Abstract
Linc-RoR was originally identified to be a regulator for induced pluripotent stem cells in humans and it has also been implicated in tumorigenesis. However, the underlying mechanism of Linc-RoR-mediated gene expression in cancer is poorly understood. The present study demonstrates that Linc-RoR plays an oncogenic role in part through regulation of c-Myc expression. Linc-RoR knockout (KO) suppresses cell proliferation and tumor growth. In particular, Linc-RoR KO causes a significant decrease in c-Myc whereas re-expression of Linc-RoR in the KO cells restores the level of c-Myc. Mechanistically, Linc-RoR interacts with heterogeneous nuclear ribonucleoprotein (hnRNP) I and AU-rich element RNA-binding protein 1 (AUF1), respectively, with an opposite consequence to their interaction with c-Myc mRNA. While Linc-RoR is required for hnRNP I to bind to c-Myc mRNA, interaction of Linc-RoR with AUF1 inhibits AUF1 to bind to c-Myc mRNA. As a result, Linc-RoR may contribute to the increased stability of c-Myc mRNA. Although hnRNP I and AUF1 can interact with many RNA species and regulate their functions, with involvement of Linc-RoR they would be able to selectively regulate mRNA stability of specific genes such as c-Myc. Together, these results support a role for Linc-RoR in c-Myc expression in part by specifically enhancing its mRNA stability, leading to cell proliferation and tumorigenesis.
INTRODUCTION
Long non-coding RNAs (lncRNAs) are a group of recently identified RNA molecules with a molecular weight of over 200 nucleotides in length, without coding capacity. Despite the non-coding nature of lncRNAs, evidence indicates that lncRNAs can play an important role in regulation of cellular pathways and disease processes. LncRNAs may function as master gene regulators through various mechanisms, and thus, the dysregulation of lncRNA expression is often associated with a variety of human diseases including cancer. For instance, a number of lncRNAs have been shown to play a role in cancer initiation, progression and metastasis as well as stem cell maintenance (1–6). This may have to do with their ability to interact with DNA, RNA or proteins such that they may serve as transcription activators; transcription repressors; guides for chromatin-modifying enzymes to be recruited to target genes; and scaffolds to bring together multiple proteins to form functional ribonucleoprotein complexes (7–10).
LincRNA regulator of reprogramming (Linc-RoR) was first identified as a regulator for reprogramming of differentiated cells to induced pluripotent stem cells (iPSCs) in humans and knockdown of Linc-RoR leads to a modest increase in apoptosis and activation of p53 pathways (11). Subsequent studies indicate that Linc-RoR may function as a key competitive endogenous RNA to link the network of microRNAs and core transcription factors, such as Oct4, Sox2, and Nanog (12). Our group demonstrates that Linc-RoR inhibits p53 translation by interacting with heterogeneous nuclear ribonucleoprotein I (hnRNP I) in response to DNA damage (13). A recent report showed that Linc-RoR functions as an oncogene in triple negative breast cancer (TNBC), promoting invasion through miR-145 and ARF6 pathway (14).
Similar to Linc-RoR, c-Myc also plays an oncogenic role. It was first identified in Burkitt's lymphoma and its activation resulted from a chromosomal translocation (15). Increased expression of c-Myc in cancer frequently correlates with poor patient survival (16). Various mechanism have been implicated in upregulation of c-Myc, including amplification, activation of transcription and posttranscriptional regulation. With regard to posttranscriptional regulation, miR-145 can suppress c-Myc by targeting its 3′-UTR (17). On the other hand, the mRNA decay factors such as AU-rich element RNA-binding protein 1 (AUF1) have been shown to induce c-Myc mRNA degradation (18). However, little is known whether Linc-RoR plays a role in regulation of c-Myc expression.
The present study demonstrates that Linc-RoR is capable of upregulating c-Myc expression, leading to tumorigenesis. In particular, we show that Linc-RoR induces c-Myc mRNA stability by facilitating the interaction of hnRNP I with c-Myc mRNA and at the same time, inhibiting the binding of AUF1 to c-Myc mRNA for degradation.
MATERIALS AND METHODS
Reagents
Sources of primary antibodies: c-Myc from Abcam (Cambridge, MA, USA); GAPDH from Protein Tech (Chicago, IL, USA); hnRNP I and AUF1 from Santa Cruz Biotechnology (Dallas, TX, USA). Secondary antibodies conjugated with IRDye 800CW or IRDye 680 were purchased from LI-COR Biosciences (Lincoln, NE, USA).
PCR primers were purchased from IDT (Coralville, IA, USA). Linc-RoR and control siRNAs were purchased from ThermoFisher Scientific (Waltham, MA, USA). hnRNP I or AUF1 siRNA mixture was purchased from Santa Cruz Biotechnology. Colon cancer tissue cDNA arrays were purchased from OriGene (Rockville, MD, USA). Breast cancer tissue microarrays (TMAs) were purchased from US Biomax (Rockville, MD, USA).
Cell culture
Colon cancer HCT-116 p53 wt and p53 null cells, breast cancer MCF-7 cell, as described previously (13,17), were grown in RPMI 1640 from Lonza (Walkersville, MD, USA), supplemented with 10% FBS from Sigma-Aldrich (St. Louis, MO, USA), 2 mM glutamine, 100 unites of penicillin/ml and 100 μg of streptomycin/ml (Lonza). Cells were incubated at 37°C and supplemented with 5% CO2 in the humidified chamber.
Transfection
Cells were transfected with siRNAs using RNAfectin reagent from Applied Biological Materials (Richmond, BC, Canada) or plasmid DNA using DNAfectin (Applied Biological Materials) following the manufacturer's protocol.
Plasmid construction
PCR reactions for cloning purpose used Phusion enzyme from ThermoFisher Scientific (Pittsburgh, PA, USA). Different fragments of Linc-RoR were cloned into pCDH-CMV-MSC-EF1-copGFP (System Biosciences) with a strategy described previously (13). For example, to clone Linc-RoR E4, we first amplified the entire exon 4 by PCR using primers RoR-E4-R1–5.1 and RoR-E4-Not1-3.2 (Supplementary Table S1) and then cloned into the designated vector at EcoR I and Not I sites using Cold Fusion kit (System Biosciences). To make miR-145 binding site mutant clones, we carried out a two-step amplification procedure as described previously (19), using primers RoR-miR145-BS1-m-5.1 and RoR-miR145-BS1-m-3.1; RoR-miR145-BS2-m-5.1 and RoR-miR145-BS2-m-3.1 (Supplementary Table S1). All PCR products were verified by DNA sequencing. RoR-E4 clone mutated at two putative binding sites was made through the gBlock method from IDT.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using Direct-zol™ RNA MiniPrep (Zymo Research) and cDNA synthesis was carried out using RevertAid™ Reverse Transcriptase (ThermoFisher) with random primers. PCR was performed using a standard SYBR Green method. We used primers RoR-E4-RT-5.1A and RoR-E4-RT-3.1A (Supplementary Table S1) to detect RoR levels, and c-Myc-RT-5.1 and c-Myc-RT-3.1 (Supplementary Table S1) to detect c-Myc levels. GAPDH or β-Actin was used as an internal control. Delta-delta Ct values were used to determine their relative expression as fold changes, as previously described (20).
In situ hybridization
In situ hybridization used to detect Linc-RoR in breast cancer tissue microarrays (TMAs) was performed according to a previously described method (21). A biotin-labeled antisense LNA probe was derived from linc-RoR exon 4 was previously described (13). The relative signal was assessed based on the intensity as - (negative), same as a negative control; + (weak positive) and ++ (strong positive).
Knockout of Linc-RoR by CRISPR/Cas9
We used a dual gRNA approach (22) to knock out exon 4 of Linc-RoR (Linc-RoR E4) by CRISPR/Cas9 system (23). To facilitate the selection of positive clones, we also generated a donor vector in such a way that targeting sequence is replaced by marker genes (GFP and PU, the puromycin resistance gene) once it is integrated into the genomic DNA by homologous recombination. Donor vector carried EF1-GFP-T2A-PU flanked by LoxP and a ∼800 bp targeting sequence derived from outside the targeting sites of Linc-RoR E4. The dual gRNA construct carrying Cas9 and donor vector were introduced into HCT-116 cells by transient transfection. The empty dual gRNA vector served as a control. One week later, the transfected cells were subject to puromycin selection; and surviving cells were sorted by FACS based on GFP signal into 96-well plates and then expanded. Initial identification of knockout clones was carried out by genomic PCR, followed by qRT-PCR, as described previously (22).
Western blot
Cells were harvested, and proteins were extracted from transfected cells and quantified as previously described (17). Samples were separated in a polyacrylamide SDS gel before transferring to PVDF membrane. Signals were detected using Odyssey systems (LI-COR).
RNA precipitation
To identify Linc-RoR binding partners, we performed RNA precipitation assay using biotin-labeled full length Linc-RoR RNA probe, followed by mass spectrometry provided by Taplin Mass Spectrometry Facility at Harvard Medical School. We also used Linc-RoR E4 RNA probe to determine its interaction with AUF1 or hnRNP I. In this case, a DNA fragment covering the entire exon 4 of Linc-RoR was amplified by PCR using a T7 promoter-containing forward primer T7-RoR-E4-5.1 and T7-RoR-E4-Not1-3.1 (see Supplementary Table S1) and then cloned into pCR8 (Invitrogen, Grand Island, NY, USA). The resultant plasmid DNA was linearized with restriction enzyme Not I which was introduced from the reverse PCR primer, and then used to synthesize RNA in vitro by T7 polymerase in the presence of biotin-labeled UTP. The precipitates were subjected to western blot using AUF1 or hnRNP I antibody. The same procedure was used for c-Myc RNA precipitation.
RNA immunoprecipitation (RIP)
To determine interaction of AUF1 or hnRNP I with Linc-RoR or c-Myc mRNA, we used AUF1 or hnRNP I antibody to pull down AUF1 or hnRNP I. Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) was used for RIP procedures according to the manufacturer's protocol. After the antibody was recovered by protein A/G beads, standard qRT-PCR was performed to detect Linc-RoR or c-Myc mRNA in the precipitates.
MTT assay
MTT assay was performed to determine the effect of Linc-RoR on cell growth as described previously (24).
Animal work
Nude (nu/nu) mice (4–5 weeks old) were purchased from Harlan Laboratories (Indianapolis, IN, USA). All animal studies were conducted in accordance with NIH animal use guidelines and a protocol approved by the UMMC Animal Care Committee. HCT-116 cells at the exponential stage were harvested and were then mixed with 50% matrigel (BD Biosciences, San Jose, CA, USA). Vector control or Linc-RoR expression vector cells (1.5 million cells/spot), and donor vector control or Linc-RoR KO cells (1.5 million cells/spot) were injected into mice s.c.. as described previously (20). Tumor growth was measured every other day 7 days after injection.
Gene network analysis
To determine whether Linc-RoR also impact other pathways, we performed network analysis of the transcriptional changes in ESCs caused by Linc-RoR knockdown (access number is GSE24182) (11) using ANAT program (25).
Statistical analysis
The continuous variables are summarized as mean and standard error of mean (S.E.M.) unless stated. The two-sample t test was used to compare the mean of a continuous variable between two samples. The satterthwaite t test was used for mean comparison when the variances in two samples were unequal. Association between two categorical variables was evaluated by using the Fisher's exact test. All P values were two-sided and P values <0.05 were considered as significant.
RESULTS
Linc-RoR is upregulated in cancer specimens and its ectopic expression promotes cell proliferation and tumor growth
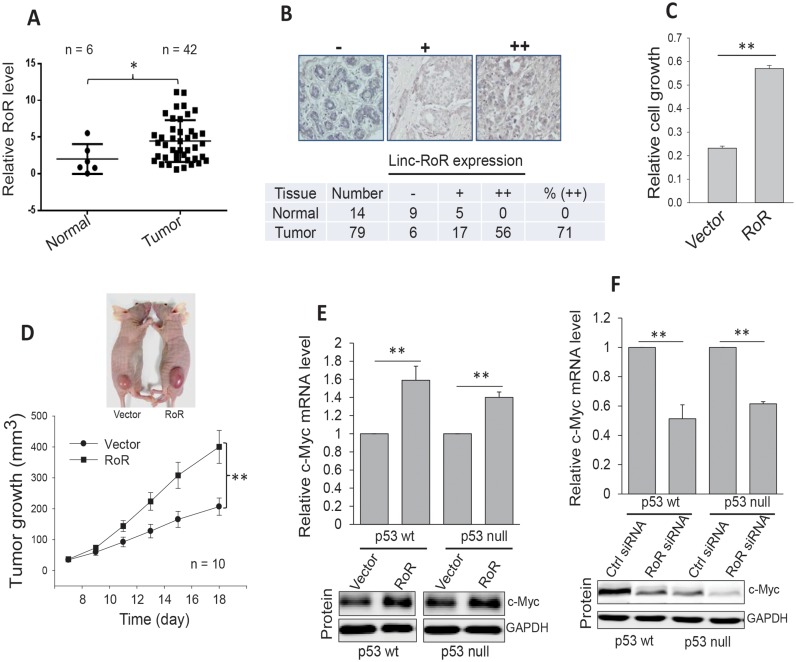
To determine the role of Linc-RoR in cancer, we first examined Linc-RoR expression using the colon cancer tissue cDNA arrays from OriGene by qPCR. We found that Linc-RoR was highly expressed in tumor specimens compared to normal tissue (Figure 1A). Furthermore in situ hybridization of breast cancer tissue microarrays (TMAs) indicated that Linc-RoR was also upregulated in tumors compared to normal tissue (Figure 1B), supporting the oncogenic role of Linc-RoR. Next, we determined the effect of Linc-RoR on cell proliferation in colon cancer HCT-116 cells carrying Linc-RoR expression vector and found that overexpression of Linc-RoR induced cell proliferation, as determined by MTT assay (Figure 1C). Furthermore, experiments with xenograft mouse model revealed that Linc-RoR also significantly induced tumor growth (Figure 1D).
Figure 1.
Linc-RoR is upregulated in cancer specimens and it promotes cell proliferation and tumor growth. (A) Linc-RoR expression in colon tissues (cDNA array from OriGene), as detected by qPCR. (B) Detection of Linc-RoR in breast cancer tissue microarrays by in situ hybridization. Top, representative images for ‘-’, ‘+’ and ‘++’. Bottom, analysis of total of 14 normal and 79 tumor samples with P < 0.01, as determined by the satterthwaite t test. (C) Ectopic expression of Linc-RoR promotes cell proliferation, as detected by MTT assay. (D) Linc-RoR promotes tumor growth in xenograft mouse model. HCT-116 cells were transfected with control or Linc-RoR and then were injected into nude mice s.c. (E) Ectopic expression of Linc-RoR induces c-Myc at mRNA and protein level both in HCT-116 p53 wt and p53 null cells. (F) Knockdown of Linc-RoR by RNAi decreases c-Myc at mRNA and protein level both in HCT-116 p53 wt and p53 null cells. Error bars represent S.E.M., n = 3 except for those as indicated. *P <0.05; **P < 0.01.
Linc-RoR upregulates c-Myc
To identify how Linc-RoR impacts cell proliferation and tumor growth, we performed network analysis for the profiling data by Loewer et al. (11) using ANAT program (25). This analysis indicated that Linc-RoR may regulate several pathways, in particular c-Myc pathway (Supplementary Figure S1). Thus, we determined the effect of Linc-RoR on c-Myc expression. Since p53 can negatively regulate c-Myc (17), to determine the possibility of p53 involvement, we carried out these experiments in both p53 wt and p53 null cells (Supplementary Figure S2A). As shown in Figure 1E, overexpression of Linc-RoR induced c-Myc in both cell lines, as detected by qRT-PCR. At the protein level, we also found a significant induction of c-Myc by Linc-RoR (Figure 1E, bottom; Supplementary Figure S2B) in both cell lines. In consistent with these results, knockdown of Linc-RoR by siRNA decreased c-Myc mRNA and protein levels (Figure 1F). These results suggest that Linc-RoR regulates c-Myc independent of p53. In addition, similar to results in HCT-116 cells, while ectopic expression of Linc-RoR increased, Linc-RoR siRNA suppressed c-Myc in MCF-7 cells (Supplementary Figure S2C). We further confirmed that p53 is a negative regulator for c-Myc (Supplementary Figure S2D).
Knockout of Linc-RoR decreases c-Myc mRNA and protein levels
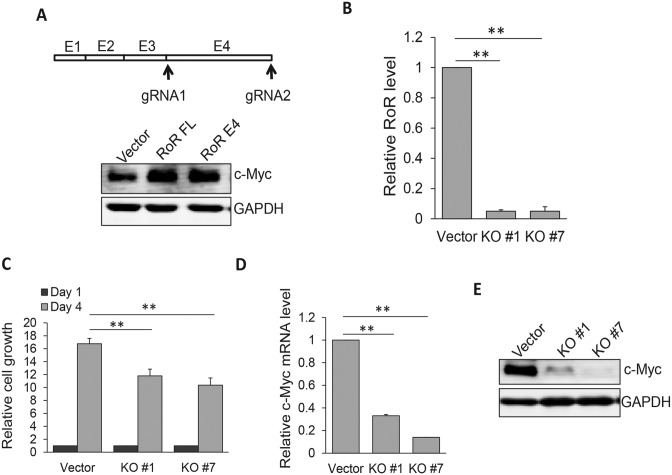
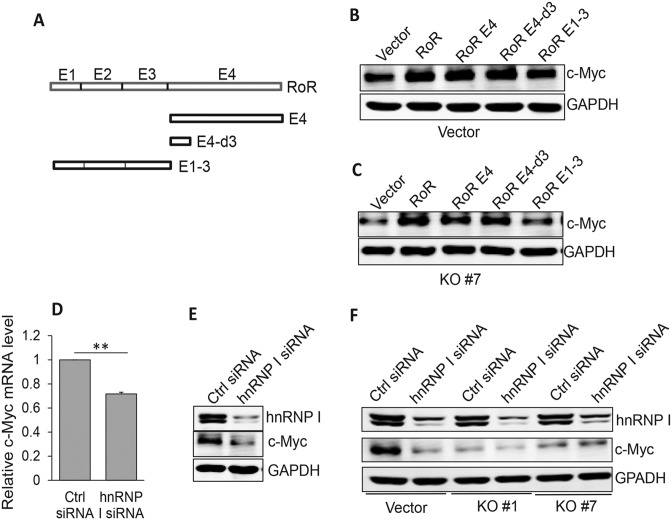
To better characterize Linc-RoR-mediated c-Myc expression, we took advantage of CRISPR/Cas9 system (26,27) to knockout Linc-RoR since RNAi is not always effective. Deletion analysis defined the active region of Linc-RoR within exon 4 (E4) because just like the full length Linc-RoR, Linc-RoR E4 was also able to induce c-Myc with a similar level (Figure 2A). Since Linc-RoR lacks an open reading frame, a small deletion or insertion after DNA breakage induced by CRISPR/Cas9 may not always cause loss of its function. Thus, we adopted a dual gRNA approach we developed recently (22). These two gRNAs were located just outside of E4 (Figure 2A; Supplementary Figure S3A) with a purpose to delete the entire exon. Both Cas9 and dual gRNAs were carried on the same vector. Screening of individual clones identified several complete knockout (KO) clones by genomic PCR and we selected two of them (Supplementary Figure S3B) for further characterization. Both clones (KO #1 and KO #7) expressed little Linc-RoR, as detected by qRT-PCR (Figure 2B). MTT assay revealed that cell proliferation rate was significantly decreased in KO cells as compared to vector control (Figure 2C). Furthermore, c-Myc mRNA level was reduced by ∼70% in KO#1 and > 80% in KO#7 clone, respectively (Figure 2D). Similarly, c-Myc protein level was also significantly decreased in these KO cells as compared to vector control (Figure 2E).
Figure 2.
Knockout of Linc-RoR E4 by CRISPR/Cas9 system decreases c-Myc mRNA and protein levels. (A) Linc-RoR E4 is sufficient to regulate c-Myc. Two gRNAs were designed to delete the entire exon 4. (B) Detection of Linc-RoR expression in KO #1 and #7 cells by qRT-PCR, as compared to vector control. (C) Suppression of cell proliferation in Linc-ROR KO cells, as determined by MTT assays. Cell proliferation was measured at day 4 after seeding as compared to day 1 as 1 (black bar). (D and E) Linc-RoR KO causes a significant reduction of c-Myc mRNA and protein level. Error bars represent S.E.M., n = 3. **P < 0.01.
Linc-RoR KO suppresses tumor growth
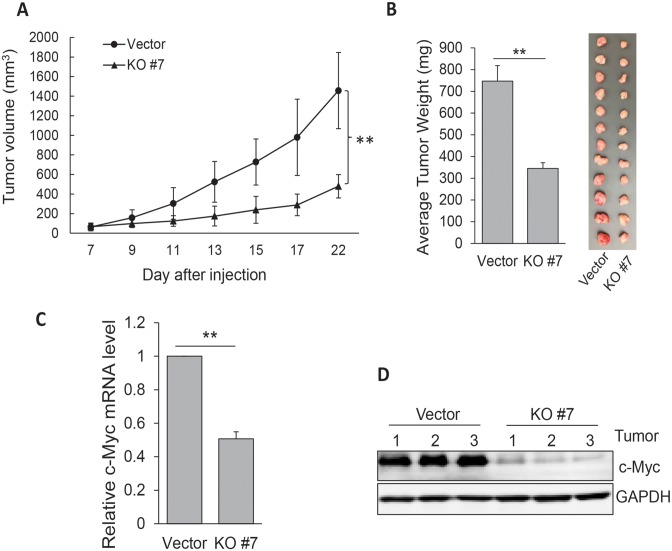
To further determine the role of Linc-RoR in tumor growth, we injected HCT-116 cells (vector control or KO#7) into nude mice. It is evident that Linc-RoR KO significantly suppressed tumor growth rate (Figure 3A) and tumor weight (Figure 3B). Importantly, tumors derived from Linc-RoR KO expressed lower levels of c-Myc mRNA (Figure 3C) and c-Myc protein (Figure 3D) than those derived from vector control, further suggesting that Linc-RoR is critical to tumor growth by regulation of c-Myc.
Figure 3.
Linc-RoR KO suppresses tumor growth and c-Myc expression. (A) Linc-RoR KO suppresses tumor growth rate. KO #7 or control cells were injected into nude mice as detailed in Materials and Methods. Error bars represent S.D.M. (B) Weight of tumors harvested at the end of experiment. (C and D) Linc-RoR KO suppresses c-Myc expression at mRNA and protein levels. Three tumors were randomly selected for RNA or protein extraction. Error bars represent S.E.M., n = 3 for (C). **P < 0.01.
Regulation of c-Myc mRNA stability by Linc-RoR through interaction with AUF1
To dissect the underlying mechanism of regulation of c-Myc by Linc-RoR, we first determined whether miR-145 plays a role in Linc-RoR-mediated c-Myc expression because there is a reciprocal repression between Linc-RoR and miR-145 (12) and c-Myc serves as a direct target for miR-145 (17). Thus, we generated a Linc-RoR E4 expression vector carrying two mutant miR-145 binding sites (Supplementary Figure S4A). In consistent with the previous report, ectopic expression of Linc-RoR E4 reduced miR-145, and mutation of two potential miR-145 binding sites in Linc-RoR E4 (Linc-RoR E4 mt) had no effect on miR-145 (Supplementary Figure S4B). Importantly, ectopic expression of both Linc-RoR E4 and Linc-RoR E4 mt induced c-Myc (Supplementary Figure S4C), suggesting that Linc-RoR can regulate c-Myc independent of miR-145.
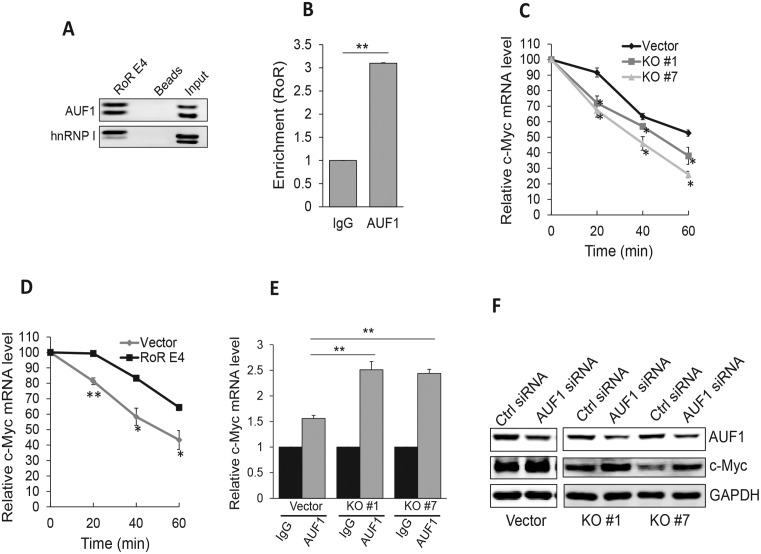
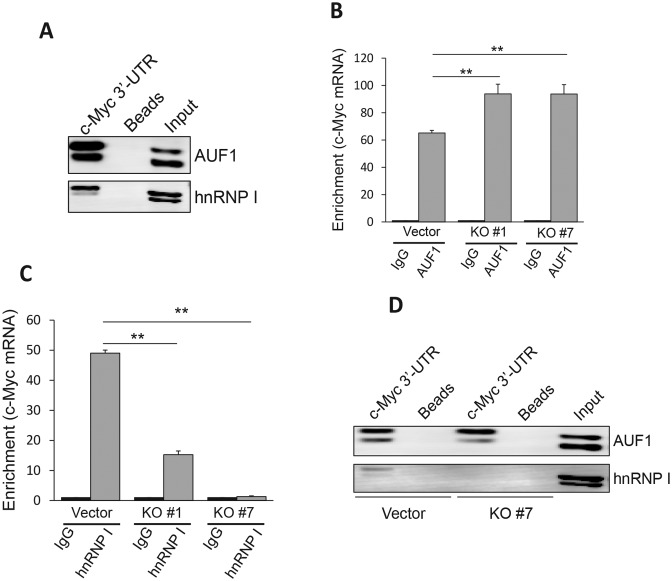
Therefore, we performed RNA precipitation using biotin-labeled Linc-RoR as a probe, and mass spectrometry analysis (Supplementary Table S2) identified a number of potential Linc-RoR binding partners. Among them was AUF1 (hnRNP D), a known factor involved in regulation of mRNA stability (28). Western blot analysis of the precipitates confirmed the interaction of AUF1 and Linc-RoR (Figure 4A). In addition, we also detected a previously identified Linc-RoR binding partner (13), hnRNP I (Figure 4A). RNA immunoprecipitation (RIP) assays with AUF1 antibody detected about a 3-fold enrichment of Linc-RoR by AUF1 antibody over IgG control (Figure 4B), providing further evidence that AUF1 is a Linc-RoR binding partner.
Figure 4.
Linc-RoR interacts with AUF1 to regulate c-Myc mRNA stability. (A) Interaction of Linc-RoR with AUF1 and hnRNP I, as detected by RNA precipitation using Linc-RoR E4 as an RNA probe, followed by western blot. (B), RIP assay using AUF1 antibody confirms that Linc-RoR interacts with AUF1. (C) Reduction of c-Myc mRNA stability in Linc-RoR KO cells as compared to control cells. Cells were treated with 2 μg/ml actinomycin D and RNA was isolated at 0, 20, 40 and 60 min, respectively. (D) Re-expression of Linc-RoR E4 restores its ability to increase c-Myc mRNA stability. KO #7 cells were firstly transfected with control or Linc-RoR E4 vector and the transfected cells were then treated with actinomycin D as in C. (E) Suppression of AUF1 by RNAi increases c-Myc mRNA. Note that this effect was more significant in KO cells than in vector control. (F) AUF1 siRNAs increase c-Myc protein, which is also more obvious in Linc-RoR KO cells than in vector control. Error bars represent S.E.M., n = 3. *P < 0.05; **P < 0.01.
Since AUF1 is known to function as a destabilizer for c-Myc mRNA (29), we determined whether Linc-RoR impacts c-Myc mRNA stability through interaction with AUF1. Thus, we treated cells with the RNA synthesis inhibitor actinomycin D (30) and then isolated total RNA at 20, 40 and 60 min, respectively. As shown in Figure 4C, the c-Myc mRNA level was significantly reduced in two KO clones as compared to control cells. Half-life for vector is more than 1 h; half-life for KO #1 and KO #7 is 50 and 40 min, respectively, suggesting that Linc-RoR KO reduces c-Myc mRNA stability. To further confirm that Linc-RoR contributes to c-Myc mRNA stability, we performed a reconstitution experiment. We introduced Linc-RoR E4 expression vector into KO #7 cells and then treated the cells with actinomycin D. While half-life for vector is ∼50 min, half-life for RoR E4 re-expression is >1 h (Figure 4D), suggesting that Linc-RoR E4 is capable of enhancing c-Myc mRNA stability.
To determine the role of Linc-RoR in the AUF1-mediated c-Myc expression, we transfected vector control or KO cells with AUF1 siRNAs. Consistent with the previous report (29), AUF1 siRNA knockdown led to ∼1.5-fold increase in c-Myc mRNA expression in control cells (Figure 4E). Importantly, Linc-RoR KO further increased the c-Myc mRNA level (about a 2.5-fold over the control siRNA) in KO cells (Figure 4E). Similarly, AUF1 siRNAs also caused a significant increase in c-Myc protein, particularly in Linc-RoR KO cells (Figure 4F), suggesting that Linc-RoR plays a role in AUF1-mediated c-Myc mRNA stability.
Linc-RoR enhances c-Myc mRNA stability by interacting with hnRNP I
Having demonstrated that Linc-RoR impacts the AUF1-mediated c-Myc mRNA stability, we did deletion analysis of Linc-RoR E4 to further define the active region responsible for regulation of c-Myc expression (Figure 5A). We found that the smallest fragment consisting of 165 bp (E4-d3) was still capable of increasing c-Myc protein level (Figure 5B). We then did reconstitution experiments by re-expression of Linc-RoR and various deletion constructs in KO cells. Similar to the results shown in Figure 5B, all constructs except for RoR E1∼3 were able to restore the KO cells to increase the c-Myc level (Figure 5C). Of interest, the Linc-RoR E4-d3 fragment contains a conserved hnRNP I binding motif (13), implying that hnRNP I might also be involved in regulation of c-Myc mRNA by interacting with Linc-RoR. RIP assay showed that Linc-RoR E4 specifically interacted with hnRNP I (Supplementary Figure S5A) To test this hypothesis, we chose HCT-116 p53 null cells because hnRNP I is also involved in regulation of p53 (13). Thus, we transfected HCT-116 p53 null cells with hnRNP I siRNAs. Both c-Myc mRNA and protein levels were reduced by hnRNP I siRNAs (Figure 5D and E), suggesting that hnRNP I can also regulate c-Myc and this regulation is independent of p53. Furthermore, we detected a significant reduction of c-Myc protein (Figure 5F) as well as mRNA (Supplementary Figure S5B) by hnRNP I siRNAs in vector control cells, but not in KO cells, suggesting the effect of hnRNP I siRNAs on c-Myc is greatly attenuated in KO cells.
Figure 5.
Linc-RoR increases c-Myc by interaction with hnRNP I. (A) Identification of active region of Linc-RoR responsible for regulation of c-Myc by deletion analysis. (B) The active region is within a 165 bp (E4-d3) fragment, as determined by ectopic expression in parental HCT-116 cells. (C) Re-expression also confirms that E4-d3 is capable of restoring its ability to enhance c-Myc expression in Linc-RoR KO cells. (D and E) Suppression of c-Myc mRNA and protein levels by hnRNP I siRNAs in HCT-116 p53 null cells. (F) Although hnRNP I siRNAs can significantly suppress c-Myc in vector control cells, no such effect is detected in Linc-RoR KO cells. Error bars represent S.E.M., n = 3. **P < 0.01.
Linc-RoR regulates the competition of AUF1 and hnRNP I for c-Myc mRNA
At this point, we identified three players, i.e. Linc-RoR, AUF1 and hnRNP I, in regulation of c-Myc. To further determine how Linc-RoR regulates c-Myc mRNA stability by interacting with AUF1 and hnRNP I, we performed RIP experiments with AUF1 or hnRNP I antibody. As expected, the enrichment of Linc-RoR was detected in control cells, but it was substantially reduced in KO cells, by AUF1 or hnRNP I antibody (Supplementary Figure S6A and S6B). We also showed that both AUF1 and hnRNP I can interact with c-Myc mRNA by RNA precipitation using c-Myc 3′-UTR as a probe (Figure 6A). Of great interest, Linc-RoR impacted the interaction of AUF1 or hnRNP I with c-Myc mRNA in an opposite way. For instance, the enrichment of c-Myc mRNA by AUF1 antibody was significantly higher in KO cells than in control cells (Figure 6B). In contrast, enrichment of c-Myc mRNA by hnRNP I antibody was decreased significantly in KO cells as compared to control cells (Figure 6C), suggesting that Linc-RoR could promote the interaction of hnRNP I with c-Myc mRNA. This was not due to alterations of AUF1 or hnRNP I levels because Linc-RoR KO had no effect on their expression (Supplementary Figure S7A) and vice versa (Supplementary Figure S7B and C). Moreover, RNA precipitation using c-Myc 3′-UTR probe also supported this notion. For example, we detected less amount of hnRNP I in KO #7 cells as compared to control cells (Figure 6D). In contrast, we detected a slight more amount of AUF1 in KO #7 cells as compared to control cells (Figure 6D), further suggesting that Linc-RoR is required for hnRNP I to interact with c-Myc mRNA, but interaction of Linc-RoR with hnRNP I inhibits the binding of AUF1 to c-Myc mRNA.
Figure 6.
Linc-RoR specifically regulates c-Myc by facilitating interaction of hnRNP I with c-Myc mRNA while inhibiting interaction of AUF1 with c-Myc mRNA. (A) Both AUF1 and hnRNP I interacts with c-Myc mRNA, as detected by RNA precipitation using c-Myc 3′-UTR as a probe, followed by western blot. (B) Linc-RoR KO enhances interaction of c-Myc mRNA with AUF1, as detected by RIP assay using AUF1 antibody. (C) Linc-RoR is required for interaction of hnRNP I with c-Myc mRNA, as detected by RIP assay using hnRNP antibody. (D) Linc-RoR KO enhances interaction of c-Myc mRNA with AUF1, but suppresses interaction of c-Myc mRNA with hnRNP I, as detected by RNA precipitation using c-Myc 3′-UTR probe. Error bars represent S.E.M., n = 3. **P < 0.01.
As RNA binding proteins, both hnRNP I and AUF1 can interact with a variety of RNA species and impact these genes in various ways (31,32). Hence, we asked whether Linc-RoR is specific to c-Myc. To test this possibility, we determined the effect of AUF1 and Linc-RoR on p21, a known target for AUF1 (33). As expected, AUF1 siRNAs were able to increase the level of p21 mRNA (Supplementary Figure S8A); however, the same level of p21 mRNA also seen in Linc-RoR KO cells (Supplementary Figure S8A), suggesting that the AUF1 siRNA-mediated increased level of p21 is not affected by Linc-RoR KO. Indeed, Linc-RoR KO alone had no effect on p21 expression (Supplementary Figure S8B). Finally, we also found that tumors derived from Linc-RoR KO cells expressed about the same level of p21 mRNA as control (Supplementary Figure S8C). These results suggest that Linc-RoR plays a specific regulatory role toward c-Myc.
DISCUSSION
c-Myc is one of the most important oncogenes and its activation can cause unregulated expression of many target genes, leading to the formation of cancer (34). Overwhelming evidence indicates that various mechanisms may contribute to the c-Myc upregulation. For example, amplification of the c-Myc gene has been often seen in cancer; similarly, transcriptional activation or posttranscriptional regulation can also lead to the increased level of c-Myc. Regulation of mRNA turnover is an important mechanism of posttranscriptional regulation of c-Myc. The present study demonstrates that Linc-RoR specifically upregulates c-Myc by in part enhancing its mRNA stability through interaction with hnRNP I and AUF1.
Linc-RoR was originally identified to be able to regulate reprogramming of iPSCs (11). Our previous study suggests the potential oncogenic role of Linc-RoR in cancer progression (13). The present study provides further evidence that Linc-RoR functions as an oncogene. For example, Linc-RoR is upregulated in colon and breast cancer specimens. Its overexpression promotes tumor cell growth both in vitro and in vivo. Furthermore, Linc-RoR KO leads to a significant reduction of tumor growth rate and overall tumor weight. Given the role of Linc-RoR in c-Myc expression, as demonstrated in this study, we suggest that upregulation of c-Myc by Linc-RoR contributes to the Linc-RoR-mediated tumorigenesis.
Our in silico analysis suggests a link between Linc-RoR and c-Myc. As a well-known oncogene, c-Myc is involved in regulation of multiple cellular processes, such as cell proliferation, apoptosis, differentiation, and stem cell self-renewal (35). It is conceivable that cells develop a complex regulatory network for precisely controlling c-Myc expression, which may in part explain why the c-Myc mRNA half-life is relatively short as compared to that of most genes. Thus, regulation of c-Myc mRNA stability is critical to c-Myc-mediated tumorigenesis. In support of this notion, the half-life of c-Myc mRNA in cancer cells is significantly longer than in normal cells (36). However, underlying mechanism is not fully understood. Although previous studies have identified AUF1 as an important destabilizer for c-Myc mRNA, little is known whether other player(s) is involved in this regulatory network.
Many labile mRNAs can be regulated in part by an AU rich element (ARE) in the 3′-untranslated region (3′-UTR). An early report suggests that ARE binding affinities of AUF1 correlate with the potency of an ARE to direct degradation of a heterologous mRNA (37), supporting a role for AUF1 in ARE-directed mRNA decay. For example, the 3′-UTR of c-Myc has been shown to be required for a high rate of mRNA turnover; in particular, a region of 140 bases of the 3′-UTR is primarily responsible for the short c-Myc RNA half-life (38). This region carries several AU-rich elements (AREs) which can be targeted by ARE-specific binding proteins (AUBP). Based on ARE site (http://rna.tbi.univie.ac.at/cgi-bin/AREsite.cgi), there are four ATTTA elements in c-Myc (Supplementary Figure S9). Our study suggests that as a member of AUBPs, AUF1 is involved in regulation of c-Myc mRNA stability.
Moreover, our study suggests that AUF1 is not the only factor involved in regulation of c-Myc mRNA stability. Like AUF1, hnRNP I can also interact with Linc-RoR. However, in contrast to AUF1, hnRNP I can enhance c-Myc mRNA stability, which is likely through its competition with AUF1 for c-Myc mRNA. Our RIP assays indicate that while more c-Myc mRNA is precipitated by AUF1 antibody in Linc-RoR KO cells than in control cells, very little c-Myc mRNA is precipitated by hnRNP I antibody in KO cells, suggesting that Linc-RoR has an opposite effect on their interactions with c-Myc mRNA. For example, hnRNP I siRNAs suppress c-Myc mRNA and protein levels. Furthermore, there is a significant reduction of c-Myc mRNA and protein levels by hnRNP I siRNAs in control cells but not in Linc-RoR KO cells. Finally, RIP assays with hnRNP I antibody indicate that interaction of c-Myc mRNA and hnRNP I is severely attenuated or lost in KO cells, suggesting that Linc-RoR is required for hnRNP I to interact with c-Myc mRNA. Therefore, these findings support a novel function of hnRNP I in regulating c-Myc mRNA stability through interaction with Linc-RoR.
Both AUF1 and hnRNP I belong to a family of RNA binding proteins that play multiple roles in the cell, including RNA processing, pre-mRNA splicing, mRNA export, localization, translation and stability (39). As an important factor for mRNA stability, AUF1 can bind and regulate many genes such as p21 in addition to c-Myc. However, Linc-RoR can impact only AUF1-mediated expression of c-Myc, but not p21. This may be due to several possibilities. Initially, we thought that Linc-RoR may directly interact with c-Myc mRNA through two 11 bp sequences complementary to Linc-RoR E4 (Supplementary Figure S9). However, mutagenesis of these two sites had no effect on binding between Linc-RoR and c-Myc mRNA (Supplementary Figure S10), suggesting that they are not crucial. Another possibility is that interaction of hnRNP I with Linc-RoR may change its conformation such that it facilitates the subsequent binding to c-Myc mRNA. However, such conformation changes may not favor its binding to p21 mRNA. Therefore, by manipulating the level or activity of hnRNP I or AUF1, we would expect that all targeted genes will be impacted. In contrast, an alteration of the Linc-RoR level would be able to specifically regulate c-Myc. This mechanism may provide the cell with more flexibility to adapt to various environmental conditions.
In summary, although several mechanisms may be involved in Linc-RoR-mediated c-Myc expression, this study focuses on the regulation of c-Myc mRNA stability by at least three players. While Linc-RoR plays a regulator role, hnRNP I and AUF1 serves as basic machinery. In other words, Linc-RoR, but not hnRNP I or AUF1, decides the specificity for the gene to be regulated. Since RNA binding proteins (e.g. hnRNP I and AUF1) usually have a broad range of substrates or targets, alterations in their levels or activity would impact a large number of genes. On the other hand, by changing levels of Linc-RoR, we would expect to see a specific set of genes or even a single gene to be regulated. For example, the level of Linc-RoR is low in normal cells. Thus, AUF1 is able to interact with ARE of c-Myc mRNA, leading to a high turnover of c-Myc mRNA. In tumor cells, Linc-RoR is upregulated such that Linc-RoR interacts with hnRNP I and facilitates the interaction of hnRNP I with c-Myc mRNA. At the same time, Linc-RoR also interacts with AUF1, which prevents its interaction with c-Myc mRNA. A consequence of both actions is an increased half-life of c-Myc such that these tumor cells become more proliferative and more aggressive.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01 CA154989 to Y.M.]; Natural Science Foundation of Zhejiang Province (LY15C050002 to X.D.) and the Key Science and Technology Project of Zhejiang Province (2014C03004 to X.D.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L., et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D., Horlings H.M., Shah N., Umbricht C., Wang P., et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D., et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W., Alvarez-Dominguez J.R., Lodish H.F. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S., et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., Wu M., Xiong J., Guo X., Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhang A., Zhou N., Huang J., Liu Q., Fukuda K., Ma D., Lu Z., Bai C., Watabe K., Mo Y.Y. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eades G., Wolfson B., Zhang Y., Li Q., Yao Y., Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol. Cancer Res.: MCR. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finver S.N., Nishikura K., Finger L.R., Haluska F.G., Finan J., Nowell P.C., Croce C.M. Sequence analysis of the MYC oncogene involved in the t(8;14)(q24;q11) chromosome translocation in a human leukemia T-cell line indicates that putative regulatory regions are not altered. Proc. Natl. Acad. Sci. U.S.A. 1988;85:3052–3056. doi: 10.1073/pnas.85.9.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sikora K., Chan S., Evan G., Gabra H., Markham N., Stewart J., Watson J. c-myc oncogene expression in colorectal cancer. Cancer. 1987;59:1289–1295. doi: 10.1002/1097-0142(19870401)59:7<1289::aid-cncr2820590710>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., Elble R., Watabe K., Mo Y.Y. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W., Wagner B.J., Ehrenman K., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., Zhu Z., Watabe K., Zhang X., Bai C., Xu M., Wu F., Mo Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Si M.L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.Y. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q., Huang J., Zhou N., Zhang Z., Zhang A., Lu Z., Wu F., Mo Y.Y. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho T.T., Zhou N., Huang J., Koirala P., Xu M., Fung R., Wu F., Mo Y.Y. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Rese. 2015;43:e17. doi: 10.1093/nar/gku1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu F., Chiocca S., Beck W.T., Mo Y.Y. Gam1-associated alterations of drug responsiveness through activation of apoptosis. Mol. Cancer Ther. 2007;6:1823–1830. doi: 10.1158/1535-7163.MCT-06-0771. [DOI] [PubMed] [Google Scholar]
- 25.Yosef N., Zalckvar E., Rubinstein A.D., Homilius M., Atias N., Vardi L., Berman I., Zur H., Kimchi A., Ruppin E., et al. ANAT: a tool for constructing and analyzing functional protein networks. Sci. Signal. 2011;4:pl1. doi: 10.1126/scisignal.2001935. [DOI] [PubMed] [Google Scholar]
- 26.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gratacos F.M., Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscipl. Rev. RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M.J., Lin S. A region within the 5′-untranslated region of hypoxia-inducible factor-1alpha mRNA mediates its turnover in lung adenocarcinoma cells. J. Biol. Chem. 2009;284:36500–36510. doi: 10.1074/jbc.M109.008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazan-Mamczarz K., Kuwano Y., Zhan M., White E.J., Martindale J.L., Lal A., Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pautz A., Linker K., Hubrich T., Korhonen R., Altenhofer S., Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J. Biol. Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- 33.Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J.L., Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nesbit C.E., Tersak J.M., Prochownik E.V. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 35.Dang C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernasconi N.L., Wormhoudt T.A., Laird-Offringa I.A. Post-transcriptional deregulation of myc genes in lung cancer cell lines. Am. J. Respir. Cell Mol. Biol. 2000;23:560–565. doi: 10.1165/ajrcmb.23.4.4233. [DOI] [PubMed] [Google Scholar]
- 37.DeMaria C.T., Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 38.Jones T.R., Cole M.D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol. Cell. Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han S.P., Tang Y.H., Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.