Abstract
Soil salinization is a resource and ecological problem in the world. Thellungiella salsuginea is becoming a new model plant because it resembles its relative species, Arabidopsis thaliana, in small genome and short life cycle. It is highly tolerant to salinity and drought stresses. Ascorbate peroxidase (APX) is an enzyme that clears H2O2 in plants. The function and molecular and regulation mechanisms of APX in T. salsuginea have rarely been reported. In this study, an APX gene, TsApx6, was cloned from T. salsuginea and its responses to abiotic stresses in transgenic Arabidopsis were studied. Under high salinity treatment, the expression of TsApx6 was significantly induced. Under drought treatment, overexpression of TsApx6 increased the survival rate and reduced leaf water loss rate in Arabidopsis. Compared to the wild type plants, high salinity treatment reduced the concentrations of MDA, H2O2 and proline but elevated the activities of APX, GPX, CAT and SOD in the TsApx6-overexpressing plants. Meanwhile, germination rate, cotyledon greening, and root length were improved in the transgenic plants compared to the wild type plants under salt and water deficit conditions. Based on these findings, TsApx6 has an important function in the resistance of plants to certain abiotic stresses. The TsApx6 promoter sequence was obtained using Genome Walking technology. Bioinformatics analysis indicated that it contains some cis-acting elements related to stress response. The treatments of salt, dehydration, and ABA induced the expression of Gus gene under the regulation of the TsApx6 promoter. Mutation analysis showed that the MBS motif present in the TsApx6 promoter might be a key negative regulatory element which has an important effect on the growth and developmental process of plants.
Introduction
The life cycle of a plant is influenced by various environmental factors. Drought and salinity are important in reducing plant productivity and limiting plant distribution. Although drought stress is more pervasive and economically damaging than salt stress, many studies on water stress focused on the effects of salt treatment. This lies in the fact that salt stress can be realized easily in a controlled condition and the responses of a plant to drought and salt stresses are associated with each other [1]. When grown under saline and drought conditions, plants produced excess reactive oxygen species (ROS), which include hydrogen peroxide (H2O2), singlet oxygen (1O2), hydroxyl radicals (•OH), and superoxide radicals (•O2-) in cells [2–4]. Reactive oxygen species have a key function to serve as signaling molecules in regulating growth, development, acclimation to abiotic stresses, and initiating pathogen defense pathways. However, over-accumulation of ROS resulted in oxidative damage to different cellular components [3,5–7]. Plants have a large network for ROS production and scavenging in various cellular compartments that are essential for regulating ROS signals in the growth, development, and defense against environmental stresses. In Arabidopsis thaliana L., such a network consists of at least 152 genes encoding ROS producing proteins, including nicotinamide adenine dinucleotide phosphate (NADPH) oxidases and ROS removing enzymes. This class of enzymes includes ascorbate peroxidases (APX, EC 1.11.1.1), glutathione peroxidase (GPX, EC 1.11.1.9), catalase (CAT, EC 1.11.1.6) and superoxide dismutases (SOD, EC 1.15.1.1) [8–11]. Ascorbate peroxidases with the strongest affinity for H2O2 are central components of the H2O2 scavenging networks and very important in the regulation of cellular ROS levels [12].
An Arabidopsis APX family, which consists of eight enzymes, uses ascorbic acid (AsA) as the substrate to reduce H2O2 into water [8,13]. Among the three cytosolic members (APX1, APX2 and APX6) in Arabidopsis, the functions of APX1 and APX2 are well studied [13–16]. As the most abundant APX member, APX1 is involved in many biological processes. This enzyme is expressed in different organs of plants, for example, root, stem, and leaf. The expression of APX1 is significantly promoted by different biotic and abiotic stresses [14,17,18]. The knockout plant that is deficient in APX1 had high accumulation of H2O2 under various growth conditions. Such a plant also had oxidized proteins under light stress, high sensitivity to oxidative stress, and growth suppression [14,17]. Cytosolic APX1 proved to be effective in protecting the chloroplasts during an excessive radiation and thylakoids and stromal/mitochondrial APXs [14]. These findings indicate that the APX1 has a function in preventing cellular components from oxidative damage and regulating cellular and intracellular ROS signals.
Under normal conditions, the expression level of Apx2 is extremely low so that it is undetectable in most plant tissues. However, upregulation of Apx2 can be promoted by wounding, heat, osmotic and oxidative stresses in roots, heat, saline and drought stresses in shoots, and heat treatment in leaves and pollen [18,19]. The expression of Apx2 gene is upregulated as high as approximately 10 folds by the drought stress and the high irradiance, about 20 folds by the treatment of H2O2, and over 20 folds by the heat shock [13,20,21]. Heat stress also significantly upregulates the expression of tomato (Solanum lycopersicum L.) Apx2 gene in pollen [19]. The lack of APX2 ensures the Arabidopsis plants more sensitive to the heat stress at the seedling stage, but makes them more tolerant to the heat stress at the reproductive stage compared to the wild-type (WT) plants [15]. These findings suggest that APX2 might have a function in the regulation of tolerance to heat at different developmental stages in plants.
Arabidopsis thaliana is a model plant in the studies of the molecular basis of tolerance to various stresses [1,22]. Because many knock-out mutants are available and genetic transformation is amenable, it is readily to dissect the functions of different stress associated genes using A. thaliana. However, it is difficult to fully understand the salt-tolerant mechanisms only by studying A. thaliana, a true glycophyte. Thellungiella salsuginea O.E. Schulz is a salt-tolerant plant species that can serve as an informative model to unravel the nature of tolerance against saline stress. As a close relative of A. thaliana, T. salsuginea shares 92% nucleotide similarity at the cDNA level with Arabidopsis. This makes it possible to use the resources of Arabidopsis, such as the information on genes and proteins, in the studies of T. salsuginea. Thellungiella salsuginea plants are extremely tolerant to salt stress, which can survive in seawater and reproduce under the salt stress as high as 500 mM NaCl treatment [23]. Thellungiella salsuginea also tolerates to ozone, drought, heat, freezing, and chilling [24]. In addition to these abiotic stresses, T. salsuginea has numerous desirable traits, for example, small genome size, short plant height and life cycle, self-fertility, and plenty of seeds. All these attributes make it suitable as a promising halophytic model plant in the genetic and genomic studies [23]. The isolation of candidate genes for salt tolerance has provided new information on the mechanisms of salt tolerance in T. salsuginea [24]. Nevertheless, the mechanisms underlying the tolerance to salt and other abiotic stresses are not fully understood.
APX6, the third cAPX of the Arabidopsis, protects mature desiccating and germinating seeds from excessive oxidative stress and maintains seed vigor in front of stress conditions [16]. Nevertheless, the expression and other functions of APX6 in the acclimation of plants to biotic and abiotic stress conditions are not known. In the present study, TsApx6 gene was cloned from T. salsuginea and the expression patterns under a high level of salt treatment was studied. The transgenic A. thaliana lines that constitutively overexpressed TsApx6 were produced. Using these transgenic lines, the concentrations of H2O2, proline and malondialdehyde (MDA), as well as the antioxidant enzyme activities under the high salt stress treatment were determined. The A. thaliana transformed plants carrying TsApx6 in the atapx6 loss-of-function mutant were also studied. In addition, the promoter regions of TsApx6 were cloned and analyzed. In the transgenic Arabidopsis plants, the Gus reporter gene, which was driven by the TsApx6 promoter, was induced by salt stress, in particular in the seedlings and flowers. By analyzing the sequences and MBS (a MYB binding site) element mutants, we identified a cis-acting element from the TsApx6 promoter, which may be a key negative regulatory element for accurately regulating the process of growth and development and responding to the environmental stresses.
Materials and Methods
Plant materials, growth conditions and abiotic stress treatments
The ecotype Shandong of T. salsuginea and the ecotype Columbia (Col-0) of A. thaliana were used throughout the study. A line of atapx6 T-DNA insertion mutant (WiscDsLox321c09) was provided by the Arabidopsis Biological Resource Center (ABRC, http://abrc.osu.edu/). All the transgenic plants were produced in the background of Col-0, except for the atapx6/35S:TsApx6-GFP line in which the TsApx6 (GenBank Accession No. AK353188) was transformed into the atapx6 homozygous mutant. Seeds were sterilized using 0.1% (w/v) HgCl2 solution and sown on the Murashige and Skoog (MS) medium (pH 5.8) supplemented with 3% (w/v) sucrose and solidified with 0.7% (w/v) agar. Germination, cotyledon greening and root growth assay were performed under various concentrations of NaCl (0, 50, 100 and 150 mM) or mannitol (0, 100, 200 and 300 mM). The surface-sterilized seeds were stratified at 4°C in the dark for 3 d prior to transferring to a growth chamber, which was set at 22°C with a 16-h light (110 μmol m-2 s-1) and 8-h dark photoperiod and a relative humidity of 70%. Alternatively, they were directly sown in pots filled with a soil-Vermiculite mixture (3:1) after stratification under the same conditions. The rates of radicle appearance and cotyledon greening were determined at 3 d and 7 d after they were cultured, respectively. To measure root growth, seedlings (3-d-old) grown on MS medium were transferred onto new MS plates supplemented with appropriate concentrations of NaCl or mannitol. After 10 d of growth on new medium, root length of seedlings was measured.
To investigate the expression profiles of TsApx6 and AtApx6 (GenBank Accession No. NM_119384) under short-term of NaCl treatment, wild-type Thellungiella and Arabidopsis seedlings at the 4–6 leaf-stage were transferred to the plastic pots containing a Vermiculite and Perlite mixture (3:1, v/v) and watered with the Hoagland nutrient solution supplemented with 300 mM NaCl for 0, 2, 4, 6, 8, 10, 12, 24, 36 or 48 h [25]. To determine the salt-tolerance of the transgenic genotypes, plants were grown in the soil and Vermiculite mixture (3:1, v/v) for four weeks before they were treated with 300 mM NaCl solution for 3 d, and the leaves were harvested from 5 plants of each line. The concentrations of H2O2, proline and MDA and the activities of APX, GPX, CAT and SOD enzymes in the transgenic and non-transgenic lines were measured using the methods that were described previously [26]. Each experiment was carried out at least in triplicate. To determine the expression of the stress/ABA-responsive genes, the WT and the 35S:TsApx6-GFP transgenic Arabidopsis plants were immersed in 300 mM NaCl for 6 h and 72 h, respectively.
For measurement of drought tolerance, watering was stopped from the 4-week-old plants for 20 days before resuming watering. Photographs were taken and the survival rates of plants were determined 3 d after recovery. Water status for the leaf tissue was determined by the water loss rate, and the detail method was described previously [26].
For GUS activities assay under abiotic stresses, NaCl, mannitol and abscisic acid (ABA) treatments were applied by immersing the seedlings one-week-old, leaves from plants three-week-old and flowers from plants five-week-old in MS nutrition solutions supplemented with 200 mM NaCl, 300 mM mannitol or 0.1 mM ABA for 10 h, respectively.
Gene expression analysis by real-time quantitative PCR
Total RNA was extracted from the plant tissues grown in the control or various stress conditions using an RNeasy kit (Qiagen, Amsterdam, Netherlands). One μg of RNA from each sample was used in the reverse transcription with a cDNA synthesis kit (Code No.: AH311-02, TransGen, Beijing, China) according to the instructions provided by the manufacturer. Real-time quantitative PCR (qPCR) was conducted on an ABI 7500 system (Applied Biosystems, New York, USA) under the default thermal cycling conditions using a TransStartTM Green qPCR SuperMix kit (TransGen) and specific primers for PCR amplification. Relative transcript abundance was determined and normalized with the reference gene Actin2 (GenBank Accession No. U41998) to minimize variation in the levels of the cDNA templates. For expression profile examination of TsApx6, AtApx6 and stress-related genes under salt stress conditions, the control sample acted as the calibrator with a nominal value of 1. When investigated the Gus expression, Actin2 itself was calibrated with a nominal value of 100. The data represent mean values derived from at least three independent repetitive amplifications and each experiment was conducted in at least three biological replicates. All calculations and analyses were performed by the 2-△△Ct method. The primers used in qPCR of TsApx6, AtApx6, Actin2 and the stress/ABA-responsive genes are shown in Table A in S1 Text.
Construction of plasmids and generation of transgenic plants
The TsApx6 and AtApx6 cDNAs were amplified using the primers Apx6 Pst I-LP and Apx6 Avr II-RP (Table A in S1 Text) for their identical primer sequences. The PCR products were digested by Pst I and Avr II and cloned into the modified binary vector pCAMBIA1302 that was digested with the same enzymes to generate 35S:TsApx6/AtApx6-GFP fusion construct. For the TsApx6 and AtApx6 promoter (containing the TsApx6 promoter whose MBS motif was mutated) and Gus fusion construct, the 5' flanking sequences were amplified from the genomic DNA with the appropriate forward and reverse primers that contained Sal I in the 5' end and Spe I in the 3' end, as shown in Table A in S1 Text (Ts6P-LP, Ts6P-RP, At6P-LP and At6P-RP). Then, the fragments generated were cloned into the appropriate sites located before the Gus reporter gene in the binary vector pCAMBIA1305.1. All the plasmids constructed were introduced into Agrobacterium tumefaciens Smith & Townsend strain GV3101 to deliver them into the wild-type Arabidopsis by means of the floral-dip method. Complementary transgenic plants of TsApx6 were produced by introducing a 35S:TsApx6-GFP construct into the atapx6 knockout mutant background. The transgenic lines were selected with 40 μg mL-1 hygromycin and confirmed by PCR amplification using the specific primers listed in Table A in S1 Text. Generation T3 plants carrying a homozygous single insertion were selected for further detailed analysis. For analyzing the phenotypic and physiological properties, 35S:TsApx6-GFP (lines 2 and 4), 35S:AtApx6-GFP (lines 8 and 12), and atapx6/35S:TsApx6-GFP (lines 1 and 6) were applied.
Isolation and bioinformatics analysis of promoter sequences
A 1,654 bp fragment located in the upstream of the ATG start codon of TsApx6 was amplified using the genomic DNA isolated from T. salsuginea by chromosome walking using a Genome Walking kit (Code No.: 6108, TaKaRa, Dalian, China) and gene specific SP primers Ts6P-1SP1, Ts6P-1SP2, Ts6P-1SP3, Ts6P-2SP1, Ts6P-2SP2, and Ts6P-2SP3 (Table A in S1 Text). The orthologous gene of TsApx6 in Arabidopsis is AtApx6 (AT4G32320). The same size upstream flanking region of AtApx6 was amplified with primers At6P-LP and At6P-RP (Table A in S1 Text) using the Arabidopsis genomic DNA as template, whose sequence was provided by The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org). Sequence alignment of the two promoters was performed against the database of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/), and the regulatory elements distributed on them were analyzed using the program PLANTCARE, which is a database of plant cis-acting regulatory elements, enhancers and repressors (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). MBS is a cis-acting regulatory element that was predicted to be a MYB binding site involved in drought inducibility. In order to confirm experimentally the function of MBS, we obtained a new promoter sequence (designated Ts6P-M0) on which the MBS motif was changed. In the new sequence, the thymine (T) of the MBS motif was deleted and one cytosine (C) was replaced by adenine (A). The specific process is shown in S1 Fig.
GUS histochemical staining and fluorometric assay
The activities of GUS in the Arabidopsis transgenic lines were determined using one-week-old seedlings, leaves from three-week-old plants, flowers, stems and siliques from five-week-old plants. Plant tissues were treated in 90% acetone for 30 min, washed with 50 mM sodium phosphate buffer (pH 7.2) for three times, and then incubated at 37°C overnight in a GUS staining solution (50 mM, pH 7.2), which consisted of 0.1% Triton X-100, 1 mM X-Gluc, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide and 10 mM ethylene diamine tetraacetic acid (EDTA, pH 8.0). The tissues stained were immersed in 70% ethanol for several hours to remove the chlorophyll before observing under a stereo fluorescence microscope (Discovery V12, Zeiss, Jena, Germany).
For quantitative fluorometric GUS assay, approximate 0.1 g of plant tissues were chilled in liquid nitrogen and ground in GUS extraction buffer (10 mM EDTA, pH 8.0, 50 mM sodium phosphate buffer, pH 7.0, 0.1% β-mercaptoethanol, 0.1% sodium dodecyl sulfonate (SDS), 0.1% Triton X-100 and 20% methanol). The extracts were centrifuged at 4000 ×g for 10 min to produce the supernatants for GUS assays. The protein concentration was determined as described previously [27]. The activity of GUS was determined using 4-methylumbelliferyl-β-D-glucuronide as the substrate following the method described by Jefferson [28]. The reaction was stopped by adding 200 mM Na2CO3. Fluorescence was quantified using a FluoroMax-4 fluorescence photometer (HORIBA Jobin Yvon, Paris, France) with the excitation and the emission filters at 365 nm and 455 nm, respectively.
Statistical analysis
All above experiments were repeated at least three times, and all the data are presented as means ± standard deviations (SD). One-way analysis of variance (ANOVA) was performed for all the data and the significance of difference in values (P < 0.05 or P < 0.01) was determined by the Fisher’s least significant difference (LSD) test using SPSS 20.0 for Windows. All figures were made with the Sigma Plot 12.5 software.
Results
Isolation and sequence analysis of TsApx6 from T. salsuginea
The TsApx6 cDNA (GenBank Accession No. AK353188) was 1,109 bp in length. It consists of a 987 bp open reading frame (ORF), encodes an APX enzyme of 328 amino acids, and has the predicted molecular mass of 36.4 kDa. Result of protein database search (http://ipsort.hgc.jp/index.html) showed that there was no nuclear localization signal, chloroplast transit, or mitochondrial targeting sequence in the N-terminus of TsAPX6 protein. This indicates that this protein might locate in the cytoplasm. Evolutionary relationship of TsAPX6 from T. salsuginea with other APXs from different plant species was detected by the phylogenetic analysis. Specially, this protein showed the highest similarity with Arabidopsis APXs and exhibited 86% identity with AtAPX6 (S2 Fig).
Expression profile analysis of TsApx6 under NaCl treatment
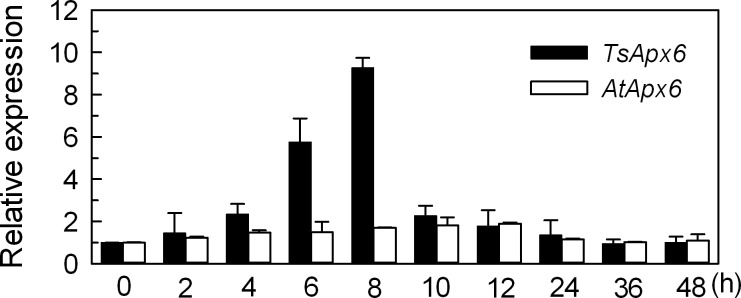
To examine whether TsApx6 expression is stress inducible, qPCR analysis was conducted using total mRNA purified from the leaves of plants (6-week-old) grown under 300 mM NaCl treatment or normal condition. The TsApx6 mRNA levels increased rapidly in the case of 300 mM NaCl treatment by about 10-fold at 8 h (P < 0.01), whereas AtApx6 mRNA showed a 1.9-fold induction at 12 h by the treatment of NaCl (P < 0.05) (Fig 1). This result suggests that TsApx6 has an important function in the response to salt stress in T. salsuginea.
Fig 1. Expression of TsApx6 and AtApx6 under the salt stress conditions as revealed by qPCR analysis.
Thellungiella plants (six-week-old) and Arabidopsis plants (four-week-old) were treated with 300 mM NaCl for 0, 2, 4, 6, 8, 10, 12, 24, 36, or 48 h, respectively. Data are shown as the means ± standard deviation (SD) from three replicates.
Expression of TsApx6 is closely associated with development of plants at the germination and post-germination stages in the presence of NaCl or mannitol
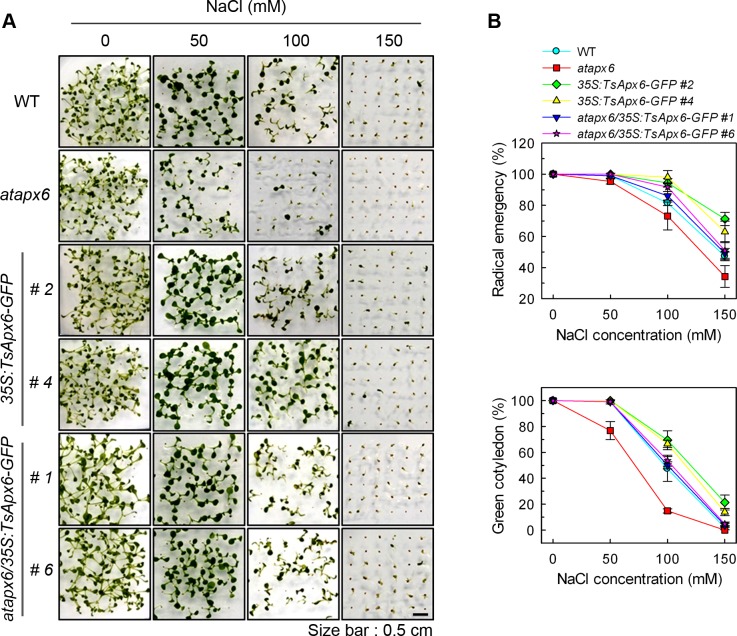
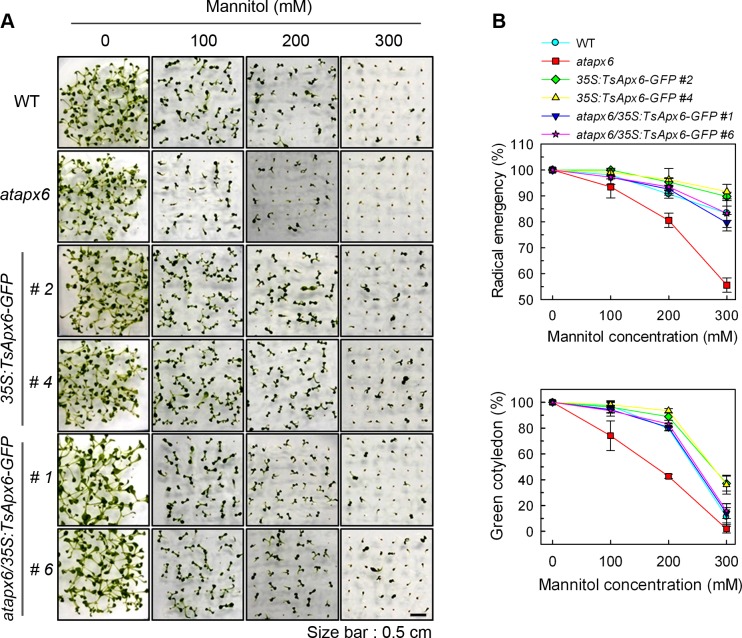
The atapx6 mutant was chosen based on its mannitol and NaCl-sensitive phenotype. Germination rates for seeds of the atapx6, WT, atapx6/35S:TsApx6-GFP, and 35S:TsApx6-GFP were compared under the treatments of 50, 100, and 150 mM of NaCl or 100, 200, and 300 mM of mannitol. On the growth media containing NaCl or mannitol, seed germination rates decreased with the increase of the concentrations of NaCl or mannitol. When treated with 100 mM NaCl, the atapx6 T-DNA insertion mutant seeds displayed a more sensitive phenotype to NaCl compared to the WT seeds (P < 0.01), 19% of the WT seeds did not germinate at 3 d after culture, and greater than 27% of the seeds from the mutant didn’t germinate normally. In contrast, both atapx6/35S:TsApx6-GFP and 35S:TsApx6-GFP transgenic seeds were highly resistant to NaCl. Over 50% of those transgenic seeds germinated under a high NaCl concentration (150 mM) (Fig 2B). Mannitol had different effects on the radicle emergence of seeds on the NaCl-containing medium. The germination rates for the wild-type and the transgenic seeds were over 80% under 300 mM mannitol treatment, with the exception of atapx6 (55%) (P < 0.01) (Fig 3).
Fig 2. Phenotypes of each line in the presence of NaCl treatment during seed germination and early growth.
(A) Sensitivity to NaCl in the WT, atapx6, and TsApx6 transgenic plants during seed germination. Surface-sterilized seeds were sown on MS media contained 0, 50, 100 or 150 mM NaCl, and incubated at 22°C for 7 d under a 16-h light and 8-h dark photoperiod. Size bar = 0.5 cm. (B) Quantification of radicle appearance at 3 d after sowing and cotyledon greening at 7 d after sowing in response to NaCl. Data represent means ± SD from four biological replicates (n = 144).
Fig 3. Phenotypes of each line in the presence of mannitol treatment during seed germination and early growth.
(A) Mannitol sensitivity of all lines at the germination stage. Surface-sterilized seeds were sown on MS medium supplemented with 0, 100, 200, or 300 mM mannitol, and incubated at 22°C for 7 d under a 16-h light and 8-h dark photoperiod. (B) Quantification of radicle emergence at 3 d after sowing and cotyledon greening at 7 d after sowing in response to mannitol. Data represent means ± SD from four biological replicates (n = 144).
Moreover, at 7 d after germination, the growth of 85% of atapx6 seedlings was fully inhibited by 100 mM NaCl treatment, and the green cotyledons failed to develop. In contrast, approximately 52% of atapx6/35S:TsApx6-GFP and 67% of 35S:TsApx6-GFP overexpressing genotypes developed green cotyledons under the same concentration of NaCl (P < 0.01) (Fig 2B). Mannitol had similar effects on NaCl treatment in terms of the cotyledon greening. Growth of almost all the atapx6 seedlings was inhibited by 300 mM mannitol treatment and only 11% of the WT plants developed true green cotyledons (P < 0.01) (Fig 3). However, over 14% of atapx6/35S:TsApx6-GFP and 36% of 35S:TsApx6-GFP overexpressing lines produced green cotyledons under the same concentration of mannitol (P < 0.01). This result indicates that TsApx6-overexpressing lines are more tolerant to salt and osmotic stresses.
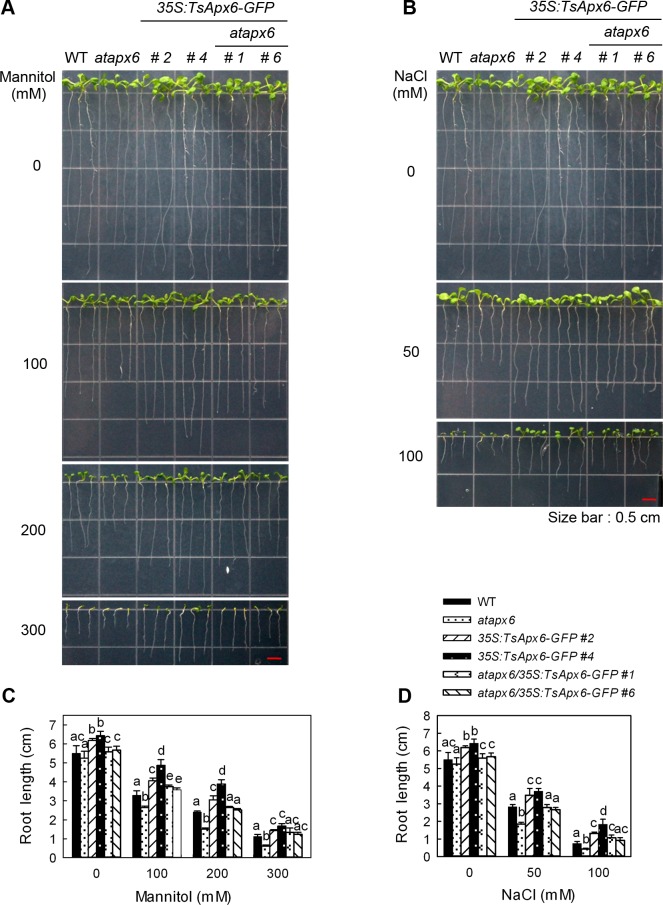
Salt and osmotic stresses produced obvious effects on the root growth on the tenth day after germination in the atapx6, WT, atapx6/35S:TsApx6-GFP, and 35S:TsApx6-GFP plants. The difference in phenotype and growth values was significant among 35S:TsApx6-GFP plants and other lines even though they were normally grown (P < 0.05) (Fig 4). The root elongation of all lines was significantly limited even by a low concentration of NaCl (50 mM) or mannitol (100 mM), indicating an increase in the sensitivity to NaCl and mannitol treatment (Fig 4). With the increase of salt or mannitol concentrations, root elongation decreased in all lines especially in the atapx6 loss-of-function mutants (Fig 4). On the MS medium supplemented with 100 mM NaCl or 300 mM mannitol, the root growth of atapx6 plants was greatly inhibited, which was different from the 35S:TsApx6-GFP roots that elongated under the same conditions (P < 0.01). The root growth of WT and atapx6/35S:TsApx6-GFP lines were in the middle between the lines of atapx6 and 35S:TsApx6-GFP (Fig 4). Thus, the atapx6 mutant was sensitive to salt and osmotic stresses in terms of root growth. In contrast, the roots of 35S:TsApx6-GFP (lines 2 and 4) overexpressing lines displayed lower sensitivity to NaCl and mannitol (P < 0.01) (Fig 4). These results indicate that the expression of TsApx6 was closely associated with the root development during germination (Figs 2 and 3) and post-germination (Fig 4) stages in Arabidopsis.
Fig 4. Root growth of all lines in response to different concentrations of NaCl or mannitol.
Surface-sterilized seeds were germinated on full-strength MS medium for 3 d and the seedlings were then moved to fresh MS medium that contained 0, 50 or 100 mM NaCl or 100, 200 or 300 mM mannitol and grown vertically for 10 d. Bars = 0.5 cm. The data represent the means ± SD (n = 45). Means followed by different letters are significantly different at P < 0.05.
Arabidopsis plants overexpressing TsApx6 are more resistant to high salinity stress than the WT genotypes
To investigate whether constitutive expression of the TsApx6 gene protects Arabidopsis from high salinity stress, the concentrations of H2O2, MDA and proline and the activities of APX, GPX, CAT and SOD in the atapx6, WT, atapx6/35S:TsApx6-GFP and 35S:TsApx6-GFP lines were measured before and after the treatment by 300 mM NaCl for 3 days. Before salt treatment, the concentration of H2O2 did not differ significantly among the WT, atapx6 mutant, and the transgenic lines (Fig 5A). After the NaCl treatment, the H2O2 concentration significantly increased in all lines particularly in the non-transgenic plants (P < 0.05). The amount of H2O2 in the atapx6 mutant was 1.31, 1.99, and 2.72 folds greater than that of the WT, atapx6/35S:TsApx6-GFP, and 35S:TsApx6-GFP genotypes, respectively (Fig 5A). This suggests that TsApx6 is involved in salt stress induced H2O2 elimination in the transgenic Arabidopsis. Malondialdehyde was highly accumulated in all plants after salt treatment for 3 days, indicating the occurrence of the lipid peroxidation caused by salt stress in all lines. However, the MDA concentration was significantly greater in the WT and atapx6 mutant than in the 35S:TsApx6-GFP overexpressing lines (P < 0.01), and the MDA concentration of atapx6/35S:TsApx6-GFP was smaller than that of the WT but greater than that of the 35S:TsApx6-GFP transgenic plants (P < 0.05) (Fig 5B). These results indicated that TsApx6 overexpressing plants had a better protection against oxidative damage and might be salt tolerant. Under the normal growth condition, a significant difference in proline levels was observed between the TsApx6-overexpressing and other plants (P < 0.05) (Fig 5C). Under the high salinity condition, the proline concentration was significantly greater in the atapx6 mutant than in the WT (P < 0.01); however, it was significantly smaller in the atapx6/35S:TsApx6-GFP and the 35S:TsApx6-GFP transgenic genotypes than in the WT (P < 0.01) (Fig 5C). In addition, we found that prior to NaCl treatment, no obvious difference for SOD and CAT activities was observed among all lines (Fig 5D and 5E). The GPX activities were greater in the 35S:TsApx6-GFP transgenic lines than in the WT and the atapx6 loss-of-function mutant (P < 0.05), and the APX activities in the 35S:TsApx6-GFP transgenic lines were significantly greater than that of the other lines (P < 0.05) (Fig 5F and 5G). Salinity stress induced the greatest activities of APX, GPX, CAT and SOD in the leaves of all NaCl treatment groups. The activities of the antioxidant enzymes displayed similar patterns in responses to salt stress (Fig 5D–5G). The activities of all the antioxidant enzymes in transgenic lines, especially 35S:TsApx6-GFP, were prominently greater than those in the atapx6 mutant and the WT (P < 0.01). These findings strongly indicated that the expression of TsApx6 in Arabidopsis resulted in the coordinated increase of antioxidant enzyme activities under the conditions of salt stress, which provided a common protection for plants against oxidative damage.
Fig 5. Effects of high salinity stress on the MDA, H2O2 and proline concentrations and APX, GPX, CAT and SOD activities in plants.
Four-week-old wild-type, atapx6 mutant, atapx6/35S:TsApx6-GFP and 35S:TsApx6-GFP plants were treated with 300 mM NaCl and the leaves were harvested after 3 d of the treatment. A significant difference (P < 0.05) among the genotypes was shown by different letters (a–d). There was no statistic difference among the groups that are not marked with letters. All data were shown as the means ± SD from three biological replicates (n = 30).
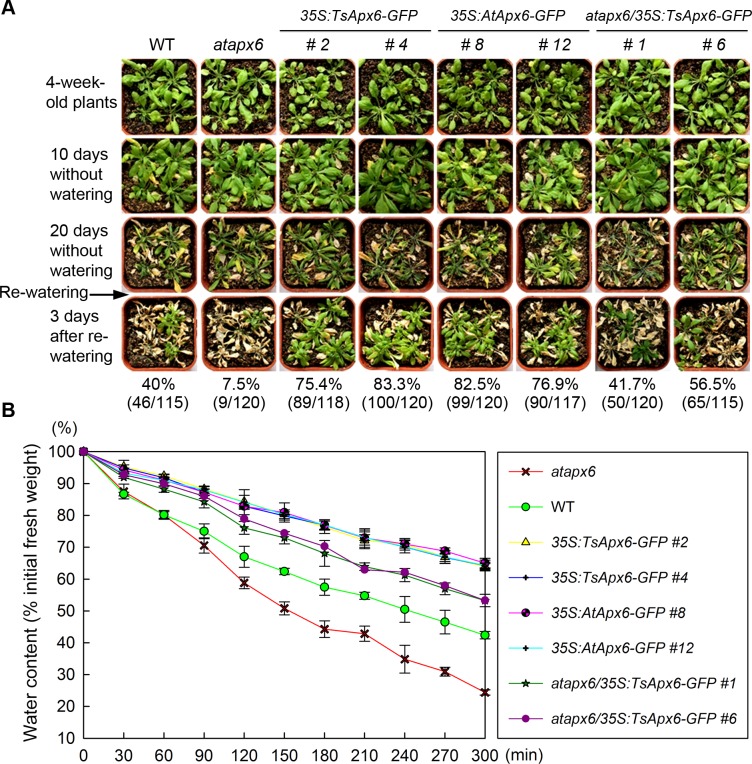
Overexpression of TsApx6 improves tolerance of Arabidopsis to drought stress
Considering the mannitol-related phenotype of the TsApx6-overexpressing plants (Fig 3), it was plausible that TsApx6 was involved in the response to drought stress. Thus, the drought tolerance of the atapx6 mutant, atapx6/35S:TsApx6-GFP, 35S:AtApx6-GFP, and 35S:TsApx6-GFP genotypes was compared with that of the WT by subjecting plants, which were grown for four weeks under well-watered condition, to drought stress by withholding watering for 20 d. During this period, most WT plants and almost all the mutant lines were seriously withered (Fig 6A). The survival rates were determined 3 d after re-watering. Significantly lower survival rate was observed in the atapx6 mutants (7.5%) compared to the WT plants (40%) (P < 0.01) (Fig 6A). However, a majority of the atapx6/35S:TsApx6-GFP, 35S:AtApx6-GFP, and 35S:TsApx6-GFP plants recovered from withering after rehydration (P < 0.05), and the survival rates of atapx6/35S:TsApx6-GFP, 35S:AtApx6-GFP, and 35S:TsApx6-GFP plants ranged from 41.7% (50 of 120 for line 1) to 56.5% (65 of 115 for line 6), 76.9% (90 of 117 for line 12) to 82.5% (99 of 120 for line 8) and 75.4% (89 of 118 for line 2) to 83.3% (100 of 120 for line 4), respectively (Fig 6A). These results suggest that the overexpression of TsApx6 enhances drought tolerance of the plants, while loss-of-function of atapx6 compromised drought tolerance. Thus, TsApx6 promotes drought tolerance of plants.
Fig 6. Estimation of drought stress resistance of the TsApx6-overexpressing plants.
(A) Drought tolerance of the WT, atapx6, atapx6/35S:TsApx6-GFP, 35S:TsApx6-GFP, and 35S:AtApx6-GFP plants. Plants (four-week-old) were subjected to drought stress by withholding water for 20 d, and the survival rates were calculated 3 d after resuming watering. (B) Measurement of leaf water loss rates. Rosette leaves detached from the plants (4-week-old) were weighed at the designated time points after their excision. Water loss was determined as the percentage of the initial fresh weight. The data represent the means ± SD of 180 leaves.
Transpiration water loss rates in the detached rosette leaves collected from the plants 4-week-old were further determined at room temperature with a humidity of 50–60%. The results indicated that the leaves from the atapx6 mutant and the WT plants exhibited a higher rate of water loss than the overexpressing genotypes (P < 0.01). The fresh weights of the atapx6 mutant and the WT leaves after 5-h of incubation were 24% and 42% of their starting values (P < 0.01) (Fig 6B). In contrast, the water contents of the Apx6 overexpressors were approximately 53% (atapx6/35S:TsApx6-GFP) to 64% (35S:TsApx6-GFP and 35S:AtApx6-GFP), and the difference between atapx6/35S:TsApx6-GFP and 35S:TsApx6/AtApx6-GFP lines was significant (P < 0.01) (Fig 6B). These results further indicate that the Apx6 provides the transgenic plants higher level of drought tolerance compared to the wild-type.
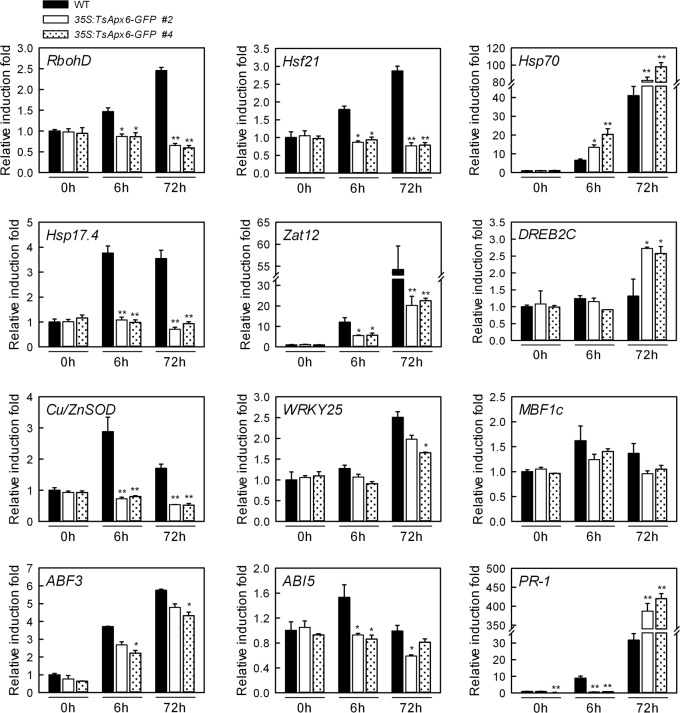
Transcriptional alterations of stress/ABA-responsive genes by TsApx6
To further study the mechanisms of the changed resistance of 35S:TsApx6-GFP overexpressing plants to stress, we assayed the expression patterns of several genes that were involved in the stress- and ABA- responsive processes under normal and salt stress conditions. After pretreatment with 300 mM NaCl for 6 h or 72 h, the expression levels of RbohD (GenBank Accession No. AF055357), Cu/ZnSOD (Accession No. EF408820), Hsp70 (Accession No. NM_112093), Hsp17.4 (Accession No. NM_114492), Hsf21 (Accession No. NM_118004), Zat12 (Accession No. NM_125374), DREB2C (Accession No. NM_129594), WRKY25 (Accession No. AF418309), MBF1c (Accession No. NM_113358), ABF3 (Accession No. AK175851), ABI5 (Accession No. NM_129185) and PR-1 (Accession No. NM_127025) were analyzed by qPCR. The transcript abundance of Hsp70, DREB2C and PR-1 was significantly higher in the TsApx6-overexpressing Arabidopsis plants than the WT under a 72-h salt stress condition (P < 0.05) (Fig 7). However, the fold changes of RbohD, Hsf21, Hsp17.4, Zat12, Cu/ZnSOD, ABF3 and ABI5 were significantly lower in the 35S:TsApx6-GFP lines than those in the WT (P < 0.05). The expression level of WRKY25 in the TsApx6-overexpressing lines was higher than that in the WT under longer duration of salt stress (P < 0.05), whereas no significant change in the MBF1c transcript was observed in both WT and 35S:TsApx6-GFP plants under salt stress conditions (Fig 7).
Fig 7. Expression of stress/ABA-responsive genes in the wild-type and the 35S:TsApx6-GFP plants subjected to salt stress.
Total RNA was purified from plants (4-week-old) treated with 300 mM NaCl for 6 h or 72 h and subjected to qPCR analysis. The expression of a gene in the WT grown under the control conditions was regarded as 1. The transcript abundance was normalized to the expression levels of Actin2 gene. Data represent means ± SD (n = 3) from three technical replicates. Asterisks indicate significant difference from the corresponding WT (*: P < 0.05; **: P < 0.01).
Isolation and bioinformatics analysis of the TsApx6 promoter
The upstream flanking region of TsApx6 1,654 bp in length was isolated from T. salsuginea by the chromosome walking method and specific primers (Table A in S1 Text). Sequence of the fragment shared about 89% similarity to the same size upstream region of AtApx6 gene (S3 Fig). Several putative cis-acting regulatory elements were identified within the amplified fragment using the online software PLANTCARE (Table C in S1 Text). They contained different elements that were associated with hormone and stress-related responses: GARE-motif (gibberellin-responsive), CGTCA-motif (MeJA-responsiveness), LTR (low-temperature), ACE (light responsiveness), and ARE (anaerobic induction) (Table 1). Among them, MBS element was specific to the TsApx6 promoter when compared to the promoter sequence of AtApx6 (S4 Fig).
Table 1. Putative cis-elements present in the promoter sequence of TsApx6.
| Putative cis-element | Motif sequence | Function |
|---|---|---|
| CGTCA-motif | CGTCA | Involved in MeJA-responsiveness |
| GARE-motif | TCTGTTG | Gibberellin-responsive element |
| LTR | CCGAAA | Involved in low-temperature |
| MBS | CGGTCA | MYB binding site involved in drought inducibility |
| TC-rich repeats | ATTTTCTTCA | Involved in defense and stress responsiveness |
| ACE | GCGACGTACC | Involved in light responsiveness |
| TGA-element | AACGAC | Auxin-responsive element |
| ARE | TGGTTT | Essential for the anaerobic induction |
Tissue-specific and stress-responsive expression patterns of TsApx6
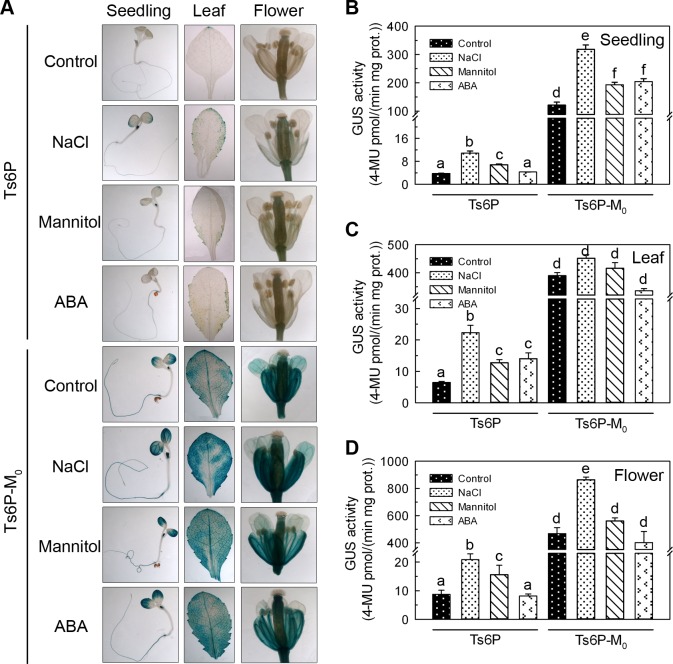
To determine the full-length TsApx6 promoter (designated Ts6P) expression pattern and to identify the effect of MBS cis-element in this promoter, histochemical staining was carried out at different developmental stages of Ts6P and Ts6P-M0 (mutation of the MBS element) plants. The activity of GUS was only weakly observed in the seedlings, leaves, flowers and stems of Ts6P plants. However, when the sequence of MBS element was mutated, the intensity was dramatically increased, and strong GUS signal was observed throughout the entire plant (Fig 8A). Interestingly, GUS activity was greater in the siliques than in the other tissues of the Ts6P plants (P < 0.01). In contrast, no significantly difference of GUS expression was observed in the whole mature Ts6P-M0 plants (Fig 8B).
Fig 8. Histochemical staining and GUS assay of transgenic Arabidopsis lines Ts6P and Ts6P-M0 in different tissues.
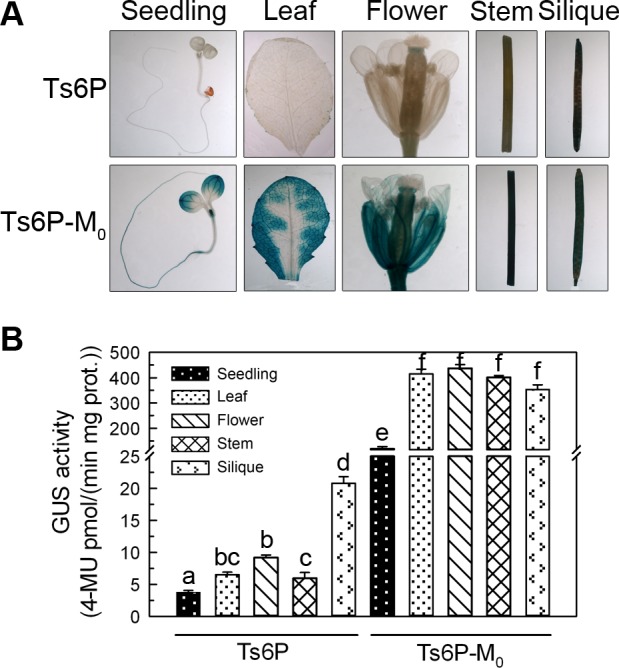
(A) Histochemical staining of seedlings, leaves, flowers, stems and siliques. (B) GUS activities of the tissues from Ts6P and Ts6P-M0 transgenic plants. A significant difference (P < 0.05) among the tissues is shown by different letters (a-f).
After treatment with 200 mM NaCl, 300 mM mannitol or 0.1 mM ABA for 10 hours separately, strong GUS activity was detected in the seedlings, leaves and flowers of the Ts6P-M0 plants, but was weekly detected in the same tissues of the Ts6P plants under the same control and stress conditions (Fig 9A). In the presence of salt stress treatment, the GUS activity in the seedlings and flowers was induced 2.89, and 2.38 folds in the Ts6P plants (P < 0.01), while it was increased about 2.63, and 1.85 folds in the same tissues of the Ts6P-M0 plants (P < 0.05) (Fig 9B and 9D). There was no significant effect of NaCl on the GUS activity in the leaves of Ts6P-M0 plants (Fig 9C). In the presence of mannitol, stronger induction of GUS activity was detected in the Ts6P plants, especially in the leaves and flowers, compared to the Ts6P-M0 plants (P < 0.05). Abscisic acid had no obvious and consistent effect on the GUS signal in the seedlings, leaves and flowers of the two lines (Fig 9B–9D). All of the results indicated that TsApx6 may take part in plant abiotic stress responses, in particularly, salt and dehydration responses.
Fig 9. Histochemical staining and GUS assay of transgenic Arabidopsis lines Ts6P and Ts6P-M0 under different stress conditions.
(A) Histochemical staining of seedlings, leaves and flowers treated with 200 mM NaCl, 300 mM mannitol and 0.1 mM ABA for 10 h, respectively. (B) GUS enzymatic activity quantification of the transgenic tissues treated with different stresses. A significant difference (P < 0.05) among the tissues is shown by different letters (a-f).
Discussion
The present study focused on an ascorbate peroxidase, which is located in cytoplasm and has important function in the physiological and metabolism systems in T. salsuginea. A strong induction of TsApx6 expression by salt stress was observed, which suggests that it might participate in the stresses tolerance. A variety of phenotypes with resistance to stresses were obtained in the TsApx6 transgenic plants. Root length, germination rate, and cotyledon greening of the TsApx6 overexpressing Arabidopsis were remarkably increased compared to the WT on the NaCl or mannitol containing MS medium. When water was withdrawn from plants (four-week-old) for 20 d, the survival rates for the transgenic plants were greater than those of the WT. The water loss of the detached rosette leaves collected from TsApx6-overexpressing lines was slower than that of the WT. Generally, malondialdehyde concentration is used as an indicator for oxidative damage that is induced by salt stress [29]. One of the mechanisms underlying tolerance to salinity is alleviating oxidative damage and mainlining the integrity of the cellular membranes under salt stress [30]. Proline is the most common osmolyte that accumulates in plants in the response to different stresses. Proline performs a wide range of protective functions, for example, osmotic adjustment, cellular structure stabilization and photosynthetic apparatus damage reduction [31]. In this study, the MDA and proline concentrations in the TsApx6-overexpressing lines were significantly lower than those in the WT plants under salt stress treatment. However, excessive accumulation of ROS results in extensive damage to plants [32], and SOD, APX, CAT and GPX are major antioxidant enzymes that are used to scavenge ROS for remedying the harm to plants [26]. The concentration of H2O2 was significantly lower and the APX, GPX, CAT and SOD activities were significantly higher in the 35S:TsApx6-GFP transgenic plants than in the WT plants in the presence of salt stress, indicating that TsApx6 might contribute to the coordinated upregulation of ROS eliminating enzymes and the lower level of H2O2. In addition, the NaCl- and mannitol-sensitive phenotype of atapx6 was sufficiently complemented by the expression of TsApx6. These results strongly suggest that TsAPX6 is a key participator in the tolerance to salt stress and water deficit.
As expected, the expression levels of RbohD, Hsf21, Hsp70, Hsp17.4, Zat12, DREB2C, Cu/ZnSOD, WRKY25, MBF1c, ABF3, ABI5 and PR-1 were up-regulated by salt stress. RbohD encodes a NADPH oxidase, which promotes ROS production especially after stress treatment [33], and Cu/ZnSOD is an important plant ROS scavenger under stress conditions [34]. In this study, the expression of RbohD and Cu/ZnSOD was significantly lower in the TsApx6 transgenic plants than in the WT, which might contribute to the lower H2O2 content and stronger salt resistance of the TsApx6-overexpressing plants. HSF21, ZAT12, DREB2C, WRKY25 and MBF1c are the key transcription factors, and they can promote many stress-responsive genes expression under different stress treatments, such as drought, salt, heat or other conditions [14,35–38]. Only DREB2C showed a significantly higher expression induction in TsApx6-overexpressing plants compared to the WT under salt stress treatment, indicating an indispensable effect on enhanced stress resistance of TsApx6-overexpressing plants. HSPs possessed molecular chaperone activities and were the key factors contributing to plants under both normal and adverse growth conditions [39]. The induction of Hsp70 in the 35S:TsApx6-GFP transgenic plant was significantly higher but the Hsp17.4 was lower than in the WT plants after NaCl treatment, suggesting that Hsp70 in the TsApx6-overexpressing plants was a significant participant in the salt stress condition. Genes ABI5 and ABF3 are vital components in the ABA signaling pathway, but the transcript levels of both genes were lower in the 35S:TsApx6-GFP transgenic lines than in the WT, indicating that TsApx6 modulated salt stress responses independent of the ABA signaling pathway. Surprisingly, the expression of PR-1, which encodes a pathogenesis-related protein and can be strongly induced in response to pathogen infections, was also much higher in TsApx6-overexpressing lines than in the WT. This suggests that TsApx6 is involved in a complicated network of regulatory stress responses [40].
A comparative study of the abiotic response between TsApx6 and AtApx6 was made in this study. The drought tolerance and dehydration rates of the 35S:TsApx6-GFP and 35S:AtApx6-GFP lines were similar. This suggests that the expression patterns of TsAPX6 and AtAPX6 were different under salt stress despite of the same biochemical function. The mRNA levels of TsApx6 significantly increased in response to the salt stress, whereas the AtApx6 transcripts showed no noticeable change. Moreover, analysis by semi-quantitative reverse transcription PCR (RT-PCR) indicated that the TsApx6 expression was higher than the AtApx6, which was almost undetectable under normal growth conditions (S5 Fig). Based on these findings, TsApx6 may play an important role in both the response to salt stress and the development of T. salsuginea, and the regulation of the APX gene expression in T. salsuginea is different from that in Arabidopsis under salt stress.
To understand the reason for the different expression patterns of TsApx6 and AtApx6 in plant development and abiotic stress response, we analyzed the promoter sequences of these genes. As expected, the two promoter sequences shared 89% sequence similarity (S3 Fig). Bioinformatics analysis showed a difference of the cis-acting elements in the two promoters (S4 Fig). The expression levels of Gus gene driven by both the TsApx6 and the AtApx6 promoters at all developmental stages were very low, but the former was slightly higher than the latter (S6 Fig), which was consistent with our previous work (S5 Fig). In addition, Gus expression in Ts6P lines showed greater inducibility than did in At6P lines under abiotic stress conditions (S7 Fig). All these demonstrated that the difference in expression patterns of the TsApx6 and AtApx6 might be attributed to the different types and distribution of the cis-acting elements in the two promoters.
MBS motif was specific to the TsApx6 promoter when compared to the promoter sequence of AtApx6 and predicted to be a MYB binding site that was involved in drought inducibility. To study the function of the MBS in detail, we constructed the Ts6P-M0 promoter-reporter vector in which the MBS motif was mutated. To our surprise, the GUS activities in the Ts6P-M0 plants were significantly higher than that of the Ts6P at different development stages and under the stress conditions. This result indicated that the MBS motif might be a negative regulatory element and indispensable to the expression regulation of TsApx6 in development and stress response. Further study on this region to identify the protein that interacts with the MBS motif will be useful to reveal the regulation mechanism of TsApx6.
In short, this work provides important understanding of the function of TsApx6 and regulatory properties of the TsApx6 promoter. TsAPX6 was an effective H2O2-scavenging enzyme and the overexpression of TsApx6 in Arabidopsis protected plants from high salinity and water deficit stresses. The expression of TsApx6 was significantly induced by the abiotic stresses, such as salt, dehydration, and ABA treatments. Moreover, the MBS motif was confirmed as a transcription silencer and it might be important in the regulation of TsApx6 expression in development and stress response in plants.
Supporting Information
(A) Electrophoresis of the mutated fragment. (B) Mutated site of the MBS motif.
(TIF)
(A) DNA electrophoresis of TsApx6. (B) Phylogenetic analysis of the DNA sequences of TsApx6 and Arabidopsis Apx family members. (C) Phylogenetic analysis of the amino acid sequences of TsApx6 and the members in the Arabidopsis Apx family.
(TIF)
(TIF)
(A) cis-acting elements predicted in AtApx6 promoter. (B) cis-acting elements predicted in TsApx6 promoter.
(TIF)
Six-week-old Thellungiella and four-week-old Arabidopsis plants were treated with 300 mM NaCl for 0, 2, 4, 6, 8, 10, 12, 24, 36, or 48h, respectively. Concentrations of RNA from different samples were accurately quantified prior to synthesis of cDNA.
(TIF)
(A) Histochemical staining of At6P seedlings, leaves, flowers, stems and siliques. (B) Histochemical staining of Ts6P. (C) Expression of Gus from At6P and Ts6P tissues. Relative expression levels were determined with respect to the expression of Actin2, whose expression level was defined as 100 relative expression units (REU). (D) GUS activities of the tissues from At6P and Ts6P transgenic plants.
(TIF)
(A) Histochemical staining of seedlings, leaves and flowers treated with 200 mM NaCl, 300 mM mannitol, and 0.1 mM ABA for 10 h, respectively. (B, D and F) Expression of Gus from At6P and Ts6P tissues treated with different stresses. Relative expression levels were determined with respect to the expression of Actin2 (= 100 REU). (C, E and G) Quantification of GUS enzymatic activity in the transgenic tissues treated with different stresses.
(TIF)
Relative expression levels were determined with respect to the expression of Actin2 (= 100 REU).
(TIF)
(Table A) Primers used for gene expression assays. (Table B) Sequence of TsApx6 promoter. (Table C) Predicted cis-acting elements of the TsApx6 promoter.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the National Natural Science Foundation of China (http://www.nsfc.gov.cn/) Grant No. 31070289 and 31270365. GZ received the funding. The funders had been used in study design, data collection and analysis, decision to publish, and preparation of the manuscript.
References
- 1.Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–73. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laloi C, Apel K, Danon A (2004) Reactive oxygen signalling: the latest news. Curr Opin Plant Biol 7: 323–8. 10.1016/j.pbi.2004.03.005 [DOI] [PubMed] [Google Scholar]
- 3.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, et al. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–9. 10.1016/j.tplants.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 4.Xu LH, Wang WY, Guo JJ, Qin J, Shi DQ, Li YL, et al. (2014) Zinc improves salt tolerance by increasing reactive oxygen species scavenging and reducing Na+ accumulation in wheat seedlings. Biol Plantarum 58: 751–7. 10.1007/s10535-014-0442-5 [DOI] [Google Scholar]
- 5.Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403. 10.1016/j.pbi.2005.05.014 [DOI] [PubMed] [Google Scholar]
- 6.Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–6. 10.1104/pp.106.082040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R (2011) Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol 14: 691–9. 10.1016/j.pbi.2011.07.014 [DOI] [PubMed] [Google Scholar]
- 8.Mittler R, Vanderauwera S, Gollery M, van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–8. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 9.Bailey-Serres J, Mittler R (2006) The roles of reactive oxygen species in plant cells. Plant Physiol 141: 311 10.1104/pp.104.900191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray PD, Huang BW, Tsuji Y (2012) Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 24: 981–90. 10.1016/j.cellsig.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naliwajski MR, Skłodowska M (2014) The oxidative stress and antioxidant systems in cucumber cells during acclimation to salinity. Biol Plantarum 58: 47–54. 10.1007/s10535-013-0378-1 [DOI] [Google Scholar]
- 12.Miao Y, Bai L, Miao C, Chen J, Song C (2005) Progress in plant glutathione peroxidase. Chin Bull Bot 22: 350–6. [Google Scholar]
- 13.Panchuk II, Volkov RA, Schoffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–53. 10.1104/pp.001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, et al. (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–81. 10.1105/tpc.104.026971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki N, Miller G, Sejima H, Harper J, Mittler R (2013) Enhanced seed production under prolonged heat stress conditions in Arabidopsis thaliana plants deficient in cytosolic ascorbate peroxidase 2. J Exp Bot 64: 253–63. 10.1093/jxb/ers335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Letnik I, Hacham Y, Dobrev P, Ben-Daniel BH, Vankova R, et al. (2014) ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol 166: 370–83. 10.1104/pp.114.245324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203. 10.1046/j.1365-313X.2003.01715.x [DOI] [PubMed] [Google Scholar]
- 18.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol 136: 2621–32. 10.1104/pp.104.046367 PMC523327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, et al. (2009) Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot 60: 3891–908. 10.1093/jxb/erp234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, et al. (2006) A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ 29: 269–81. 10.1111/j.1365-3040.2005.01419.x [DOI] [PubMed] [Google Scholar]
- 21.Volkov RA, Panchuk II, Mullineaux PM, Schoffl F (2006) Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol Biol 61: 733–46. 10.1007/s11103-006-0045-4 [DOI] [PubMed] [Google Scholar]
- 22.Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19: 371–9. 10.1016/j.tplants.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Lai J, Sun S, Li Y, Liu Y, Liang L, et al. (2008) Comparison analysis of transcripts from the halophyte Thellungiella halophila. J Integr Plant Biol 50: 1327–35. 10.1111/j.1744-7909.2008.00740.x [DOI] [PubMed] [Google Scholar]
- 24.Higashi Y, Ohama N, Ishikawa T, Katori T, Shimura A, Kusakabe K, et al. (2013) HsfA1d, a protein identified via FOX hunting using Thellungiella salsuginea cDNAs improves heat tolerance by regulating heat-stress-responsive gene expression. Mol Plant 6: 411–22. 10.1093/mp/sst024 [DOI] [PubMed] [Google Scholar]
- 25.Cho SK, Ryu MY, Song C, Kwak JM, Kim WT (2008) Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–914. 10.1105/tpc.108.060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li ZQ, Li JX, Li HJ, Shi ZH, Zhang GF (2015) Overexpression of TsApx1 from Thellungiella salsuginea improves abiotic stress tolerance in transgenic Arabidopsis thaliana. Biol Plantarum 59: 497–506. 10.1007/s10535-015-0533-y [DOI] [Google Scholar]
- 27.Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–54. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 28.Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez JA, Almansa MS (2002) Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol Plant 115: 251–7. 10.1034/j.1399-3054.2002.1150211.x [DOI] [PubMed] [Google Scholar]
- 30.Stevens J, Senaratna T, Sivasithamparam K (2006) Salicylic acid induces salinity tolerance in tomato (Lycopersicon esculentum cv. Roma): Associated changes in gas exchange, water relations and membrane stabilisation. Plant Growth Regul 49: 77–83. 10.1007/s10725-006-0019-1 [DOI] [Google Scholar]
- 31.Nounjan N, Nghia PT, Theerakulpisut P (2012) Exogenous proline and trehalose promote recovery of rice seedlings from salt-stress and differentially modulate antioxidant enzymes and expression of related genes. J Plant Physiol 169: 596–604. 10.1016/j.jplph.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 32.Li XY, Liu X, Yao Y, Li YH, Liu S, He CY, et al. (2013) Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int J Mol Sci 14: 12827–42. 10.3390/ijms140612827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–67. 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- 34.Xu X, Zhou Y, Wei S, Ren D, Yang M, Bu H, et al. (2009) Molecular cloning and expression of a Cu/Zn-Containing superoxide dismutase from Thellungiella halophila. Mol Cells 27: 423–8. 10.1007/s10059-009-0060-9 [DOI] [PubMed] [Google Scholar]
- 35.Shim D, Hwang JU, Lee J, Lee S, Choi Y, An G, et al. (2009) Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21: 4031–43. 10.1105/tpc.109.066902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62: 4731–48. 10.1093/jxb/err210 [DOI] [PubMed] [Google Scholar]
- 37.Li S, Fu Q, Huang W, Yu D (2009) Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep 28: 683–93. 10.1007/s00299-008-0666-y [DOI] [PubMed] [Google Scholar]
- 38.Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139: 1313–22. 10.1104/pp.105.070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Lin S, Liu Q, Huang J, Zhang W, Lin J, et al. (2014) Expression and interaction of small heat shock proteins (sHsps) in rice in response to heat stress. Biochim Biophys Acta 1844: 818–28. 10.1016/j.bbapap.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 40.Jiang L, Wu J, Fan S, Li W, Dong L, Cheng Q, et al. (2015) Isolation and characterization of a novel pathogenesis-related protein gene (GmPRP) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. PLoS One 10: e0129932 10.1371/journal.pone.0129932 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Electrophoresis of the mutated fragment. (B) Mutated site of the MBS motif.
(TIF)
(A) DNA electrophoresis of TsApx6. (B) Phylogenetic analysis of the DNA sequences of TsApx6 and Arabidopsis Apx family members. (C) Phylogenetic analysis of the amino acid sequences of TsApx6 and the members in the Arabidopsis Apx family.
(TIF)
(TIF)
(A) cis-acting elements predicted in AtApx6 promoter. (B) cis-acting elements predicted in TsApx6 promoter.
(TIF)
Six-week-old Thellungiella and four-week-old Arabidopsis plants were treated with 300 mM NaCl for 0, 2, 4, 6, 8, 10, 12, 24, 36, or 48h, respectively. Concentrations of RNA from different samples were accurately quantified prior to synthesis of cDNA.
(TIF)
(A) Histochemical staining of At6P seedlings, leaves, flowers, stems and siliques. (B) Histochemical staining of Ts6P. (C) Expression of Gus from At6P and Ts6P tissues. Relative expression levels were determined with respect to the expression of Actin2, whose expression level was defined as 100 relative expression units (REU). (D) GUS activities of the tissues from At6P and Ts6P transgenic plants.
(TIF)
(A) Histochemical staining of seedlings, leaves and flowers treated with 200 mM NaCl, 300 mM mannitol, and 0.1 mM ABA for 10 h, respectively. (B, D and F) Expression of Gus from At6P and Ts6P tissues treated with different stresses. Relative expression levels were determined with respect to the expression of Actin2 (= 100 REU). (C, E and G) Quantification of GUS enzymatic activity in the transgenic tissues treated with different stresses.
(TIF)
Relative expression levels were determined with respect to the expression of Actin2 (= 100 REU).
(TIF)
(Table A) Primers used for gene expression assays. (Table B) Sequence of TsApx6 promoter. (Table C) Predicted cis-acting elements of the TsApx6 promoter.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.