Abstract
As the formation of ribonucleoprotein complexes is a major mechanism of angiotensin II type 1 receptor (AT1R) regulation, we sought to identify novel AT1R mRNA binding proteins. By affinity purification and mass spectroscopy, we identified TIA-1. This interaction was confirmed by colocalization of AT1R mRNA and TIA-1 by FISH and immunofluorescence microscopy. In immunoprecipitates of endogenous TIA- 1, reverse transcription-PCR amplified AT1R mRNA. TIA-1 has two binding sites within AT1R 3′-UTR. The binding site proximal to the coding region is glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-dependent whereas the distal binding site is not. TIA-1 functions as a part of endoplasmic reticulum (ER) stress response leading to stress granule (SG) formation and translational silencing. We and others have shown that AT1R expression is increased by ER stress-inducing factors. In unstressed cells, TIA-1 binds to AT1R mRNA and decreases AT1R protein expression. Fluorescence microscopy shows that ER stress induced by thapsigargin leads to the transfer of TIA-1 to SGs. In FISH analysis AT1R mRNA remains in the cytoplasm and no longer colocalizes with TIA-1. Thus, release of TIA-1-mediated suppression by ER stress increases AT1R protein expression. In conclusion, AT1R mRNA is regulated by TIA-1 in a ER stress-dependent manner.
INTRODUCTION
Angiotensin II is central hormonal regulator of the function and structure of cardiovascular system. It is among the most powerful vasoconstrictors and inducers of aldosterone secretion and it exerts these effects via the angiotensin II type 1 receptor (AT1R). Accordingly, the regulation of AT1R expression is a critical mechanism that modulates the activity of renin-angiotensin system (RAS). Apart from hemodynamic effects, angiotensin II (Ang II) induces extracellular matrix production, cellular hypertrophy and apoptosis (1,2).
Through their influence on protein expression patterns, RNA-binding proteins (RBPs) regulate the response of AT1R to stress and hormones. Messenger RNA (mRNA) levels do not directly translate into protein levels as mRNA localization, half-life, and translation are regulated post-transcriptionally by RBPs or small interfering RNAs. Ang II forms a negative feedback loop by two post-transcriptional mechanisms. First, Ang II increases AUF1 expression and its binding to AT1R mRNA. The binding of AUF1 to mRNA results in its destabilization and decreased protein expression (3). Second, Ang II activates the phosphorylation of calreticulin by cellular Src kinase. Phosphorylated calreticulin in turn increases the decay of AT1R mRNA (4,5).
AT1R expression has been shown to be positively regulated for example in hyperinsulinemia, hyperthyroidism and during oxidative stress. Post-transcriptional regulation of AT1R by insulin links hyperinsulinemia, a hallmark of type 2 diabetes, to hypertension. The post-transcriptional regulation mediated by insulin results from HuR translocation from nucleus to cytoplasm where it stabilizes AT1R mRNA via 3′-untranslated region (3′-UTR) leading to increased receptor expression (6). Hyperthyroidism may lead to hypertension, due to increased RAS activity. Thyroid hormone has been shown to induce an increase in the poly-A tail length of AT1R mRNA leading to increased AT1R expression (7). Reactive oxygen species (ROS) are key mediators of signaling pathways that underlie vascular inflammation in atherogenesis, starting from the initiation of fatty streak development, through lesion progression, to ultimate plaque rupture. In atherosclerotic lesions, AT1R is upregulated (8). Oxidative stress results in a wide variety of coordinated adaptive responses that result in increased AT1R expression. Oxidative stress by H2O2 dissociates GAPDH from AT1R mRNA, leading to reduced translational suppression by GAPDH, and thus increased AT1R protein expression (9).
Our previous studies, focused on the post-transcriptional regulation of AT1R, have used affinity purification using AT1R mRNA as bait to identify RBPs (6,9,10). In this study, we started from an observation that TIA-1 interacts with AT1R mRNA under unstressed condition. TIA-1 is an RBP that shuttles between cytoplasm and nucleus and functions downstream of the endoplasmic reticulum (ER) stress-induced phosphorylation of eIF2α to promote recruitment of mRNA into stress granules (SG) (11). TIA-1 binds to AT1R mRNA in unstressed cells whereas ER stress dissociates TIA-1 from AT1R mRNA and thus AT1R mRNA avoids sequestration into SGs and translational suppression. Rather than being suppressed, AT1R expression is increased under ER stress.
MATERIALS AND METHODS
Cell culture, luciferase assay and protein extraction
HEK293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), ampicillin/streptomycin and glutamine. Human coronary artery VSMC cells were purchased from Lonza and early passage cells were grown in smooth muscle growth medium-2 with 5% FBS and supplements. To induce ER stress the culture media of subconfluent VSMC was replaced with serum free media containing 1 μM thapsigargin or Dimethyl sulfoxide (DMSO) as control. At the appropriate time points the culture media was aspirated and the plates washed once with phosphate buffered saline (PBS), followed by subsequent assay dependent treatments. Expression constructs were transiently transfected into HEK293 cells using a standard Fugene 6 protocol (Roche). Silencing GAPDH and negative control silencing RNAs (siRNAs) were obtained from Ambion. Custom made siRNAs for TIA-1, including cacaacaaauuggccagua (sense) and uacuggccaauuugguugug (antisense), were obtained from Qiagen. The siRNAs were transfected using Lipofectamine2000 reagent (Invitrogen) according to manufacturer's instructions. Cells were harvested 24–72 h after transfection and firefly luciferase activities were measured using the Dual Luciferase Assay System (Promega). The luciferase activity was normalized to the activity of the cotransfected renilla luciferase plasmid. Cytoplasmic protein lysates were prepared in TX100 lysis buffer as described earlier (10). Recombinant human GAPDH-protein was obtained from Abcam.
DNA constructs
Expression construct for TIA-1 was cloned form HeLa cell library and constructed as described earlier (10). To create a construct for maltose-binding protein (MBP)-TIA-1 fusion protein production, TIA-1 cDNA was inserted into pMAL-c4x using EcoR1 and HindIII restriction sites and the ligated vector was transformed to OneShot Top10 chemically competent Escherichia coli cells (Invitrogen). Other constructs used in the experiments have been described earlier (6,9).
RNA probe preparation
cDNA was used as a template for polymerase chain reaction (PCR) reactions whereby T7 RNA polymerase promoter sequence was added to the 5′-end of all fragments and a 30 bases long poly-A tail was included in the 3′-oligos for production of polyadenylated RNA probes for affinity purification. The following 3′-UTR fragments were produced: 1–847 (full length 3′-UTR), 1–100, 100–300, 1–600, 300–600 and 600–887. The primer sequences used for the PCR were described earlier (9). The PCR products were run on agarose gel and stained with ethidium bromide. The PCR products were excised and purified from the gel with a PCR purification kit (NucleoSpin, Macherey-Nagel). RNA transcripts were synthesized from the PCR-generated templates according to manufacturer's instructions (MEGAscript, Ambion).
RNA affinity purification
RNA affinity purification was done as described earlier (10). Briefly, 2 μg of polyadenylated RNA probes were incubated with polystyrene latex beads with dC10T30 oligonucleotides covalently linked to the surface (Oligotex, Qiagen). The RNA-coated beads were incubated with cytoplasmic extracts in the RNA binding buffer (5 mM Hepes, pH 7.9, 7.5 mM KCl, 0.5 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 0.5 mM dithiotreitol (DTT), 0.1 mg/ml yeast tRNA, 0.1 mg/ml bovine serum albumin, BSA). The beads were then extensively washed with RNA binding buffer without BSA. Proteins bound to the RNA probe were resolved on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected either to western blotting or to coomassie blue staining according to manufacturer's instructions (Bio-Rad) for mass spectrometry.
Mass spectrometry
Coomassie blue stained protein bands of interest were cut out the SDS-PAGE gel and the proteins identified by mass spectrometry as described earlier (6,9,10).
Western blotting
Affinity purified proteins or cell lysates were electrophoresed in SDS-PAGE and transferred into nitrocellulose membrane (Hybond ECL). Immunodetection was performed as described earlier (6). In brief, the membrane was incubated with specific antibodies against the protein of interest followed by incubation with either fluorescently (Li-Cor and Invitrogen) or biotin labeled (Dako Cytomation) secondary antibodies. The fluorescently labeled secondary antibodies were detected with Odyssey infrared Imaging System (Li-Cor). The biotinylated secondary antibodies were detected on X-ray film using streptavidin-HRP conjugate (GE Healthcare) followed by enhanced chemiluminecence (Amersham ECL Western Blotting Detection Reagent, GE Healthcare). Primary antibodies used, were as followed: monoclonal anti-β-tubulin (Upstate), monoclonal anti-pEIF2α (Abcam), polyclonal anti-eIF2α (Abcam), monoclonal anti-GRP78 (Abcam), monoclonal anti-MBP (New England Biolabs), monoclonal anti-α-smooth muscle actin (Sigma Aldrich) polyclonal anti-GAPDH (Trevigen), polyclonal anti-TIA-1 and monoclonal anti-TIA-1/TIAR (Santa-Cruz) and polyclonal anti-TIA-1 (BioVision).
Protein immunorecipitation (IP)
The assay was performed essentially as described earlier (9). In short, TIA-1 was immunoprecipitated from HEK293 cell lysates with polyclonal anti-TIA-1 antibody. Pre-immuno IgG or anti-cyclooxygenase 2 (Cox-2) antibodies were used as controls. The samples were precleared with protein G-sepharose beads before adding the anti-TIA-1 or control antibodies. Protein G-sepharose beads were added to bind the antibodies followed by extensive washing of the beads with 100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% NP-40, 1 mM DTT, 100 U/ml RNase inhibitor, 2 mM vanadyl ribonucleoside complexes and protein and phosphatase inhibitor cocktails. Proteins were digested with 0.1% SDS and proteinase K followed by RNA extraction with phenol-chloroform-isoamyl alcohol mixture. RNA was precipitated with 10 μg of yeast tRNA as carrier. The precipitated RNA was reverse transcribed using poly-dT oligos followed by PCR with primers targeting the AT1R coding region.
Production and purification of MBP-TIA-1 proteins
The TIA-1 fusion protein production and purification was done according to manufacturer's instructions (New England Biolabs). The vector for fusion protein production was extracted and transformed to E. coli K12 TB1 strain for protein production. To produce MBP-TIA-1 fusion protein, the TIA-1-pMAL-c4x transformed E. coli cells were grown to OD600 of 0.5 and the recombinant protein production was initiated by 0.3 mM IPTG. After 2 h the cells were harvested by centrifugation, resuspended in column buffer (20 mM Tris–HCL, 200 mM NaCl, 1 mM EDTA) and stored overnight in −20°C. The cells were thawed and sonicated on ice before centrifugation. The supernatant was diluted 1:5 in column buffer and loaded to an amylose resin column. The column was washed with column buffer 12× the column volume. The fusion protein was eluted in 3 ml fractions with column buffer containing 10 mM maltose. After the elution the protein concentrations of the fractions were measured with Protein Assay (Bio-Rad) and the protein containing fractions were pooled. Finally, the proteins were concentrated with Amicon Ultra centrifugal filters (Millipore).
Lentiviral silencing of TIA-1 in VSMCs
VSMCs were cultured in 6 well plates (5 × 104 cells/well). One day after seeding, the cells were transduced with lentiviral particles packed with pGIPZ shRNA constructs against TIA-1 or negative controls (Thermo Scientific) at MOI 10, together with 4 μg/ml polybrene in 1 ml of serum free medium. Four hours post-infection 1 ml of medium with serum was added to the cultures, and 24 h post-infection the medium was replaced with virus-free medium. To kill any uninfected cells, puromycin was added to the cells (2 μg/ml) 48 h post-infection. The puromycin selective medium was replaced daily for 4 days. The cells were grown to confluency in puromycin-free medium and seeded to 6 cm plates for qPCR and western blots or 24-well plates for ligand binding assay.
Real-time PCR (qPCR)
In RNA expression experiments, normal or shRNA silenced VSMCs were grown subconfluent on 6 cm cell culture plates. Normal VSMCs were exposed to 1 μM thapsigargin or dimethyl sulfoxide (DMSO) as controls and the stimulations were stopped at different time points. The total RNA was isolated from the cultures with NucleospinRNA II (Macherey-Nagel). A total of 1 μg of total RNA from each sample was used for the first strand cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen). The cDNA synthesis was carried out according to manufacturer's instructions using oligo-dT primers. qPCR was performed in LightCycler (Roche) using Maxima SYBR Green (Fermentas) according to manufacturer's instructions. In short, AT1R and β actin (ActB) or β tubulin (TubB) mRNAs were amplified using specific primers obtained from Invitrogen. ActB was used as a control for normalization in thapsigargin experiments whereas TubB was used with the shRNA silenced samples. The primers used in the PCR were as follows: gcaccaggtgtatttgatatag (AT1R sense), ctgtttccaaatattcccacc (AT1R antisense), gacgacatggagaaaatctg (ActB sense), atgatctgggtcatcttctc (ActB antisense), gtactacaatgaagccacag (TubB sense) and ccagactgaccaaatacaaag (TubB antisense). The relative concentrations were calculated from the first 35 cycles against a standard curve.
Ligand-binding assay
TIA-1-silenced or negative control-transduced VSMCs and untreated VSMCs were grown to subconfluency on 24-well plates and either exposed to ER stress by 1 μM thapsigargin for various timepoints or left untreated. To study the expression of AT1R on cell surface the cultures were subjected to ligand binding assay as described earlier (12). In short, the cells were washed twice with PBS containing 0.1% BSA followed by incubation with 0.2 nM 125I-[Sar1,lle8]Ang II (PerkinElmer) at +37°C for 1 h in the absence (total count) or presence of 1 μM Ang II as competitor. The cells were then washed twice with ice-cold PBS containing 0.1% BSA and lysed in 0.5 N NaOH. The lysates were mixed with Optiphase Hisafe 3 (PerkinElmer) and the activity of the radiolabeled ligand was measured with liquid scintillation counter (Wallac 1414 WinSpectral). AT1R binding was calculated as the difference between the total count and the count from samples incubated with unlabeled Ang II.
Immunofluorescence staining and RNA-fluorescence in situ hybridization
Subconfluent VSMC grown on coverslips were exposed to 1 μM thapsigargin for 1 h to induce ER stress or to DMSO as controls. After the stimulation the culture media was aspirated and the cells were washed once with PBS. The cells were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature (RT) followed by two washes with PBS. After fixation the samples were subjected to either immunofluorescence (IF) staining or to RNA fluorescence in situ hybridization (FISH). The antibodies used in the experiments was as follows: monoclonal anti-GAPDH (Novus Biologicals), polyclonal anti-GAPDH and anti-TIA-1 (Santa-Cruz), monoclonal anti-PABP1 (Abcam).
For IF, the fixed cells were permeabilized with 0.25% Triton X-100 (Tx-100) (Bio-Rad) in PBS for 15 min at RT followed by three washes with PBS. The samples were blocked with blocking buffer [5% BSA, 5% normal donkey serum (NDS), 0.3 M glycine in PBS] for 1 h at RT. Subsequently the samples were incubated with primary antibodies diluted in antibody buffer (2.5% BSA, 2.5% NDS, 0.1% Tween-20 in PBS) overnight at +4°C in a humidified chamber. The samples were washed quickly with washing buffer [0.1% Tween-20 in PBS (PBST)] followed by a single 5 min and two 10 min washes. Next the samples were incubated with fluorescently labeled secondary antibodies (Alexa Fluor, Invitrogen) diluted in the antibody buffer for 1 h at RT followed by a series of washes as before with PBST. Finally the samples were rinsed with PBS and the nuclei were stained with 4 μM Hoechst 33258 (Sigma Aldrich) in PBS. After rinsing the samples twice with PBS and once with H2O, the coverslips were mounted to microscope slides with ProLong Gold (Invitrogen) and visualized with Zeiss LSM 780 confocal microscope.
For RNA FISH, the cells were permeabilized with 70% ethanol overnight at +4°C. After permeabilization the samples were incubated for 5 min in wash buffer [10% formamide in 2× saline-sodium citrate (SSC)] followed by hybridization with fluorescently labeled RNA probes designed against AT1R mRNA coding region (Stellaris RNA FISH, Biosearch technologies). The samples were hybridized with 250 nM probes in hybridization buffer (100 mg/ml dextran sulfate and 10% formamide in 2× SSC) for 4 h at +37°C in a humidified chamber. Following hybridization the samples were washed twice with washing buffer for 30 min at +37°C in a humidified chamber. Next the samples were rinsed once with PBS and proceeded to blocking and subsequent IF staining as describes above. The samples were visualized using Zeiss Axioplan 2 microscope.
Statistical analysis
Data are presented as means ± SD. Statistical analysis was performed using Student's paired one-tailed t-test. P < 0.05 were considered significant. Quantification of western blot band intensities were calculated using the quantification option of Image Studio version 3.1 imaging software (Li-Cor) used with the Odyssey Infrared Imaging System for detection of western blots.
RESULTS
TIA-1 interacts with AT1R mRNA
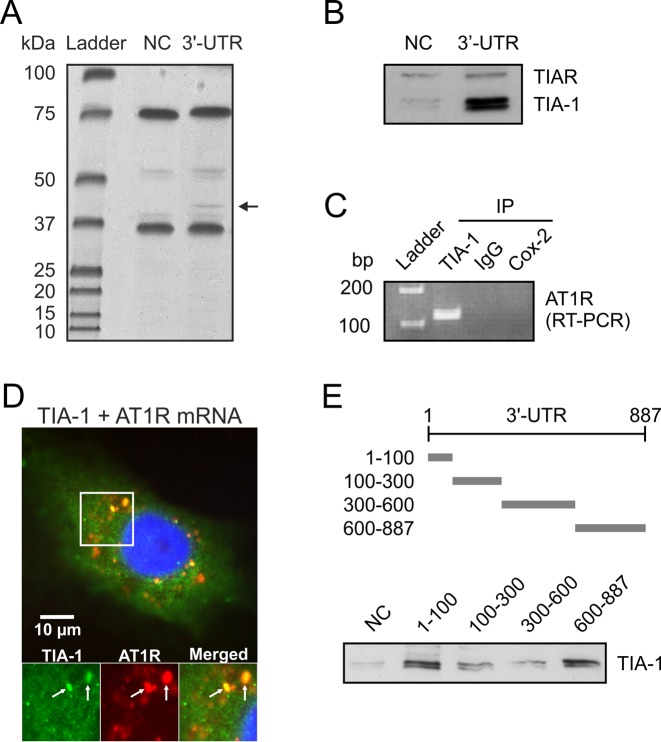
Our research group has searched for AT1R mRNA-binding proteins to understand the post-transcriptional mechanisms of AT1R regulation. We assayed the existence of such RBPs by employing affinity purification using 3′-UTR of AT1R mRNA as bait. Previously, we used this technique to identify p100, GAPDH and HuR as AT1R 3′-UTR binding proteins (6,9,10). In the present study, a polyadenylated probe corresponding to nucleotides 1–847 of 3′-UTR was transcribed and bound to poly-T beads, and then incubated with cytoplasmic lysates followed by extensive washing. A band migrating at ∼40 kDa was identified (Figure 1A). In a negative control experiment using fragments of luciferase coding region, binding was undetectable. Protein was excised from the gel and digested ‘in gel’ by trypsin. The generated peptides were extracted and subjected to mass spectrometric analysis. Database (MSDB) search with the combined MALDI-TOF peptide mass fingerprint data identified (Mascot score 566) the protein as human TIA-1.
Figure 1.
Cytoplasmic association of TIA-1 with AT1R mRNA in VSMCs. (A) Proteins binding to AT1R 3′-UTR were affinity purified from VSMC lysates using an in vitro transcribed AT1R 3′-UTR as bait. A probe consisting of luciferase coding region was used as the negative control (NC). At 40 kDa a protein was present in 3′-UTR lane but not in control lane and this protein (arrow) was then excised, digested and identified by mass spectrometry. The details of the purification are given under ‘Materials and Methods’ section. (B) A western blot of proteins isolated by affinity purification from HEK293 cell lysates was performed and binding of TIA-1 to AT1R 3′-UTR was confirmed by antibody recognizing both TIA-1 and TIAR. (C) Binding of endogenous TIA-1 with endogenous AT1R mRNA was detected by PCR assay with AT1R specific primers of reverse transcribed material obtained by IP from cytoplasmic fractions. Changes in the level of AT1R mRNA associated with TIA-1 were evaluated by measuring its abundance in the IP mixture. Immunoprecipitation was performed with TIA-1-specific antibody, preimmuno IgG or Cox-2-specific antibody. PCR products were visualized after electrophoresis in 1% agarose gels stained with ethidium bromide. (D) The colocalization of TIA-1 and AT1R mRNA in VSMCs was examined by in situ hybridization using a fluorescently labeled AT1R mRNA-specific probe, followed by immunocytochemistry with the anti-TIA-1 antibodies. The distribution of TIA-1 is shown in green (Figure 1D left panel), AT1R mRNA is shown in red (Figure 1D middle panel) and the overlap is shown in yellow (Figure 1D right panel). The colocalization of the signals is indicated (arrows). (E) Mapping of the TIA-1 binding site. Upper panel, a schematic illustration of AT1R mRNA 3′-UTR probes used for mapping of TIA-1 interaction sites. Lower panel, western blot detection of TIA-1 from affinity purifications with probes consisting of various segments of AT1R 3′-UTR from HEK293 cell lysates.
To directly test if TIA-1 interacts with AT1R mRNA, we performed protein separation from HEK293 cell lysates by RNA affinity purification with full length AT1R 3′-UTR probe. The samples were immunoblotted for TIA-1 and endogenous TIA-1 was detected (Figure 1B). In this blot, we utilized antibody recognizing also TIAR (TIA-1 related protein) but no specific TIAR binding to 3′-UTR was detected. Ribonucleoprotein (RNP) IP was performed to study the association of TIA-1 with endogenous AT1R mRNA. The relative enrichment of AT1R mRNA in RNP IP reaction was tested by reverse transcription, followed by conventional PCR amplification and then visualized on agarose gels. RNP IP assays revealed a strong enrichment of AT1R mRNA with TIA-1 in anti-TIA-1 antibody reactions relative to that of control IP reactions (IgG and Cox-2) (Figure 1C). These results indicate that TIA-1 forms RNP complex with endogenous AT1R mRNA.
To test whether TIA-1 and AT1R mRNA are colocalized within VSMCs, we performed in situ hybridization using a fluorescently labeled AT1R mRNA-specific probe, followed by immunocytochemistry with the anti-TIA-1 antibodies. These double-staining results (Figure 1D) show a clear colocalization of TIA-1 and AT1R mRNA within VSMCs. The distribution of TIA-1 is shown in green (Figure 1D left panel), AT1R mRNA is shown in red (Figure 1D middle panel) and the overlap is shown in yellow (Figure 1D right panel). These data are consistent with the colocalization of TIA-1 with AT1R mRNA in the cytoplasm of VSMCs. A small fraction of TIA-1 colocalized with a P-body marker GW182 (data not shown).
TIA-1 has two binding sites within AT1R 3′-UTR
Next, we sought to identify the AT1R 3′-UTR regions involved in interaction with TIA-1. To this end, four polyadenylated fragments spanning the different mRNA regions of AT1R 3′-UTR were used in affinity purification. Probes corresponding to 3′-UTR bases 1–100, 100–300, 300–600 and 600–887 were transcribed and poly-A tail was included as a part of the primer used in the PCR-reaction. Schematic representations of the probes are described in Figure 1E (upper panel). Synthesized mRNAs with poly-A were bound to poly-T beads, incubated with cytoplasmic lysates followed by extensive washing. A western blot of proteins isolated by RNA affinity purification was performed and TIA-1 expression was detected by polyclonal anti-TIA-1 antibodies. We mapped the interaction to fragments of 1–100 and 600–887 of 3′-UTR (Figure 1E, lower panel). These findings indicate that TIA-1 interacts with two binding sites, one in the proximity of the coding region and the other in the distal part of the 3′-UTR of AT1R mRNA.
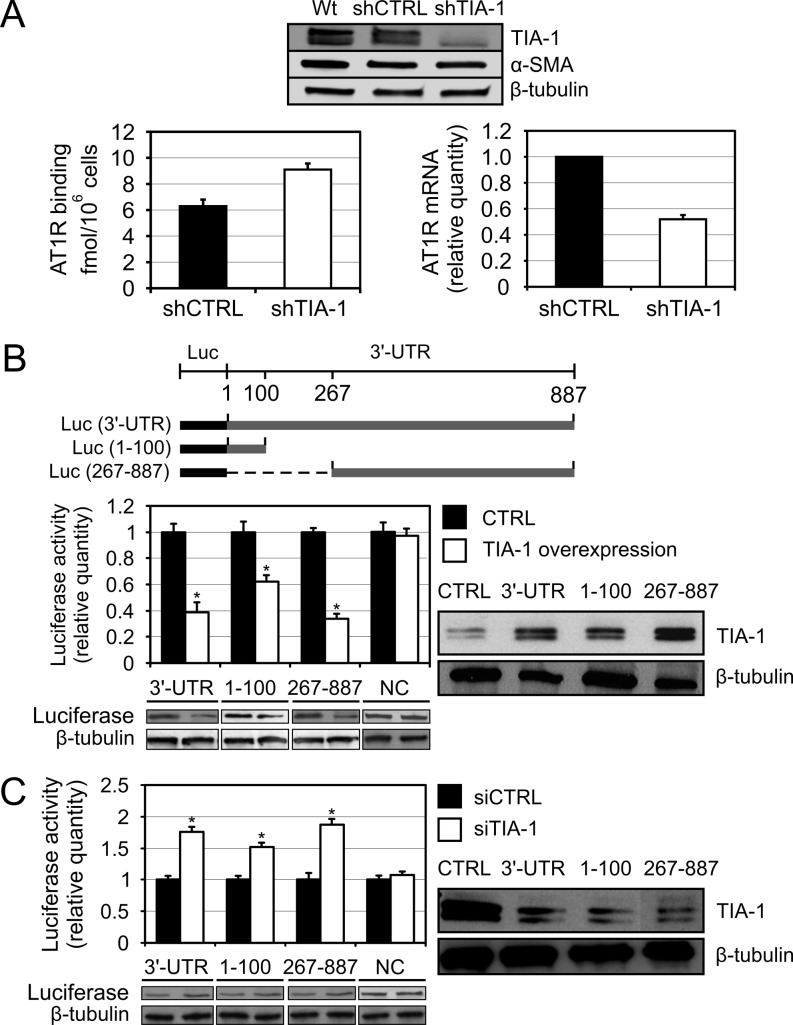
TIA-1 is a negative regulator of AT1R protein expression
To test the functional consequences of the association between TIA-1 and AT1R mRNA we used RNA interference. Silencing of TIA-1 by transducing coronary artery VSMCs with TIA-1-directed shRNA effectively lowered TIA-1 abundance (Figure 2A, upper panel). Depletion of TIA-1 increased AT1R protein expression as observed by ligand binding utilizing radiolabeled Ang II to measure changes in cell surface AT1R expression (Figure 2A, lower left panel). While increasing AT1R protein expression, TIA-1 silencing decreased the AT1R mRNA expression as measured by qPCR (Figure 2A, lower right panel). This suggests that in the absence of stress, endogenous TIA-1 represses the protein expression of AT1R while having an opposite effect on the mRNA expression.
Figure 2.
TIA-1 negatively regulates AT1R protein expression via mRNA 3′-UTR. (A) The effect of TIA-1 silencing on endogenous AT1R protein and mRNA expression. VSMC were transduced with a lentiviral shRNA construct against TIA-1 (shTIA-1) or with a non-silencing control (shCTRL). The western blot at the top show successful TIA-1 silencing and α-smooth muscle actin (α-SMA) expression to confirm the VSMC phenotype of the cells after virus infection. β-tubulin was used as a loading control. The bottom left panel shows AT1R protein expression as quantified by ligand binding using radiolabeled Ang II to measure changes in the cell surface receptor expression. The bottom right panel show AT1R mRNA expression as quantified by qPCR. The results are shown as relative quantity to shCTRL AT1R mRNA expression and the results are normalized to β-tubulin expression. Results represent the mean ± SD of an average of three independent experiments. P < 0.05 versus the shCTRL. (B) TIA-1 effect on AT1R 3′-UTR-mediated reporter gene expression. Upper panel, a schematic illustration of luciferase constructs with AT1R 3′-UTR fragments used in the experiment. Lower panel, HEK293 cells were cotransfected with the various luciferase-3′-UTR constructs together with renilla luciferase and either empty vector (CTRL) or with TIA-1 expression plasmid. A construct with inverted luciferase coding region as 3′ UTR was used as negative control (NC). Cells were lysed and assayed for firefly and renilla luciferase activities. Results represent the mean ± SD of an average of three independent experiments. *P < 0.05 versus samples transfected with the empty expression vector. Western blots showing the luciferase expression and β-tubulin as loading control are shown below. The anti-TIA-1 western blots showing the overexpression of TIA-1 and β-tubulin expression as loading control are shown on the right. (C) Effect on reporter gene expression following TIA-1 silencing. HEK293 cells were transfected with the same luciferase constructs as in Figure 2B together with a non-silencing control (siCTRL) or with siTIA-1 expression construct and assayed for firefly and renilla luciferase activities in similar manner. Results represent the mean ± SD of an average of three independent experiments. *P < 0.05 versus samples transfected with the siCTRL. Western blots showing the luciferase expression and β-tubulin as loading control are shown below. The western blots showing TIA-1 silencing and a β-tubulin control blot are shown on the right.
To investigate if TIA-1 regulates AT1R mRNA through region where it binds more extensively, we prepared the heterologous reporter construct of luciferase fused with AT1R 3′-UTR. TIA-1 overexpression did not influence luciferase activity of a construct lacking AT1R 3′-UTR. As shown in Figure 2B, TIA-1 overexpression in HEK293 cells decreased luciferase activity of a vector containing at least one TIA-1 binding site fused to the 3′-end of a luciferase coding region. Conversely, TIA-1 silencing in HEK293 cells using a specific TIA-1-directed siRNA resulted in a significant increase in the luciferase activity of a fusion construct with either 5′ end (1–100) or 3′-end (267–887) TIA-1 binding sites of the 3′-UTR (Figure 2C). These findings indicate that there are TIA-1-dependent negative regulatory elements within the 3′-UTR of AT1R mRNA. These results are consistent with the notion that TIA-1 reduces AT1R protein expression through binding sites within the 3′-UTR.
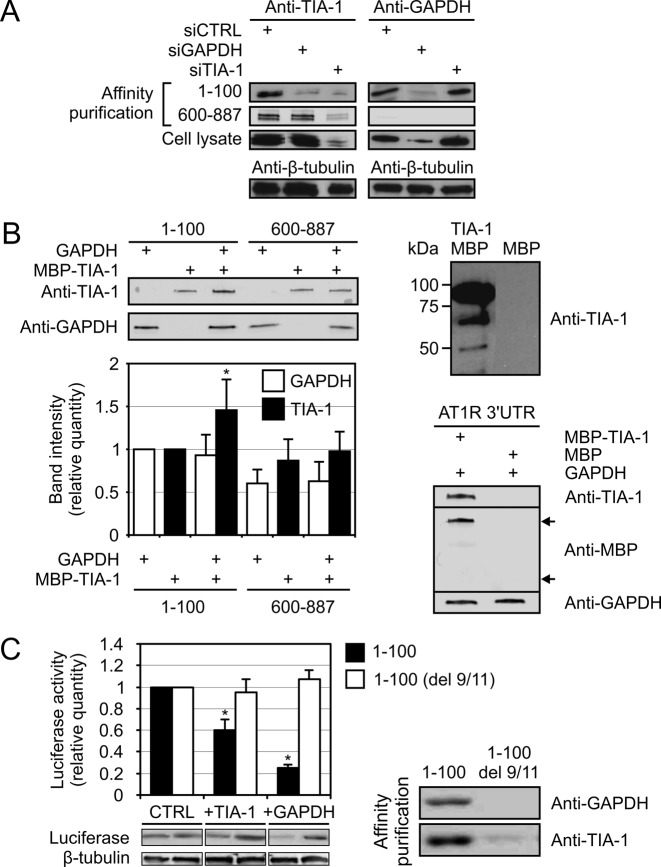
GAPDH-dependent and -independent binding of TIA1 to AT1R mRNA
Our prior data with GAPDH showed that TIA-1 and GAPDH have an overlapping binding site within the 3′-UTR proximal to the coding region (9). Accordingly, we postulated that both GAPDH and TIA-1 interact either directly or through mRNA. To test if the binding of TIA-1 to AT1R mRNA is influenced by GAPDH, polyadenylated RNAs spanning the 1–100 and 600–887 regions of AT1R 3′-UTR were synthesized and their interaction with TIA-1 and GAPDH was studied after incubating them with HEK293 cytoplasmic lysates and affinity purifying the resulting RNP complexes using poly-T-coated beads. Western blot analysis was used to identify TIA-1 and GAPDH present in the complexes. As shown in Figure 3A (left lanes), both GAPDH and TIA-1 are present in the complexes. In GAPDH-depleted HEK293 lysates, TIA-1 was no longer present in the 1–100 RNP complexes whereas in the complexes containing the 600–887-region the binding of TIA-1 was unaffected by GAPDH-depletion. In contrast, depletion of TIA-1 did not have an effect on GAPDH binding to the 1–100 region whereas with the 600–887-region there was no binding of GAPDH to the RNA in any of the samples. These results suggest that GAPDH is required for TIA-1 binding to mRNA at the 1–100 region of AT1R 3′-UTR. We confirmed this data with purified GAPDH protein and recombinant MBP-TIA-1 fusion protein. In this affinity purification experiment MBP-TIA-1 and GAPDH were added separately and together to AT1R 3′-UTR. TIA-1 binding to 1–100 of AT1R 3′-UTR significantly increased if GAPDH was present, whereas at the distal site (600–887) TIA-1 binding was not dependent on GAPDH (Figure 3B, upper left panel). When GAPDH was present, quantification of the western blot band intensities showed an ∼1.5-fold binding of TIA-1 to the 1–100-region relative to samples containing TIA-1 alone. At the distal 600–887-region there was no statistically significant change (Figure 3B, lower left panel). To confirm that the mRNA binding of MBP-TIA-1-fusion protein was mediated by TIA-1, affinity purification with AT1R 3′-UTR was performed in samples containing GAPDH and either MBP-TIA-1 or MBP. Western blot analysis of the samples showed that while MBP-TIA-1 bound to the RNA, MBP alone did not (Figure 3B, lower right panel). To examine the possibility that TIA-1 and GAPDH would interact directly, we tried co-immunoprecipitation under various conditions. We could not demonstrate co-immunoprecipitation of GAPDH and TIA-1 in mRNA-depleted environment neither with purified proteins nor with HEK293 cell lysates (data not shown). Nor did strong overexpression of GAPDH and TIA-1 in the HEK293 lysates improve the yield of immunoprecipitation (data not shown). This data indicates that TIA-1 and GAPDH form complexes with AT1R mRNA but the two proteins do not interact directly.
Figure 3.
GAPDH-dependent and -independent binding of TIA-1 to AT1R mRNA. (A) GAPDH-dependent TIA-1 binding at the 1–100 binding site. Left panel, affinity purification with the 1–100 and 600–887 fragments of AT1R 3′-UTR were performed from siCTRL-, siTIA-1- or siGAPDH-transfected HEK293 cells as indicated. The upper lane shows TIA-1 detection by western blot from the affinity purified samples from the 1–100 binding site and the second lane from the 600–887 binding site. The third lane show TIA-1 expression in whole cell lysates and the bottom lane β-tubulin expression, used as a loading control. Right panel, same experimental setup was used as in the left panel but the samples were probed for GAPDH. (B) Validation of direct TIA-1 and GAPDH interaction on AT1R 3′-UTR. Affinity purification was performed with 1–100 or 600–887 fragments of AT1R 3′-UTR from samples containing purified recombinant MBP-TIA-1, GAPDH or both, as indicated (left panel). Binding of TIA-1 and GAPDH to the probes were visualized by western blotting (upper left panel). The band-intensities from the western blot were quantified (lower left panel). Results represent the mean ± SD of an average of three independent experiments. *P < 0.05 versus the result from 1–100 region with only TIA-1 added. A western blot of purified MBP-TIA-1 detected by anti-TIA-1 antibody is shown in the upper right panel. To confirm that the binding of MBP-TIA-1 to RNA is mediated by TIA-1 and not MBP the full length AT1R 3′-UTR was used in affinity purification containing recombinant GAPDH and either MBP-TIA-1 or MBP only. The binding of the proteins to the probe were visualized by western blotting (lower right panel). The upper arrow indicate the location of MBP-TIA-1-fusion protein and the lower arrow the location of MBP. (C) Effect of TIA-1 and GAPDH on the expression of a reporter gene with mutated GAPDH binding site. HEK293 cells were transfected with a luciferase expression construct fused with 1–100 fragment of AT1R 3′-UTR or with a fragment lacking the GAPDH binding motif (del 9/11). The cells were cotransfected with TIA-1 or GAPDH expression vectors or with an empty control vector (CTRL). Following transfections, the cells were lysed and the luciferase activities measured (Left panel). Results represent the mean ± SD of an average of three independent experiments. *P < 0.05 versus samples transfected with the empty expression vector. Western blots showing the luciferase expression and β-tubulin as loading control are shown below. Binding of TIA-1 and GAPDH to the wild-type (1–100) and mutated (del 9/11) RNA was studied by affinity purification from HEK293 lysates. The affinity purified proteins visualized by western blot are shown in the right panel.
GAPDH-dependent TIA-1 binding site
To seek further evidence for GAPDH-dependent TIA-1 binding to mRNA, we used an altered version of a chimeric construct in which luciferase was fused with a mutated 1–100 of AT1R 3′-UTR that does not bind to GAPDH nor respond to GAPDH silencing or overexpression (9). Initially, this construct unresponsive to GAPDH was identified in random, saturating mutagenesis designed to identify critical binding site for GAPDH-RNA interaction. Inactive 1–100 of AT1R 3′-UTR had bases at sites 9 and 11 removed (del 9/11). We hypothesized that TIA-1 does not affect the luciferase activity when a necessary mediator protein, GAPDH, is unable to link TIA-1 to AT1R 3′-UTR. To test this, HEK293 cells were transfected with a luciferase expression construct fused with either the 1–100 fragment of AT1R 3′-UTR or with 3′-UTR lacking functional GAPDH binding motif (del 9/11). The cells were cotransfected with either TIA-1 or GAPDH expression constructs or with an empty vector as control. Following the transfections the cells were lysed and the luciferase activity measured. Both TIA-1 and GAPDH overexpression led to a decreased luciferase activity of the wild-type construct (Figure 3C, left panel). In line with our expectations, the construct lacking the binding site for GAPDH (del 9/11) did not respond to neither GAPDH nor TIA-1 overexpression. Binding of TIA-1 and GAPDH to the 1–100 and mutated (del 9/11) fragment of AT1R 3′-UTR was studied by affinity purification from HEK193 lysates followed by western blot detection. Both TIA-1 and GAPDH bound to the 1–100 fragment whereas almost no binding of TIA-1 was observed with the del 9/11 fragment lacking the GAPDH binding site (Figure 3C, right panel). Taken together, our data argues for the proximal binding site within AT1R 3′-UTR to require GAPDH for both binding and function.
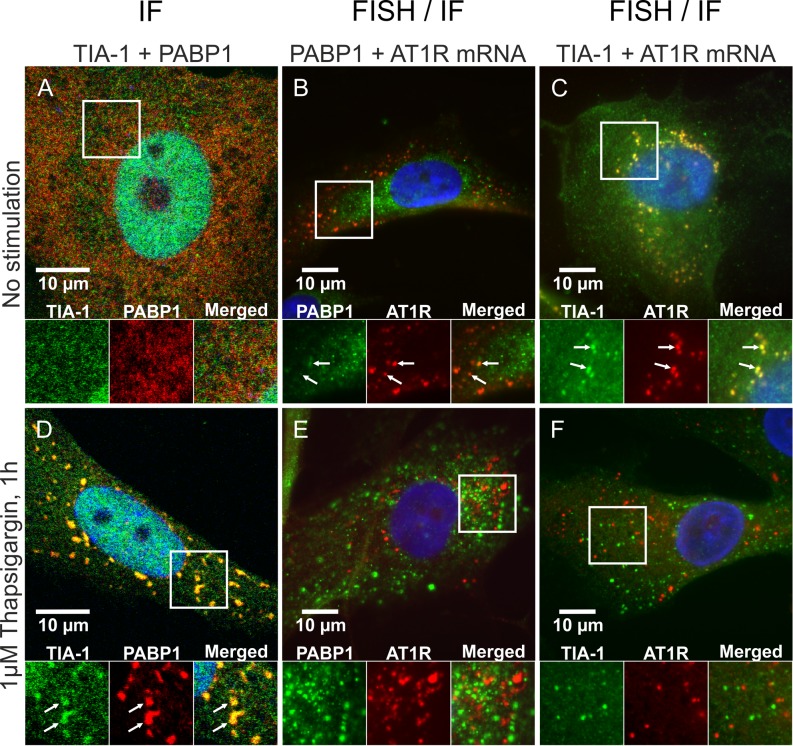
Interaction of AT1R mRNA and TIA-1 in stressed cells
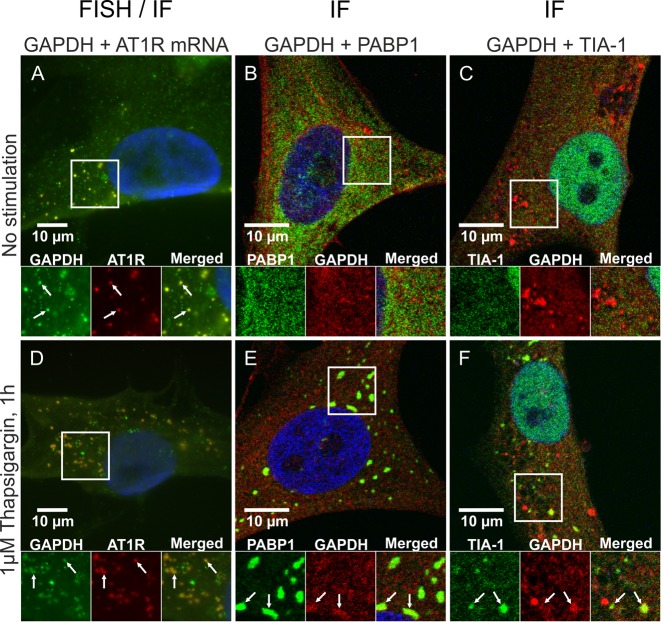
To explore the interaction of TIA-1 with AT1R mRNA in stressed cells, we exposed coronary artery VSMC to thapsigargin, an ER stressor that induces the formation of SG. VSMCs were fixed, permeabilized and blocked before staining for TIA-1 and another well-established SG marker, poly-A binding protein 1 (PABP1) (11,13). In the unstressed cells TIA-1 was primarily nuclear, showing only a diffuse cytoplasmic expression (Figure 4A), whereas in thapsigargin-treated VSMCs TIA-1 accumulated at multiple cytoplasmic foci, which represented SGs (Figure 4D). The nuclear localization of TIA-1 is detectable in Figure 4A and D as the nuclear envelope is permeabilized by the 0.25% TX-100 used for the immunofluorescent staining. Colocalization of PABP1 confirms that TIA-1 is accumulated in SGs. The localization of AT1R mRNA in stressed cells was studied in FISH experiment using a probe against the coding region of AT1R mRNA and immunostaining of the SG marker PABP1 or TIA-1. Superposition of FISH of AT1R mRNA combined with immunostaining of PABP1 show that no AT1R mRNA accumulation to SGs occurred during ER stress (Figure 4B and E). Superposition with TIA-1 immunostaining, in turn, shows that in unstressed coronary artery VSMCs TIA-1 and AT1R mRNA colocalize but in stressed cells they do not (Figure 4C and F). These observations suggest that the distribution of AT1R mRNA is unaffected by ER stress. AT1R mRNA and TIA-1 colocalize in unstressed conditions, but when cells are stressed TIA-1 is recruited to SGs while AT1R mRNA is not. Thus, ER stress releases TIA-1 from AT1R mRNA. The FISH experiments do not show the nuclear TIA-1 in Figure 4C and F as the cells are permebilized with 70% ethanol leaving the nuclear proteins undetected by the antibodies.
Figure 4.
AT1R mRNA dissociates from TIA-1 in response to ER stress. VSMCs were exposed to vehicle (A–C) or 1 μM thapsigargin for 1 h (D–F). After stimulations the cells were fixed and immunofluorescently stained against TIA-1 and PABP1 (A and D). Alternatively the cells were in situ hybridized with a fluorescently labeled RNA probe against AT1R mRNA and immunofluorescently stained against PABP1 (B and E) or TIA-1 (C and F). The subcellular localization of the mRNA and proteins (FISH/IF) was visualized with a fluorescence microscope whereas colocalization of the proteins (IF) was visualized with a confocal laser scanning microscope. Colocalization of the signals are indicated (arrows). The nuclear localization of TIA-1 is visible in the IF stainings (A and D) as the cells are permeabilized with 0.25% TX-100. In the FISH combined with IF stainings against TIA-1 (C and F) the nuclear localization of TIA-1 is not shown as the cells are permeabilized with 70% ethanol leaving the nuclear proteins undetected by the antibodies. The details of the staining procedures are given under ‘Materials and Methods’ section.
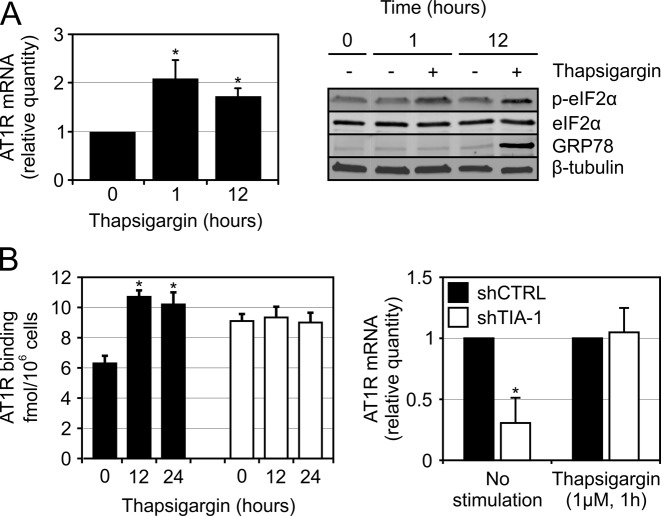
ER stress increases endogenous AT1R mRNA and protein expression
So far, we have shown that in unstressed cells cytoplasmic TIA-1 binds to AT1R mRNA and suppresses AT1R protein expression. However, the major biological function of TIA-1 is related to ER stress where it sequesters mRNAs in SGs. Prior reports suggest that ER stress increases AT1R protein expression (6,9,14–16). To directly test whether ER stress upregulates AT1R protein expression, we treated coronary artery VSMCs with a well-known ER stress-inducing agent, thapsigargin and measured mRNA and ligand binding of AT1R. RNA was purified from thapsigargin stimulated and unstimulated VSMCs, reverse-transcribed, quantified by qPCR and normalized against β actin mRNA. ER stress-induced SG assembly is initiated by phosphorylation of eIF2α which inhibits the formation of functional preinitiation complexes (11,17). Successful induction of ER stress by thapsigargin was thus monitored by western blotting, showing an increase in both eIF2α phosphorylation and ER chaperone GRP78 (Figure 5A, right panel). In thapsigargin treated cells the AT1R mRNA levels were ∼2-fold relative to untreated cells normalized against the β actin expression (Figure 5A, left panel). Similarly, AT1R ligand binding using radiolabeled Ang II to detect the cell surface expressed AT1R shows that thapsigargin increased the protein expression of AT1R (Figure 5B, left panel). To test whether TIA-1 silencing has an effect on ER stress-induced increase in AT1R mRNA-levels as seen in Figure 5A, the control and TIA-1-silenced VSMCs were exposed to thapsigargin and the change in AT1R mRNA expression was quantified by qPCR. Under unstressed conditions there was a significant decrease in AT1R mRNA levels in TIA-1 silenced cells compared to the controls, consistent with the results seen in Figure 2A. In the thapsigargin treated cells, however, there was no statistically significant difference in the AT1R mRNA levels between the control and TIA-1-silenced cells (Figure 5B, right panel). These results are consistent with earlier observations that AT1R escapes the stress-induced transcriptional silencing (6,9,14–16). Further, the thapsigargin effect was dependent on TIA-1 as TIA-1 silenced VSMCs had increased baseline AT1R protein expression but no longer responded to the thapsigargin treatment.
Figure 5.
ER stress increases AT1R mRNA and protein expression. (A) ER stress effect on AT1R mRNA expression. VSMCs were treated with 1 μM thapsigargin and at the indicated time points the cells were harvested and the total RNA was extracted. One microgram of total RNA from each time point was reverse transcribed to cDNA using oligo-dT primers. The cDNAs were analyzed by qPCR using gene specific primers for AT1R and β actin. The results are normalized against the β actin values and represent the mean ± SD values relative to untreated samples from same time points of an average of three independent experiments. *P < 0.05 versus the results at 0 h. As shown on right, the induction of ER stress was confirmed by visualizing the increase of phosphorylated eIF2α (p-eIF2α) by western blot using a specific antibody against serine 51 phosphorylated eIF2α (upper lane). The expression of total eIF2 α is shown in the second lane. As another indicator of ER stress induction, the increased expression of the ER chaperone GRP78 is shown in the third lane. β tubulin was used as a loading control. (B) Effect of ER stress and TIA-1 on endogenous AT1R expression. The same TIA-1 and control silenced VSMCs as in Figure 2A were subjected to thapsigargin-induced ER stress. At the indicated time points endogenous AT1R expression was measured by ligand binding using radiolabeled Ang II to measure changes in the cell surface receptor expression (left panel). Results represent the mean ± SD of an average of three independent experiments. *P < 0.05 versus the shCTRL at 0 h. To study if TIA-1 silencing has an effect on ER stress induced upregulation of AT1R mRNA expression as seen in Figure 5A, the TIA-1 and control silenced VSMCs were subjected to 1 μM thapsigargin for 1 h and the AT1R mRNA expression was quantified by qPCR as in Figure 5A (right panel). The results are normalized against the control silenced samples (shCTRL) and represent the mean ± SD of an average of three independent experiments. *P < 0.05 versus the shCTRL.
GAPDH interaction with AT1R mRNA 3′-UTR is not regulated by ER stress
TIA-1 interaction with the proximal 3′-UTR binding site in AT1R mRNA is GAPDH-dependent. To know if GAPDH has a role in ER stress response, we hybridized fluorescently labeled RNA probe with AT1R mRNA in VSMCs and followed GAPDH localization by immunofluorescence. The colocalization of GAPDH with AT1R mRNA remained unaltered by ER stress (Figure 6A and D). This occurred despite the fact that in thapsigargin-stressed cells, GAPDH colocalized with both PABP1 (Figure 6B and E) and TIA-1 (Figure 6C and F) in SGs. This result identified GAPDH as a novel component of SGs. The abundant GAPDH appears to have different pools of protein. One pool is taken to SGs by ER stress whereas the other pool is bound to AT1R mRNA.
Figure 6.
GAPDH binding to AT1R mRNA is unaffected by ER stress. VSMCs were exposed to vehicle (A–C) or 1 μM thapsigargin for 1 h (D–F). After stimulations the cells were fixed and in situ hybridized with a fluorescently labeled RNA probe against AT1R mRNA and immunofluorescently stained against GAPDH (A and D). Alternatively the cells were immunofluorescently dual labeled against GAPDH and PABP1 (B and E) or TIA-1 (C and F). The subcellular localization of the mRNA and proteins (FISH/IF) was visualized with a fluorescent microscope whereas the colocalization of the proteins (IF) was visualized with a confocal laser scanning microscope. Colocalization of the signals is indicated (arrows). The details of the staining procedures are given under ‘Materials and Methods’ section.
DISCUSSION
Our earlier studies have systematically analyzed the mechanisms of post-transcriptional regulation of AT1R. Since post-transcriptional regulation is governed by the formation of RNP complexes, we sought to identify RNPs that bind to AT1R mRNA. The binding of TIA-1 to AT1R mRNA in unstressed cells or cell lysates was supported by affinity purification of TIA-1 with AT1R 3′-UTR and colocalization of TIA-1 and AT1R mRNA by immunofluorescence microscopy as well as immunoprecipitation of AT1R mRNA from TIA-1 lysates. In affinity purification of cell lysates with different fragments of AT1R 3′-UTR, TIA-1 was found to have two binding sites. TIA-1 and GAPDH cooperate in binding to the binding site proximal to coding region, whereas TIA-1 binding to the distal binding site appears to be direct as shown by in vitro binding of purified GAPDH with MBP-TIA-1. In unstressed cells, TIA-1 associated with AT1R mRNA and decreased the protein expression of the receptor. The better-known function for TIA-1 is its role in the ER stress-induced translational silencing. SG assembly is regulated by one or more RBPs including TIA-1 (11,13). Interestingly, ER stress induces the dissociation of TIA-1 from AT1R mRNA. TIA-1 goes into the newly formed SGs whereas AT1R mRNA remains in the cytoplasm escaping the ER stress-induced translational suppression.
Angiotensin II and its most important receptor AT1R are part of system responsible for the adaptation of cardiovascular system to new hemodynamic environments (1). Hemodynamic and structural changes due to various stresses such as hypertension, atherosclerosis or myocardial infarction are partly mediated by AT1R (8,18–22). AT1R is upregulated by different oxidative and ER stress-inducing factors (6,9,14,15). Oxidative stress is associated with a wide variety of pathophysiologic events such as inflammation in atherosclerosis. Exposure to nonlethal level of oxidative stress leads to dissociation of GAPDH from AT1R 3′-UTR that results in a loss of GAPDH-mediated translational suppression and increase in AT1R expression both in kidney and vascular cells (9). ER stress is activated by stress-sensing serine/threonine kinases that phosphorylate eIF2α resulting in translational stalling and recruitment of e.g. TIA-1 to initiate the assembly of SGs (23).
Oxidative and ER stress are closely linked (24,25). For example, accumulation of ROS induces ER stress and unfolded protein response (UPR) by disrupting the ER Ca2+ homeostasis. Accumulation of cytoplasmic Ca2+ increases mitochondrial ROS production (26). We have previously reported GAPDH to bind the 3′ UTR of AT1R mRNA and to suppress its translation (9). This regulation was shown to be ROS dependent as H2O2 dissociated GAPDH from the AT1R mRNA, leading to increased AT1R protein expression.
During cellular stress, mRNAs are directed to SGs where they remain translationally inactive until the stress has been relieved and the mRNAs can be restored to the translational machinery (27). However, some mRNAs escape SGs and are translated despite ER stress (28,29). TIA-1 is part of stress-regulatory machinery that reprograms translation from a normal state to a stress state in order to focus its limited translational capacity to selectively express stress response genes. Although TIA-1 is predominantly in the nucleus, cytoplasmic TIA-1 binds to AT1R mRNA in unstressed cells. Triggering of the ER stress response by thapsigargin leads to TIA-1 translocation into SGs and loss of colocalization with AT1R mRNA. ER stress dissociates TIA-1 from AT1R 3′-UTR, TIA-1-mediated suppression is relieved and AT1R mRNA avoids transportation to SGs.
We found that GAPDH is required for TIA-1 binding to AT1R 3′-UTR. This is supported by affinity purification data in which silencing of GAPDH decreased the presence of TIA-1 in RNP complexes pulled down by 1–100 fragment of AT1R 3′-UTR. Secondly, partial deletion of GAPDH binding site within 1–100 of AT1R 3′-UTR blocks the binding of GAPDH as well as TIA-1. SGs comprise a number of RBPs including regulators of mRNA stability and translation such as TTP, TIA-1 and CPEB (11,30,31). We found GAPDH to be a novel SG protein that is translocated into SGs by ER stress. In contrast to TIA-1, GAPDH remains bound to AT1R mRNA while a separate pool of GAPDH is translocated to SGs by ER stress.
In summary, the ER stress-dependent association of TIA-1 with AT1R mRNA was validated using several approaches. In unstressed cells, TIA-1 decreases AT1R protein expression. In stressed cells, TIA-1 dissociates from AT1R mRNA and is targeted to SGs. AT1R protein expression is increased as AT1R mRNA avoids the ER stress-induced translational suppression and as TIA-1-mediated suppression is released. These discoveries provide comprehensive and valuable insight into the RNP complexes that govern AT1R expression at the post-transcriptional level.
Acknowledgments
We thank Ms Susanna Saarinen for expert technical assistance.
FUNDING
Finnish Cultural Foundation; Paavo Nurmi Foundation; Maud Kuistila Memorial Foundation; Ida Montin Foundation (to M.B.); Finnish Foundation for Cardiovascular Research; Sigrid Juselius Foundation (to K.K.); Finnish Medical Foundation; Instrumentarium Research Foundation (to J.L.). Funding for open access charge: Foundation for cardiovascular research.
Conflict of interest statement. None declared.
REFERENCES
- 1.de Gasparo M., Catt K.J., Inagami T., Wright J.W., Unger T. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacol. Rev. 2000;52:415–472. [PubMed] [Google Scholar]
- 2.Mehta P.K., Griendling K.K. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am. J. Physio.l Cell Physiol. 2007;292:C82–C97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 3.Pende A., Giacche M., Castigliola L., Contini L., Passerone G., Patrone M., Port J.D., Lotti G. Characterization of the binding of the RNA-binding protein AUF1 to the human AT(1) receptor mRNA. Biochem. Biophys. Res. Commun. 1999;266:609–614. doi: 10.1006/bbrc.1999.1862. [DOI] [PubMed] [Google Scholar]
- 4.Mueller C.F., Wassmann K., Berger A., Holz S., Wassmann S., Nickenig G. Differential phosphorylation of calreticulin affects AT1 receptor mRNA stability in VSMC. Biochem. Biophys. Res. Commun. 2008;370:669–674. doi: 10.1016/j.bbrc.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Nickenig G., Michaelsen F., Muller C., Berger A., Vogel T., Sachinidis A., Vetter H., Bohm M. Destabilization of AT(1) receptor mRNA by calreticulin. Circ. Res. 2002;90:53–58. doi: 10.1161/hh0102.102503. [DOI] [PubMed] [Google Scholar]
- 6.Paukku K., Backlund M., De Boer R.A., Kalkkinen N., Kontula K.K., Lehtonen J.Y. Regulation of AT1R expression through HuR by insulin. Nucleic Acids Res. 2012;40:5250–5261. doi: 10.1093/nar/gks170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diniz G.P., Takano A.P., Bruneto E., Silva F.G., Nunes M.T., Barreto-Chaves M.L. New insight into the mechanisms associated with the rapid effect of T(3) on AT1R expression. J. Mol. Endocrinol. 2012;49:11–20. doi: 10.1530/JME-11-0141. [DOI] [PubMed] [Google Scholar]
- 8.Schieffer B., Schieffer E., Hilfiker-Kleiner D., Hilfiker A., Kovanen P.T., Kaartinen M., Nussberger J., Harringer W., Drexler H. Expression of angiotensin II and interleukin 6 in human coronary atherosclerotic plaques: potential implications for inflammation and plaque instability. Circulation. 2000;101:1372–1378. doi: 10.1161/01.cir.101.12.1372. [DOI] [PubMed] [Google Scholar]
- 9.Backlund M., Paukku K., Daviet L., De Boer R.A., Valo E., Hautaniemi S., Kalkkinen N., Ehsan A., Kontula K.K., Lehtonen J.Y. Posttranscriptional regulation of angiotensin II type 1 receptor expression by glyceraldehyde 3-phosphate dehydrogenase. Nucleic Acids Res. 2009;37:2346–2358. doi: 10.1093/nar/gkp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paukku K., Kalkkinen N., Silvennoinen O., Kontula K.K., Lehtonen J.Y. p100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Res. 2008;36:4474–4487. doi: 10.1093/nar/gkn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akishita M., Ito M., Lehtonen J.Y., Daviet L., Dzau V.J., Horiuchi M. Expression of the AT2 receptor developmentally programs extracellular signal-regulated kinase activity and influences fetal vascular growth. J. Clin. Invest. 1999;103:63–71. doi: 10.1172/JCI5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatt S.R., Lokhandwala M.F., Banday A.A. Vascular oxidative stress upregulates angiotensin II type I receptors via mechanisms involving nuclear factor kappa B. Clin. Exp. Hypertens. 2014;36:367–373. doi: 10.3109/10641963.2014.943402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D., Saldeen T., Romeo F., Mehta J.L. Oxidized LDL upregulates angiotensin II type 1 receptor expression in cultured human coronary artery endothelial cells: the potential role of transcription factor NF-kappaB. Circulation. 2000;102:1970–1976. doi: 10.1161/01.cir.102.16.1970. [DOI] [PubMed] [Google Scholar]
- 16.Nickenig G., Roling J., Strehlow K., Schnabel P., Bohm M. Insulin induces upregulation of vascular AT1 receptor gene expression by posttranscriptional mechanisms. Circulation. 1998;98:2453–2460. doi: 10.1161/01.cir.98.22.2453. [DOI] [PubMed] [Google Scholar]
- 17.Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 18.Le T.H., Kim H.S., Allen A.M., Spurney R.F., Smithies O., Coffman T.M. Physiological impact of increased expression of the AT1 angiotensin receptor. Hypertension. 2003;42:507–514. doi: 10.1161/01.HYP.0000092000.07559.57. [DOI] [PubMed] [Google Scholar]
- 19.Billet S., Bardin S., Verp S., Baudrie V., Michaud A., Conchon S., Muffat-Joly M., Escoubet B., Souil E., Hamard G., et al. Gain-of-function mutant of angiotensin II receptor, type 1A, causes hypertension and cardiovascular fibrosis in mice. J. Clin. Invest. 2007;117:1914–1925. doi: 10.1172/JCI28764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D.Y., Zhang Y.C., Philips M.I., Sawamura T., Mehta J.L. Upregulation of endothelial receptor for oxidized low-density lipoprotein (LOX-1) in cultured human coronary artery endothelial cells by angiotensin II type 1 receptor activation. Circ. Res. 1999;84:1043–1049. doi: 10.1161/01.res.84.9.1043. [DOI] [PubMed] [Google Scholar]
- 21.Schieffer B., Wirger A., Meybrunn M., Seitz S., Holtz J., Riede U.N., Drexler H. Comparative effects of chronic angiotensin-converting enzyme inhibition and angiotensin II type 1 receptor blockade on cardiac remodeling after myocardial infarction in the rat. Circulation. 1994;89:2273–2282. doi: 10.1161/01.cir.89.5.2273. [DOI] [PubMed] [Google Scholar]
- 22.Nio Y., Matsubara H., Murasawa S., Kanasaki M., Inada M. Regulation of gene transcription of angiotensin II receptor subtypes in myocardial infarction. J. Clin. Invest. 1995;95:46–54. doi: 10.1172/JCI117675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedersha N., Ivanov P., Anderson P. Stress granules and cell signaling: more than just a passing phase. Trends Biochem. Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokouchi M., Hiramatsu N., Hayakawa K., Okamura M., Du S., Kasai A., Takano Y., Shitamura A., Shimada T., Yao J., et al. Involvement of selective reactive oxygen species upstream of proapoptotic branches of unfolded protein response. J. Biol. Chem. 2008;283:4252–4260. doi: 10.1074/jbc.M705951200. [DOI] [PubMed] [Google Scholar]
- 25.van der Vlies D., Makkinje M., Jansens A., Braakman I., Verkleij A.J., Wirtz K.W., Post J.A. Oxidation of ER resident proteins upon oxidative stress: effects of altering cellular redox/antioxidant status and implications for protein maturation. Antioxid. Redox Signal. 2003;5:381–387. doi: 10.1089/152308603768295113. [DOI] [PubMed] [Google Scholar]
- 26.Malhotra J.D., Kaufman R.J. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid. Redox Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 27.Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Anderson P., Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unsworth H., Raguz S., Edwards H.J., Higgins C.F., Yague E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24:3370–3380. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- 30.Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W.F., Blackwell T.K., Anderson P. MK2-induced tristetraprolin:14–3–3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]