Abstract
Background
Organic cation transporters transfer solutes with a positive charge across the plasma membrane. The novel organic cation transporter 1 (OCTN1) and 2 (OCTN2) transport ergothioneine and carnitine, respectively. Mutations in the SLC22A5 gene encoding OCTN2 cause primary carnitine deficiency, a recessive disorders resulting in low carnitine levels and defective fatty acid oxidation. Variations in the SLC22A4 gene encoding OCTN1 are associated with rheumatoid arthritis and Crohn disease.
Methods
Here we evaluate the functional properties of the OCTN1 transporter using chimeric transporters constructed by fusing different portion of the OCTN1 and OCTN2 cDNAs. Their relative abundance and subcellular distribution was evaluated through western blot analysis and confocal microscopy.
Results
Substitutions of the C-terminal portion of OCTN1 with the correspondent residues of OCTN2 generated chimeric OCTN transporters more active than wild-type OCTN1 in transporting ergothioneine. Additional single amino acid substitutions introduced in chimeric OCTN transporters further increased ergothioneine transport activity. Kinetic analysis indicated that increased transport activity was due to an increased Vmax, with modest changes in Km toward ergothioneine.
Conclusions
Our results indicate that the OCTN1 transporter is tolerant to extensive amino acid substitutions. This is in sharp contrast to the OCTN2 carnitine transporter that has been selected for high functional activity through evolution, with almost all substitutions reducing carnitine transport activity.
General significance
The widespread tolerance of OCTN1 to amino acid substitutions suggests that the corresponding SLC22A4 gene may have derived from a recent duplication of the SLC22A5 gene and might not yet have a defined physiological role.
Keywords: Ergothioneine transport, OCTN1, SLC22A4, OCTN2, SLC22A5, Organic cation transporters
Introduction
Organic cation transporters (OCTs) transfer solutes with a positive charge across the plasma membrane [1]. They are encoded by the SLC22 family of membrane transporters that also includes organic anion (OATs) and novel organic cation transporters (OCTNs) [1, 2]. The novel organic cation transporter 1 (OCTN1) and the novel organic cation transporter 2 (OCTN2) are encoded by the SLC22A4 and SLC22A5 genes, respectively [2]. OCTN1 was identified in 1997 from a human fetal liver library [3]. OCTN2 was subsequently identified in 1998 for its homology to OCTN1 [4, 5], since their sequence is 88% homologous and 77% identical [6]. The regions less conserved between OCTN1 and OCTN2 are the intracellular loop between transmembrane domains 6 and 7 and the intracellular C-terminal domain [6].
OCTN2 operates a sodium-dependent transport of carnitine and a sodium-independent organic cation transport [6]. Defects in the OCTN2 transporter cause primary carnitine deficiency [7]. Carnitine is essential to transfer long-chain fatty acids from the cytosol to mitochondria for subsequent β-oxidation [7]. Therefore, the lack of carnitine impairs the ability to use fat as energy source during periods of fasting or stress and this can result in an acute metabolic decompensation early in life with hypoketotic hypoglycemia, Reye syndrome, and sudden infant death, or in a more insidious presentation, later in life, with skeletal or cardiac myopathy [8]. OCTN2 is expressed in polarized cells of intestine, kidney, placenta and mammary gland, and also in other tissues such as heart, testis, skeletal muscle and brain, allowing carnitine absorption and distribution within the entire organism [9].
OCTN1 encodes a 551-amino acid protein with 12 transmembrane domains and one nucleotide binding site motif [3]. OCTN1 is widely expressed in bone marrow, gut, heart, kidney, lung, placenta, prostate, skeletal muscle, trachea, fetal but not adult liver, and other organs that are critical for transfer of many endogenous small organic cations as well as a wide array of drugs and environmental toxins [2, 3, 10]. Little is known about the physiological role of OCTN1. When expressed in mammalian human embryonic kidney (HEK) 293 cells, OCTN1 operates a pH-dependent tetraethylammonium transport [4, 11]. Expression of OCTN1 in Xenopus oocytes increased the transport of compounds such as [3H]pyrilamine, [3H]quinidine, and [3H]verapamil and zwitterionic L-[3H]carnitine [12]. When expressed in human and rodent cells, OCTN1 mediates a Na+-independent and pH-dependent transport of the prototypical organic cation tetraethylammonium, while interacts with carnitine with very low affinity [13-15]. OCTN1 affinity for carnitine has been linked to the presence of functional variants of the amino acid in position 503 of the SLC22A4 gene: individuals homozygous for the more prevalent 503L showed a Km of 507 μM toward carnitine, while in individuals homozygous for the 503F variant the Km was 1360 μM [16, 17]. The affinity of OCTN1 toward carnitine is significantly lower than that of the human high affinity carnitine transporter OCTN2 (Km toward carnitine 2-5 μM) [18, 19] and OCTN1 probably is not a physiologically relevant carnitine transporter.
In 2005, Grundemann and collaborators identified ergothioneine as a specific substrate (apparent Km 21 μM in HEK-293 cells) for OCTN1 using a metabolomic approach [20]. Ergothioneine is a water soluble thiol compound of dietary origin [21]. Ergothioneine is synthesized in Actinomycetales bacteria and in various non-yeast-like fungi, while it cannot be synthetized in higher plants or any animal species [22]. Animals assimilate ergothioneine through plant and animal foods rich in ergothioneine [22], with major dietary sources being mushrooms, some meat products (kidney and liver) and plant products such as black and red beans and oat bran [22]. Ergothioneine has antioxidant and cytoprotectant properties and might modulate inflammation and autoimmune disease [21]. Variations in the SLC22A4 gene encoding OCTN1 have been associated with autoimmune disorders such as rheumatoid arthritis and Crohn disease [16, 23]. However, the role of ergothioneine in inflammation is somewhat controversial and there is no clear-cut phenotype in humans due to defective or excessive activity of OCTN1 [21, 22]. Mice in which the gene encoding for OCTN1 has been inactivated have greater susceptibility to intestinal inflammation under the ischemia/reperfusion model, but no defined phenotype in the absence of harsh experimental treatments [24]. Conversely, patients with mildly active rheumatoid arthritis have significantly higher erythrocytic and monocytic levels of ergothioneine, with the 503F variant of OCTN1 associated with Crohn disease having 50% greater transport efficiency than the more prevalent 503L variant [21, 25]. However, the exact role of ergothioneine in inflammation and auto-immune diseases still needs to be clarified and the gene encoding for OCTN1 might just be casually located close to the pathological gene [26].
Substitution of amino acid residues within OCTN1 with those in corresponding positions of OCTN2 (that does not transport ergothioneine) can increase ergothioneine transport by OCTN1 while retaining carnitine transport activity [27]. Amino acids in multiple transmembrane domains (7-10) seem important for substrate discrimination and transport activity [27]. However, a systematic approach to understand the role of different domains of the OCTN1 transporter was not completed.
Here we evaluate ergothioneine transport by chimeric OCTN transporters constructed by fusing together portions of the OCTN1 and OCTN2 transporters expressed in Chinese Hamster Ovary (CHO) cells. Our results show that, unlike the OCTN2 carnitine transporter, the ergothioneine transport activity of OCTN1 can be increased by amino acid substitutions, with some artificially created transporters having much higher ergothioneine transport activity than the natural occurring OCTN1 transporter.
Experimental Procedures
Construction of Chimeric OCTN1-OCTN2 Expression Vectors
The human OCTN1 and OCTN2 cDNAs, kindly provided by Dr. Vadivel Ganapathy, Medical College of Georgia, Augusta, GA, were amplified by PCR using Pfu high-fidelity polymerase and primers to add a EcoRI site to the 5′ and a BamHI site to the 3′, as previously described for OCTN2 [28]. The 3′ primer removed the physiologic STOP codon of these cDNAs. The PCR product was digested with EcoRI and BamHI and ligated in the corresponding restriction sites of pEGFP-N2 (Clontech Laboratories, Inc., Mountain View, CA). The resulting plasmid had the OCTN cDNAs fused in frame with the green fluorescent protein (GFP) under control of the Cytomegalovirus promoter. The final vector was sequenced to exclude PCR artifacts. These plasmids were transfected into CHO cells by lipofectamine-2000 according to the manufacturer instructions (Invitrogen™, Grand Island, NY). Cells were selected with 0.8 mg/ml G418 (Invitrogen™, Grand Island, NY) for three weeks and resistant cells were isolated. The presence of green fluorescence was used to confirm successful transfection. For the construction of chimeric plasmids, unique restriction sites within each cDNA were identified and are shown in Figure 1.
Figure 1. Schematic of the ergothioneine transporter hOCTN1 and the chimeric OCTN transporters.
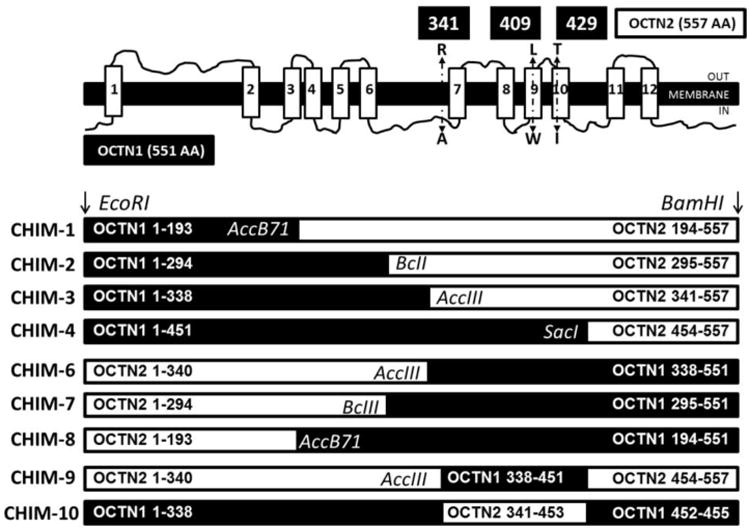
Top figure. Schematic of the OCTN1 ergothioneine transporter with transmembrane domains. The indicated substitutions, R341A, L409W, and T429I, introduced back OCTN1 residues in the portion of OCTN2 cDNA of chimeric transporters CHIM-2 and CHIM-3, in order to evaluate their effect on ergothioneine transport. Bottom figure. Chimeric OCTN transporters constructed using unique restriction sites as indicated.
Chimeric transporters were constructed fusing the N-terminus of OCTN1 with the C-terminus of OCTN2 (CHIM1, CHIM2, CHIM3, CHIM4) and the mirror image of most of these transporters fusing the N-terminus of OCTN2 with the C-terminus of OCTN1 (CHIM6, CHIM7, CHIM8). Our OCTN1 cDNA contained a phenylalanine in position 503 (503F), instead of a leucine. The 503F variant has higher ergothioneine transport activity when compared to 503L [25]. In initial experiments, cells transfected with the green florescent protein alone were used and found to be equivalent to wild-type CHO cells in terms of ergothioneine transport activity (data not shown). Analysis of these cells by confocal microscopy indicated a diffuse cytoplasmic distribution of GFP (data not shown) and Western blot analysis indicated the presence of a single band of approximately 25 kDa.
Site-directed mutagenesis
The amino acid substitutions indicated at the top of figure 1 were introduced in chimeric OCTN transporters CHIM2 and CHIM3 by site-directed mutagenesis using the Quick Change system (Agilent Technologies Inc., Santa Clara, CA) following the manufacturer's instructions. The final plasmids were sequenced to confirm the presence of the mutation and the absence of PCR artifacts and stably (not transiently) transfected into CHO cells. The plasmid contained the neomycin-resistance gene and cells were selected for resistance to G418 (Geneticin, 0.8 mg/ml) for 3 weeks before testing. A mass culture including different clones was then used for functional and imaging studies.
Western Blot Analysis
Cells were scraped in ice-cold phosphate buffered saline (PBS), and centrifuged for 10 minutes at 8,500 g at 4°C. To partially enrich the membrane fraction, cell pellets were re-suspended in ice cold hypotonic buffer (25 mM NaCl; 0.5 mM EDTA; 25 mM TRIS-HCl pH 7.2; 1× protease inhibitor), manually disrupted with 30 gauge syringe, and centrifuged at 21,000 g for 15 minutes at 4°C. Cell pellets were re-suspended in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl; 1 mM EDTA; 50 mM TRIS-HCl pH 7.2; 1× protease inhibitor, 10% Triton) and incubated for 10 minutes on ice. Each sample was then centrifuged at 21,000 g for 30 minutes at 4°C and supernatants were collected. Proteins in the supernatant were estimated by the BCA Protein Assay Kit (Thermo Fisher Scientific Inc., Waltham, MA). Sample buffer (0.25 mM 2-Mercaptoethanol; 10% glycerol; 63 mM M TRIS-HCl pH 6.8; 2% Sodium dodecyl sulfate, 0.0025% Bromophenol blue) was added to the membrane proteins without boiling. For each sample, equal concentration of protein was separated by electrophoresis on a 12% SDS-PAGE gel (Mini-PROTEAN® TGX™ Precast Gels, BioRad, Hercules, CA). Proteins were then transferred to nitrocellulose overnight. The membrane was blocked in PBS containing 5% milk and 0.02% NaN3 for 6 hours. Anti-GFP (Invitrogen™, Grand Island, NY) rabbit antibodies (1:1,000) were added to the membrane overnight at 4°C. After washing five times with 0.5% Tween20 in PBS, the membrane was blocked in PBS containing 5% milk and 0.02% NaN3 for 30 minutes at room temperature and incubated with anti-rabbit (Invitrogen™, Grand Island, NY) antibodies (1:5,000) in PBS with 5% milk and 0.02% NaN3 for one hour at room temperature. After washing the membrane three times with 0.5% Tween20 in PBS and two times with PBS, Li-Cor Odessey Infrared Imager was used to visualize labeled proteins. Blots were then stripped by incubation for 10 minutes in mild stripping buffer (0.2 M glycine, 3.5 mM Sodium dodecyl sulfate and 1% Tween20, pH 2.2), 10 minutes with PBS twice and 5 minutes with 0.1% Tween20 in Tris buffered saline (0.2 M Tris, 1.5 M NaCl, pH 7.6) for two times (http://www.abcam.com/ps/pdf/protocols/Stripping%20for%20reprobing.pdf). The blot was then blocked again and incubated with α1 Na, K ATPase rabbit polyclonal antibodies (1:1000) (Cell Signaling Technology, Inc, Danvers, MA) as control for normalization.. Digital files were analyzed with Image Studio™ Lite to estimate the intensity of each band.
Confocal Microscopy
Subcellular localization of wild-type and chimeric OCTN transporters conjugated with the GFP was analyzed with confocal microscopy (Olympus FVX laser scanning confocal IX70 microscope using a 60× 1.4NA PLANAPO oil objective). Cells were seeded on glass bottom (No. 0 cover-glass) micro-well dishes (P35G-0-14-C, MatTek Corporation, Ashland, MA), covered with medium and evaluated in vivo after labeling with Bodipy-ceramide (Molecular Probes, Eugene, OR) to visualize the Golgi/endoplasmic reticulum in red. Images of the cells were obtained at 1 μm sections. Contrast microscopy was used on the same cells to define cell borders and intracellular structures. Multiple cells were scanned and representative cells were selected. Images were then digitally superimposed [29].
Cell Culture and Ergothioneine Transport
Chinese Hamster Ovary (CHO) cells were grown in Ham F12 medium supplemented with 6% fetal bovine serum. Ergothioneine transport was measured at 37°C with the cluster-tray method as described previously for carnitine [6, 28, 30]. Cells were grown to confluence in 24-well plates (Corning® Costar® cell culture plates, Sigma-Aldrich Corp., St. Louis, MO) and depleted of intracellular amino acids by incubation for 60 min in Earle's balanced salt solution containing 5.5 mM D-glucose and supplemented with 0.1% bovine serum albumin. Ergothioneine (1 μM, 0.1 μCi/ml) was then added to the cells for 60 min. The transport reaction was stopped by rapidly washing the cells four times with ice-cold 0.1 M MgCl2. Intracellular ergothioneine was normalized for intracellular water content and transport velocity was expressed as nmol/ml cell water/h [6, 28, 30]. Saturable ergothioneine transport was calculated by subtracting non-specific uptake, measured in the presence of 1 mM unlabeled substrate, from all points. Values are reported as means ± SE of 9-12 independent determinations. Initial experiments indicated that ergothioneine transport increased linearly with time up to 4 hours of incubation (data not shown).
Transport activity among different cell lines was compared using the Student's t-test. The transport activity of cells that transported significantly more ergothioneine than CHO cells was also compared to the activity of cells expressing the wild-type OCTN1 transporter and a second symbol was used to indicate whether a significant difference was found.
Kinetic constants for ergothioneine transport were determined by measuring the transport of ergothioneine at different concentrations (1-100 μM). Data were analyzed by nonlinear regression analysis according to a Michaelis-Menten equation [30]. Nonlinear parameters are expressed as means ± SD.
Ergothioneine transport in the absence of sodium was measured substituting methylglucamine for sodium, so that the sum of methylglucamine and sodium remained constant at 140 mM [28]. In previous studies, we used multiple carnitine concentrations to define the Km of OCTN2 toward sodium (KNa) [6, 28]. However, we noted that the value obtained from the intercept of multiple regressions was identical to the value obtained at the lowest concentration of substrate [6, 28]. This is because, in a random bireactant system, such as the carnitine/Na+ or ergothioneine/Na+ co-transporter, the apparent Km toward the co-substrate (KNa) approaches the true Km (KNa) as the concentration of the substrate (carnitine or ergothioneine) decreases to near zero [6, 28, 31]. Since our previous studies indicated experimentally that a concentration of carnitine of 0.5 μM gave already values of apparent KNa indistinguishable from those calculated from the intersection of multiple curves, in this study we used a low concentration of ergothioneine (1 μM) to obtain an apparent KNa that approaches closely true KNa [6, 28].
Results
Western Blot Analysis
Expression of wild-type and chimeric OCTN transporters was assessed by Western blot analysis (using an antibody directed against GFP) and by confocal microscopy visualizing directly GFP. CHO cells and CHO cells expressing the GFP alone were included as internal controls and had no OCTN band, but a band of about 27 kDa (corresponding to the green fluorescent protein) was seen in CHO-GFP cells (Fig. 2A, lane 2). OCTN1, OCTN2 and chimeric OCTN transporters had an apparent molecular weight of 92,000 (Fig. 2A), which corresponds to the predicted size of 62 kDa of the unglycosylated OCTN1 and OCTN2 with the addition of the green fluorescent protein (about 27 kDa). In a previous report with native gel electrophoresis, we observed substantial glycosylation of the OCTN2 carnitine transporter that was abolished by tunicamycin and substitutions of N57, N64 and N91 [32]. In the experiments reported here, we did not test directly glycosylation of wild-type and chimeric transporters and the apparent molecular weight might differ from the one that we previously saw using native gels [32].
Figure 2. Western blot analysis of CHO cells expressing OCTN1, OCTN2, and chimeric OCTN transporters without (A) and with additional point mutations (B).
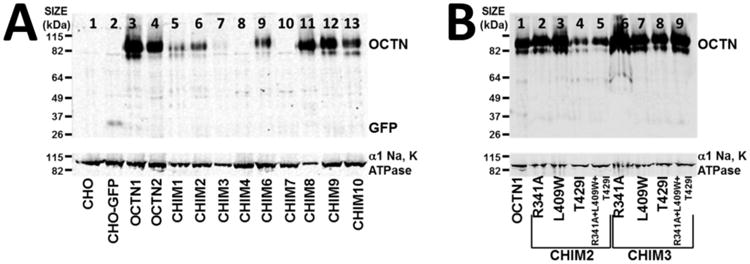
Cell lysates (20 μg of protein) from CHO cells expressing wild-type transporters OCTN1 and OCTN2 and chimeric OCTN transporters were separated by gel electrophoresis and recognized by an anti-GFP polyclonal antibody. CHO cells and CHO cells expressing GFP alone were included as an internal control. α1 Na, K ATPase was used as control for membrane protein loading. The migration of known proteins (OCTN, α1 Na, K ATPase and GFP) is indicated on the right side of the blots. The experiment was repeated 4 times with similar results.
While all transporters had a similar size, there was variation in the intensity of the bands measured in the wild-type OCTN1, OCTN2 and chimeric transporters, despite the fact that an equal concentration of protein was loaded in each lane (Fig. 3).
Figure 3. Relative intensity of the chimeric OCTN transporters and wild-type ergothioneine (OCTN1) and carnitine (OCTN2) transporters.
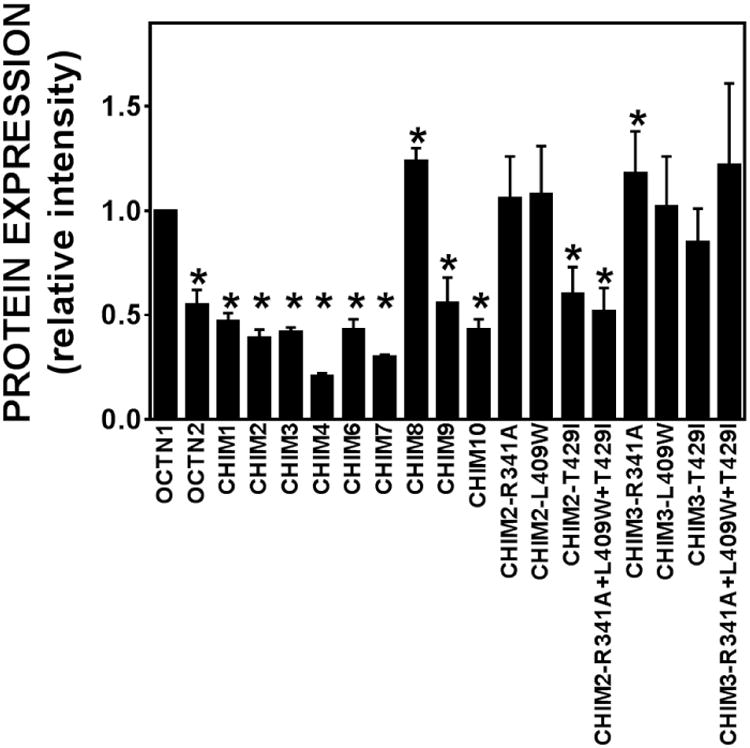
The intensity of each OCTN band in Fig. 2 was normalized to the α1 Na, K ATPase signal and then to the wild-type OCTN1 transporter. Values are means ± SD of 4 independent determinations. *p<0.05 versus OCTN1 using Student's t-test.
Normalization of the relative intensity to that of OCTN1 indicated that all chimeric OCTN transporters (CHIM1, CHIM2, CHIM3, CHIM4, CHIM6, CHIM7, CHIM9, and CHIM10) had signal intensity significantly lower than that of OCTN1, with the exception of CHIM8 whose signal intensity was higher (Fig. 3).
Confocal Microscopy
Subcellular localization of wild-type and chimeric OCTN transporters conjugated with the GFP was evaluated by confocal microscopy visualizing in green the membrane transporter and in red the endoplasmic reticulum (Fig. 4).
Figure 4. Subcellular distribution of normal and chimeric OCTN transporters tagged with the green fluorescent protein.
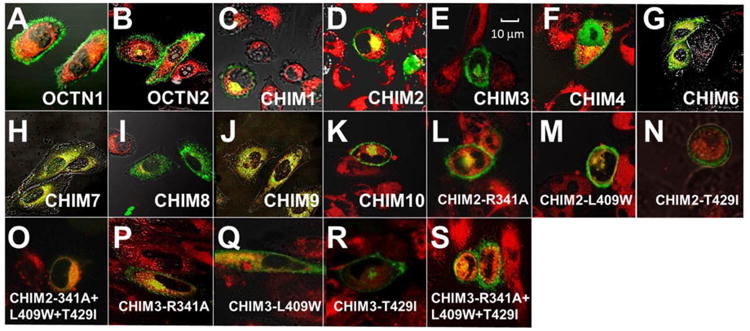
CHO cells stably transfected with OCTN1, OCTN2 and chimeric OCTN transporters were seeded on glass-bottom dishes. Live cells were visualized by Olympus FVX laser scanning confocal IX70 microscope using a 60× oil objective at 1 μm sections. Cells were labeled in vivo with Bodipy-ceramide to visualize the Golgi. Phase contrast microscopy was used to define the cell borders and intracellular structures. Images were digitally superimposed to show the location of green transporters compared to the red Golgi and plasma membrane. Co-localization of membrane transporters with the Golgi is seen as a yellow signal. Relative cell size can be estimated from the reference bar in panel E.
Wild-type OCTN1 and OCTN2 transporters mostly localized on the cell membrane (panels A, B). CHIM1 and CHIM2 were located predominantly on the plasma membrane, with minimal amounts retained in the cytoplasm (panels C, D). CHIM3 and CHIM4 localized to the plasma membrane, with a good amount of transporters retained in the cytoplasm (panels E, F). Substitution of the N-terminus of OCTN1 with correspondent portion of OCTN2 (CHIM6, CHIM7 and CHIM8) resulted in predominant localization of chimeric transporters to the cytoplasm (panels G-I). CHIM9, which contains the N- and C-terminus of OCTN2 and only amino acid residues 338-451 of OCTN1 (Fig. 1), was found mostly in the cytoplasm, around the nuclear envelope (panel J). CHIM10, which retained the N-terminus and C-terminus portions of OCTN1, localized predominantly on the plasma membrane, with a small amount retained intracellularly (panel K).
Overall, these data suggest that, in the chimeric OCTN transporters, the presence of the N-terminus of OCTN1 might be more effective for plasma membrane localization. There was an overall correlation between localization to the plasma membrane and transporter abundance (by Western blot analysis), with transporters retained inside the cell having lower abundance as compared to wild-type OCTN1. The only exception was CHIM9 that despite an intracellular localization had signal intensity similar to that of OCTN1. This latter finding may correlate with the presence in CHIM9 of both N- and C-terminus from the wild-type OCTN2 transporter.
Ergothioneine Transport by CHO Cells Transfected with Wild-Type and Chimeric OCTN Transporters
Ergothioneine is a specific substrate of the OCTN1 transporter and measurement of its initial rate of entry was used to assess the function of wild-type and chimeric transporters (Fig. 5A) [20].
Figure 5. Ergothioneine transport by wild-type and chimeric OCTN transporters. A.
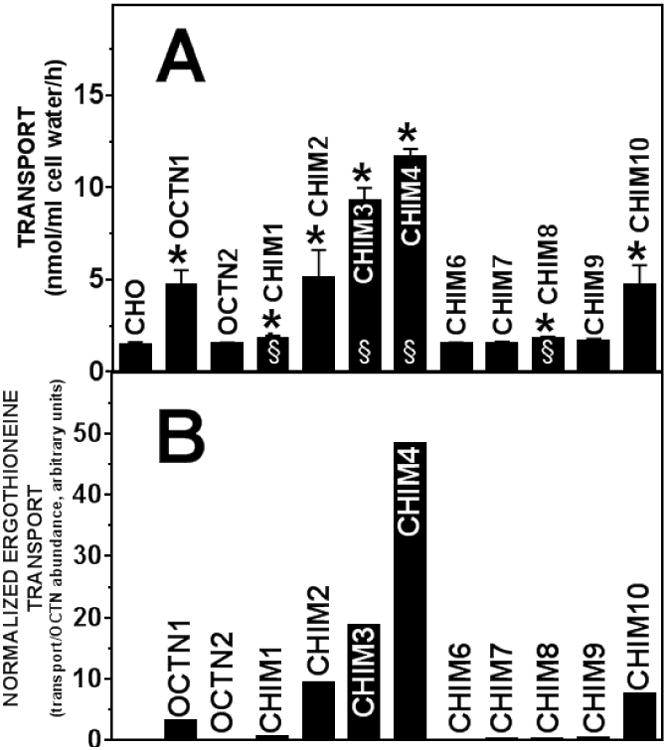
Ergothioneine (1 μM) transport was measured for 60 minutes at 37°C in CHO cells stably transfected with cDNA of the indicated transporter. Saturable ergothioneine transport was calculated by subtracting non-specific uptake, measured in the presence of 1 mM unlabeled substrate, from all points. Values are means ± SD of 9-12 independent determinations. *p<0.01 versus CHO; §p<0.01 versus OCTN1 using Student's t-test (only cells whose transport activity was significantly higher that CHO cells were compared to OCTN1). B. Values of ergothioneine transport in panel A were corrected for the baseline transport value measured in untransfected CHO cells and normalized by the amount of protein relative to OCTN1 (Fig. 3).
Transfection of CHO cells with the OCTN1 cDNA significantly increased ergothioneine transport as compared to untransfected CHO cells, while no significant increase was observed with the OCTN2 carnitine transporter. Chimeric OCTN transporters CHIM6, CHIM7, and CHIM9 failed to increase ergothioneine transport above the levels measured in untransfected CHO cells, while only minimal increase was observed for CHIM8. Since these transporters were localized mostly intracellularly (Fig. 4), we postulated that the lack of increase in ergothioneine transport was at least in part due to their abnormal subcellular distribution. Chimeric OCTN transporters CHIM2, CHIM3, CHIM4 and CHIM10, all localized on the plasma membrane (Fig. 4) and transported ergothioneine as or better than the wild-type OCTN1 transporter. Despite plasma membrane localization (Fig. 4), CHIM1 failed to transport ergothioneine. These findings suggest that amino acid residues 194-294 of OCTN1, contained in CHIM2, CHIM3, CHIM4 and CHIM10, but not in CHIM1, are necessary for ergothioneine transport by OCTN1.
While CHIM2 and CHIM10 were similar in activity to wild-type OCTN1, ergothioneine transport in CHIM3 and CHIM4 was significantly higher as compared to OCTN1. Since both CHIM3 and CHIM4 had similar subcellular distribution (Fig. 4) to wild-type OCTN1, the difference in ergothioneine transport is likely due to the specific combination of amino acid residues 295-451 of OCTN1 (Fig. 1) with the C-terminus of OCTN2.
When ergothioneine transport activity was normalized to the relative amount of OCTN protein, CHIM2, CHIM3, CHIM4, and CHIM10 all had higher intrinsic ergothioneine transport activity as compared to wild-type OCTN1 (Fig. 5B), with the intrinsic activity progressively increasing from CHIM2 to CHIM4.
Residues 341, 409, and 429 of the OCTN1 transporter and ergothioneine transport
In the OCTN2 transporter, three substitutions (R341A, L409W and T429I) that introduced OCTN1 residues for corresponding residues of OCTN2 completely abolished carnitine transport [6]. These residues are located before (R341A) or within (L409W and T429I) transmembrane domains (top Fig. 1). R341A, L409W, and T429I were substituted in the portion of OCTN2 cDNA in chimeric transporters CHIM2 and CHIM3 and their effect on protein abundance (Fig. 2B), subcellular localization (Fig. 4), and ergothioneine transport (Fig. 6) was evaluated.
Figure 6. Ergothioneine transport by wild-type and chimeric OCTN transporters.
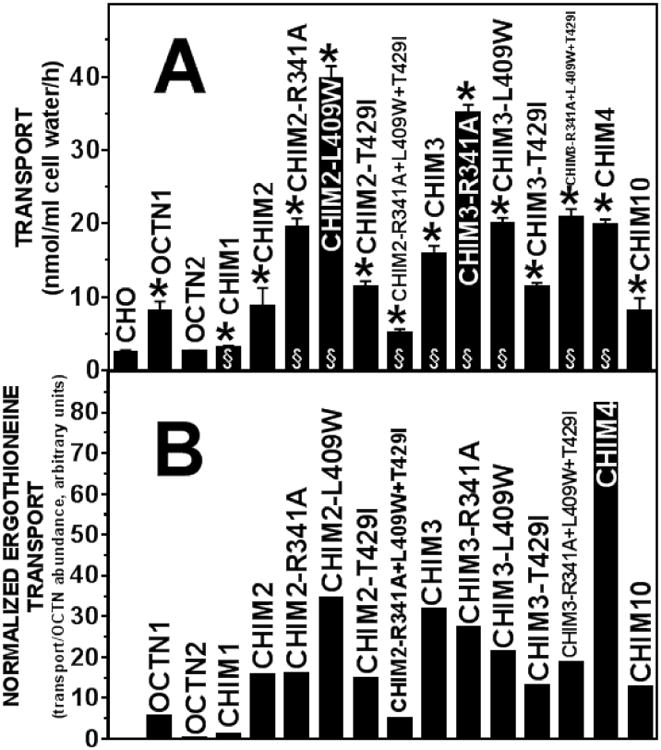
A Ergothioneine (1 μM) transport was measured for 60 minutes at 37°C in CHO cells stably transfected with the indicated transporter. Substitutions R341A, L409W, and T429I introduced back OCTN1 residues in the portion of OCTN2 cDNA of chimeric transporters CHIM-2 and CHIM-3. Saturable ergothioneine transport was calculated by subtracting nonspecific uptake, measured in the presence of 1 mM unlabeled substrate, from all points. Values are means ± SD of 9-12 independent determinations (SD not shown when smaller than the size of the symbols). *p<0.01 versus CHO; §p<0.01 versus OCTN1 using Student's t-test (only cells whose transport activity was significantly higher that CHO cells were compared to OCTN1). B. Values of ergothioneine transport in panel A were corrected for the baseline transport value measured in untransfected CHO cells and normalized by the amount of protein relative to OCTN1 (Fig. 3).
The R341A and L409W substitutions increased protein abundance when added to CHIM2, restoring protein levels similar to those seen with OCTN1, while the T429I and the combination of the three substitutions (R341A+L409W+T429I) showed similar protein abundance to that of CHIM2 (Fig. 2B, Fig. 3). The same substitutions in chimeric transporter CHIM3 (Fig. 2B) restored protein levels similar to OCTN1 and significantly increased the signal intensity above the level of OCTN1 only for CHIM3-R341A (Fig. 3).
All the substitutions tested in both CHIM2 and CHIM3 localized on the plasma membrane, with only minimal amounts retained in the cytoplasm (Fig 4, panels L-S).
Addition of R341A and L409W to CHIM2 increased ergothioneine transport above the levels measured in CHIM2 or OCTN1 (Fig. 6A). These same two substitutions increased ergothioneine transport in CHIM3 as well. No significant difference as compared to wild-type OCTN1 was observed for T429I, in both CHIM2 and CHIM3. The combination of the three substitutions (R341A+L409W+T429I) in CHIM2 reduced ergothioneine transport below the levels of wild-type OCTN1, while in CHIM3 reproduced ergothioneine transport measured with CHIM4. Since all these chimeric OCTN transporters localized on the plasma membrane, as OCTN1 (Fig. 4, panels L-S), the different levels of transport might be due to either increased efficiency of transmembrane solute transfer or different kinetic properties of these chimeric OCTN transporters as compared to wild-type OCTN1.
After semi-quantitative normalization of transport activity to the amount of OCTN protein, most chimeric transporters had increased intrinsic ergothioneine transport activity as compared to wild-type OCTN1 (Fig. 6B). The most dramatic increase in intrinsic activity was seen with CHIM4 that contains the N-terminus of OCTN1 and the C-terminus of OCTN2 (Fig.1). CHIM2-L409W and CHIM3 containing or not point mutations (CHIM3-R341A, CHIM3-L409W, and CHIM3-R341A+L409W+T429I) also had a dramatic increase in intrinsic activity (Fig. 6B), while CHIM2, CHIM2-R341A, CHIM2-T429I, CHIM3-T429I, and CHIM10 had about twice the intrinsic transport activity of OCTN1. CHIM1 and CHIM2-R341A+L409W+T429I were ineffective ergothioneine transporters, with intrinsic transport activity lower of that of OCTN1.
Kinetics of Ergothioneine Transport in CHO Cells Expressing Wild-Type and Chimeric OCTN Transporters
Kinetic analysis of ergothioneine transport by CHO cells expressing wild-type and chimeric OCTN transporters is shown in Fig. 7.
Figure 7. Kinetics of ergothioneine transport in CHO cells transfected with normal and chimeric OCTN cDNAs.
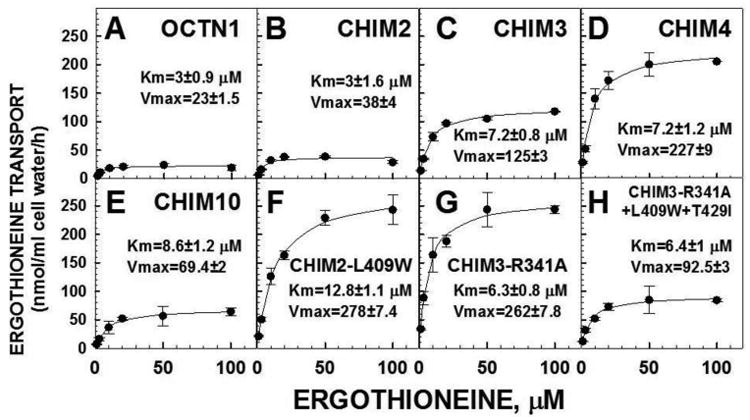
Ergothioneine (1-100 μM) transport was measured for 60 minutes at 37°C in the presence of 140 mM sodium. Non-specific ergothioneine transport, measured in the presence of 1 mM unlabeled substrate, was subtracted from each point. Data are means ± SE of triplicates (SE not shown when smaller than the size of the symbols). Black lines represent the best fit of data to a Michaelis-Menten equation. Parameters for ergothioneine transport are expressed as means ± SD. Units for Vmax are nmol/ml cell water/h. All panels have the same scale to allow direct comparison.
CHO cells overexpressing the OCTN1 cDNA transported ergothioneine with a Km of 3±0.9 μM and a Vmax of 23±1.5 nmol/ml cell water/h. CHIM2 retained the same Km toward ergothioneine (3±1.6 μM), with a moderate increase in the Vmax value (38±4 nmol/ml cell water/h). The progressive addition of OCTN1 residues to the chimeric transporters (panels C, D) increased modestly the Km toward ergothioneine up to 7.2 μM in CHIM3 and CHIM4, but determined a much larger increase in the Vmax. Chimeric transporter CHIM10, in which only residues 339-451 of OCTN1 were substituted by correspondent residues of OCTN2 (Fig. 1), had a Km toward ergothioneine similar to that observed with chimeric transporters CHIM3 and CHIM4 (8.6±1.2 μM) but the Vmax value significantly decreased compared to CHIM3 and CHIM4.
Increased ergothioneine transport activity by CHIM2-L409W, CHIM3-R341A and CHIM3-R341A+L409W+T429I was due to a higher Vmax for CHIM2-L409W and CHIM3-R341A and occurred is spite of a lower affinity toward ergothioneine, with the Km increasing up to 12.8±1.1 μM (Fig. 7, panels F-H). The Vmax was lower in the triple mutant CHIM3-R341A+L409W+T429I, with a relative normalization of the Km (6.4±1 μM).
Sodium-Dependence of Ergothioneine Transport in CHO Cells Expressing normal and Chimeric OCTN Transporters
The binding of a co-substrate can affect the affinity of a transporter toward the substrate [28, 31] and the extracellular sodium concentration can affect the Km of OCTN2 toward carnitine [28]. In the case of OCTN2 (carnitine transport), sodium at low concentrations lowers the Km of OCTN2 toward carnitine, suggesting binding to a site close to the carnitine binding site, while higher sodium concentrations provide the electrochemical gradient to transfer the carnitine-sodium complex inside the cell and increase the Vmax [6]. In order to exclude that the changes in the Vmax and Km toward ergothioneine measured for CHIM3, CHIM4 and CHIM10, were caused by abnormal interaction with the co-transported sodium, kinetics of sodium stimulated ergothioneine transport were obtained (Fig. 8, panels A-E).
Figure 8. Sodium dependence of ergothioneine transport by CHO cells expressing normal and chimeric OCTN transporters.
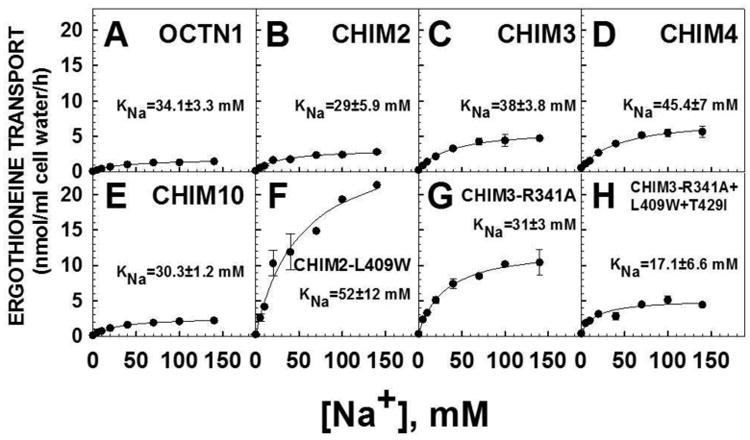
Ergothioneine (1 μM, 0.1 μCi/ml) transport was measured for 60 minutes at 37°C in the presence of increasing concentrations of sodium (0-140 mM). Cells were washed 3 times with a sodium-free solution prior to transport. Points are means ± SE of triplicates (SE not shown when smaller than the size of the symbols). Lines are the best fitting to a rectangular hyperbola. KNa is the concentration of sodium at which half-maximal stimulation of ergothioneine transport is measured. All panels have the same scale to allow direct comparison.
Half-maximal stimulation of ergothioneine transport was obtained at a sodium concentration of 34±3.3 mM in CHO cells expressing the normal OCTN1 cDNA. A similar value of KNa (the concentration of sodium at which half-maximal stimulation of ergothioneine transport was observed) was measured with all chimeric transporters CHIM2, CHIM3, CHIM4 and CHIM10.
Kinetics of sodium stimulated ergothioneine transport were also obtained for CHIM2-L409W, CHIM3-R341A, CHIM3-R341A+L409W+T429I (panels F-H). No significant modification of the KNa value was observed in these chimeric transporters as compared to wild-type transporter OCTN1. Thus, as for all chimeric and substituted transporters, the changes in the Vmax and Km toward ergothioneine were not caused by an abnormal interaction with the co-substrate sodium.
Discussion
Organic anion and cation transporters (OATs and OCTs) transfer solutes with a negative and positive charge across the plasma membrane [1]. This family of transporters also includes the novel organic cation transporter 1 (OCTN1) [3] and 2 (OCTN2) [4]. Organic cation transporters exhibiting significant homology to mammalian OCTs are present in Drosophila (organic cation transporter-like, Orct) [33] and in Caenorhabditis elegans (organic cation transporter from Caenorhabditis elegans, CeOCT1) [34]. Phylogenetic analysis suggests that the organic ion transporter family trifurcate into three branches: OATs, OCTs and a third cluster including OCTNs, fly-like putative transporters 1 and 2 (Flipts 1 and 2) and Drosophila putative transporters Orct and SD08136, which are related, but distinct and phylogenetically near-equidistant [35]. The OCTN lineage, as well as OAT and OCT lineages, formed after the divergence of vertebrates and invertebrates [36]. These lineages then expanded through independent tandem duplications to produce multiple gene pairs [36]. Flipts 1 and 2 and OAT2 are the only unpaired family members [37]. Since pair members are functionally alike, such pairing may confer the benefits of redundancy or broader substrate specificity [36] and pair members may have been hold together due to recent gene duplication or the presence of shared regulatory elements, such as locus control regions [37, 38]. The pair hOCTN1 (encoded by SLC22A4, chr5:131,630,145-131,679,899) and hOCTN2 (encoded by the SLC22A5 gene, chr5:131,705,401-131,731,306) is juxtaposed on chromosome 5 [4, 5]. Defects in the OCTN2 transporter cause primary carnitine deficiency [7], while variations in the OCTN1 ergothioneine transporter have been associated with autoimmune disorders, even though the putative mechanism at the basis of this association is unclear [16, 21-24]. The SLC22A4 gene is part of a 250 kb haplotype on chromosome 5 associated with an increased risk of Crohn disease in Europeans [17]. The association with the disease might be due to variations in SLC22A4 or one of the nearby genes, such as IRF1 which, unlike OCTN1, is actually overexpressed in lower gastrointestinal biopsies of individuals with Crohn disease [17].
In this manuscript, we evaluate chimeric OCTN transporters, generated by swapping corresponding portions of OCTN1 and OCTN2 (Fig. 1), for their capacity to transport ergothioneine, the putative physiological substrate of OCTN1 (Figs. 5A, 6A) [20]. In our experiments, we used cells stably transfected with normal or variant OCTN cDNA. Despite stable transfection, changes in activity (and probably of protein expression) can occur during the course of the experiments. With this limitation, we attempted to normalize transport activity in transfected cells to the amount of OCTN protein (Figs. 5B, 6B). Unexpectedly, some of our chimeric OCTN transporters were able to transport ergothioneine above the level measured for wild-type OCTN1, with or without normalization for OCTN content (Figs. 5, 6).
Our results indicate that the presence of the N-terminal portion of OCTN1 in chimeric OCTN transporters is essential for proper membrane localization. Chimeric OCTN transporters in which the N-terminus of OCTN1 was substituted by correspondent amino acid residues of OCTN2 (CHIM6, CHIM7, CHIM8 and CHIM9) localized intracellularly (Fig. 4) and consequently failed to transport ergothioneine (Fig. 5A). By contrast, CHIM2, CHIM3, CHIM4 and CHIM10, which retained the N-terminus of OCTN1, localized to the plasma membrane and were able to transport ergothioneine as or better than wild-type OCTN1. Since wild-type OCTN1 and OCTN2 have normal membrane localization, as do CHIM9 and CHIM10 that retain both termini from the same OCTN transporter, it seems that the two ends need to be compatible to allow membrane maturation, with the N-terminus of OCTN2 not being compatible with the C-terminus of OCTN1. There is no available crystal structure of the OCTN1 or any other mammalian organic cation transporter. In the high-affinity phosphate transporter, PiPT, which is structurally related to the SLC22 family of human transporters, the N- and C-terminal domains face one another across the central pore [39] and abnormal interactions between juxtaposed transmembrane domains in addition to the N- and C-terminus might be responsible for decreased protein stability or membrane maturation. Despite retaining the N-terminus of OCTN1 and having similar protein abundance to that of CHIM2, CHIM3, CHIM4 and CHIM10, CHIM1 failed to transport ergothioneine. This finding suggested that, in the OCTN1 ergothioneine transporter, amino acid residues 194-294 are crucial for proper functioning of this transporter, since these residues were present in CHIM2, CHIM3, CHIM4 and CHIM10, but not in CHIM1.
The portion of OCTN1 transporter comprising amino acids 295-451 includes alanine 341, next to transmembrane domain 7, tryptophan 409, within trans-membrane domain 9, and isoleucine 429, within trans-membrane domain 10 (top Fig. 1). These residues, when substituted into OCTN2, completely abolished carnitine transport [6]. Introduction of A341 and W409 for the corresponding residue of OCTN2 in both CHIM2 and CHIM3 significantly increased ergothioneine transport, with CHIM2-L409W and CHIM3-R341A having the highest transport activity among OCTN transporters (Fig. 6A). Both CHIM2-L409W and CHIM3-R341A localized to the plasma membrane (Fig. 4) and had similar (CHIM2-L409W) or significantly increased (CHIM3-R341A) protein abundance as compared to wild-type OCTN1 (Fig. 3).
After transport activity was corrected by the amount of OCTN protein, most chimeric transporters showed increased intrinsic ergothioneine transport activity as compared to wild-type OCTN1 (Figs. 5B, 6B). However, the most dramatic increase in intrinsic activity was seen with CHIM4 that contains the longest stretch of N-terminal OCTN1 residues with the C-terminus of OCTN2 (Fig. 6B). CHIM2-L409W and CHIM3 containing or not point mutations (CHIM3-R341A, CHIM3-L409W, and CHIM3-R341A+L409W+T429I) also had a dramatic increase in intrinsic activity (Fig. 6B). Since the R341A, L409W, T429I substitutions are those that completely abolished carnitine transport by the OCTN2 transporter [6], overall these data confirm an essential role of residues 341, 409 and 429 in the specific activity of both OCTN1 and OCTN2.
Kinetic analysis of ergothioneine transport in CHIM2-L409W and CHIM3-R341A showed a significantly increase in the Vmax, with values ten folds higher than in wild-type OCTN1, with minimal changes in the Km (Fig.7). Similarly, there was no significant change in the sodium-dependence of the transporter (Fig. 8), suggesting that the increased ergothioneine transport activity of chimeric transporters was due either to an increased number of membrane transporters or a more efficient transmembrane solute transfer. Western blot analysis indicated that both transporters (CHIM2-L409W and CHIM3-R341A) had a substantial increase in abundance as compared to wild type OCTN1 (Fig. 3).
It is surprising that OCTN1 appears very tolerant to domain swapping and single amino acid substitutions. This is in sharp contrast with OCTN2, encoded by the SLC22A5 gene, in which almost any substitution decreased carnitine transport and all chimeric transporters were less efficient that wild-type OCTN2 [6]. OCTN2 has a very well defined physiological function (carnitine transport) that has been selected through evolution and whose deficiency causes a well-defined human disease [8]. By contrast, the physiological function of OCTN1 seems less well defined and its removal from animal models does not cause an overt phenotype [24]. The widespread tolerance of OCTN1 to amino acid substitutions with residues of the OCTN2 carnitine transporter suggests that the corresponding SLC22A4 gene may have derived from a recent duplication of the SLC22A5 gene [37, 38], and might not have been subject to evolutionary constraints.
Supplementary Material
Highlights.
The SLC22A4 gene encodes the OCTN1 (ergothioneine) transporter.
Variations in the SLC22A4 gene have been associated with autoimmune disorders.
Chimeric OCTN1/OCTN2 transporters transported ergothioneine more efficiently than wild-type OCTN1.
Single amino acid substitutions also increased OCTN1 activity.
Unlike the OCTN2 carnitine transporter, the OCTN1 ergothioneine transporter is tolerant to extensive amino acid substitutions.
Acknowledgments
This work was supported in part by NIH grant DK 53824. We thank Dr. Vadivel Ganapathy for providing the cDNAs for OCTN1 and OCTN2 and Dr. Bijina Balakrishnan for the contribution provided during the revision phase.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koepsell H. The SLC22 family with transporters of organic cations, anions and zwitterions. Molecular aspects of medicine. 2013;34:413–435. doi: 10.1016/j.mam.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Tamai I. Pharmacological and pathophysiological roles of carnitine/organic cation transporters (OCTNs: SLC22A4, SLC22A5 and Slc22a21) Biopharmaceutics & drug disposition. 2013;34:29–44. doi: 10.1002/bdd.1816. [DOI] [PubMed] [Google Scholar]
- 3.Tamai I, Yabuuchi H, Nezu J, Sai Y, Oku A, Shimane M, Tsuji A. Cloning and characterization of a novel human pH-dependent organic cation transporter, OCTN1. FEBS Lett. 1997;419:107–111. doi: 10.1016/s0014-5793(97)01441-5. [DOI] [PubMed] [Google Scholar]
- 4.Tamai I, Ohashi R, Nezu J, Yabuuchi H, Oku A, Shimane M, Sai Y, Tsuji A. Molecular and functional identification of sodium ion-dependent, high affinity human carnitine transporter OCTN2. J Biol Chem. 1998;273:20378–20382. doi: 10.1074/jbc.273.32.20378. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Prasad PD, Leibach FH, Ganapathy V. cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem Biophys Res Commun. 1998;246:589–595. doi: 10.1006/bbrc.1998.8669. [DOI] [PubMed] [Google Scholar]
- 6.Amat di San Filippo C, Wang Y, Longo N. Functional domains in the carnitine transporter OCTN2, defective in primary carnitine deficiency. J Biol Chem. 2003;278:47776–47784. doi: 10.1074/jbc.M307911200. [DOI] [PubMed] [Google Scholar]
- 7.Rose EC, di San Filippo CA, Ndukwe Erlingsson UC, Ardon O, Pasquali M, Longo N. Genotype-phenotype correlation in primary carnitine deficiency. Hum Mutat. 2012;33:118–123. doi: 10.1002/humu.21607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longo N, Amat di San Filippo C, Pasquali M. Disorders of carnitine transport and the carnitine cycle. Am J Med Genet C Semin Med Genet. 2006;142C:77–85. doi: 10.1002/ajmg.c.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pochini L, Scalise M, Galluccio M, Indiveri C. OCTN cation transporters in health and disease: role as drug targets and assay development. J Biomol Screen. 2013;18:851–867. doi: 10.1177/1087057113493006. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi D, Aizawa S, Maeda T, Tsuboi I, Yabuuchi H, Nezu J, Tsuji A, Tamai I. Expression of organic cation transporter OCTN1 in hematopoietic cells during erythroid differentiation. Exp Hematol. 2004;32:1156–1162. doi: 10.1016/j.exphem.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Burckhardt G, Wolff NA. Structure of renal organic anion and cation transporters. Am J Physiol Renal Physiol. 2000;278:F853–866. doi: 10.1152/ajprenal.2000.278.6.F853. [DOI] [PubMed] [Google Scholar]
- 12.Yabuuchi H, Tamai I, Nezu J, Sakamoto K, Oku A, Shimane M, Sai Y, Tsuji A. Novel membrane transporter OCTN1 mediates multispecific, bidirectional, and pH-dependent transport of organic cations. J Pharmacol Exp Ther. 1999;289:768–773. [PubMed] [Google Scholar]
- 13.Wu X, George RL, Huang W, Wang H, Conway SJ, Leibach FH, Ganapathy V. Structural and functional characteristics and tissue distribution pattern of rat OCTN1, an organic cation transporter, cloned from placenta. Biochim Biophys Acta. 2000;1466:315–327. doi: 10.1016/s0005-2736(00)00189-9. [DOI] [PubMed] [Google Scholar]
- 14.Xuan W, Lamhonwah AM, Librach C, Jarvi K, Tein I. Characterization of organic cation/carnitine transporter family in human sperm. Biochem Biophys Res Commun. 2003;306:121–128. doi: 10.1016/s0006-291x(03)00930-6. [DOI] [PubMed] [Google Scholar]
- 15.Tamai I, Ohashi R, Nezu JI, Sai Y, Kobayashi D, Oku A, Shimane M, Tsuji A. Molecular and functional characterization of organic cation/carnitine transporter family in mice. J Biol Chem. 2000;275:40064–40072. doi: 10.1074/jbc.M005340200. [DOI] [PubMed] [Google Scholar]
- 16.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 17.Lamhonwah AM, Tein I. Novel localization of OCTN1, an organic cation/carnitine transporter, to mammalian mitochondria. Biochem Biophys Res Commun. 2006;345:1315–1325. doi: 10.1016/j.bbrc.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Tein I, De Vivo DC, Bierman F, Pulver P, De Meirleir LJ, Cvitanovic-Sojat L, Pagon RA, Bertini E, Dionisi-Vici C, Servidei S, et al. Impaired skin fibroblast carnitine uptake in primary systemic carnitine deficiency manifested by childhood carnitine-responsive cardiomyopathy. Pediatr Res. 1990;28:247–255. doi: 10.1203/00006450-199009000-00020. [DOI] [PubMed] [Google Scholar]
- 19.Treem WR, Stanley CA, Finegold DN, Hale DE, Coates PM. Primary carnitine deficiency due to a failure of carnitine transport in kidney, muscle, and fibroblasts. N Engl J Med. 1988;319:1331–1336. doi: 10.1056/NEJM198811173192006. [DOI] [PubMed] [Google Scholar]
- 20.Grundemann D, Harlfinger S, Golz S, Geerts A, Lazar A, Berkels R, Jung N, Rubbert A, Schomig E. Discovery of the ergothioneine transporter. Proc Natl Acad Sci U S A. 2005;102:5256–5261. doi: 10.1073/pnas.0408624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheah IK, Halliwell B. Ergothioneine; antioxidant potential, physiological function and role in disease. Biochimica et biophysica acta. 2012;1822:784–793. doi: 10.1016/j.bbadis.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Ey J, Schomig E, Taubert D. Dietary sources and antioxidant effects of ergothioneine. Journal of agricultural and food chemistry. 2007;55:6466–6474. doi: 10.1021/jf071328f. [DOI] [PubMed] [Google Scholar]
- 23.Maeda T, Hirayama M, Kobayashi D, Miyazawa K, Tamai I. Mechanism of the regulation of organic cation/carnitine transporter 1 (SLC22A4) by rheumatoid arthritis-associated transcriptional factor RUNX1 and inflammatory cytokines. Drug metabolism and disposition: the biological fate of chemicals. 2007;35:394–401. doi: 10.1124/dmd.106.012112. [DOI] [PubMed] [Google Scholar]
- 24.Kato Y, Kubo Y, Iwata D, Kato S, Sudo T, Sugiura T, Kagaya T, Wakayama T, Hirayama A, Sugimoto M, Sugihara K, Kaneko S, Soga T, Asano M, Tomita M, Matsui T, Wada M, Tsuji A. Gene knockout and metabolome analysis of carnitine/organic cation transporter OCTN1. Pharm Res. 2010;27:832–840. doi: 10.1007/s11095-010-0076-z. [DOI] [PubMed] [Google Scholar]
- 25.Taubert D, Grimberg G, Jung N, Rubbert A, Schomig E. Functional role of the 503F variant of the organic cation transporter OCTN1 in Crohn's disease. Gut. 2005;54:1505–1506. doi: 10.1136/gut.2005.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huff CD, Witherspoon DJ, Zhang Y, Gatenbee C, Denson LA, Kugathasan S, Hakonarson H, Whiting A, Davis CT, Wu W, Xing J, Watkins WS, Bamshad MJ, Bradfield JP, Bulayeva K, Simonson TS, Jorde LB, Guthery SL. Crohn's disease and genetic hitchhiking at IBD5. Molecular biology and evolution. 2012;29:101–111. doi: 10.1093/molbev/msr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacher P, Giersiefer S, Bach M, Fork C, Schomig E, Grundemann D. Substrate discrimination by ergothioneine transporter SLC22A4 and carnitine transporter SLC22A5: gain-of-function by interchange of selected amino acids. Biochim Biophys Acta. 2009;1788:2594–2602. doi: 10.1016/j.bbamem.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Meadows TA, Longo N. Abnormal sodium stimulation of carnitine transport in primary carnitine deficiency. J Biol Chem. 2000;275:20782–20786. doi: 10.1074/jbc.M000194200. [DOI] [PubMed] [Google Scholar]
- 29.Amat di San Filippo C, Pasquali M, Longo N. Pharmacological rescue of carnitine transport in primary carnitine deficiency. Hum Mutat. 2006;27:513–523. doi: 10.1002/humu.20314. [DOI] [PubMed] [Google Scholar]
- 30.Scaglia F, Wang Y, Longo N. Functional characterization of the carnitine transporter defective in primary carnitine deficiency. Arch Biochem Biophys. 1999;364:99–106. doi: 10.1006/abbi.1999.1118. [DOI] [PubMed] [Google Scholar]
- 31.Segel IH. Enzyme Kinetics. John Wiley & Son; New York: pp. 274–345. Place Published, 1975. [Google Scholar]
- 32.Filippo CA, Ardon O, Longo N. Glycosylation of the OCTN2 carnitine transporter: study of natural mutations identified in patients with primary carnitine deficiency. Biochim Biophys Acta. 2011;1812:312–320. doi: 10.1016/j.bbadis.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor CA, Stanley KN, Shirras AD. The Orct gene of Drosophila melanogaster codes for a putative organic cation transporter with six or 12 transmembrane domains. Gene. 1997;201:69–74. doi: 10.1016/s0378-1119(97)00429-0. [DOI] [PubMed] [Google Scholar]
- 34.Wu X, Fei YJ, Huang W, Chancy C, Leibach FH, Ganapathy V. Identity of the F52F12.1 gene product in Caenorhabditis elegans as an organic cation transporter. Biochimica et biophysica acta. 1999;1418:239–244. doi: 10.1016/s0005-2736(99)00020-6. [DOI] [PubMed] [Google Scholar]
- 35.Eraly SA, Nigam SK. Novel human cDNAs homologous to Drosophila Orct and mammalian carnitine transporters. Biochemical and biophysical research communications. 2002;297:1159–1166. doi: 10.1016/s0006-291x(02)02343-4. [DOI] [PubMed] [Google Scholar]
- 36.Eraly SA, Monte JC, Nigam SK. Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiological genomics. 2004;18:12–24. doi: 10.1152/physiolgenomics.00014.2004. [DOI] [PubMed] [Google Scholar]
- 37.Eraly SA, Hamilton BA, Nigam SK. Organic anion and cation transporters occur in pairs of similar and similarly expressed genes. Biochemical and biophysical research communications. 2003;300:333–342. doi: 10.1016/s0006-291x(02)02853-x. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, Harju S, Peterson KR. Locus control regions: coming of age at a decade plus. Trends in genetics : TIG. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen BP, Kumar H, Waight AB, Risenmay AJ, Roe-Zurz Z, Chau BH, Schlessinger A, Bonomi M, Harries W, Sali A, Johri AK, Stroud RM. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013;496:533–536. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


