Abstract
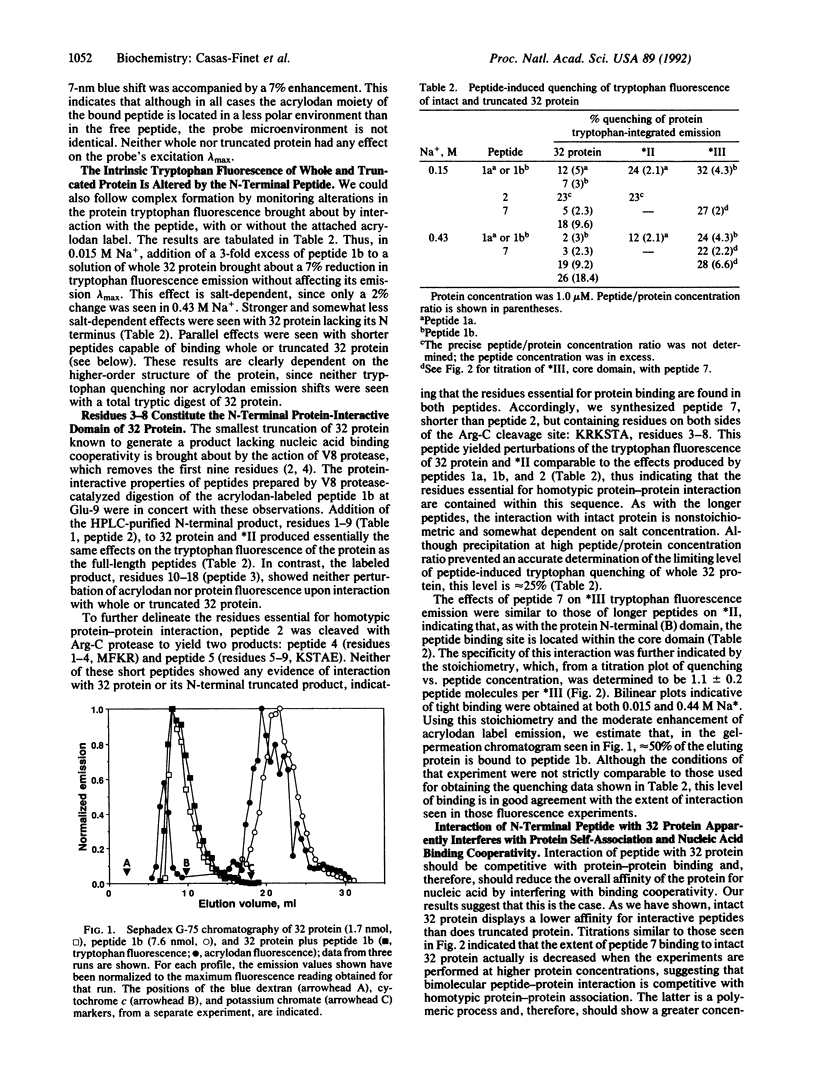
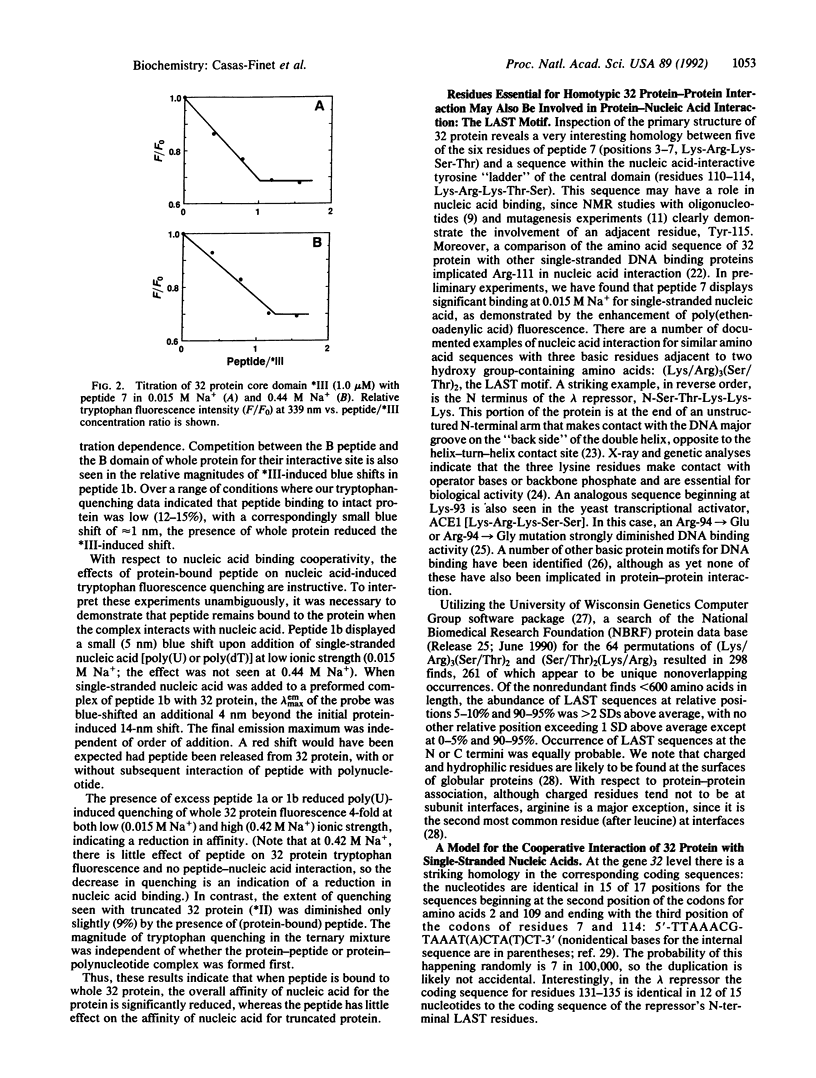
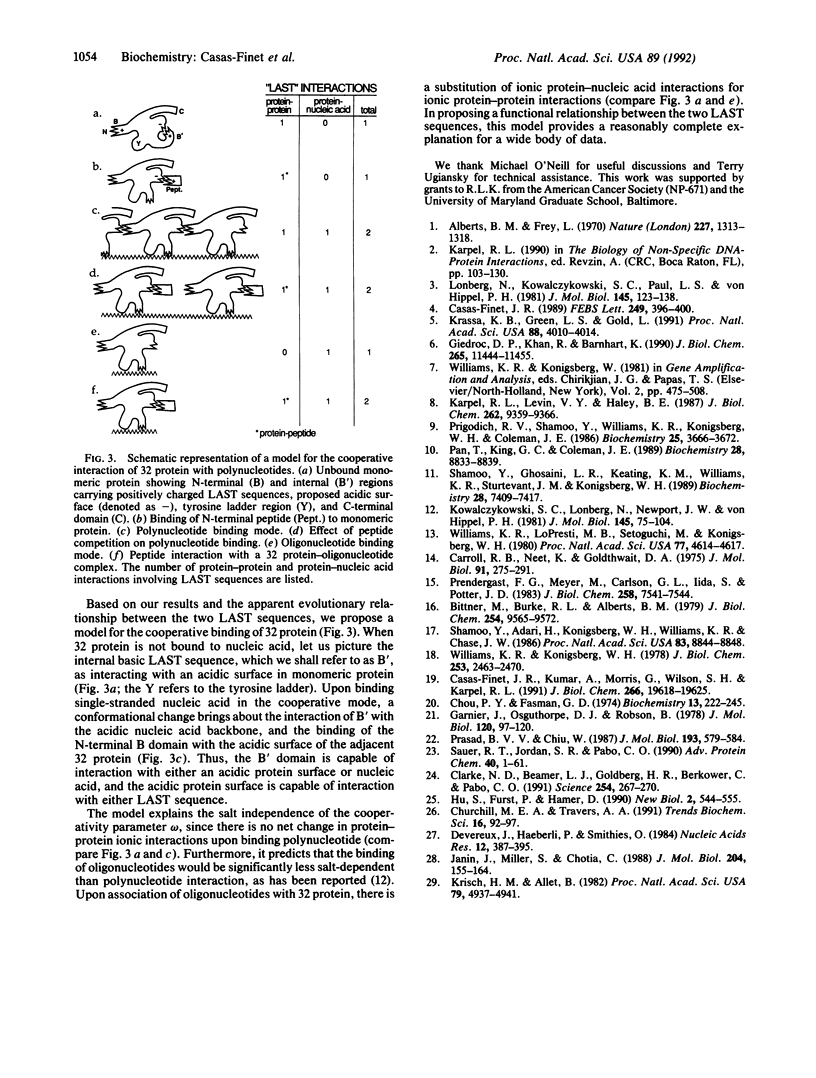
To identify the functional residues of the N-terminal B region of bacteriophage T4 gene 32 protein involved in its cooperative binding to single-stranded nucleic acids, a process dependent on homotypic protein-protein interaction, we have studied the interaction of the protein with synthetic peptides containing different portions of this domain. Gel-permeation chromatography showed that a 6-acryloyl-2-dimethylaminonaphthalene (acrylodan)-labeled fluorescent peptide corresponding to the first 17 residues of gene 32 protein formed a complex with whole protein. The fluorescence was blue-shifted 14 nm upon interaction with intact protein, and somewhat less so (7-11 nm) with cleavage products of the protein lacking B domains. The intrinsic tryptophan fluorescence of whole and truncated protein was quenched by this peptide and by the nonderivatized peptide. The peptide bound tightly to truncated protein at both 0.015 and 0.44 M Na+, with a stoichiometry of 1:1. Similar tryptophan quenching or acrylodan blue shifts were obtained with peptides corresponding to residues 1-9 and 3-8, but not residues 1-4, 5-9, or 5-17, indicating that the essential amino acids are contained within positions 3-8, Lys-Arg-Lys-Ser-Thr-Ala. Several other DNA binding proteins contain a LAST motif with documented involvement of these residues in nucleic acid interaction. The amino acid and coding sequence of residues 110-114, a region proposed to be involved in nucleic acid binding, is virtually identical to that of residues 3-7. Based on these observations, we have formulated a model for the cooperative interactions of gene 32 protein with single-stranded nucleic acids.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Bittner M., Burke R. L., Alberts B. M. Purification of the T4 gene 32 protein free from detectable deoxyribonuclease activities. J Biol Chem. 1979 Oct 10;254(19):9565–9572. [PubMed] [Google Scholar]
- Carroll R. B., Neet K., Goldthwait D. A. Studies of the self-association of bacteriophage T4 gene 32 protein by equilibrium sedimentation. J Mol Biol. 1975 Jan 25;91(3):275–291. doi: 10.1016/0022-2836(75)90380-0. [DOI] [PubMed] [Google Scholar]
- Casas-Finet J. R. Binding properties of T4 gene 32 protein fragments carrying partially cleaved terminal domains. FEBS Lett. 1989 Jun 5;249(2):396–400. doi: 10.1016/0014-5793(89)80666-0. [DOI] [PubMed] [Google Scholar]
- Casas-Finet J. R., Kumar A., Morris G., Wilson S. H., Karpel R. L. Spectroscopic studies of the structural domains of mammalian DNA beta-polymerase. J Biol Chem. 1991 Oct 15;266(29):19618–19625. [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Clarke N. D., Beamer L. J., Goldberg H. R., Berkower C., Pabo C. O. The DNA binding arm of lambda repressor: critical contacts from a flexible region. Science. 1991 Oct 11;254(5029):267–270. doi: 10.1126/science.254.5029.267. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Giedroc D. P., Khan R., Barnhart K. Overexpression, purification, and characterization of recombinant T4 gene 32 protein22-301 (g32P-B). J Biol Chem. 1990 Jul 15;265(20):11444–11455. [PubMed] [Google Scholar]
- Hu S., Fürst P., Hamer D. The DNA and Cu binding functions of ACE1 are interdigitated within a single domain. New Biol. 1990 Jun;2(6):544–555. [PubMed] [Google Scholar]
- Janin J., Miller S., Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988 Nov 5;204(1):155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- Karpel R. L., Levin V. Y., Haley B. E. Photoaffinity labeling of T4 bacteriophage 32 protein. J Biol Chem. 1987 Jul 5;262(19):9359–9366. [PubMed] [Google Scholar]
- Kowalczykowski S. C., Lonberg N., Newport J. W., von Hippel P. H. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J Mol Biol. 1981 Jan 5;145(1):75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- Krassa K. B., Green L. S., Gold L. Protein-protein interactions with the acidic COOH terminus of the single-stranded DNA-binding protein of the bacteriophage T4. Proc Natl Acad Sci U S A. 1991 May 1;88(9):4010–4014. doi: 10.1073/pnas.88.9.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisch H. M., Allet B. Nucleotide sequences involved in bacteriophage T4 gene 32 translational self-regulation. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4937–4941. doi: 10.1073/pnas.79.16.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonberg N., Kowalczykowski S. C., Paul L. S., von Hippel P. H. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. III. Binding properties of two specific proteolytic digestion products of the protein (G32P*I and G32P*III). J Mol Biol. 1981 Jan 5;145(1):123–138. doi: 10.1016/0022-2836(81)90337-5. [DOI] [PubMed] [Google Scholar]
- Pan T., King G. C., Coleman J. E. Comparison of cooperative and isolated site binding of T4 gene 32 protein to ssDNA by 1H NMR. Biochemistry. 1989 Oct 31;28(22):8833–8839. doi: 10.1021/bi00448a023. [DOI] [PubMed] [Google Scholar]
- Prasad B. V., Chiu W. Sequence comparison of single-stranded DNA binding proteins and its structural implications. J Mol Biol. 1987 Feb 5;193(3):579–584. doi: 10.1016/0022-2836(87)90268-3. [DOI] [PubMed] [Google Scholar]
- Prendergast F. G., Meyer M., Carlson G. L., Iida S., Potter J. D. Synthesis, spectral properties, and use of 6-acryloyl-2-dimethylaminonaphthalene (Acrylodan). A thiol-selective, polarity-sensitive fluorescent probe. J Biol Chem. 1983 Jun 25;258(12):7541–7544. [PubMed] [Google Scholar]
- Prigodich R. V., Shamoo Y., Williams K. R., Chase J. W., Konigsberg W. H., Coleman J. E. 1H NMR (500 MHz) identification of aromatic residues of gene 32 protein involved in DNA binding by use of protein containing perdeuterated aromatic residues and by site-directed mutagenesis. Biochemistry. 1986 Jun 17;25(12):3666–3672. doi: 10.1021/bi00360a029. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Jordan S. R., Pabo C. O. Lambda repressor: a model system for understanding protein-DNA interactions and protein stability. Adv Protein Chem. 1990;40:1–61. doi: 10.1016/s0065-3233(08)60286-7. [DOI] [PubMed] [Google Scholar]
- Shamoo Y., Adari H., Konigsberg W. H., Williams K. R., Chase J. W. Cloning of T4 gene 32 and expression of the wild-type protein under lambda promoter PL regulation in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8844–8848. doi: 10.1073/pnas.83.23.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoo Y., Ghosaini L. R., Keating K. M., Williams K. R., Sturtevant J. M., Konigsberg W. H. Site-specific mutagenesis of T4 gene 32: the role of tyrosine residues in protein-nucleic acid interactions. Biochemistry. 1989 Sep 5;28(18):7409–7417. doi: 10.1021/bi00444a039. [DOI] [PubMed] [Google Scholar]
- Williams K. R., Konigsberg W. Structural changes in the T4 gene 32 protein induced by DNA polynucleotides. J Biol Chem. 1978 Apr 10;253(7):2463–2470. [PubMed] [Google Scholar]
- Williams K. R., LoPresti M. B., Setoguchi M., Konigsberg W. H. Amino acid sequence of the T4 DNA helix-destabilizing protein. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4614–4617. doi: 10.1073/pnas.77.8.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]



