. Utilizing a variety of techniques including candidate gene studies, the use of genotyping arrays such as Immunochip, and genome wide association studies (GWAS), both human leukocyte antigen (HLA) and non-HLA susceptibility loci associated with JIA have been described. Several of these polymorphisms (e.g. HLA class II, PTPN22, STAT4) are shared with other common autoimmune conditions; other novel polymorphisms that have been identified may be unique to JIA.
Associations with oligoarticular and RF-negative polyarticular JIA are the best characterized. A strong association between HLA DRB1:11:03/04 and DRB1:08:01, and a protective effect of DRB1:15:01 have been described. HLA DPB1:02:01 has also been associated with oligoarticular and RF-negative polyarticular JIA. Besides PTPN22, STAT4 and PTPN2 variants, IL2, IL2RA, IL2RB, as well as IL6 and IL6R loci also harbor variants associated with oligoarticular and RF-negative polyarticular JIA. RF-positive polyarticular JIA is associated with many of the shared epitope encoding HLA DRB1 alleles, as well as PTPN22, STAT4 and TNFAIP3 variants. ERA is associated with HLA B27. Most other associations between JIA categories and HLA or non-HLA variants need confirmation. The formation of International Consortia to ascertain and analyze large cohorts of JIA categories, validation of reported findings in independent cohorts, and functional studies will enhance our understanding of the genetic underpinnings of JIA.
Keywords: Juvenile idiopathic arthritis, Genetics, Association, Immunochip, Candidate gene, Genome-wide association studies
1. Introduction
Juvenile Idiopathic Arthritis (JIA), also previously termed Juvenile Rheumatoid Arthritis (JRA) or Juvenile Chronic Arthritis (JCA), is the most common chronic arthropathy of childhood and affects hundreds of thousands of children in the United States and around the world [1]. It is a serious disorder of childhood with potentially devastating consequences for the individual and society. JIA has the potential to disrupt growth, and adversely affect the joints resulting in permanent joint damage and long-term functional limitation and disability. Most children with JIA continue to have active disease years after onset, and in the majority of cases, the disease persists into adulthood, dispelling the notion that children can “outgrow” JIA [2,3]. Furthermore, JIA is associated with a substantial economic burden [4].
2. Overview of JIA genetics
JIA is believed to be a complex genetic trait influenced by both genetic and environmental factors [5]. There is substantial evidence for genetic contribution to JIA. Twin and affected sibling pair studies have supported a role for genetic susceptibility to JIA. Twin studies have shown that the monozygotic twin concordance rates for JIA range between 25 and 40%, a risk that is substantially greater than the population prevalence of 1 in 1000 [6–8]. In the largest twin study from the JIA affected sibling pair registry, 14 pairs of twins concordant for JIA were analyzed [9]. Of the 14 pairs of twins, one was discordant for gender and the rest were same-sex twins. All 13 pairs were concordant for onset and course. Twelve were concordant for presence/absence of anti-nuclear antibodies. The first twins to develop JIA did so a mean of 5.5 months before the second twins. In contrast, among the 104 non-twin affected sibling pairs in the registry, the difference in age at onset between the first and second sibling was 37 months, which was statistically different compared to twins. DNA was available on 11 twin pairs and all 11 were found to be monozygotic, in contrast to one third that were expected to be monozygotic based on the occurrence of twinning in the USA. Together, these twin studies provide strong evidence for genetic factors contributing to the susceptibility to JIA.
Affected sibling pair studies have also been carried out in JIA. Examination of the phenotypes of siblings affected with JIA has shown that sibling pairs were significantly concordant for sex, onset type and course type of JIA [10]. This was confirmed in a larger analysis of 164 sibling pairs [11]. Siblings were more likely to develop disease at the onset age, rather than calendar year. With the exception of number of joints at onset among children with polyarticular JIA, other clinical features did not differ between sporadic and multiple JIA cases.
Family studies can also provide evidence for genetic contribution to JIA. Although traditional multiplex families with numerous affected cases of JIA have been only rarely described, innovative approaches using a probabilistic record-linking of records in a JIA registry to the Utah Population Database have resulted in the identification of extended multiplex pedigrees with multiple affected individuals with JIA [12,13]. This approach identified 22 founders who had a significantly increased number of descendants with JIA (5–13 descendants) compared to what would be expected based on the prevalence of JIA. This study demonstrated that siblings of probands with JIA have an 11.6-fold increase in the risk of JIA (range 4.9–27.5, p < 2.6 × 10−8) compared to the general population [13]. Similarly, first cousins have a 5.8-fold increase in the prevalence of JIA (range 2.5–13.8, p < 6.07 × 10−5) compared to the general population. These observations also support a role for genetic factors in the predisposition to JIA.
In addition to clustering of JIA in some families, there is also evidence for familial autoimmunity in JIA. A case–control study of 110 families of probands with JIA and 45 healthy control families demonstrated that the prevalence of autoimmunity was three fold higher among relatives of JIA probands [14]. Of the 110 families of JIA probands, 81 families had at least one relative with a history of an autoimmune disorder, compared with only 15 of 45 families of control probands (OR 5.6 [2.5–12.7], p < 4 × 10−6). The prevalence of autoimmunity was higher among first degree relatives compared to second degree relatives. Thyroid autoimmunity was the most prevalent. In all, 52.3% of the relatives of the JIA probands were women, compared with 50.6% of the relatives of the controls. However, when those with autoimmune disorders were compared, 80.6% of the relatives of JIA patients and 80.0% of the relatives of controls were women. This is consistent with the female preponderance of autoimmunity. A follow up study demonstrated that the prevalence of autoimmunity was significantly higher among maternal aunts and maternal grandmothers compared to paternal aunts and paternal grandmothers, suggesting a maternal parent of origin effect in JIA [15]. These findings support the hypothesis that clinically distinct autoimmune phenotypes share common genetic susceptibility factors. The successful identification of variants that predispose to multiple autoimmune disorders, such as STAT4 and PTPN22, strongly supports this hypothesis. This was the basis for the success of the ImmunoChip consortium in the identification of shared susceptibility variants across many autoimmune phenotypes.
3. JIA categories
Genetic studies for JIA are complicated by the heterogeneity of the condition. There are seven categories of JIA as defined by International League of Associations for Rheumatology (ILAR) classification criteria [16]. By definition, all patients with JIA have a chronic arthropathy with symptoms beginning at <16 years of age, however each category of JIA varies in its clinical symptoms and associated laboratory studies [16]. Systemic-onset JIA is characterized by fever and rash in addition to arthritis; polyarticular JIA is characterized by arthritis in five or more joints in the first six months of disease, and can be rheumatoid factor (RF)-negative or RF-positive. Polyarticular RF-positive JIA is phenotypically similar to rheumatoid arthritis (RA) in adults. Oligoarticular JIA is characterized by arthritis involving one to four joints in the first six months of disease. In many genetic studies, subjects with polyarticular RF-negative JIA and oligoarticular JIA are grouped together since these categories of JIA are phenotypically similar with the exception of the number of joints involved, thus suggesting that they may have a similar genetic basis. Additionally, the ILAR classification of JIA includes enthesitis-related arthritis (ERA), psoriatic arthritis, and unclassified arthritis. While the various JIA categories are characterized by different clinical features, natural histories, and immunogenetic associations, they all share in common chronic inflammation of the synovium.
Over the past two decades there have been numerous studies which have identified potential susceptibility loci for JIA. In some cases these studies have included subjects with all JIA categories, and in others they have included subjects limited to particular JIA categories. Oligoarticular and polyarticular RF-negative categories of JIA have been the most often investigated for genetic associations. Identified polymorphisms also vary by ethnic groups. A variety of techniques have been employed to identify potential variants including candidate gene studies of single nucleotide polymorphisms (SNP) and genome wide association studies (GWAS) including the use of high density genotyping arrays such as Immunochip. In this review we will describe the known HLA and non-HLA susceptibility loci for specific subtypes of JIA including oligoarticular JIA/polyarticular RF-negative JIA, polyarticular RF-positive JIA, systemic JIA, ERA and psoriatic JIA.
4. Genetics of oligoarticular JIA/polyarticular RF-negative JIA
Oligoarticular JIA is the most common category of JIA, affecting up to 40% of all patients with JIA. Patients with oligoarticular JIA have 4 or fewer joints affected during the first six months of the disease. After 6 months following onset, if the arthritis is confined to 4 or fewer joints, the disease is referred to as “persistent” oligoarticular JIA, whereas when more than 4 joints are involved, it is referred to as “extended” oligoarticular JIA [16]. Oligoarticular JIA has its peak incidence between 2 and 4 years of age and it has a female/male ratio of 3:1. One of its distinctions from other forms of arthritis is that it is associated with asymptomatic chronic anterior uveitis, a form of inflammatory eye disease which is more common in patients who are anti-nuclear antibody (ANA) positive. Like oligoarticular JIA, polyarticular RF-negative JIA also tends to predominantly affect girls, and has an early age of onset. Children with polyarticular RF-negative JIA are frequently ANA positive and they have an increased risk of chronic anterior uveitis. Thus, these two categories of JIA, which account for ~70% of all JIA cases, are phenotypically similar with the exception in the number of joints involved, and the symmetric involvement typically noted in polyarticular RF-negative JIA.
4.1. HLA associations with oligoarticular JIA/polyarticular RF-negative JIA
The major histocompatibility complex (MHC) on chromosome 6 has more than 200 genes most of which are essential to the immune system. The genes encoding human leukocyte antigen (HLA) are also located in the MHC region. HLA genes are extremely polymorphic and have been shown to be associated with multiple autoimmune diseases including RA, celiac disease, type 1 diabetes mellitus (type 1 DM) and many others. Numerous associations between HLA alleles and JIA categories have been reported in multiple populations. Class I allele HLA:A2 is associated with oligoarticular and polyarticular RF negative JIA [17]. Susceptibility to oligoarticular JIA is also associated with HLA DRB1:01, DRB1:08, DRB1:11, DRB1:13, DPB1:02 and DQB1:04 [18]. Interestingly, HLA DRB1*04 and DRB1*07 are seen less frequently in children with oligoarticular JIA than controls, suggesting that they are protective against oligoarticular JIA [19]. Polyarticular RF negative JIA is associated with DRB1:08 and DPB1:03 [20].
A large study of 820 children with JIA and 273 healthy controls, reported by Hollenbach et al., in 2010 utilized high resolution HLA typing for class I and class II loci [21]. An HLA DRB1 effect which was similar between children with oligoarticular JIA and a subset of younger patients with polyarticular JIA was reported by the authors. HLA DRB1:11:03/11:04 conferred susceptibility to both oligoarticular JIA and younger polyarticular JIA patients, but not older patients with polyarticular RF-negative JIA. However, HLA DRB1:08:01 conferred increased risk of JIA in younger and older patients with both oligoarticular and polyarticular RF-negative JIA. HLA DRB1:15:01 conferred a very strong protective effect to both oligoarticular and polyarticular RF-negative JIA subjects.
This study also confirmed the age-specific effects of susceptibility conferred by HLA alleles previously reported by Murray et al who also reported that presence of multiple risk alleles resulted in an earlier onset of JIA (Fig. 1) [22]. Younger age at the onset of polyarticular JIA is mediated by DRB1:08:01 and DRB1:11:03/11:04. Younger age at the onset of polyarticular RF-negative JIA and both persistent and extended oligoarticular JIA was found to be mediated by DPB1:02:01 in the absence of the predisposing alleles DRB1:08:01 and 11:03/11:04. Some HLA alleles were associated with a reduced risk of JIA. Whereas HLA DRB1:15:01 was shown to reduce risk across the whole JIA cohort, HLA DRB1:04:01 and DRB1:0701 were protective against persistent oligoarticular JIA only. In addition to the HLA DRB1 associations, a modest association was observed between HLA DPB1:02:01 and JIA. Surprisingly, the disease predisposition mediated by DPB1:02:01 in individuals without any risk associated DRB1 alleles was large enough to overcome even the very strong protective effect observed for DRB1:15:01. Associations noted with HLA DQB1 variants were believed to be due to linkage disequilibrium. A summary of the HLA alleles and their effect on JIA risk are shown in Fig. 2.
Fig. 1.
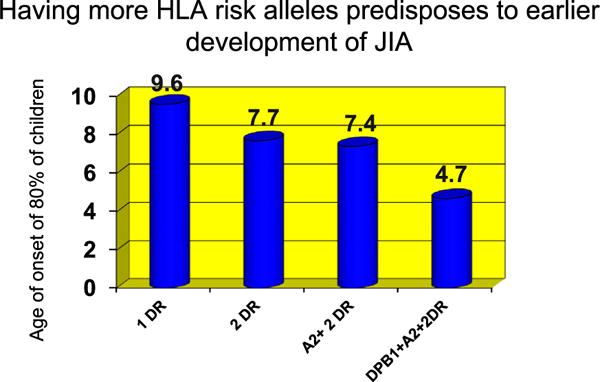
Age of onset of 80% of children with JIA with various combinations of HLA risk alleles. 80% of children with any 2 HLA DR risk alleles, HLA DPB1 and A2 alleles develop JIA by age 4.7 years. Adapted from Murray KJ, Moroldo MB, Donnelly P, Prahalad S, Passo MH, Giannini EH, et al. Age-specific effects of juvenile rheumatoid arthritis associated HLA alleles. Arthritis Rheum 1999; 42:1843–53.
Fig. 2.
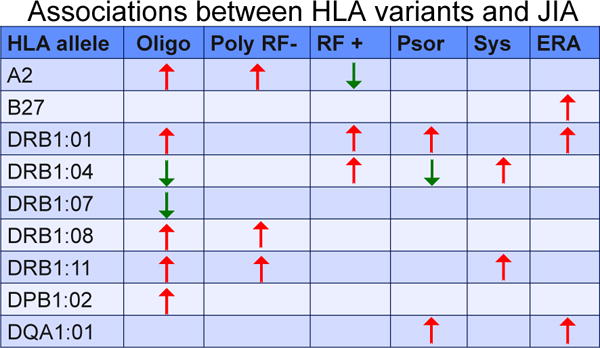
Summary of associations between selected HLA alleles and different categories of JIA. Some alleles that predispose to the risk of a category might be protective against another JIA category. From Murray KJ, Moroldo MB, Donnelly P, Prahalad S, Passo MH, Giannini EH, et al. Age-specific effects of juvenile rheumatoid arthritis-associated HLA alleles. Arthritis Rheum 1999; 42:1843–53, Thomson W, Barrett JH, Donn R, Pepper L, Kennedy LJ, Ollier WE, et al. Juvenile idiopathic arthritis classified by the ILAR criteria: HLA associations in UK patients. Rheumatology (Oxford) 2002; 41:1183–9, and Hollenbach JA, Thompson SD, Bugawan TL, Ryan M, Sudman M, Marion M, et al. Juvenile idiopathic arthritis and HLA class I and class II interactions and age-at-onset effects. Arthritis Rheum 2010; 62:1781–91.
In addition to these candidate gene studies over several decades, evidence of the crucial role played by genetic variants in the HLA region is provided by the recent GWAS studies of JIA. HLA variants demonstrated the strongest associations in the GWAS studies by Hinks et al and Thompson et al in 2010. [23,24], This was also confirmed in the ImmunoChip study in which the SNP rs7775055 located in the class II HLA region (DQ) was associated with oligoarticular and polyarticular RF-negative JIA with an odds ratio of 6.01 (95% CI 5.30–6.81; p < 3.1 × 10−174) [25]. Together, these studies confirm the critical role played by HLA variants in predis posing to risk of JIA.
4.2. Non-HLA associations with oligoarticular JIA/polyarticular RF-negative JIA
While numerous case control association studies between non-HLA variants and JIA have been published, most suffered from relatively modest sample sizes and heterogeneity of subjects included in the studies with most studies including several categories of JIA. Some associations were well established (PTPN22, STAT4, TNFAIP3) however many others had not been replicated. The best characterized loci demonstrating associations with oligoarticular and polyarticular RF-negative JIA were identified as a result of the international JIA Immunochip consortium which uncovered for the first time many loci at genome wide levels of significance, and others at suggestive levels of significance.
The Immunochip Consortium was established with a goal to investigate shared loci identified in GWAS across multiple autoimmune disorders [26]. The Immunochip contains about 200,000 SNPs, including dense coverage of the MHC region, and ~180 loci that have shown genome wide evidence of association with one or more of 12 autoimmune diseases [26]. The Immunochip consortium has been successful at identifying loci associated with many autoimmune disorders including celiac disease [27], inflammatory bowel disease [28], RA [29], and AS [30].
The International JIA Immunochip consortium published the results of the analysis in 2816 cases with oligoarticular and polyarticular RF-negative JIA and 13,056 controls (Fig. 3) [25]. In addition to confirming the 3 loci that have previously been associated at genome-wide level of significance (HLA, PTPN22 and PTPN2), 14 loci, several of which had been previously associated with JIA reached genome-wide level of significance for the first time [Table 1]. An additional 11 loci reached suggestive levels of significance. Of the 25 loci described in the Immunochip study 6 were replicated utilizing cases and controls from the Childhood Arthritis Risk Factor Identification Study (CLARITY); the replicated loci include C5orf56-IRF1 (rs4705862), ERAP2-LNPEP (rs27290), PRR5L (rs4755450), RUNX1 (rs9979383), RUNX3 (rs4648881), and UBE2L3 (rs2266959) [31]. Many of the loci are shared with RA, T1DM and celiac disease. The Immunochip study by Hinks et al also highlighted crucial pathways including IL-2 pathway in JIA pathogenesis.
Fig. 3.
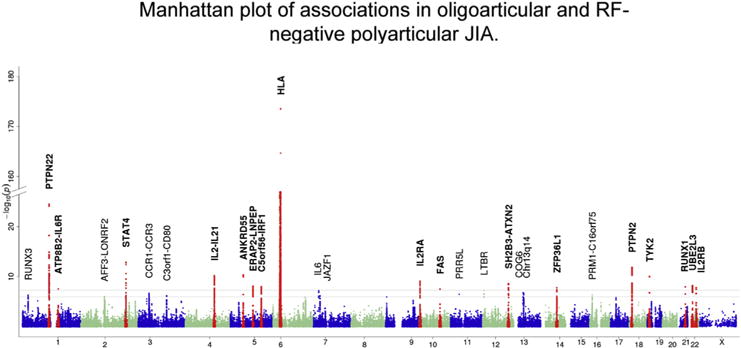
Manhattan plot of association statistics for oligoarticular and RF-negative polyarticular juvenile idiopathic arthritis risk loci. The upper black line indicates the threshold for genome-wide significance (P < 5 × 10−8). Loci reaching this threshold are highlighted in bold, and individual SNPs mapping to these loci are shown in red. The lower gray line indicates the threshold for suggestive association (5 × 10 −8 < P < 1 × 10−6). Reproduced from Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet 2013; 45:664–9.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Genome-wide significant associations in oligoarticular and RF-negative polyarticular JIA.
| Locus | Chr | SNP | Best p value | MAF control | MAF case | OR | SNP position |
|---|---|---|---|---|---|---|---|
| PTPN22* | 1 | rs6679677 | 3.2 × 10−25 | 0.10 | 0.14 | 1.59 | Intergenic |
| ATP8B2-IL6R | 1 | rs11265608 | 2.8 10−8 | 0.10 | 0.12 | 1.33 | Intergenic |
| STAT4 | 2 | rs10174238 | 1.3 × 10−13 | 0.23 | 0.28 | 1.29 | Intron |
| IL2-IL21 | 4 | rs1479924 | 6.2 × 10−11 | 0.29 | 0.24 | 0.79 | Intergenic |
| ANKRD55 | 5 | rs71624119 | 4.4 × 10−11 | 0.25 | 0.20 | 0.78 | Intron |
| ERAP2-LNPEP | 5 | rs27290 | 7.5 × 10−9 | 0.44 | 0.47 | 1.32 | Intron |
| C5orf56-IRF1 | 5 | rs4705862 | 1.0 × 10−8 | 0.44 | 0.39 | 0.84 | Intergenic |
| HLA DQB1-DQA2* | 6 | rs7775055 | 3.1 × 10−174 | 0.02 | 0.12 | 6.01 | Intergenic |
| IL2RA | 10 | rs7909519 | 8.0 × 10−10 | 0.11 | 0.08 | 0.72 | Intron |
| FAS | 10 | rs7069750 | 2.9 × 10−8 | 0.44 | 0.48 | 1.18 | Intron |
| SH2B3-ATXN2 | 12 | rs3184504 | 2.6 × 10−9 | 0.49 | 0.54 | 1.20 | Coding |
| ZFP36L1 | 14 | rs12434551 | 1.6 × 10−8 | 0.47 | 0.43 | 0.77 | Intergenic |
| PTPN2* | 18 | rs2847293 | 1.4 × 10−12 | 0.17 | 0.20 | 1.31 | Intergenic |
| TYK2 | 19 | rs34536443 | 1.0 × 10−10 | 0.05 | 0.03 | 0.56 | Coding |
| RUNX1 | 21 | rs9979383 | 1.1 × 10−8 | 0.37 | 0.33 | 0.78 | Intergenic |
| UBE2L3 | 22 | rs2266959 | 6.2 × 10−9 | 0.19 | 0.22 | 1.24 | Intron |
| IL2RB | 22 | rs2284033 | 1.6 × 10−8 | 0.44 | 0.39 | 0.84 | Intron |
Chr: chromosome; SNP: Single nucleotide polymorphism; MAF: Minor allele frequency; OR: Odds ratio.
indicates associations that had been described previously at genome wide levels of significance.
From Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet 2013; 45:664–9.
Prior to the report of the ImmunoChip associations, the strongest non-HLA genetic association identified by means of candidate gene case control studies with JIA was protein tyrosine phosphatase nonreceptor 22 (PTPN22), a gene located on chromosome 1p13.3-13.1 which encodes lymphoid protein tyrosine kinase, an enzyme which negatively regulates T cells. It is thought that functional mutations in the PTPN22 gene lead to T cell activation and the subsequent promotion of autoimmune disease. Mutations in PTPN22 have been associated with multiple autoimmune conditions including rheumatoid arthritis, systemic lupus erythematosus (SLE), autoimmune thyroid disease and T1DM [32]. Thus, PTPN22 variants appear to be generally predisposing to risk of autoimmunity. Interestingly, there are some studies that suggest that those variants that increases the risk of autoimmunity might confer protection against tuberculosis [33,34].
Several studies have confirmed associations between PTPN22 variants and JIA [35,36]. An association between a missense SNP (rs2476601, 1858C→T) in PTPN22 and JIA was first reported by Hinks et al in 2005 [37]. In this study the PTPN22-1858T allele had a frequency of 15% in JIA cases versus 10.3% in controls (p = 0.0005). In a brief communication Viken et al., also reported an increased odds of the same SNP of PTPN22 in patients with JIA compared to controls (OR = 1.58, p = 0.001) [38]. These findings have been replicated in a JIA GWAS study and in candidate gene studies limited to distinct ethnic groups [39,40]. A recent meta-analysis of PTPN22 associations in JIA confirmed the strong association between this variant and JIA (OR 1.44, p < 0.0001) [35]. Subtype meta-analyses of the PTPN22 variant revealed associations between oligoarticular and polyarticular RF-negative JIA that remained significant after multiple hypothesis correction (p 0.0007, and p < 0.0005, respectively). Thus the association between PTPN22 and oligoarticular and polyarticular RF-negative JIA has been validated in multiple studies.
Aside from HLA and PTPN22, the other genetic association to reach genome wide significance in early GWAS studies was PTPN2, which like PTPN22 encodes a protein tyrosine phosphatase involved in T cell regulation. A novel association between polymorphisms in PTPN2 and oligoarticular JIA and polyarticular RF-negative JIA was described in 2010, by Thompson et al. [24]. This association was confirmed in replication cohorts. Other novel polymorphisms identified in this study were mutations in the conserved oligomeric golgi 6 (COG6) gene and polymorphisms in the angiopoetin-1 (ANGPT1) gene. Both COG6 and ANGTP1 polymorphisms have been associated with other autoimmune conditions including autoimmune thyroiditis [41,42].
The gene encoding the signal transducer and activator of transcription factor 4 (STAT 4) is also strongly associated with JIA and other autoimmune conditions including RA, SLE, type I DM, Sjogren’s syndrome and inflammatory bowel disease [43]. STAT4 is thought to play an important role in T cell differentiation. A case control study by Prahalad et al., demonstrated an association of the G→T substitution in intron 3 of STAT4 (rs7574865) with JIA [44]. This finding has been replicated in several other studies [23,39,44,45]. Variation in the C12orf30 region on chromosome 12 has been associated with JIA and several other autoimmune conditions, including Type I DM [24,44,45].
Multiple studies have demonstrated associations between polymorphisms in the TNFA gene and JIA [46–50]. TNFA encodes the pro-inflammatory cytokine tumor necrosis factor-alpha; it is located on chromosome 6 in the MHC region. While mixed results have been reported for various TNF polymorphisms, the TNFA G-308A allele (rs1800629) has been found to be associated with JIA in several studies, including a meta-analysis [35]. In addition to investigations of susceptibility, there has also been an investigation of response to therapy associated with polymorphisms in TNFA. Schmeling et al found that patients with polyarticular RF negative JIA with the genotype TNFA-308GG responded better to therapy with the anti-TNF agent etanercept than patients with the –308GA or AA genotype [51].
IL2RA which encodes the interleukin receptor alpha (also known as CD25) is another gene with strong associations with RA and other autoimmune conditions as well as JIA. SNP rs2104286 was significantly associated with JIA in a 2009 study [52]. This is an intriguing gene for JIA pathogenesis because of the role of interleukin 2 in promoting inflammation in autoimmune disease via binding to the IL2 receptor. The TRAF1-C5 locus on chromosome 9, which is a well described risk factor for RA, has also been associated with JIA in case control and GWAS studies [53]. TRAF1 encodes tumor necrosis factor receptor-associated factor 1, a regulator in the TNF pathway.
A GWAS study used the Affymetrix GeneChip 100K array identified several SNPs in the VTCN gene to be associated with JIA [54]. It should be noted that 97% of the 279 subjects in the discovery cohort and 80% of the 321 subjects in the validation cohort had oligoarticular or polyarticular RF-negative JIA (the rest had polyarticular RF-positive JIA). This was followed by fine mapping of 10 VTCN1 variants in a third cohort of 654 children with JIA (of whom only 64% had oligoarticular and polyarticular RF-negative JIA). VTCN1 encodes a costimulatory molecule, B7-H1, expressed on activated T and B cells, monocytes and dendritic cells and it may play a role in T cell inhibition and attenuation of the inflammatory response. Notably, VTCN1 variants were not associated at genome wide significance in the Immunochip study of JIA. VTCN1 rs10923223 polymorphism has also been associated with disease course in JIA [53]. In support of a critical role for this gene in inflammatory arthritis, variants in VTCN1 were found to be associated with RA in a Dutch cohort [55].
Macrophage migration inhibitory factor (MIF) is a protein which is a cofactor in T-cell activation and promotes pro-inflammatory activity include the secretion of pro-inflammatory cytokines. Polymorphisms in the MIF gene have been associated with various forms of JIA including oligoarticular JIA and polyarticular RF negative JIA [36,56,57]. The MIF-173*C allele has been reported to be associated with susceptibility to JIA in multiple ethnic groups [36]. One study found that children with oligoarticular JIA who carry the MIF-173*C allele are more likely to relapse within three months of intra-articular glucocorticoid injections, however there was no difference in long-term outcomes based on MIF polymorphisms [58].
In a 2014 study Reinards et al performed a genetic association study which identified a polymorphism in the CD226 (DNAM1) gene in association with JIA [59]. DNAX accessory molecule-1 is a type 1 membrane protein belonging to the Ig-supergene family. It is expressed on T and NK cells and is involved in co-stimulation of these cells. This association was not seen on a follow-up Immunochip study but the association was confirmed in a meta-analysis [59]. Other polymorphisms associated with JIA are located in the IL2-IL21 region, the AFF3 gene and ANKRD55 [45,59,60].
Genes which may be protective against JIA have also been identified. CCR5 is a chemokine which recruits T helper cells to the synovium, promoting synovitis and joint inflammation. Synovial T-cells from subjects with JIA/RA express high levels of the chemokine receptor CCR5. A 32-bp deletion in the gene encoding CCR5 has been implicated in protection against RA. A meta-analysis of published association studies confirmed a protective role for CCR5-Δ32 in RA [61]. Subsequent investigation in a JIA cohort has shown that two functional CCR5 variants protect against early-onset JIA [62]. This was confirmed in an independent cohort from the UK, and accompanying meta-analysis by Hinks et al. [63] Variations in the TNFAIP3 gene are associated with both protection from or increased risk of RA and JIA [44]. The T allele at TNFAIP3 SNP rs10499194 demonstrates protective effect (OR 0.74, p < 0.004) while the minor allele (A) at TNFAIP3 SNP rs6920220 conferred an increased risk of JIA in the cohort studied (OR 1.30, p < 0.02).
4.3. Summary of genetic associations with oligoarticular JIA/polyarticular RF-negative JIA
The cumulative data from genetic studies of oligoarticular and polyarticular JIA allow us to draw several conclusions. While some discovered genetic associations are very strong (e.g., HLA variants generally have high odds ratios), these remain the minority, and most other associations are of modest magnitude. The risk alleles identified in genomic screens are relatively common in the general population, have only a modest effect on risk, and together explain only a small part of the variance in disease risk. Whereas the actual causal variants for most risk loci identified to date remain to be determined, some themes have emerged: most variants are located in intronic or intergenic regions, many risk loci and variants are associated with more than one autoimmune disease, and many genes are associated with discrete biological pathways [64]. Finally, it is clear that identifying the numerous risk factors for complex rheumatic diseases in general, and pediatric rheumatic diseases in particular requires international collaboration to investigate large cohorts. The Immunochip study is a great example of a successful multinational collaboration.
5. Genetics of polyarticular RF-positive JIA
Polyarticular RF- positive JIA, which can considered the childhood onset of seropositive adult rheumatoid arthritis, represents about 10% of all cases of JIA. Patients are typically diagnosed in late childhood/adolescence. As with adult RA, patients have a chronic symmetric, erosive polyarticular arthritis. To have polyarticular RF-positive disease, the ILAR classification requires the involvement of 5 or more joints [16]. Also, by definition, patients with polyarticular RF + positive JIA must have two positive tests for IgM rheumatoid factor (RF). Although anti-cyclic-citrullinated peptide (anti-CCP) antibody status is not part of the ILAR criteria, children with childhood-onset RA also commonly have anti-CCP antibodies. In order to improve the sensitivity of diagnosing childhood-onset RA, we have proposed prioritization of RF/anti-CCP positivity over specific exclusions (e.g. number of joints involved), along with inclusion of anti-CCP, in future revisions of the JIA classification criteria [65]. Thus, some of the following genetic associations described might include all children with RF/CCP positive JIA, albeit a substantial majority of them would fall under the polyarticular RF-positive JIA category defined by the ILAR criteria.
5.1. HLA associations with polyarticular RF-positive JIA
An association between seropositive rheumatoid arthritis (RA) in adults and certain HLA-DR molecules has been known for a long time. In 1987 the Shared epitope (SE) hypothesis was proposed by Gregersen et al, based on the observation that different RA associated HLA-DR molecules shared some common amino acid motifs [66]. There have been seven studies of polyarticular RF-positive JIA and HLA [67–73]. Earlier studies had small number of subjects (13–52), and used lower resolution HLA-DR typing. Cases with polyarticular RF-positive JIA had a significantly higher frequency of SE compared to controls. In an investigation of 149 Non-Hispanic White children with polyarticular RF-positive JIA, 76% carried 1 or 2 SE alleles compared with 46% of 373 controls (OR = 3.81, 95% CI: 2.4–6.0; p < 1 × 10−7) [73]. Also notable was that a significantly greater proportion of cases carried 2 copies of the SE allele compared to controls (33% vs 5.4%, OR = 8.6 (4.7–15.8); p < 1 × 10−7). When individual SE encoding HLA DRB1 alleles were examined, HLA DRB1:01:01, 04:01, 04:04, 04:05, 04:08, and 10:01 were confirmed to be associated with polyarticuar RF-positive JIA (Fig. 4). Significantly more patients than controls were heterozygous for HLA DRB1 04:01/04:04 compared to controls (10 of 149 patients vs 1 of 373 controls; OR 28.8). These firmly establish the role for SE encoding HLA DRB1 alleles in conferring a risk of susceptibility to polyarticular RF-positive JIA.
Fig. 4.
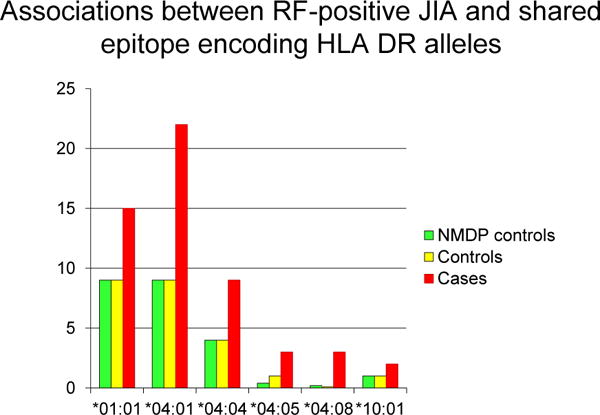
Frequencies of shared epitope encoding HLA DRB1 alleles among children with RF-positive JIA, healthy controls and external controls reported in the National Marrow Donor Program Registry. Adapted from Prahalad S, Conneely KN, Jiang Y, Sudman M, Wallace CA, Brown MR, et al. Susceptibility to childhood-onset rheumatoid arthritis: investigation of a weighted genetic risk score that integrates cumulative effects of variants at five genetic Loci. Arthritis Rheum 2013; 65:1663–7.
In addition, this study evaluated if there was a hierarchy of the SE alleles in polyarticular RF-positive JIA by classifying alleles into high risk (S2: KRAA at positions 71–74 of the DRβ1 chain e.g., 04:01 and 13:03), intermediate risk (S3:Q/R-RRAA at positions 70–74; e.g., 01:01, 01:02, 04:04, 04:05, 04:08, 10:01, 14:02, 14:06), or low risk (L; the other DRB1 alleles) as proposed by Du Montcel et al. [74] Genotype specific risk estimates with L/L as the reference group suggested a hierarchy of risk. For subjects with a single copy of S2 (S2/L genotype) or S3 (S3/L genotype) the OR was 2.2 and 2.6 compared to L/L respectively. For those with two copies of S2 (S2/S2) or S3 (S3/S3), the OR was 7.7 and 7.2 respectively. However, for those with a copy of S2 and S3 (S2/S3 genotype) the OR was substantially higher at 22.3. This suggests that individuals with DRβ1 chains containing both the KRAA and Q/R-RRAA sequences are especially prone to polyarticular RF-positive JIA.
While most genetic studies of JIA are confined to Non-Hispanic White cohorts, small cohorts of Hispanic subjects (n = 25), and African American cases (n = 21) with polyarticular RF-positive JIA have been investigated for association with SE encoding HLA DRB1 alleles. For African American cases, the OR was 1.84 (0.84–3.82; p < 0.08) and for Hispanic cases the OR was 2.88 (1.59–5.21, p < 0.0001) compared to appropriate controls from the National Marrow Donor Program Registry. Together these studies confirm that the variants in the HLA DRB1 gene play a major role in shaping RF-positive disease in childhood similar to that seen in adults with RA. Recently Raychaudhuri et al have shown that the association between seropositive RA and MHC can be explained by just five amino acid positions in three HLA proteins [75]. Similar studies of children with polyarticular RF-positive JIA, and comparisons with adult RA would be of great interest.
5.2. Non-HLA associations with polyarticular RF-positive JIA
Unlike oligoarticular JIA and polyarticular RF negative JIA, there have only been limited investigations of non-HLA risk loci for association with polyarticular RF-positive JIA. In a study of 155 children with polyarticular RF-positive JIA and 684 healthy autoimmunity free controls, subjects were genotyped for 5 variants in the PTPN22, TRAF1/C5, STAT4 and TNFAIP3 loci, all of which have been associated with RA [76]. Polyarticular RF-positive JIA was associated with TNFAIP3 rs10499194 (OR: 0.60 (0.44–0.83)), PTPN22 rs 2476601 (1.61 (1.11–2.31)), and STAT4 rs7574865 (OR 1.41 (1.06–1.87)) variants. It was notable that the association between TNFAIP3 rs10499194 and polyarticular RF-positive JIA in this study [76] was more pronounced than with adult RA, as the confidence intervals of the odds ratios did not overlap with those of a meta-analysis of RA [77]. Using the largest cohort of patients with polyarticular RF-positive JIA, this study demonstrated that TNFAIP3, STAT4 and PTPN22 variants were associated with RF-positive JIA in a similar magnitude and direction as had been observed in adult RA. The association between RF-positive polyarticular JIA and PTPN22 C1858T variant has been confirmed in a meta-analysis as well [35].
In addition to studies of association between individual HLA and non-HLA variants with polyarticular RF-positive JIA, there has been an investigation of a cumulative weighted genetic risk score (wGRS) with polyarticular RF-positive JIA. Using HLA-DRB1 and PTPN22, TRAF1/C5, STAT4 and TNFAIP3 genotypes in 149 cases with polyarticular RF-positive JIA and 373 healthy controls, a wGRS was computed for each subject [76]. Weights were based on natural log of the published OR for each allele investigated. The wGRS was significantly different between cases and controls (3.32 (2.47–4.45); p < 2 × 10 −16). Even after excluding the HLA there was a significant difference (2.18 (1.39–342); p < 9.7 × 10 −4). Examination of the wGRS quintiles suggested a pattern of increasing disease risk with each wGRS quintile. Logistic regression confirmed that individuals in the third to the fifth quintiles of the wGRS had three to twelve fold increase in the odds of disease compared to the baseline. The mean age of onset did not differ between individuals in the top and bottom quintiles of the wGRS, similar to a study in adults with RA [78]. However, there was a significantly greater proportion of males with polyarticular RF-positive JIA in the upper quintiles compared to lower quintiles of wGRS. There was also an interaction between wGRS and sex such that the wGRS had a greater effect size in males, suggesting that higher wGRS appear to increase the risk of polyarticular RF-positive JIA especially among males who generally have a lower risk.
5.3. Summary of genetic associations with polyarticular RF-positive JIA
Polyarticular RF-positive JIA in childhood is strongly associated with HLA variants similar to other JIA categories as well as adult RA. It is notable that the HLA alleles encoding the SE are associated with Polyarticular RF-positive JIA in similar magnitudes and direction as adult RA. There also does not appear to be a difference in the proportion of children who carry two copies of SE in general, the frequency of heterozygosity for S2/S3P alleles was higher in a cohort of patients with childhood onset RA compared to adults with RA [73]. The non-HLA associations appear to be similar between RA and polyarticular RF-positive JIA as well. Analysis focusing on rare variants with large effects or higher cumulative load of genetic risk factors might be influencing the onset of RF/CCP positive inflammatory arthritis in childhood.
6. Genetics of systemic onset JIA
In North America and Europe, systemic onset JIA accounts for about 10% of all cases of JIA. It affects males and females equally, which is a distinguishing feature from other forms of JIA (except ERA) which have a female predominance. Systemic onset JIA occurs throughout the pediatric age spectrum from toddlerhood to adolescence. This is in contrast to oligoarticular JIA and RF-negative polyarticular JIA which tend to have its onset in early childhood (<10 years of age). Perhaps the most striking features which distinguish systemic onset JIA from other categories of JIA is the presence of systemic symptoms at disease onset with spiking fevers and a characteristic rash; hepatosplenomegaly, lymphadenopathy and serositis are also common manifestations. Patients with systemic onset JIA have elevated inflammatory markers including ESR, CRP and ferritin. Macrophage activation syndrome is a well described complication of systemic JIA but is infrequently seen with other forms of JIA. Newer treatments include biologics which block the active cytokines in systemic onset JIA including interleukin 1 (IL-1) and interleukin 6 (IL-6). These distinguishing clinical features have led some to argue that systemic onset JIA should be considered an auto-inflammatory syndrome as opposed to a subcategory of JIA, wherein most categories appear to be autoimmune disorders [79].
6.1. HLA associations with systemic onset JIA
Associations with MHC Class II have been described for systemic onset JIA. These include HLA DRB1:04:05 and DQB1:04:01 [80]. In a Mexican population HLA-DRB1:01 and HLA-DRB1:04 were strongly associated with JIA [81]. A major limitation of studying the genetics of the systemic JIA has hitherto been the rarity of this condition. However the International Childhood Arthritis Genetics Consortium (INCHARGE) performed a genome wide meta-analysis if systemic JIA, obtained from 988 children with systemic JIA and 514 healthy control subjects as well as 7370 additional healthy controls. Since the cohort had been derived from many international sites, the dataset was analyzed after dividing the cohort into 9 strata by country of origin [82]. Meta-analysis of the MHC locus identified two strong association signals. The first was around HLA DRB1 (OR 1.5, p < 1.6 × 10−10). The second signal of association was located between BTNL2 and HLA-DRA (OR 2.2; p < 7.1 × 10 −15, OR 2.2). Univariate regression analysis suggested that these two markers likely represented independent sources of risk to systemic JIA. This meta-analysis of systemic JIA also identified a novel genomic region on chromosome 1 as a systemic JIA susceptibility locus adjacent to LOC284661, which encodes a long, intergenic noncoding RNA. Data from this large multinational genome wide meta-analysis strongly implicates the class II MHC molecule HLA DR in the pathogenesis of systemic JIA.
6.2. Non-HLA associations with systemic onset JIA
Several studies have identified potential genetic risk factors for the development of systemic onset JIA. The first study suggesting a genetic association for systemic onset JIA was a 1998 study from the United Kingdom (UK) which examined polymorphisms in the 5′ flanking region of the IL6 gene among 92 children with systemic JIA; a G/C polymorphism was identified at position –174 [83]. The authors found a lower frequency of the CC (vs GG or GC) in patients with systemic onset JIA compared to controls. In in vitro studies, cells transfected with the CC construct demonstrated lower expression of IL-1 when stimulated with LPS. Among 102 healthy controls, CG homozygotes had fasting plasma levels of IL-6 twice as high as those homozygous for the C allele. The authors concluded that the CC genotype may have a protective effect for systemic onset JIA.
In 1999 a Japanese group examined polymorphisms in the 5′ flanking region of the TNFA gene in 111 JIA patients, including 50 subjects with systemic onset JIA, and found a significantly higher frequency of the compared –1031C, –863A and –857T alleles in subjects with systemic JIA to controls [80]. Subjects with both TNFA-857T allele and the DRB1:04:05 allele had an increased OR of 3.84 for systemic JIA. In 2005 a Spanish group reported on the association between the TNFA -308 allele and systemic onset JIA [84]. In 2001 Donn et al identified a 5′ flanking region polymorphism of the MIF gene which encodes the macrophage migration inhibitory factor in patients with systemic onset JIA vs controls (MIF-173*C) [85]. The presence of the MIF-173*C allele introduced an AP-4 transcription factor binding site which was thought to alter MIF expression. The presence of the MIF-173*C allele has also been associated with worse outcome in systemic onset JIA [86].
A 2006 study from the UK identified increased prevalence of a polymorphism in the interleukin-10 (IL10) gene family among patients with systemic onset JIA [87]. The allele was associated with low expression of IL-10; IL-10 has been shown to suppress the release of pro-inflammatory cytokines such as IL-6 and tumor necrosis factor (TNF) alpha. Variants in the IL1 gene family have also been associated with systemic onset JIA [88]. Using a tagged SNP approach the investigators identified three IL-1 ligand cluster SNPs (r6712572, rs2071374 and rs1688075) and one Il-1 receptor cluster SNP (rs12712122) that were seen more frequently in systemic onset JIA patients vs controls in a case–control association analysis. More recently Wakil et al described the association of a mutation in the LACC1 gene in a familial form of systemic onset JIA affecting 5 Saudi Arabian families [89]. LACC1 encodes the enzyme laccase domain-containing 1 and has previously been identified as a risk variant for leprosy and Crohn’s disease [90].
Since it has been suggested that systemic onset JIA should be considered an autoinflammatory condition, it is not surprising that genetic variants associated with inherited forms of auto-inflammatory syndromes have been associated with systemic JIA [91,92]. These include SNPs in the interleukin 1 ligand cluster, TNFRSF1A and MVK genes.
6.3. Summary of genetic associations with systemic onset JIA
Systemic onset JIA is a distinct category of juvenile arthritis, felt by some to be an autoinflammatory (vs an autoimmune) condition. The genetics of systemic onset JIA is distinct from other subtypes of JIA, and there does appear to be some overlap between genetic susceptibility to systemic onset JIA and other traditional auto-inflammatory syndromes. Improved understanding of the genetics of systemic onset JIA may help clarify these relationships further. However, emerging information suggests that the class II MHC molecule HLA DR might play a role in susceptibility to systemic JIA as well. Non HLA associations demonstrating the involvement of genes encoding several pro-inflammatory cytokines suggest therapeutic targets for systemic JIA many of which are already in clinical use.
7. Genetics of enthesitis-related arthritis (ERA)
As defined by the ILAR classification, ERA (previously termed juvenile ankylosing spondylitis or seronegative arthritis and enthesitis (SEA) syndrome) is diagnosed in a child with arthritis AND enthesitis or in a child with arthritis OR enthesitis as well as two of the following: sacroiliac joint tenderness and/or inflammatory back pain, HLAB27 positive, family history of one first or second degree relative with HLAB27 associated disease, acute anterior uveitis or onset of arthritis in a boy after 6 years of age [16]. There are very few reports of investigations focused on the genetics of ERA in contrast to the efforts towards unraveling the genetics of ankylosing spondylitis in adults.
7.1. HLA associations with enthesitis-related arthritis
There is a well described between HLA B27 and ERA; HLA B27 has been reported in 60–90% of patients with ERA [93]. In a study of HLA B27 in 410 Norwegian children with JIA, the prevalence of HLA B27 was 72% among children with ERA category of JIA, but was found in 5–21% of children with other JIA categories, which likely reflects the higher prevalence of HLA B27 in Scandinavian populations [94]. HLA B27 positive boys tended to have an older age of onset and had clinical signs of sacroiliitis. HLA B27 was negatively associated with long term remission status, although this could reflect the association with features such as sacroiliitis. Patients with ERA who are HLA B27 positive are more likely to have a chronic course with the eventual development of axial disease. In a study of Latvian patients with ERA the most common HLA B27 allele identified was HLAB:27:05 [95]. Although the association between ERA and HLA B27 is the best characterized, there has been an investigation of associations between class II HLA alleles and ERA in a small cohort of 34 children [72]. ERA was found to be associated with HLA DRB1:01 (OR 3.6), DQA1:01:01 (OR 2.8), and DQB1:05 (OR 3.5), and a haplotype carrying these alleles (OR 4.9, 95% CI, 2 to 12.1). This study also found that DRB1:07 was protective against ERA (OR 0.3), as was DPB1:02:01 (OR 0.1). In another study of 45 Norwegian children with ERA, presence of HLA DRB1:01 was a predictor of failure to achieve disease remission [96]. This study also found that HLA DPB1:02 was protective, similar to findings reported by Thomson et al. [72].
7.2. Non-HLA associations with enthesitis-related arthritis
Several non-HLA polymorphisms have been associated with ERA. Endoplasmic reticulum aminopeptidase 1 (ERAP1) is a susceptibility locus previously associated with ankylosing spondylitis, a related condition to ERA which is diagnosed primarily in adults. In 2011 Hinks and colleagues genotyped SNPs in the ERAP1 gene in 1054 JIA cases (including 74 cases with ERA) and 5200 controls and found a strong association between the ERAP1 gene and the ERA category of JIA [97].
In a 2010 study, Hinks and colleagues investigated type I DM and celiac disease susceptibility loci and their association with JIA [52]. They identified a polymorphism in the IL12A gene which was associated with ERA, but not with other subtypes of JIA. A subsequent study identified a micro RNA polymorphism (miR-146a rs2910164) which was significantly associated with ERA [98]. The same authors investigated polymorphisms in the IL1 gene cluster and toll like receptors 2 and 4 genes but did not find an association with ERA [99,100]. At least two studies have identified a high frequency of MEFV gene mutations among Mediterranean patients with ERA but without the classical symptoms of Familial Mediterranean Fever which is associated with homozygous mutations in MEFV [101,102].
While AS in adults has been associated with IL23R variants, which have also been implicated in inflammatory bowel disease and psoriasis [93], ERA was not associated with IL23R in the study by Hinks et al., although the small size of the ERA cohort (n = 63) might have resulted in the study being underpowered [97].
7.3. Summary of genetic associations with ERA
The relationship between HLA B27 and ERA is well described. In addition, several non-HLA polymorphisms have been identified which appear to confer an increased risk of ERA. The impact of HLA and non-HLA polymorphisms on the risk of ERA is not well known and may deserve further study. There are only limited studies of the genetics of ERA, and similar to efforts in other JIA categories with limited prevalence, multicenter studies of genetics of ERA are needed.
8. Psoriatic JIA
The ILAR criteria define psoriatic JIA as the presence of both arthritis and psoriasis, or the presence of arthritis, with at least two of the following criteria: family history of psoriasis in a first degree relative, dactylitis, and nail pitting or onycholysis [16]. It has been proposed that psoriatic JIA might actually comprise two distinct subgroups, an older onset phenotype with male predominance, enthesitis and axial involvement, and an early-onset subphenotype resembling early onset oligoarticular and polyarticular JIA, including female predominance and ANA positivity [103]. Psoriatic JIA is among the least investigated categories of JIA in genetic association studies. The following studies summarize the published literature.
8.1. HLA associations with psoriatic JIA
A small cohort of 28 children with juvenile psoriatic arthritis, which can be considered to be equivalent to psoriatic JIA, was described by Hamilton et al in 1990 [104]. HLA genetic associations, uncorrected for multiple comparisons, suggested an increase in HLA A2 in patients (71% of patients compared to 45% among 276 controls) and HLA B27 (32% among cases vs. 8% among controls). The prevalence of HLA B27 was 11% among children with psoriatic JIA. In a subsequent study of 70 children with, only HLA B27 was found to statistically significantly elevated among cases with psoriatic JIA (20% vs 6% among 310 controls) [105]. Other associations, including with HLA A2, were not statistically significant.
In their investigation of associations between class II HLA genes and different ILAR categories of JIA, Thomson et al included 37 cases with psoriatic JIA [72]. An association was found between psoriatic JIA and DRB1:01 (OR 2.7), DQA1:01:01 (OR 4.2) and DQB1:05 (OR 4.4). All three HLA variants were also associated with ERA. DRB1:01 was protective against psoriatic JIA (OR 0.3), as was DQA1:03 (OR 0.3), and DQB1:03 (OR 0.5).
8.2. Non-HLA associations with psoriatic JIA
Since psoriatic JIA is also considered to be an auto-inflammatory phenotype, variants predisposing to other hereditary periodic fever syndromes have been investigated for an association in cohorts of children with psoriatic JIA. In a study of 950 children with JIA and 728 healthy controls, Day et al investigate 41 SNPs across NLRP3, NOD2, MEFV and PSTPIP1 loci [106]. After correction for multiple testing, two genotype associations remained significant in the group of patients with psoriatic JIA category, including MEFV SNP rs 224204 (p 0.25) and NLRP3 SNP rs3806265 (p = 0.04). These have not been reported in adult GWAS and have not been replicated in other pediatric cohorts. Hinks et al investigated an association between a SNP in IL23R and JIA in 1244 cases of JIA and 5200 healthy controls. In this cohort there were 93 subjects with psoriatic JIA. An association was found between IL23R SNP (rs11209026) and psoriatic JIA (OR 0.4 95% CI 0.16–0.98, p = 0.04) [97].
8.3. Summary of genetic associations with psoriatic JIA
The heterogeneity of psoriatic JIA, the relative rarity of this condition and lack of well-powered studies limit our ability to understand this subphenotype of JIA. However, both HLA and non-HLA associations have been reported with this phenotype. In contrast, the genetics of psoriasis in adults have been successful in identifying IL-23 and NF-kB pathways as being associated with the pathogenesis of psoriasis [107]. Multinational collaborations of psoriatic JIA to replicate these findings, and to query if there are specific pediatric associations are needed.
9. Conclusions
Juvenile idiopathic arthritis is a complex heterogeneous entity with multiple disease phenotypes included under that broad umbrella of “JIA”. While earlier studies often grouped all children with JIA in genetic studies, more recent studies have focused on the differences in categories and have reported data based on JIA categories, allowing us to understand the differences in genetic association between various categories. For oligoarticular and RF-negative polyarticular JIA, there are numerous associations with genome wide levels of significance. For most categories of JIA, there are only limited number of studies which have largely not been confirmed in independent cohorts. Given the clinical heterogeneity, it is not surprising that genetic susceptibility and associated polymorphisms also vary according to the JIA category. As the number of the known associated variants grows it will become essential to determine the magnitude of the contribution of each variant singly and in combination, to the development of the different JIA categories. Furthermore, the identification of variants would improve the understanding of the pathogenesis of different JIA categories, as well as allow us to develop better diagnostic and therapeutic options. Genetic studies can play a major role in refining future JIA classification criteria, by allowing us to incorporate genomic information in addition to clinical and family history variables that currently play a major role in classification of JIA. Finally, in the era of personalized medicine, it is essential to understand the contribution of genetics to treatment response and outcome. In order to accomplish this, it is likely that multinational collaborative studies will be needed to assess genetic risk between the distinct JIA phenotypes and among various ethnic groups.
Acknowledgments
Dr. Prahalad is supported by grants from The National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01-AR060893), The Marcus Foundation Inc. and The Arthritis Foundation. Dr. Hersh is supported by a grant from the The National Institute of Arthritis and Musculoskeletal and Skin Diseases (K23-AR066064). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Arthritis and Musculoskeletal and Skin Diseases or the National Institutes of Health. Dr. Prahalad has served on an advisory board for Novartis but this had no influence on the contents of this publication.
References
- 1.Oen KG, Cheang M. Epidemiology of chronic arthritis in childhood. Semi Arthritis Rheum. 1996;26:575–591. doi: 10.1016/s0049-0172(96)80009-6. [DOI] [PubMed] [Google Scholar]
- 2.Fantini F, Gerloni V, Gattinara M, Cimaz R, Arnoldi C, Lupi E. Remission in juvenile chronic arthritis: a cohort study of 683 consecutive cases with a mean 10 year followup. J Rheumatol. 2003;30:579–584. [PubMed] [Google Scholar]
- 3.Minden K, Niewerth M, Listing J, Biedermann T, Bollow M, Schontube M, et al. Long-term outcome in patients with juvenile idiopathic arthritis. Arthritis Rheum. 2002;46:2392–2401. doi: 10.1002/art.10444. [DOI] [PubMed] [Google Scholar]
- 4.Minden K, Niewerth M, Listing J, Biedermann T, Schontube M, Zink A. Burden and cost of illness in patients with juvenile idiopathic arthritis. Ann Rheum Dis. 2004;63:836–842. doi: 10.1136/ard.2003.008516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glass DN, Giannini EH. Juvenile rheumatoid arthritis as a complex genetic trait. Arthritis Rheum. 1999;42:2261–2268. doi: 10.1002/1529-0131(199911)42:11<2261::AID-ANR1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 6.Ansell BM, Bywaters EG, Lawrence JS. Familial aggregation and twin studies in Still’s disease. Juvenile chronic polyarthritis, Rheumatology. 1969;2:37–61. [PubMed] [Google Scholar]
- 7.Baum J, Fink C. Juvenile rheumatoid arthritis in monozygotic twins: a case report and review of the literature. Arthritis Rheum. 1968;11:33–36. doi: 10.1002/art.1780110104. [DOI] [PubMed] [Google Scholar]
- 8.Savolainen A, Saila H, Kotaniemi K, Kaipianen-Seppanen O, Leirisalo-Repo M, Aho K. Magnitude of the genetic component in juvenile idiopathic arthritis. Ann Rheum Dis. 2000;59:1001. doi: 10.1136/ard.59.12.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prahalad S, Ryan MH, Shear ES, Thompson SD, Glass DN, Giannini EH. Twins concordant for juvenile rheumatoid arthritis. Arthritis Rheum. 2000;43:2611–2612. doi: 10.1002/1529-0131(200011)43:11<2611::AID-ANR33>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Moroldo MB, Tague BL, Shear ES, Glass DN, Giannini EH. Juvenile rheumatoid arthritis in affected sibpairs. Arthritis Rheum. 1997;40:1962–1966. doi: 10.1002/art.1780401107. [DOI] [PubMed] [Google Scholar]
- 11.Moroldo MB, Chaudhari M, Shear E, Thompson SD, Glass DN, Giannini EH. Juvenile rheumatoid arthritis affected sibpairs: extent of clinical phenotype concordance. Arthritis Rheum. 2004;50:1928–1934. doi: 10.1002/art.20292. [DOI] [PubMed] [Google Scholar]
- 12.Prahalad S, O’Brien E, Fraser AM, Kerber RA, Mineau GP, Pratt D, et al. Familial aggregation of juvenile idiopathic arthritis. Arthritis Rheum. 2004;50:4022–4027. doi: 10.1002/art.20677. [DOI] [PubMed] [Google Scholar]
- 13.Prahalad S, Zeft AS, Pimentel R, Clifford B, McNally B, Mineau GP, et al. Quantification of the familial contribution to juvenile idiopathic arthritis. Arthritis Rheum. 2010;62:2525–2529. doi: 10.1002/art.27516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prahalad S, Shear ES, Thompson SD, Giannini EH, Glass DN. Increased prevalence of familial autoimmunity in simplex and multiplex families with juvenile rheumatoid arthritis. Arthritis Rheum. 2002;46:1851–1856. doi: 10.1002/art.10370. [DOI] [PubMed] [Google Scholar]
- 15.Zeft A, Shear ES, Thompson SD, Glass DN, Prahalad S. Familial autoimmunity: maternal parent-of-origin effect in juvenile idiopathic arthritis. Clin Rheumatol. 2008;27:241–244. doi: 10.1007/s10067-007-0778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–392. [PubMed] [Google Scholar]
- 17.Hall PJ, Burman SJ, Laurent MR, Briggs DC, Venning HE, Leak AM, et al. Genetic susceptibility to early onset pauciarticular juvenile chronic arthritis: a study of HLA and complement markers in 158 British patients. Ann Rheum Dis. 1986;45:464–474. doi: 10.1136/ard.45.6.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vicario JL, Martinez-Laso J, Gomez-Reino JJ, Gomez-Reino FJ, Regueiro JR, Corell A, et al. Both HLA class II and class III DNA polymorphisms are linked to juvenile rheumatoid arthritis susceptibility. Clin Immunol Immunopathol. 1990;56:22–28. doi: 10.1016/0090-1229(90)90165-m. [DOI] [PubMed] [Google Scholar]
- 19.Paul C, Schoenwald U, Truckenbrodt H, Bettinotti MP, Brunnler G, Keller E, et al. HLA-DP/DR interaction in early onset pauciarticular juvenile chronic arthritis. Immunogenetics. 1993;37:442–448. doi: 10.1007/BF00222468. [DOI] [PubMed] [Google Scholar]
- 20.Arnaiz-Villena A, Gomez-Reino JJ, Gamir ML, Regueiro JR, Vicario JL, Gomez-Reino FJ, et al. DR, C4, and Bf allotypes in juvenile rheumatoid arthritis. Arthritis Rheum. 1984;27:1281–1285. doi: 10.1002/art.1780271110. [DOI] [PubMed] [Google Scholar]
- 21.Hollenbach JA, Thompson SD, Bugawan TL, Ryan M, Sudman M, Marion M, et al. Juvenile idiopathic arthritis and HLA class I and class II interactions and age-at-onset effects. Arthritis Rheum. 2010;62:1781–1791. doi: 10.1002/art.27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray KJ, Moroldo MB, Donnelly P, Prahalad S, Passo MH, Giannini EH, et al. Age-specific effects of juvenile rheumatoid arthritis-associated HLA alleles. Arthritis Rheum. 1999;42:1843–1853. doi: 10.1002/1529-0131(199909)42:9<1843::AID-ANR8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Hinks A, Eyre S, Ke X, Barton A, Martin P, Flynn E, et al. Overlap of disease susceptibility loci for rheumatoid arthritis and juvenile idiopathic arthritis. Ann Rheum Dis. 2010;69:1049–1053. doi: 10.1136/ard.2009.110650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson SD, Sudman M, Ramos PS, Marion MC, Ryan M, Tsoras M, et al. The susceptibility loci juvenile idiopathic arthritis shares with other autoimmune diseases extend to PTPN2, COG6, and ANGPT1. Arthritis Rheum. 2010;62:3265–3276. doi: 10.1002/art.27688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet. 2013;45:664–669. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.International Genetics of Ankylosing Spondylitis C. Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiaroni-Clarke RC, Munro JE, Chavez RA, Pezic A, Allen RC, Akikusa JD, et al. Independent confirmation of juvenile idiopathic arthritis genetic risk loci previously identified by immunochip array analysis. Pediatr Rheumatol Online J. 2014;12:53. doi: 10.1186/1546-0096-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol. 2014;10:602–611. doi: 10.1038/nrrheum.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boechat AL, Ogusku MM, Sadahiro A, dos Santos MC. Association between the PTPN22 1858C/T gene polymorphism and tuberculosis resistance. Infect Genet Evol: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013;16:310–313. doi: 10.1016/j.meegid.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 34.Gomez LM, Anaya JM, Martin J. Genetic influence of PTPN22 R620W polymorphism in tuberculosis. Hum Immunol. 2005;66:1242–1247. doi: 10.1016/j.humimm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Kaalla MJ, Broadaway KA, Rohani-Pichavant M, Conneely KN, Whiting A, Ponder L, et al. Meta-analysis confirms association between TNFA-G238A variant and JIA, and between PTPN22-C1858T variant and oligoarticular, RF-polyarticular and RF-positive polyarticular JIA. Pediatr Rheumatol Online J. 2013;11:40. doi: 10.1186/1546-0096-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YH, Bae SC, Song GG. The association between the functional PTPN22 1858 C/T and MIF-173 C/G polymorphisms and juvenile idiopathic arthritis: a meta-analysis. Inflamm Res: official journal of the European Histamine Research Society [et al] 2012;61:411–415. doi: 10.1007/s00011-012-0447-5. [DOI] [PubMed] [Google Scholar]
- 37.Hinks A, Barton A, John S, Bruce I, Hawkins C, Griffiths CE, et al. Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum. 2005;52:1694–1699. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- 38.Viken MK, Amundsen SS, Kvien TK, Boberg KM, Gilboe IM, Lilleby V, et al. Association analysis of the 1858C>T polymorphism in the PTPN22 gene in juvenile idiopathic arthritis and other autoimmune diseases. Genes Immun. 2005;6:271–273. doi: 10.1038/sj.gene.6364178. [DOI] [PubMed] [Google Scholar]
- 39.Fan ZD, Wang FF, Huang H, Huang N, Ma HH, Guo YH, et al. STAT4 rs7574865 G/T and PTPN22 rs2488457 G/C polymorphisms influence the risk of developing juvenile idiopathic arthritis in Han Chinese patients. PloS One. 2015;10:e0117389. doi: 10.1371/journal.pone.0117389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dimopoulou DG, Zervou MI, Trachana M, Myrthianou E, Pratsidou-Gertsi P, Kardassis D, et al. Investigation of juvenile idiopathic arthritis susceptibility loci: results from a Greek population. Hum Immunol. 2013;74:1194–1198. doi: 10.1016/j.humimm.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 41.Ungar D. Golgi linked protein glycosylation and associated diseases. Semin Cell Dev Biol. 2009;20:762–769. doi: 10.1016/j.semcdb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Figueroa-Vega N, Sanz-Cameno P, Moreno-Otero R, Sanchez-Madrid F, Gonzalez-Amaro R, Marazuela M. Serum levels of angiogenic molecules in autoimmune thyroid diseases and their correlation with laboratory and clinical features. J Clin Endocrinol Metab. 2009;94:1145–1153. doi: 10.1210/jc.2008-1571. [DOI] [PubMed] [Google Scholar]
- 43.Liang YL, Wu H, Shen X, Li PQ, Yang XQ, Liang L, et al. Association of STAT4 rs7574865 polymorphism with autoimmune diseases: a meta-analysis. Mol Biol Rep. 2012;39:8873–8882. doi: 10.1007/s11033-012-1754-1. [DOI] [PubMed] [Google Scholar]
- 44.Prahalad S, Hansen S, Whiting A, Guthery SL, Clifford B, McNally B, et al. Variants in TNFAIP3, STAT4, and C12orf30 loci associated with multiple autoimmune diseases are also associated with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:2124–2130. doi: 10.1002/art.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellis JA, Chavez RA, Pezic A, Ponsonby AL, Akikusa JD, Allen RC, et al. Independent replication analysis of genetic loci with previous evidence of association with juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2013;11:12. doi: 10.1186/1546-0096-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee YH, Bae SC, Song GG. TNF promoter -308 A/G and -238 A/G polymorphisms and juvenile idiopathic arthritis: a meta-analysis. Mol Biol Rep. 2012;39:8497–8503. doi: 10.1007/s11033-012-1704-y. [DOI] [PubMed] [Google Scholar]
- 47.Jimenez-Morales S, Velazquez-Cruz R, Ramirez-Bello J, Bonilla-Gonzalez E, Romero-Hidalgo S, Escamilla-Guerrero G, et al. Tumor necrosis factor-alpha is a common genetic risk factor for asthma, juvenile rheumatoid arthritis, and systemic lupus erythematosus in a Mexican pediatric population. Hum Immunol. 2009;70:251–256. doi: 10.1016/j.humimm.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 48.Mourao AF, Caetano-Lopes J, Costa P, Canhao H, Santos MJ, Pinto P, et al. Tumor necrosis factor-alpha -308 genotypes influence inflammatory activity and TNF-alpha serum concentrations in children with juvenile idiopathic arthritis. J Rheumatol. 2009;36:837–842. doi: 10.3899/jrheum.080615. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- 49.Ozen S, Alikasifoglu M, Bakkaloglu A, Duzova A, Jarosova K, Nemcova D, et al. Tumour necrosis factor alpha G–>A -238 and G–>A -308 polymorphisms in juvenile idiopathic arthritis. Rheumatology (Oxford) 2002;41:223–227. doi: 10.1093/rheumatology/41.2.223. [DOI] [PubMed] [Google Scholar]
- 50.Zeggini E, Thomson W, Kwiatkowski D, Richardson A, Ollier W, Donn R, et al. Linkage and association studies of single-nucleotide polymorphism-tagged tumor necrosis factor haplotypes in juvenile oligoarthritis. Arthritis Rheum. 2002;46:3304–3311. doi: 10.1002/art.10698. [DOI] [PubMed] [Google Scholar]
- 51.Schmeling H, Horneff G. Tumour necrosis factor alpha promoter polymorphisms and etanercept therapy in juvenile idiopathic arthritis. Rheumatol Int. 2007;27:383–386. doi: 10.1007/s00296-006-0208-2. [DOI] [PubMed] [Google Scholar]
- 52.Hinks A, Ke X, Barton A, Eyre S, Bowes J, Worthington J, et al. Association of the IL2RA/CD25 gene with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:251–257. doi: 10.1002/art.24187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albers HM, Kurreeman FA, Houwing-Duistermaat JJ, Brinkman DM, Kamphuis SS, Girschick HJ, et al. The TRAF1/C5 region is a risk factor for polyarthritis in juvenile idiopathic arthritis. Ann Rheum Dis. 2008;67:1578–1580. doi: 10.1136/ard.2008.089060. [DOI] [PubMed] [Google Scholar]
- 54.Hinks A, Barton A, Shephard N, Eyre S, Bowes J, Cargill M, et al. Identification of a novel susceptibility locus for juvenile idiopathic arthritis by genome-wide association analysis. Arthritis Rheum. 2009;60:258–263. doi: 10.1002/art.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daha NA, Lie BA, Trouw LA, Stoeken G, Schonkeren JJ, Ding B, et al. Novel genetic association of the VTCN1 region with rheumatoid arthritis. Ann Rheum Dis. 2012;71:567–571. doi: 10.1136/annrheumdis-2011-200574. [DOI] [PubMed] [Google Scholar]
- 56.Berdeli A, Ozyurek AR, Ulger Z, Gurses D, Levent E, Salar K, et al. Association of macrophage migration inhibitory factor gene -173 G/C polymorphism with prognosis in Turkish children with juvenile rheumatoid arthritis. Rheum Int. 2006;26:726–731. doi: 10.1007/s00296-005-0062-7. [DOI] [PubMed] [Google Scholar]
- 57.Donn R, Alourfi Z, Zeggini E, Lamb R, Jury F, Lunt M, et al. A functional promoter haplotype of macrophage migration inhibitory factor is linked and associated with juvenile idiopathic arthritis. Arthritis Rheum. 2004;50:1604–1610. doi: 10.1002/art.20178. [DOI] [PubMed] [Google Scholar]
- 58.Vivarelli M, D’Urbano LE, Insalaco A, Lunt M, Jury F, Tozzi AE, et al. Macrophage migration inhibitory factor (MIF) and oligoarticular juvenile idiopathic arthritis (o-JIA): association of MIF promoter polymorphisms with response to intra-articular glucocorticoids. Clin Exp Rheumatol. 2007;25:775–781. [PubMed] [Google Scholar]
- 59.Reinards TH, Albers HM, Brinkman DM, Kamphuis SS, van Rossum MA, Girschick HJ, et al. CD226 (DNAM-1) is associated with susceptibility to juvenile idiopathic arthritis. Ann Rheum Dis ( 2014 doi: 10.1136/annrheumdis-2013-205138. http://dx.doi.org/10.1136/annrheumdis-2013-205138. [DOI] [PubMed]
- 60.Hinks A, Eyre S, Ke X, Barton A, Martin P, Flynn E, et al. Association of the AFF3 gene and IL2/IL21 gene region with juvenile idiopathic arthritis. Genes Immun. 2010;11:194–198. doi: 10.1038/gene.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prahalad S. Negative association between the chemokine receptor CCR5-Delta32 polymorphism and rheumatoid arthritis: a meta-analysis. Genes Immun. 2006;7:264–268. doi: 10.1038/sj.gene.6364298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prahalad S, Bohnsack JF, Jorde LB, Whiting A, Clifford B, Dunn D, et al. Association of two functional polymorphisms in the CCR5 gene with juvenile rheumatoid arthritis. Genes Immun. 2006;7:468–475. doi: 10.1038/sj.gene.6364317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hinks A, Martin P, Flynn E, Eyre S, Packham J, Childhood Arthritis Prospective Study UCBSG et al. Association of the CCR5 gene with juvenile idiopathic arthritis. Genes Immun. 2010;11:584–589. doi: 10.1038/gene.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferrell EG, Ponder LA, Minor LS, Angeles-Han ST, Kennedy CW, Rouster-Stevens KA, et al. Limitations in the classification of childhood-onset rheumatoid arthritis. J Rheumatol. 2014;41:547–553. doi: 10.3899/jrheum.130563. [DOI] [PubMed] [Google Scholar]
- 66.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 67.Clemens LE, Albert E, Ansell BM. HLA studies in IgM rheumatoid-factor-positive arthritis of childhood. Ann Rheum Dis. 1983;42:431–434. doi: 10.1136/ard.42.4.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forre O, Dobloug JH, Hoyeraal HM, Thorsby E. HLA antigens in juvenile arthritis. Genetic basis for the different subtypes. Arthritis Rheum. 1983;26:35–38. doi: 10.1002/art.1780260106. [DOI] [PubMed] [Google Scholar]
- 69.Nepom BS, Nepom GT, Mickelson E, Schaller JG, Antonelli P, Hansen JA. Specific HLA-DR4-associated histocompatibility molecules characterize patients with seropositive juvenile rheumatoid arthritis. J Clin Invest. 1984;74:287–291. doi: 10.1172/JCI111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vehe RK, Begovich AB, Nepom BS. HLA susceptibility genes in rheumatoid factor positive juvenile rheumatoid arthritis. J Rheumatol Suppl. 1990;26:11–15. [PubMed] [Google Scholar]
- 71.Barron KS, Silverman ED, Gonzales JC, Owerbach D, Reveille JD. DNA analysis of HLA-DR, DQ, and DP alleles in children with polyarticular juvenile rheumatoid arthritis. J Rheumatol. 1992;19:1611–1616. [PubMed] [Google Scholar]
- 72.Thomson W, Barrett JH, Donn R, Pepper L, Kennedy LJ, Ollier WE, et al. Juvenile idiopathic arthritis classified by the ILAR criteria: HLA associations in UK patients. Rheumatology (Oxford) 2002;41:1183–1189. doi: 10.1093/rheumatology/41.10.1183. [DOI] [PubMed] [Google Scholar]
- 73.Prahalad S, Thompson SD, Conneely KN, Jiang Y, Leong T, Prozonic J, et al. Hierarchy of risk of childhood-onset rheumatoid arthritis conferred by HLA-DRB1 alleles encoding the shared epitope. Arthritis Rheum. 2012;64:925–930. doi: 10.1002/art.33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.du Montcel ST, Michou L, Petit-Teixeira E, Osorio J, Lemaire I, Lasbleiz S, et al. New classification of HLA-DRB1 alleles supports the shared epitope hypothesis of rheumatoid arthritis susceptibility. Arthritis Rheum. 2005;52:1063–1068. doi: 10.1002/art.20989. [DOI] [PubMed] [Google Scholar]
- 75.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prahalad S, Conneely KN, Jiang Y, Sudman M, Wallace CA, Brown MR, et al. Susceptibility to childhood-onset rheumatoid arthritis: investigation of a weighted genetic risk score that integrates cumulative effects of variants at five genetic Loci. Arthritis Rheum. 2013;65:1663–1667. doi: 10.1002/art.37913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chibnik LB, Keenan BT, Cui J, Liao KP, Costenbader KH, Plenge RM, et al. Genetic risk score predicting risk of rheumatoid arthritis phenotypes and age of symptom onset. PLoS One. 2011;6:e24380. doi: 10.1371/journal.pone.0024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossi-Semerano L, Kone-Paut I. Is still’s disease an autoinflammatory syndrome? Int J Inflam. 2012;2012:480373. doi: 10.1155/2012/480373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Date Y, Seki N, Kamizono S, Higuchi T, Hirata T, Miyata K, et al. Identification of a genetic risk factor for systemic juvenile rheumatoid arthritis in the 5′-flanking region of the TNFalpha gene and HLA genes. Arthritis Rheum. 1999;42:2577–2582. doi: 10.1002/1529-0131(199912)42:12<2577::AID-ANR10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 81.Silva-Ramirez B, Cerda-Flores RM, Rubio-Perez N, Vargas-Alarcon G, Perez-Hernandez N, Granados-Arriola J, et al. Association of HLA DRB1 alleles with juvenile idiopathic arthritis in Mexicans. Clin Exp Rheumatol. 2010;28:124–127. [PubMed] [Google Scholar]
- 82.Ombrello MRE, Tachmazidou I, Grom AA, Foll D, Martini A, et al. Genome Wide Association Meta-Analysis Implicates HLA-DRB1, The BTNL2/HLA-DRA Region, And A Novel Susceptibility Locus On Chromosome 1 In Systemic Juvenile Idiopathic Arthritis. Arthritis Rheum. 2013 Oct;65 Abstract Supplement 2013. [Google Scholar]
- 83.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Modesto C, Patino-Garcia A, Sotillo-Pineiro E, Merino J, Garcia-Consuegra J, Merino R, et al. TNF-alpha promoter gene polymorphisms in Spanish children with persistent oligoarticular and systemic-onset juvenile idiopathic arthritis. Scand J Rheumatol. 2005;34:451–454. doi: 10.1080/03009740510026652. [DOI] [PubMed] [Google Scholar]
- 85.Donn RP, Shelley E, Ollier WE, Thomson W, British Paediatric Rheumatology Study G A novel 5′-flanking region polymorphism of macrophage migration inhibitory factor is associated with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2001;44:1782–1785. doi: 10.1002/1529-0131(200108)44:8<1782::AID-ART314>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 86.De Benedetti F, Meazza C, Vivarelli M, Rossi F, Pistorio A, Lamb R, et al. Functional and prognostic relevance of the -173 polymorphism of the macrophage migration inhibitory factor gene in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2003;48:1398–1407. doi: 10.1002/art.10882. [DOI] [PubMed] [Google Scholar]
- 87.Fife MS, Gutierrez A, Ogilvie EM, Stock CJ, Samuel JM, Thomson W, et al. Novel IL10 gene family associations with systemic juvenile idiopathic arthritis. Arthritis Res Ther. 2006;8:R148. doi: 10.1186/ar2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stock CJ, Ogilvie EM, Samuel JM, Fife M, Lewis CM, Woo P. Comprehensive association study of genetic variants in the IL-1 gene family in systemic juvenile idiopathic arthritis. Genes Immun. 2008;9:349–357. doi: 10.1038/gene.2008.24. [DOI] [PubMed] [Google Scholar]
- 89.Wakil SM, Monies DM, Abouelhoda M, Al-Tassan N, Al-Dusery H, Naim EA, et al. Association of a mutation in LACC1 with a monogenic form of systemic juvenile idiopathic arthritis. Arthritis Rheumatol. 2015;67:288–295. doi: 10.1002/art.38877. [DOI] [PubMed] [Google Scholar]
- 90.Grant AV, Alter A, Huong NT, Orlova M, Van Thuc N, Ba NN, et al. Crohn’s disease susceptibility genes are associated with leprosy in the Vietnamese population. J Infect Dis. 2012;206:1763–1767. doi: 10.1093/infdis/jis588. [DOI] [PubMed] [Google Scholar]
- 91.Hinks A, Martin P, Thompson SD, Sudman M, Stock CJ, Thomson W, et al. Autoinflammatory gene polymorphisms and susceptibility to UK juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2013;11:14. doi: 10.1186/1546-0096-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lotfy HM, Kandil ME, Issac MS, Salah S, Ismail NA, Abdel Mawla MA. MEFV mutations in Egyptian children with systemic-onset juvenile idiopathic arthritis. Mol Diagn Ther. 2014;18:549–557. doi: 10.1007/s40291-014-0105-4. [DOI] [PubMed] [Google Scholar]
- 93.Colbert RA. Classification of juvenile spondyloarthritis: enthesitis-related arthritis and beyond. Nat Rev Rheumatol. 2010;6:477–485. doi: 10.1038/nrrheum.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berntson L, Nordal E, Aalto K, Peltoniemi S, Herlin T, Zak M, et al. HLA-B27 predicts a more chronic disease course in an 8-year followup cohort of patients with juvenile idiopathic arthritis. J Rheumatol. 2013;40:725–731. doi: 10.3899/jrheum.121257. [DOI] [PubMed] [Google Scholar]
- 95.Stanevicha V, Eglite J, Zavadska D, Sochnevs A, Lazareva A, Guseinova D, et al. HLA B27 allele types in homogeneous groups of juvenile idiopathic arthritis patients in Latvia. Pediatr Rheumatol Online J. 2010;8:26. doi: 10.1186/1546-0096-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Flato B, Hoffmann-Vold AM, Reiff A, Forre O, Lien G, Vinje O. Long-term outcome and prognostic factors in enthesitis-related arthritis: a case-control study. Arthritis Rheum. 2006;54:3573–3582. doi: 10.1002/art.22181. [DOI] [PubMed] [Google Scholar]
- 97.Hinks A, Martin P, Flynn E, Eyre S, Packham J, Childhood Arthritis Prospective Study C et al. Subtype specific genetic associations for juvenile idiopathic arthritis: ERAP1 with the enthesitis related arthritis subtype and IL23R with juvenile psoriatic arthritis. Arthritis Res Ther. 2011;13:R12. doi: 10.1186/ar3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singh S, Rai G, Aggarwal A. Association of microRNA-146a and its target gene IRAK1 polymorphism with enthesitis related arthritis category of juvenile idiopathic arthritis. Rheumatol Int. 2014;34:1395–1400. doi: 10.1007/s00296-014-3001-7. [DOI] [PubMed] [Google Scholar]
- 99.Aggarwal A, Srivastava R, Singh S, Kumar Dubey P. IL1 gene polymorphisms in enthesitis related arthritis category of juvenile idiopathic arthritis (ERA-JIA) Clin Rheumatol. 2012;31:607–611. doi: 10.1007/s10067-011-1898-8. [DOI] [PubMed] [Google Scholar]
- 100.Myles A, Aggarwal A. Lack of association of single nucleotide polymorphisms in toll-like receptors 2 and 4 with enthesitis-related arthritis category of juvenile idiopathic arthritis in Indian population. Rheumatol Int. 2013;33:417–421. doi: 10.1007/s00296-012-2396-2. [DOI] [PubMed] [Google Scholar]
- 101.Comak E, Dogan CS, Akman S, Koyun M, Gokceoglu AU, Keser I. MEFV gene mutations in Turkish children with juvenile idiopathic arthritis. Eur J Pediatr. 2013;172:1061–1067. doi: 10.1007/s00431-013-2003-x. [DOI] [PubMed] [Google Scholar]
- 102.Gulhan B, Akkus A, Ozcakar L, Besbas N, Ozen S. Are MEFV mutations susceptibility factors in enthesitis-related arthritis patients in the eastern Mediterranean? Clin Exp Rheumatol. 2014;32:S160–S164. [PubMed] [Google Scholar]
- 103.Stoll ML, Punaro M. Psoriatic juvenile idiopathic arthritis: a tale of two subgroups. Curr Opin Rheumatol. 2011;23:437–443. doi: 10.1097/BOR.0b013e328348b278. [DOI] [PubMed] [Google Scholar]
- 104.Hamilton ML, Gladman DD, Shore A, Laxer RM, Silverman ED. Juvenile psoriatic arthritis and HLA antigens. Ann Rheum Dis. 1990;49:694–697. doi: 10.1136/ard.49.9.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ansell B, Beeson M, Hall P, Bedford P, Woo P. HLA and juvenile psoriatic arthritis. Br J Rheumatol. 1993;32:836–837. doi: 10.1093/rheumatology/32.9.836. [DOI] [PubMed] [Google Scholar]
- 106.Day TG, Ramanan AV, Hinks A, Lamb R, Packham J, Wise C, et al. Autoinflammatory genes and susceptibility to psoriatic juvenile idiopathic arthritis. Arthritis Rheum. 2008;58:2142–2146. doi: 10.1002/art.23604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]


