Abstract
Background
Experiments in isolated perfused heart have shown that heart rate is lower and sinoatrial node (SAN) action potential duration is longer in streptozotocin (STZ)–induced diabetic rat compared to controls. In sino-atrial preparations the pacemaker cycle length and sino-atrial conduction time are prolonged in STZ heart. To further clarify the molecular basis of electrical disturbances in the diabetic heart the profile of mRNA encoding a wide variety of proteins associated with the generation and transmission of electrical activity has been evaluated in the SAN of STZ-induced diabetic rat heart.
Methodology/Principal Findings
Heart rate was measured in isolated perfused heart with an extracellular suction electrode. Expression of mRNA encoding a variety of intercellular proteins, intracellular Ca2+-transport and regulatory proteins, cell membrane transport proteins and calcium, sodium and potassium channel proteins were measured in SAN and right atrial (RA) biopsies using real-time reverse transcription polymerase chain reaction techniques. Heart rate was lower in STZ (203±7 bpm) compared to control (239±11 bpm) rat. Among many differences in the profile of mRNA there are some worthy of particular emphasis. Expression of genes encoding some proteins were significantly downregulated in STZ-SAN: calcium channel, Cacng4 (7-fold); potassium channel, Kcnd2 whilst genes encoding some other proteins were significantly upregulated in STZ-SAN: gap junction, Gjc1; cell membrane transport, Slc8a1, Trpc1, Trpc6 (4-fold); intracellular Ca2+-transport, Ryr3; calcium channel Cacna1g, Cacna1h, Cacnb3; potassium channels, Kcnj5, Kcnk3 and natriuretic peptides, Nppa (5-fold) and Nppb (7-fold).
Conclusions/Significance
Collectively, this study has demonstrated differences in the profile of mRNA encoding a variety of proteins that are associated with the generation, conduction and regulation of electrical signals in the SAN of STZ-induced diabetic rat heart. Data from this study will provide a basis for a substantial range of future studies to investigate whether these changes in mRNA translate into changes in electrophysiological function.
Introduction
Cardiovascular complications are common in patients with Type 1 and Type 2 diabetes mellitus and these complications lead to an increased risk of mortality [1]. Although vascular diseases including coronary artery disease and hypertension increase the risk of mortality these patients are also at increased risk of developing cardiac abnormalities that are independent of cardiovascular complications [2]. In addition to mechanical dysfunction disturbances in electrical activity, manifesting as arrhythmias, have also been widely reported in diabetic heart. Patients with diabetes have an increased risk of arrhythmias including QT interval and QRS prolongation which is associated with sudden death [3] Atrial fibrillation, bradyarrhythmias, bundle branch block and atrioventricular block are more prevalent in diabetic patients [4]. In vivo biotelemetry studies performed in experimental models of diabetes have demonstrated disturbances in the electrocardiogram which are variously associated with bradycardia, prolongation of PQ, QRS and QT intervals [5–7]. Action potential duration is frequently prolonged in the diabetic heart and prolongation can occur to different extents in different regions of the heart including the SAN [6,8,9]. Very little experimental data is available about the effects of diabetes on SAN electrophysiology. It is known that SAN conduction and pacemaker cycle length are prolonged and that the SAN action potential duration may be prolonged in diabetic heart [10,11]. A variety of ionic currents including L-type and T-type Ca2+ current, hyperpolarization-activated "funny" current, Na+/Ca2+ exchange current and various K+ currents are important in the generation of the SAN action potential [12]. Sarcoplasmic reticulum (SR) Ca2+ signaling may also contribute to the generation and decay of the SAN action potential [12]. Disturbances of one or more of these ionic conductances would undoubtedly have implications for the generation and conduction of electrical signals in the SAN which in turn may underlie some of the electrical disturbances that have been frequently reported in diabetic heart [5–7]. To further clarify the molecular basis of electrical disturbances in the SAN of diabetic heart the profile of mRNA that encodes a wide variety of proteins that are associated with the generation and conduction of electrical activity in the pacemaker has been evaluated in the STZ-induced diabetic rat heart.
Materials and Methods
Experimental protocol
Forty male Wistar rats aged 8 weeks were divided into 2 subgroups. All animals received normal rat chow and drinking water ad libitum. One subgroup of rats received STZ/citrate buffer (60 mg/kg, intraperitoneal) whilst the other subgroup received citrate buffer alone. Experiments began 10 weeks after STZ treatment. Blood glucose was measured 5 days following STZ treatment to confirm diabetes. Body weight, heart weight and blood glucose were measured immediately prior to experiments. All animal experimentation was carried out in accordance with the Animals (Scientific Procedures) Act 1986 and conforms to the Recommendation from the Declaration of Helsinki and the Guiding Principles in the Care and Use of Animals. Approval for this project was obtained from the Animal Ethics Committee, College of Medicine & Health Sciences, United Arab Emirates University.
Measurement of heart rate and action potentials
Rats were killed using a guillotine. Hearts were then rapidly removed, mounted in Langendorff mode and perfused retrogradely at a constant flow of 8 ml.g heart-1 min-1 and at physiological temperature (36–37°C) with a normal Tyrode containing: 140 mM NaCl; 5 mM KCl; 1 mM MgCl2; 10 mM glucose; 5 mM HEPES; 1.8 mM CaCl2 and adjusted to pH 7.4 with NaOH and continuously bubbled with oxygen. To measure heart rate action potentials were recorded in spontaneously beating hearts with a purpose built extracellular suction electrode with a tip ~ 2 mm in diameter according to previously described techniques [13]. Recordings were made in the region of the left ventricle. Signals from the electrode were collected at 400 Hz, amplified (ADInstuments, ML136 Bioamp) and conveyed via a Powerlab (ADInstruments, PL410) for display on a PC. Data were analyzed with ADInstruments software version v 4.21 (ADInstruments, Australia).
Expression of mRNA
Expression of genes encoding a range of cardiac muscle proteins was assessed using modifications of previously described techniques [14–16]. After sacrifice hearts were removed rapidly from the rats and placed in a dish containing: NaCl 140 mM; KCl 5.4 mM; MgCl2 1 mM; HEPES 5 mM; D-glucose 5.5 mM; CaCl2 1.8 mM and adjusted to pH 7.4 with NaOH. The ventricles and the left atrium were removed and the right atrium was opened to expose the SAN and crista terminalis (Fig 1a and 1b). The SAN artery was used to identify the SAN. The SAN was exposed and 2 mm biopsy samples of SAN were carefully collected from 20 STZ (STZ-SAN) and 20 control (CON-SAN) hearts according to previously described techniques [17,18]. Samples of right atrial (RA) tissue were also collected. Once dissected, the samples were immediately placed in RNAlater (AM7021, Life Technologies, Carlsbad, CA, USA) and kept overnight at room temperature to allow thorough penetration of the tissue. Tissue samples were then frozen at -20°C pending further processing. The samples were homogenized at 6500 rpm for 2 runs of 20 seconds each with a 15 second gap (Preceylls 24, Berlin Technology, USA). Isolation of total RNA from the tissue was performed using the SV Total RNA Isolation System (Promega, Madison, USA) according to the manufacturer’s instructions. The concentration and purity of the RNA samples was determined by measuring the absorbance at 260 nm and the ratio of absorbance at 260 nm and 280 nm (ND-1000, NanoDrop). A two-step RT-PCR procedure was used to generate cDNA. Total RNA (500 ng) was converted into cDNA in a 25 μl PCR reaction with 10 x RT Buffer 2.0 μl, 25 x dNTP Mix (100 mM) 0.8 μl, 10 x RT Random Primers 2.0 μl, MultiScribe™ Reverse Transcriptase 1.0 μl, RNase inhibitor 1.0 μl, and Nuclease-free H2O (High Capacity cDNA Reverse Transcription Kit (4374966, Applied Biosystems, USA). Reverse transcription was carried out using the following parameter values: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min on the Veriti thermal cycler (Applied Biosystems, USA). Gene Expression Assays were performed using custom TaqMan Low Density Arrays (Format 32, 4346799, Applied Biosystems, USA). The TaqMan assays are pre-loaded in each reaction well of the array in triplicate for each RNA sample. As in previous studies 18S ribosomal RNA was used as an endogenous control [19]. Expression of 18S was not significantly different (P>0.05) between the samples of STZ-SAN, CON-SAN, STZ-RA and CON-RA. 100 ng of cDNA (RNA-equivalent) was loaded together with 2 × TaqMan Gene Expression Master Mix (No AmpErase UNG, Applied Biosystems, USA) for a total of 100 μL per port. Two SAN samples were combined and two RA samples were combined for each real-time RT-PCR assay. Real-time RT-PCR was performed in a Fast ABI Prism 7900HT Sequence Detection System (Applied Biosystems, USA). The PCR thermal cycling parameters were run in standard mode as follows: 50°C for 2 min, 94.5°C for 10 min, followed by 40 cycles of 97°C for 30 sec and 59.7 for 1 min. Results were initially analyzed using ABI Prism 7900HT SDS, v2.4. All remaining calculations and statistical analysis were performed by the SDS RQ Manager 1.1.4 software using the 2−ΔΔCt method with a relative quantification RQmin/RQmax confidence set at 95%. A list of the target genes and the proteins encoded by these genes is shown in Table 1.
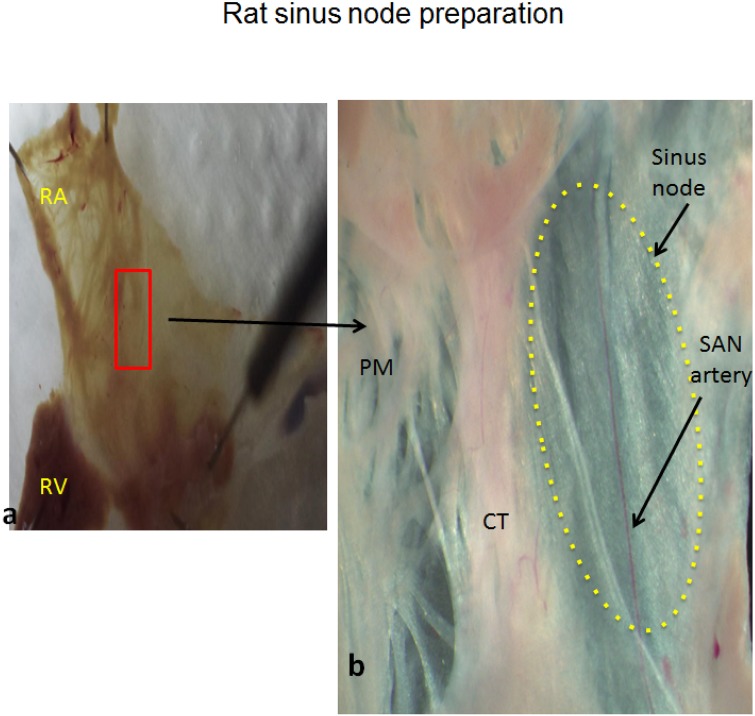
Fig 1. a & b –Gross anatomy of the sinus node in a typical control heart showing the location where tissue samples were collected.
Table 1. Target genes and proteins.
| Genes | Proteins | Protein descriptions |
|---|---|---|
| Cardiac muscle and associated regulatory proteins | ||
| Myh6 | MHC-α | Myosin Heavy Chain, Cardiac Muscle Alpha Isoform |
| Myl1 | MLC1 | Myosin light chain 3, skeletal muscle isoform |
| Myl2 | MLC-2 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform |
| Tnnc1 | TN-C | Troponin C Type 1 |
| Intercellular proteins | ||
| Gja1 | Cx43 | Connexin43 |
| Gja5 | Cx40 | Connexin40 |
| Gja7 | Cx45 | Connexin45 |
| Gjd3 | Cx31.9 | Connexin31.9 |
| Cell membrane transport | ||
| Atp1a1 | Na/K ATPase,α1 | ATPase, Na+/K+ Transporting, Alpha 1 Polypeptide |
| Atp1a2 | Na/K ATPase,α2 | ATPase, Na+/K+ Transporting, Alpha 2 Polypeptide |
| Atp1a3 | Na/K ATPase,α3 | ATPase, Na+/K+ Transporting, Alpha 3 Polypeptide |
| Atp1b1 | Na/K ATPase,β1 | ATPase, Na+/K+ Transporting, Beta 1 Polypeptide |
| Atp2b1 | Na/K ATPase,β2 | ATPase, Ca++ Transporting, Plasma Membrane 1 |
| Slc8a1 | NCX1 | Solute Carrier Family 8 (Sodium/Calcium Exchanger), Member 1 |
| Trpc1 | TRPC1 | Transient receptor potential channel 1 |
| Trpc3 | TRPC3 | Transient receptor potential channel 3 |
| Trpc6 | TRPC6 | Transient receptor potential channel 6 |
| Intercellular Ca2+ transport and Ca2+ regulation | ||
| Atp2a2 | SERCA2 | Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase 2 |
| Calm1 | Calm1 | Calmodulin1 |
| Calm3 | Calm3 | Calmodulin3 |
| Casq2 | Casq2 | Calsequestrin 2 |
| Itpr1 | IP3R1 | Inositol 1,4,5-Trisphosphate Receptor, Type 1 |
| Itpr2 | IP3R2 | Inositol 1,4,5-Trisphosphate Receptor, Type 2 |
| Itpr3 | IP3R3 | Inositol 1,4,5-Trisphosphate Receptor, Type 3 |
| Pln | PLB | Phospholamban |
| Ryr2/ RYR2 | RYR2 | Ryanodine Receptor 2 |
| Ryr3/ RYR3 | RYR3 | Ryanodine Receptor 3 |
| Hyperpolarization-activated cyclic nucleotide-gated channels | ||
| Hcn1 | HCN1 | Hyperpolarization-activated cyclic nucleotide-gated channels 1 |
| Hcn2 | HCN2 | Hyperpolarization-activated cyclic nucleotide-gated channels 2 |
| Hcn3 | HCN3 | Hyperpolarization-activated cyclic nucleotide-gated channels 3 |
| Hcn4 | HCN4 | Hyperpolarization-activated cyclic nucleotide-gated channels 4 |
| Calcium channels | ||
| Cacna1c | Cav1.2 | Voltage-Dependent, L Type, Alpha 1C Subunit |
| Cacna1d | Cav1.3 | Voltage-Dependent, L Type, Alpha 1D Subunit |
| Cacna1g | Cav3.1 | Voltage-Dependent, T Type, Alpha 1G Subunit |
| Cacna1h | Cav3.2 | Voltage-Dependent, T Type, Alpha 1H Subunit |
| Cacna2d1 | Cavα2δ1 | Voltage-Dependent, Alpha 2/Delta Subunit 1 |
| Cacna2d2 | Cavα2δ2 | Voltage-Dependent, Alpha 2/Delta Subunit 2 |
| Cacna2d3 | Cavα2δ3 | Voltage-Dependent, Alpha 2/Delta Subunit 3 |
| Cacnb1 | Cavβ1 | Voltage-Dependent, Beta 1 Subunit |
| Cacnb2 | Cavβ2 | Voltage-Dependent, Beta 2 Subunit |
| Cacnb3 | Cavβ3 | Voltage-Dependent, Beta 3 Subunit |
| Cacng4 | Cavγ4 | Voltage-Dependent, Gamma Subunit 4 |
| Cacng7 | Cavγ7 | Voltage-Dependent, Gamma Subunit 7 |
| Sodium channels | ||
| Scn1a | Nav1.1 | Voltage Gated, Type I Alpha Subunit |
| Scn1b | Navβ1 | Voltage Gated, Type I Beta Subunit |
| Scn2b | Navβ2 | Voltage Gated, Type II Beta Subunit |
| Scn3a | Nav1.3 | Voltage Gated, Type III Alpha Subunit |
| Scn3b | Navβ3 | Voltage Gated, Type III Beta Subunit |
| Scn4a | Nav1.4 | Voltage Gated, Type IV Alpha Subunit |
| Scn5a | Nav1.5 | Voltage Gated, Type V Alpha Subunit |
| Scn7a | Nav2.1 | Voltage Gated, Type VII Alpha Subunit |
| Potassium channels | ||
| Kcna2 | Kv1.2 | Voltage Gated Shaker Related Subfamily A, Member 2 |
| Kcna3 | Kv1.3 | Voltage Gated Shaker Related Subfamily A, Member 3 |
| Kcna4 | Kv1.4 | Voltage Gated Shaker Related Subfamily A, Member 4 |
| Kcna5 | Kv1.5 | Voltage Gated Shaker Related Subfamily A, Member 5 |
| Kcna6 | Kv1.6 | Voltage Gated Shaker Related Subfamily A, Member 6 |
| Kcnb1 | Kv2.1 | Voltage Gated Shab Related Subfamily B, Member 1 |
| Kcnd1 | Kv4.1 | Voltage Gated Shal Related Subfamily D, Member 1 |
| Kcnd2 | Kv4.2 | Voltage Gated Shal Related Subfamily D, Member 2 |
| Kcnd3 | Kv4.3 | Voltage Gated Shal Related Subfamily D, Member 3 |
| Kcne4 | MIRP3 | Minimum Potassium Ion Channel-Related Peptide 3 |
| Kcnh2 | ERG-1 | Ether-A-Go-Go-Related Protein 1 |
| Kcnip2 | KChIP2 | Kv Channel Interacting Protein 2 |
| Kcnj11 | Kir6.2 | Inwardly Rectifying Subfamily J, Member 11 |
| Kcnj12 | Kir2.2 | Inwardly Rectifying Subfamily J, Member 12 |
| Kcnj14 | Kir2.4 | Inwardly Rectifying Subfamily J, Member 14 |
| Kcnj2 | Kir2.1 | Inwardly Rectifying Subfamily J, Member 2 |
| Kcnj3 | Kir3.1 | Inwardly Rectifying Subfamily J, Member 3 |
| Kcnj5 | Kir3.4 | Inwardly Rectifying Subfamily J, Member 5 |
| Kcnj8 | Kir6.1 | Inwardly Rectifying Subfamily J, Member 8 |
| Kcnk1 | TWIK1 | Two Pore Domain Subfamily K, Member 1 |
| Kcnk2 | TREK1 | Two Pore Domain Subfamily K, Member 2 |
| Kcnk3 | K2P3.1 | Two Pore Domain Subfamily K, Member 3 |
| Kcnk5 | K2P5.1 | Two Pore Domain Subfamily K, Member 5 |
| Kcnk6 | TWIK2 | Two Pore Domain Subfamily K, Member 6 |
| Kcnn1 | SK1 | Calcium Activated Intermediate/Small Conductance Subfamily N Alpha, Member 1 |
| Kcnn2 | SK2 | Calcium Activated Intermediate/Small Conductance Subfamily N Alpha, Member 2 |
| Kcnn3 | SK3 | Calcium Activated Intermediate/Small Conductance Subfamily N Alpha, Member 3 |
| Kcnq1 | Kv7.1 | Voltage Gated KQT-Like Subfamily Q, Member 1 |
| Miscellaneous proteins | ||
| Abcc8 | SUR1 | ATP-binding cassette transporter sub-family C member 8 |
| Abcc9 | SUR2 | ATP-binding cassette, sub-family C member 9 |
| Nppa | ANP | Atrial natriuretic peptide |
| Nppb | BNP | Brain natriuretic peptide |
| Pias3 | KChAP | Protein Inhibitor of activated STAT, 3 |
Expression of protein
Protein expression was measured using previously described SDS-PAGE and Western blotting techniques [20]. SAN from STZ and control rats were dissected, rinsed with ice-cold saline and homogenised in 100 mM potassium phosphate buffer (pH7.4) containing 1 mM EDTA and 0.1mM phenylmethylsulfonyl fluoride at 6500 rpm for 2 runs of 20 seconds each with a 15 second gap (Preceylls 24 homogeniser, Berlin Technology, USA). Protein concentration was measured using Bio-Rad reagent. The supernatant was used for SDS-PAGE and Western Blotting. Briefly, 10–20 ug protein was electrophoretically separated onto 7.5% or 12% (depending on the molecular weight of the protein to be separated) polyacrylamide gels and transferred onto nitrocellulose membranes. The expression of the specific proteins was checked by immunoreaction with their specific antibodies by Western blot analysis. β-actin was used as a loading control. The blots were developed using the Pierce Western Blot kit. Densitometric analysis of the protein bands was performed using the Typhoon FLA 9500, GE Healthcare Bio-Sciences AB (Uppsala, Sweden). The ratio of specific protein signal to that of actin control were used to calculate fold change.
Statistics
Results were expressed as the mean ± S.E.M. of ‘n’ observations. Statistical comparisons were performed using one-way ANOVA and Bonferroni post hoc for multiple comparisons or Independent Samples t-test, as appropriate (SPSS v. 20). P< 0.05 was considered to indicate a significant difference.
Results
Body and heart weight were significantly (P<0.01) reduced and heart weight / body weight ratio was increased in STZ compared to control rat. Blood glucose was increased 5-fold in STZ rat compared to control (Table 2).
Table 2. General characteristics of streptozotocin-induced diabetic rats.
| Control | Streptozotocin | |
|---|---|---|
| Body weight (g) | 322.50±22.98 | 207.00±51.99** |
| Heart weight (g) | 1.15±0.12 | 0.85±0.13** |
| Heart weight / Body weight ratio | 3.59±0.45 | 4.19±0.48** |
| Blood glucose (mg/dl) | 96.92±15.03 | 514.33±54.60** |
Data are mean ± SEM, n = 12 hearts,
** P<0.01
Heart rate and action potential
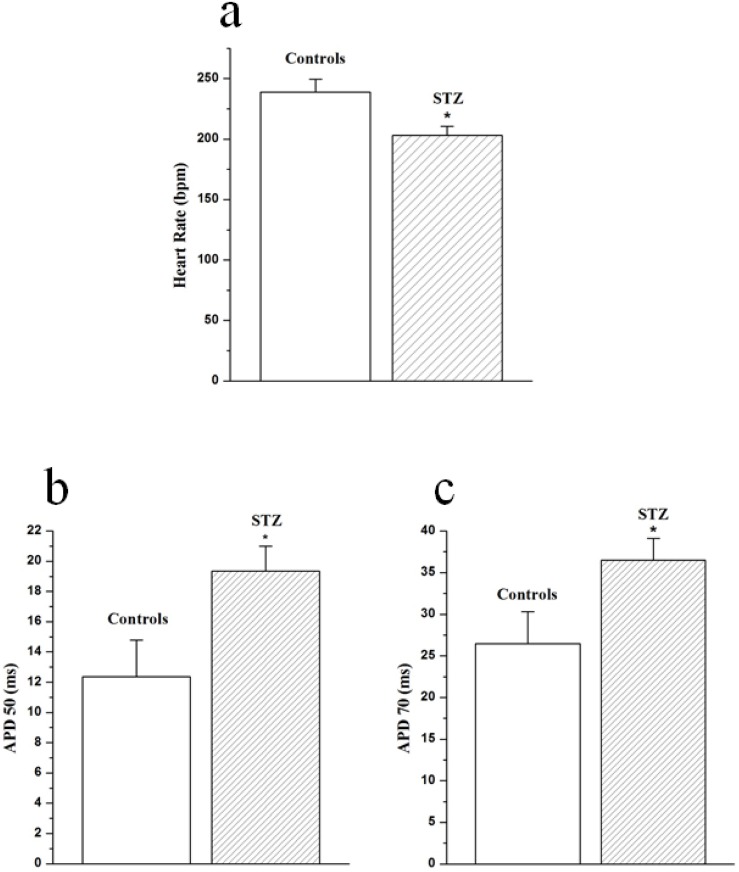
Heart rate and action potential duration data are shown in Fig 2. Heart rate was significantly (P<0.05) reduced in STZ (203±7 bpm, n = 12) compared to control (239±11 bpm, n = 12) heart (Fig 2a). Time to peak action potential was not significantly (P>0.05) altered in STZ (4.9±0.3 ms) compared to control (4.7±0.3 ms) heart. Action potential duration (APD) at 50% repolarization was significantly prolonged in STZ (19.3±1.7 ms) compared to control (12.4±2.4 ms) heart (Fig 2b). APD at 70% repolarization was also significantly prolonged in STZ (36.5±2.6 ms) compared to control (26.4±3.9 ms) heart (Fig 2c).
Fig 2. Effects of STZ-induced diabetes on heart rate (a), action potential duration at 50% (b) and 70% (c) recovery from peak action potential.
Data are mean ± SEM, n = 12 hearts.
Expression of mRNA
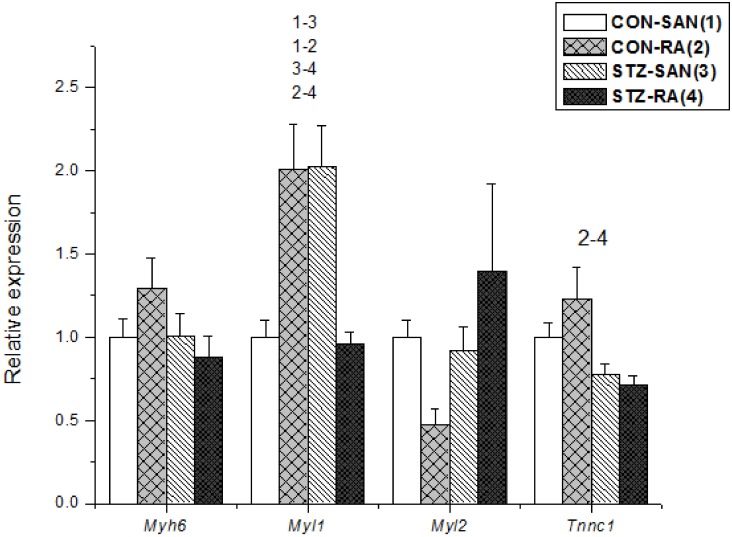
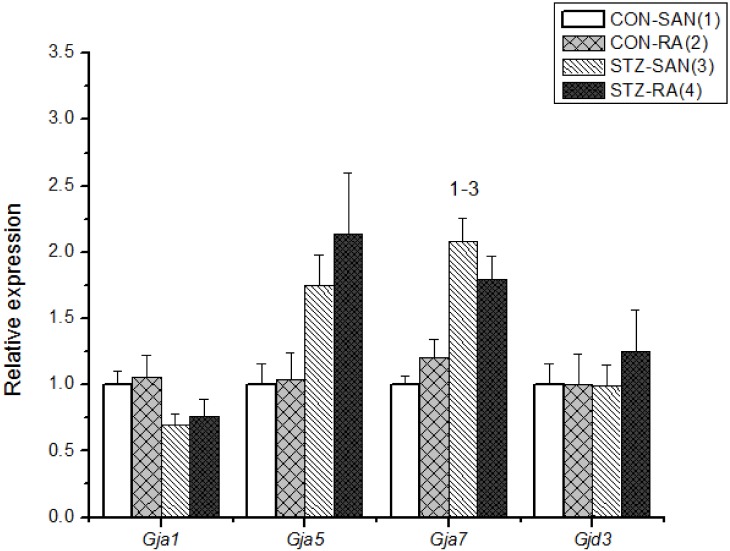
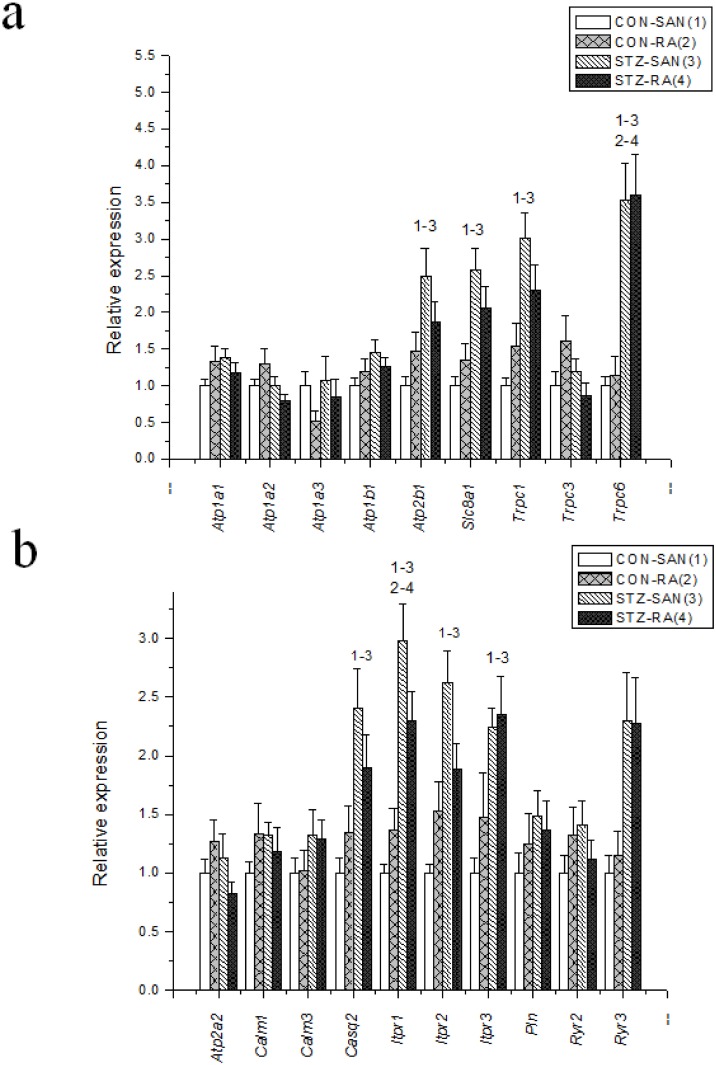
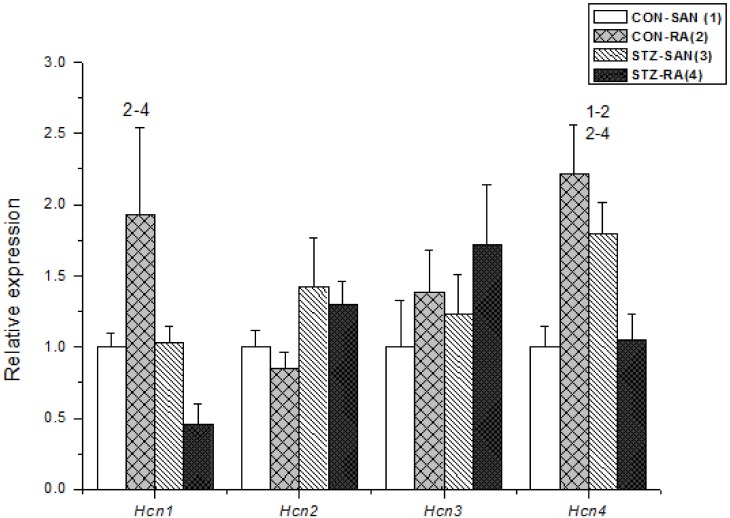
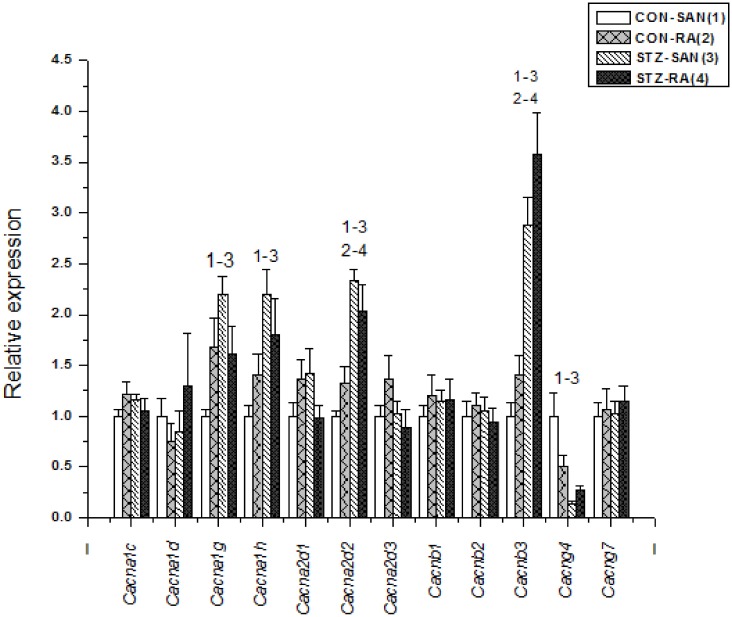
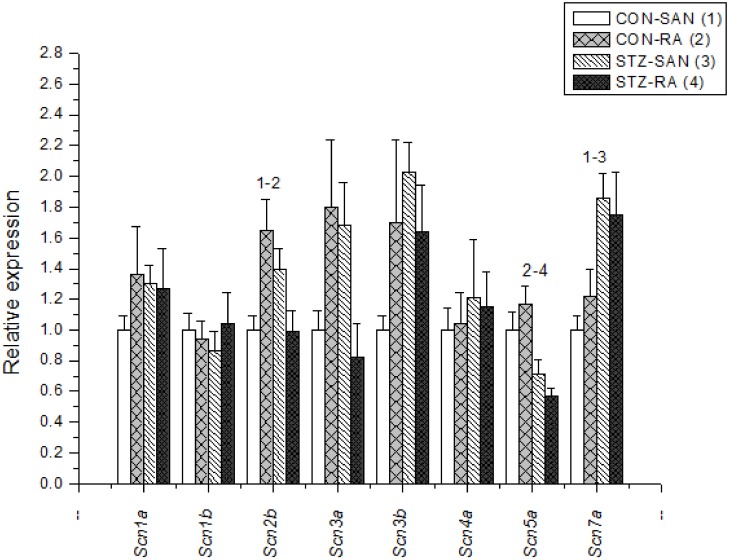
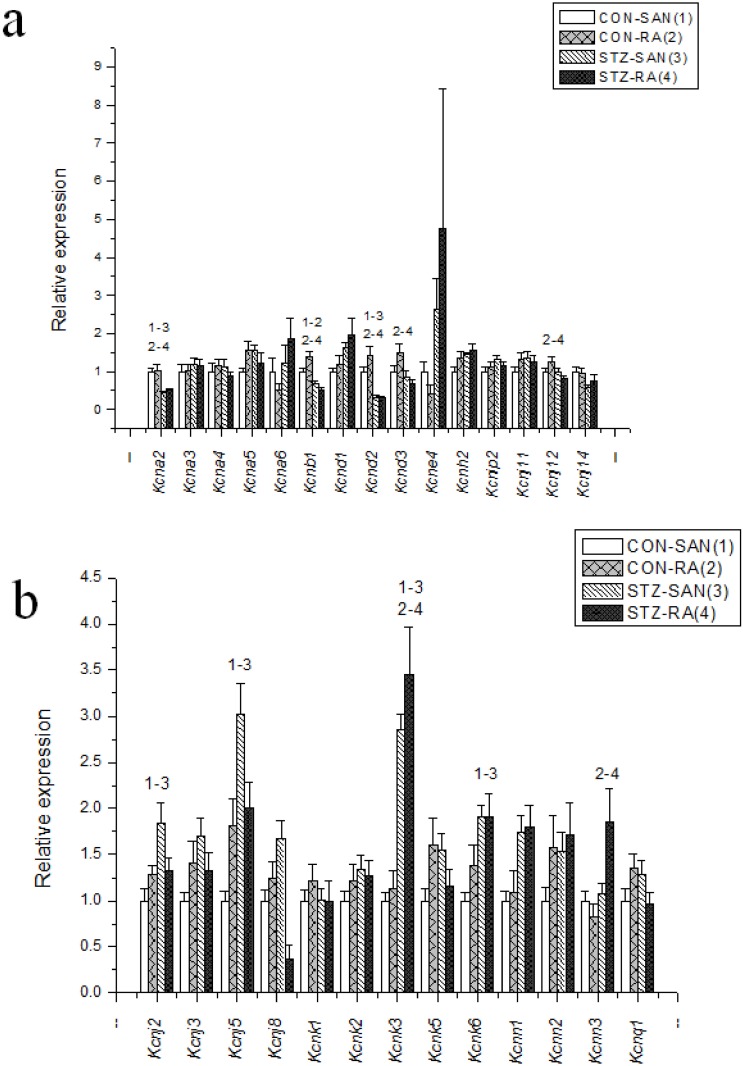
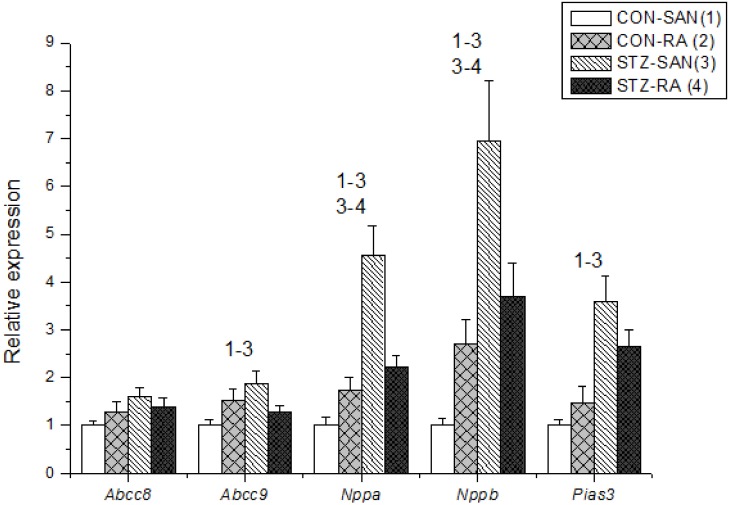
Expression of mRNA for cardiac muscle proteins are shown in Fig 3. Expression of Myl1 was significantly (P<0.05) upregulated in STZ-SAN compared to CON-SAN whereas Myl1 was significantly downregulated STZ-RA compared to CON-RA. Expression of mRNA for intercellular proteins are shown in Fig 4. Expression of Gja7 was upregulated in STZ-SAN compared to CON-SAN. Expression of mRNA for cell membrane transport and intracellular Ca2+ transport are shown in Fig 5a and 5b, respectively. Among the cell membrane transport and Ca2+ transport proteins expression of Atp2b1, Slc8a1, Trpc1, Trpc6, Casq2 and Itpr1-3 were significantly upregulated and Ryr3 was modestly upregulated in STZ-SAN compared to CON-SAN. Expression of mRNA for hyperpolarization-activated cyclic nucleotide-gated channel proteins are shown in Fig 6. Hcn4 was modestly upregulated in STZ-SAN compared to CON-SAN however, the difference was not significant. Expression of mRNA for calcium channel proteins are shown in Fig 7. Expression of Cacna1g, Cacna1h, Cacna2d and Cacnb3 were upregulated whilst Cacng4 (7-fold) was downregulated in STZ-SAN compared to CON-SAN. Expression of mRNA for sodium channel proteins are shown in Fig 8. Expression of Scn7a was upregulated in STZ-SAN compared to CON-SAN. Expression of mRNA for potassium channel proteins are shown in Fig 9a and 9b. Expression of Kcna2 and Kcnd2 were downregulated whilst Kcnj2, Kcnj5, Kcnk3 and Kcnk6 were upregulated in STZ-SAN compared to CON-SAN. Expression of Kcnd2, Kcnd3 and Kcnj12 were downregulated and Kcnk3 and Kcnn3 were upregulated in STZ-RA compared to CON-RA. Expression of mRNA for miscellaneous cardiac proteins are shown in Fig 10. Expression of Abcc9, Nppa, Nppb and Pias3 were upregulated in STZ-SAN compared to CON-SAN.
Fig 3. Expression of genes encoding various cardiac muscle proteins.
Data are mean ± SEM, n = 4–10 samples from STZ and control rat each containing SANs from 2 hearts.
Fig 4. Expression of genes encoding various intercellular proteins.
Data are mean ± SEM, n = 6–10 samples from STZ and control rat each containing SANs from 2 hearts.
Fig 5. (a) Expression of genes encoding various intracellular Ca2+-transport and regulatory proteins and (b) membrane transport proteins.
Data are mean ± SEM, n = 6–10 samples from STZ and control rat each containing SANs from 2 hearts.
Fig 6. Expression of genes encoding various hyperpolarization-activated cyclic-nucleotide-gated channels.
Data are mean ± SEM, n = 5–10 samples from STZ and control rat each containing SANs from 2 hearts.
Fig 7. Expression of genes encoding various calcium channel proteins.
Data are mean ± SEM, n = 5–10 samples from STZ and control rat each containing SANs from 2 hearts.
Fig 8. Expression of genes encoding various sodium channel proteins.
Data are mean ± SEM, n = 8–10 samples from STZ and control rat each containing SANs from 2 hearts.
Fig 9. a and b—Expression of genes encoding various potassium channel proteins.
Data are mean ± SEM, n = 6–10 samples from STZ and control rat each containing SANs from 2 hearts.
Fig 10. Expression of genes encoding miscellaneous cardiac proteins.
Data are mean ± SEM, n = 8–10 samples from STZ and control rat each containing SANs from 2 hearts.
Expression of protein
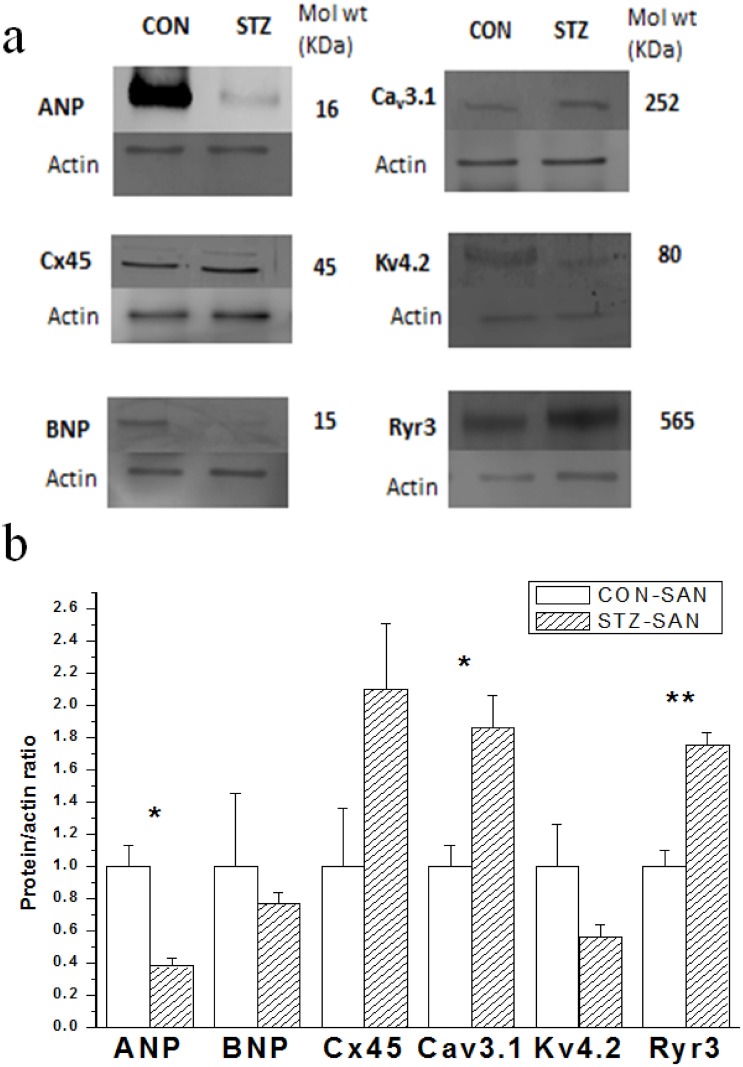
Western blot techniques were used to compare expression of selected proteins in STZ and control SAN and typical results are shown in Fig 11a. Expression of ANP was significantly reduced whilst Cav3.1 and Ryr3 were increased in STZ compared to control SAN (Fig 11b).
Fig 11. Typical Western blots comparing expression of various proteins from STZ and control SAN are shown in Fig 11a.
The blots shown are representative of 3 individual samples from STZ and control rats each containing SANs from 2 hearts. The Protein/actin ratio for the different proteins are shown in Fig 11b. Data are mean ± SEM, n = 3 samples from STZ and control rat each containing SANs from 2 hearts. * P<0.05, ** P<0.01
Discussion
Spontaneous heart rate was lower in STZ compared to control hearts. Previous in vivo biotelemetry experiments have also demonstrated reduced heart rate in STZ rat [5,21]. Prolongation of SAN action potentials in isolated perfused heart and prolonged pacemaker cycle length and sino-atrial conduction time in isolated sino-atrial preparations have been demonstrated in STZ rat heart [9,10]. Collectively, data from various isolated preparations suggest that the reduced heart rate in STZ rat could, at least in part, may be attributed to intrinsic defects in the electrophysiological function of the SAN. An early step in elucidating the mechanisms that underlie low heart rate in STZ rat is to study the profile of genes associated with proteins that are involved in the generation and propagation of the SAN action potential. Of particular interest were genes encoding Gja7 (2-fold), Trpc1 (3-fold), Trpc6 (4-fold), Ryr3 (2-fold), Cacna1g (2-fold), Cacnb3 (3-fold), Kcnj5 (3-fold), Nppa (5-fold) and Nppb (7-fold) which were upregulated and Cacng4 (3-fold) which was downregulated in STZ-SAN compared to CON-SAN.
Gja7 (Cx45) was upregulated in STZ-SAN compared to CON-SAN. Cx45 protein was also moderately increased in STZ-SAN compared to CON-SAN. Connexin proteins, pore forming subunits of gap junctions, play an important role in ensuring efficient cell-to-cell communication and the maintenance of cardiac rhythmicity [22]. At least five connexins (Cx30.2, Cx37, Cx40, Cx43 and Cx45) are prominently expressed in the heart and each shows regional and cell type specific expression [22,23]. Gja7 (Cx45) is expressed in the SAN and previous studies have demonstrated upregulation of genes encoding connexin proteins, and in particular Cx45, in STZ-SAN [10,24–26]. It is known that action potential duration and SAN conduction and pacemaker cycle length can be prolonged in diabetic heart and this may be associated with remodeling of connexin proteins [10,11].
Trpc1 (TRPC1) and Trpc6 (TRPC6) were upregulated in STZ-SAN compared to CON-SAN. The transient receptor potential (TRP) channels are a large family of non-selective and non-voltage gated ion channels that are widely expressed in human tissue including the heart and vasculature [27,28]. TRPC1 and TRPC6 are mechano-sensitive, non-selective cation channels that are expressed in mouse ventricular muscle [29]. In the cardiovascular system the TRPC family has been found to play a role in vascular and cardiac disease [30]. Upregulation of TRPC channels is involved in the development of cardiac hypertrophy and heart failure [27,31,32]. TRPC6 is induced in heart hypertrophy and inhibition of TRPC6 has been shown to suppress agonist-induced hypertrophic responses [33–35]. Whilst the physiological role of the TRP channels in the SAN is unclear, upregulation of Trpc1 and Trpc6 in STZ-SAN if associated with altered entry of Na+ or Ca2+ through non-selective TRP channels might have implications for the generation of the pacemaker or action potential in diabetic SAN cells [31,36,37].
Cacna1g (Cav3.1) was upregulated in STZ-SAN compared to CON-SAN however, expression of Cacna1g was not altered in STZ-RA compared to CON-RA. Cav3.1 protein was also increased in STZ-SAN compared to CON-SAN. Previous studies have reported upregulation of Cacna1g in ventricle from the Goto-Kakizaki rat, an experimental model of type 2 diabetes mellitus, and the Zucker diabetic fatty rat [38,39]. The protein encoded by Cacna1g represents the alpha 1G subunit, also known as Cav3.1, of the T-type calcium channel. In the heart T-type Ca2+ channels are found in the SAN and conduction cells [40]. In mice disruption of the gene encoding Cav3.1channels abolishes T-type Ca2+ current in isolated cells from the SAN and atrioventricular node without affecting L-type Ca2+ current. Inactivation of Cacna1g slowed the heart rate in vivo and prolonged the SAN recovery time and slowed pacemaker activity of individual SAN cells through a reduction of the slope of the diastolic depolarization [41]. Upregulation of Cacna1g (Cav3.1) might be expected to increase T-type Ca2+ current and hence, the slope of the pacemaker potential and heart rate in STZ rat.
Cacnb3 (Cavβ3) was upregulated in STZ-SAN compared to CON-SAN. Cacnb3 (Cavβ3) encodes expression of the beta 3 subunit (Cavβ3) of the L-type Ca2+ channel. Previous studies have demonstrated that overexpression of the beta subunits (1–4) in adult cultured heart cells increased whole-cell L-type Ca2+ current density [42]. In the SAN L-type Ca2+ current contributes to the pacemaker potential and generates the upstroke of the action potential therefore upregulation of Cacnb3 (Cavβ3) might be expected to increase L-type Ca2+ current, the slope of the pacemaker potential and hence heart rate.
Ryr3 (RYR3) was upregulated in STZ-SAN compared to CON-SAN. RY3 protein was also increased in STZ-SAN compared to CON-SAN. Various studies have shown that sub-sarcolemmal diastolic Ca2+ release from the sarcoplasmic reticulum (SR) may contribute to the generation of electrical activity in SAN cells [43,44]. Upregulation of the SR Ca2+ release channel (ryandodine receptor) would be expected to facilitate increased release of Ca2+ from the SR and hence, might influence spontaneous electrical activity in the SAN cell.
Cacng4 (Cavγ4) was downregulated (3-fold) in STZ-SAN compared to CON-SAN however, expression of Cacng4 was not altered in STZ-RA compared to CON-RA. The cardiac voltage-gated L-type Ca2+ channel is the Ca2+ channel that is required for excitation-contraction coupling and that also contributes to the plateau phase of the cardiac action potential and pacemaker activity in nodal cells [45,46]. The protein encoded by Cacng4 (Cavγ4) represents one of the gamma subunits of the L-type Ca2+ channel. It has been reported that the gamma subunits expressed in heart (gamma 4, 6, 7 and 8) form macromolecular complexes with Cav1.2 and can differentially modulate its function [45]. Downregulation of Cacng4 might have implications for the modulation and hence, function of the L-type Ca2+ channel and L-type Ca2+ current which in turn are important for generation of the pacemaker potential and the upstroke of the SAN action potential.
Nppa (ANP) and Nppb (BNP) were upregulated in STZ-SAN compared to CON-SAN however, neither Nppa or Nppb were significantly altered in STZ-RA compared to CON-RA. ANP protein was also significantly reduced in STZ-SAN compared to CON-SAN. Natriuretic peptides are a family of related peptides that include atrial natriuretic peptide (ANP) and B-type natriuretic peptide (BNP) that are secreted from the cardiac atria and ventricles [47]. ANP and BNP decrease blood pressure and cardiac hypertrophy and BNP acts locally to reduce ventricular fibrosis and they are both involved in the pathogenic mechanisms leading to major cardiovascular diseases, including heart failure, coronary heart diseases, hypertension and left ventricular hypertrophy [47–49]. Previous studies have demonstrated the expression of ANP, albeit at low levels, in the SAN node [50,51]. ANP plays a key role in cardiac electrophysiology, modulating the autonomic nervous system and regulating the function of various cardiac ion channels [52]. Previous studies have demonstrated increases in ANP and BNP in blood plasma and atrial tissues and varying effects of ANP and BNP on the amplitude and kinetics of shortening and [Ca2+]i in ventricular myocytes from STZ-induced diabetic rat [53,54]. BNP has been shown to increase heart rate and electrical conduction velocity in isolated hearts and in the SAN and also increase spontaneous action potential frequency in isolated SAN myocytes [55–57]. Upregulation of Nppa and Nppb in the SAN may be associated with mechanisms that compensate for the low heart rate seen in the STZ-induced diabetic heart [53,54]. Interestingly, expression of Nppa and Nppb were increased whilst ANP protein was reduced in STZ-SAN compared to CON-SAN. Previous studies have demonstrated increased ANP and BNP levels in plasma and atria in STZ rats compared to controls [53,54].
Collectively, this study has demonstrated differences in the profile of mRNA encoding a variety of proteins that are associated with the generation, conduction and regulation of electrical signals in the SAN of STZ-induced diabetic rat heart. Data from this study will provide a basis for a substantial range of studies to investigate whether these changes in mRNA translate into changes in electrophysiological function.
Acknowledgments
Grant from UAE University.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by UAE University and LABCO, a partner of SIGMA-ALDRICH. UAE University provided a grant to support this research project. LABCO did not provide any funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Laing SP, Swerdlow AJ, Slater SD, Burden AC, Morris A, Waugh NR et al. (2003) Mortality from heart disease in a cohort of 23,000 patients with insulin-treated diabetes. Diabetologia 46(6):760–765. [DOI] [PubMed] [Google Scholar]
- 2.Piccini JP, Klein L, Gheorghiade M, Bonow RO (2004) New insights into diastolic heart failure: role of diabetes mellitus. Am J Med 116 Suppl 5A:64S–75S. [DOI] [PubMed] [Google Scholar]
- 3.Movahed MR (2007) Diabetes as a risk factor for cardiac conduction defects: a review. Diabetes Obes Metab 9(3):276–281. [DOI] [PubMed] [Google Scholar]
- 4.Aksnes TA, Schmieder RE, Kjeldsen SE, Ghani S, Hua TA, Julius S (2008) Impact of new-onset diabetes mellitus on development of atrial fibrillation and heart failure in high-risk hypertension (from the VALUE Trial). Am J Cardiol 101(5):634–638. 10.1016/j.amjcard.2007.10.025 [DOI] [PubMed] [Google Scholar]
- 5.Howarth FC, Jacobson M, Shafiullah M, Adeghate E (2005) Long-term effects of streptozotocin-induced diabetes on the electrocardiogram, physical activity and body temperature in rats. Exp Physiol 90(6):827–835. [DOI] [PubMed] [Google Scholar]
- 6.Howarth FC, Jacobson M, Qureshi MA, Shafiullah M, Hameed RS, Zilahi E et al. (2009) Altered gene expression may underlie prolonged duration of the QT interval and ventricular action potential in streptozotocin-induced diabetic rat heart. Mol Cell Biochem 328(1–2): 57–65. 10.1007/s11010-009-0074-9 [DOI] [PubMed] [Google Scholar]
- 7.Howarth FC, Jacobson M, Shafiullah M, Adeghate E (2008) Long-term effects of type 2 diabetes mellitus on heart rhythm in the Goto-Kakizaki rat. Exp Physiol 93(3):362–369. [DOI] [PubMed] [Google Scholar]
- 8.Yuill KH, Tosh D, Hancox JC (2010) Streptozotocin-induced diabetes modulates action potentials and ion channel currents from the rat atrioventricular node. Exp Physiol 95(4):508–517. 10.1113/expphysiol.2009.050286 [DOI] [PubMed] [Google Scholar]
- 9.Howarth FC, Al Sharhan R, Al Hammadi A, Qureshi MA (2007) Effects of streptozotocin-induced diabetes on action potentials in the sinoatrial node compared with other regions of the rat heart. Mol Cell Biochem 300(1–2):39–46. [DOI] [PubMed] [Google Scholar]
- 10.Howarth FC, Nowotny N, Zilahi E, El Haj MA, Lei M (2007) Altered expression of gap junction connexin proteins may partly underlie heart rhythm disturbances in the streptozotocin-induced diabetic rat heart. Mol Cell Biochem 2007; 305(1–2):145–151. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe M, Yokoshiki H, Mitsuyama H, Mizukami K, Ono T, Tsutsui H (2012) Conduction and refractory disorders in the diabetic atrium. Am J Physiol 303(1):H86–H95. [DOI] [PubMed] [Google Scholar]
- 12.Monfredi O, Dobrzynski H, Mondal T, Boyett MR, Morris GM (2010) The anatomy and physiology of the sinoatrial node—a contemporary review. Pacing Clin Electrophysiol 33(11):1392–1406. 10.1111/j.1540-8159.2010.02838.x [DOI] [PubMed] [Google Scholar]
- 13.Howarth FC, Qureshi MA (2006) Effects of carbenoxolone on heart rhythm, contractility and intracellular calcium in streptozotocin-induced diabetic rat. Mol Cell Biochem 289(1–2):21–29. [DOI] [PubMed] [Google Scholar]
- 14.Salem KA, Qureshi MA, Sydorenko V, Parekh K, Jayaprakash P, Iqbal T et al. (2013) Effects of exercise training on excitation-contraction coupling and related mRNA expression in hearts of Goto-Kakizaki type 2 diabetic rats. Mol Cell Biochem 380(1–2):83–96. 10.1007/s11010-013-1662-2 [DOI] [PubMed] [Google Scholar]
- 15.Salem KA, Adrian TE, Qureshi MA, Parekh K, Oz M, Howarth FC (2012) Shortening and intracellular Ca2+ in ventricular myocytes and expression of genes encoding cardiac muscle proteins in early onset type 2 diabetic Goto-Kakizaki rats. Exp Physiol 97(12):1281–1291. 10.1113/expphysiol.2012.066639 [DOI] [PubMed] [Google Scholar]
- 16.Gaber EM, Jayaprakash P, Qureshi MA, Parekh K, Oz M, Adrian TE et al. (2014) Effects of a sucrose-enriched diet on the pattern of gene expression, contraction and Ca(2+) transport in Goto-Kakizaki type 2 diabetic rat heart. Exp Physiol 99(6):881–893. 10.1113/expphysiol.2013.077594 [DOI] [PubMed] [Google Scholar]
- 17.Yanni J, Tellez JO, Maczewski M, Mackiewicz U, Beresewicz A, Billeter R et al. (2011) Changes in ion channel gene expression underlying heart failure-induced sinoatrial node dysfunction. Circ Heart Fail 4(4):496–508. 10.1161/CIRCHEARTFAILURE.110.957647 [DOI] [PubMed] [Google Scholar]
- 18.Yanni J, Tellez JO, Sutyagin PV, Boyett MR, Dobrzynski H (2010) Structural remodelling of the sinoatrial node in obese old rats. J Mol Cell Cardiol 48(4):653–662. 10.1016/j.yjmcc.2009.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Souza A, Bucchi A, Johnsen AB, Logantha SJ, Monfredi O, Yanni J et al. (2014) Exercise training reduces resting heart rate via downregulation of the funny channel HCN4. Nat Commun 5:3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raza H, John A, Howarth FC (2015) Increased oxidative stress and mitochondrial dysfunction in zucker diabetic rat liver and brain. Cell Physiol Biochem 35(3):1241–1251. 10.1159/000373947 [DOI] [PubMed] [Google Scholar]
- 21.Howarth FC, Adeghate E, Jacobson M (2009) Heart rate and QT interval in streptozotocin-induced diabetic rat. Journal of Medical Sciences 2(3):108–118. [Google Scholar]
- 22.Jansen JA, van Veen TA, de Bakker JM, van Rijen HV (2010) Cardiac connexins and impulse propagation. J Mol Cell Cardiol 48(1):76–82. 10.1016/j.yjmcc.2009.08.018 [DOI] [PubMed] [Google Scholar]
- 23.Duffy HS, Fort AG, Spray DC (2006) Cardiac connexins: genes to nexus. Adv Cardiol 42:1–17. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Dobrzynski H, Tellez J, Niwa R, Billeter R, Honjo H et al. (2006) Extended atrial conduction system characterised by the expression of the HCN4 channel and connexin45. Cardiovasc Res 72(2):271–281. [DOI] [PubMed] [Google Scholar]
- 25.Jones SA, Lancaster MK, Boyett MR (2004) Ageing-related changes of connexins and conduction within the sinoatrial node. J Physiol 560(Pt 2):429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verheijck EE, van Kempen MJ, Veereschild M, Lurvink J, Jongsma HJ, Bouman LN (2001) Electrophysiological features of the mouse sinoatrial node in relation to connexin distribution. Cardiovasc Res 52(1):40–50. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe H, Murakami M, Ohba T, Ono K, Ito H (2009) The pathological role of transient receptor potential channels in heart disease. Circ J 73(3):419–427. [DOI] [PubMed] [Google Scholar]
- 28.Bon RS, Beech DJ (2013) In pursuit of small molecule chemistry for calcium-permeable non-selective TRPC channels—mirage or pot of gold? Br J Pharmacol 170(3):459–474. 10.1111/bph.12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward ML, Williams IA, Chu Y, Cooper PJ, Ju YK, Allen DG (2008) Stretch-activated channels in the heart: contributions to length-dependence and to cardiomyopathy. Prog Biophys Mol Biol 97(2–3):232–249. 10.1016/j.pbiomolbio.2008.02.009 [DOI] [PubMed] [Google Scholar]
- 30.Rowell J, Koitabashi N, Kass DA (2010) TRP-ing up heart and vessels: canonical transient receptor potential channels and cardiovascular disease. J Cardiovasc Transl Res 3(5):516–524. 10.1007/s12265-010-9208-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishida M, Kurose H (2008) Roles of TRP channels in the development of cardiac hypertrophy. Naunyn Schmiedebergs Arch Pharmacol 378(4):395–406. 10.1007/s00210-008-0321-8 [DOI] [PubMed] [Google Scholar]
- 32.Guinamard R, Bois P (2007) Involvement of transient receptor potential proteins in cardiac hypertrophy. Biochim Biophys Acta 1772(8):885–894. [DOI] [PubMed] [Google Scholar]
- 33.Vindis C, D'Angelo R, Mucher E, Negre-Salvayre A, Parini A, Mialet-Perez J (2010) Essential role of TRPC1 channels in cardiomyoblasts hypertrophy mediated by 5-HT2A serotonin receptors. Biochem Biophys Res Commun 391(1):979–983. 10.1016/j.bbrc.2009.12.001 [DOI] [PubMed] [Google Scholar]
- 34.Nishida M, Watanabe K, Nakaya M, Kurose H (2010) [Mechanism of cardiac hypertrophy via diacylglycerol-sensitive TRPC channels]. Yakugaku Zasshi 130(3):295–302. [DOI] [PubMed] [Google Scholar]
- 35.Nishida M, Watanabe K, Sato Y, Nakaya M, Kitajima N, Ide T et al. (2010) Phosphorylation of TRPC6 channels at Thr69 is required for anti-hypertrophic effects of phosphodiesterase 5 inhibition. J Biol Chem 285(17):13244–13253. 10.1074/jbc.M109.074104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harper MT, Londono JE, Quick K, Londono JC, Flockerzi V, Philipp SE et al. (2013) Transient receptor potential channels function as a coincidence signal detector mediating phosphatidylserine exposure. Sci Signal 6(281):ra50 10.1126/scisignal.2003701 [DOI] [PubMed] [Google Scholar]
- 37.Sun YH, Li YQ, Feng SL, Li BX, Pan ZW, Xu CQ et al. (2010) Calcium-sensing receptor activation contributed to apoptosis stimulates TRPC6 channel in rat neonatal ventricular myocytes. Biochem Biophys Res Commun 394(4):955–961. 10.1016/j.bbrc.2010.03.096 [DOI] [PubMed] [Google Scholar]
- 38.Salem KA, Qureshi MA, Sydorenko V, Parekh K, Jayaprakash P, Iqbal T et al. (2013) Effects of exercise training on excitation-contraction coupling and related mRNA expression in hearts of Goto-Kakizaki type 2 diabetic rats. Mol Cell Biochem 380(1–2):83–96. 10.1007/s11010-013-1662-2 [DOI] [PubMed] [Google Scholar]
- 39.Howarth FC, Qureshi MA, Hassan Z, Isaev D, Parekh K, John A et al. (2012) Contractility of ventricular myocytes is well preserved despite altered mechanisms of Ca(2+) transport and a changing pattern of mRNA in aged type 2 Zucker diabetic fatty rat heart. Mol Cell Biochem 361(1–2):267–280. 10.1007/s11010-011-1112-y [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Wang F, Zhang X, Qi Z, Tang M, Szeto C et al. (2012) Beta-Adrenergic stimulation increases Cav3.1 activity in cardiac myocytes through protein kinase A. PLoS ONE 7(7):e39965 10.1371/journal.pone.0039965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangoni ME, Traboulsie A, Leoni AL, Couette B, Marger L, Le Quang K et al. (2006) Bradycardia and slowing of the atrioventricular conduction in mice lacking CaV3.1/alpha1G T-type calcium channels. Circ Res 98(11):1422–1430. [DOI] [PubMed] [Google Scholar]
- 42.Colecraft HM, Alseikhan B, Takahashi SX, Chaudhuri D, Mittman S, Yegnasubramanian V et al. (2002) Novel functional properties of Ca(2+) channel beta subunits revealed by their expression in adult rat heart cells. J Physiol 541(Pt 2):435–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinogradova TM, Brochet DX, Sirenko S, Li Y, Spurgeon H, Lakatta EG (2010) Sarcoplasmic reticulum Ca2+ pumping kinetics regulates timing of local Ca2+ releases and spontaneous beating rate of rabbit sinoatrial node pacemaker cells. Circ Res 107(6):767–775. 10.1161/CIRCRESAHA.110.220517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maltsev VA, Vinogradova TM, Bogdanov KY, Lakatta EG, Stern MD (2004) Diastolic calcium release controls the beating rate of rabbit sinoatrial node cells: numerical modeling of the coupling process. Biophys J 86(4):2596–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Katchman A, Morrow JP, Doshi D, Marx SO (2011) Cardiac L-type calcium channel (Cav1.2) associates with gamma subunits. FASEB J 25(3):928–936. 10.1096/fj.10-172353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess DL, Davis CF, Gefrides LA, Noebels JL (1999) Identification of three novel Ca(2+) channel gamma subunit genes reveals molecular diversification by tandem and chromosome duplication. Genome Res 9(12):1204–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potter LR, Yoder AR, Flora DR, Antos LK, Dickey DM (2009) Natriuretic peptides: their structures, receptors, physiologic functions and therapeutic applications. Handb Exp Pharmacol (191):341–366. 10.1007/978-3-540-68964-5_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volpe M, Rubattu S, Burnett J Jr (2014) Natriuretic peptides in cardiovascular diseases: current use and perspectives. Eur Heart J 35(7):419–425. 10.1093/eurheartj/eht466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGrath MF, de Bold ML, de Bold AJ (2005) The endocrine function of the heart. Trends Endocrinol Metab 16(10):469–477. [DOI] [PubMed] [Google Scholar]
- 50.Skepper JN (1989) An immunohistochemical study of the sinoatrial node and atrioventricular conducting system of the rat for atrial natriuretic peptide distribution. Histochemical Journal 21, 72–78. [DOI] [PubMed] [Google Scholar]
- 51.Benvenuti LA, Aiello VD, Higuchi ML, Palomino SA (1997) Immunohistochemical expression of atrial natriuretic peptide (ANP) in the conducting system and internodal atrial myocardium of human hearts. Acta Histochem 99(2):187–193. [DOI] [PubMed] [Google Scholar]
- 52.Perrin MJ, Gollob MH (2012) The role of atrial natriuretic peptide in modulating cardiac electrophysiology. Heart Rhythm 9(4):610–615. 10.1016/j.hrthm.2011.11.019 [DOI] [PubMed] [Google Scholar]
- 53.Howarth FC, Al Shamsi N, Al Qaydi M, Al Mazrouei M, Qureshi A, Chandranath SI et al. (2006) Effects of brain natriuretic peptide on contraction and intracellular Ca2+ in ventricular myocytes from the streptozotocin-induced diabetic rat. Ann N Y Acad Sci 1084:155–165. [DOI] [PubMed] [Google Scholar]
- 54.Howarth FC, Adem A, Adeghate EA, Al Ali NA, Al Bastaki AM, Sorour FR et al. (2005) Distribution of atrial natriuretic peptide and its effects on contraction and intracellular calcium in ventricular myocytes from streptozotocin-induced diabetic rat. Peptides 26(4):691–700. [DOI] [PubMed] [Google Scholar]
- 55.Vespasiani G, Bruni M, Meloncelli I, Clementi L, Amoretti R, Branca S et al. (1996) Validation of a computerized measurement system for guided routine evaluation of cardiovascular autonomic neuropathy. Computer Methods & Programs In Biomedicine 51:211–216. [DOI] [PubMed] [Google Scholar]
- 56.Azer J, Hua R, Vella K, Rose RA (2012) Natriuretic peptides regulate heart rate and sinoatrial node function by activating multiple natriuretic peptide receptors. J Mol Cell Cardiol 53(5):715–724. 10.1016/j.yjmcc.2012.08.020 [DOI] [PubMed] [Google Scholar]
- 57.Springer J, Azer J, Hua R, Robbins C, Adamczyk A, McBoyle S et al. (2012) The natriuretic peptides BNP and CNP increase heart rate and electrical conduction by stimulating ionic currents in the sinoatrial node and atrial myocardium following activation of guanylyl cyclase-linked natriuretic peptide receptors. J Mol Cell Cardiol 52(5):1122–1134. 10.1016/j.yjmcc.2012.01.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.