Abstract
DNA double-strand breaks (DSBs) and their repair can cause extensive epigenetic changes. As a result, DSBs have been proposed to promote transcriptional and, ultimately, physiological dysfunction via both cell-intrinsic and cell-non-autonomous pathways. Studying the consequences of DSBs in higher organisms has, however, been hindered by a scarcity of tools for controlled DSB induction. Here, we describe a mouse model that allows for both tissue-specific and temporally controlled DSB formation at ∼140 defined genomic loci. Using this model, we show that DSBs promote a DNA damage signaling-dependent decrease in gene expression in primary cells specifically at break-bearing genes, which is reversed upon DSB repair. Importantly, we demonstrate that restoration of gene expression can occur independently of cell cycle progression, underlining its relevance for normal tissue maintenance. Consistent with this, we observe no evidence for persistent transcriptional repression in response to a multi-day course of continuous DSB formation and repair in mouse lymphocytes in vivo. Together, our findings reveal an unexpected capacity of primary cells to maintain transcriptome integrity in response to DSBs, pointing to a limited role for DNA damage as a mediator of cell-autonomous epigenetic dysfunction.
INTRODUCTION
The accumulation of DNA damage is a conserved hallmark of cancer and aging (1). Of all DNA lesions, DNA double-strand breaks (DSBs) are arguably the most harmful. Defects in DSB repair can result in cell cycle arrest, apoptosis or genomic aberrations and have been linked to both disease progression and a premature onset of aging phenotypes (2,3). Consistent with the latter, DSB induction was found to be sufficient to promote a subset of age-related pathologies in mice (4). In addition to the often detrimental effects of mutations and chromosomal abnormalities, DSBs cause significant changes in the chromatin environment both at and beyond the break site, raising the intriguing possibility that DSB repair contributes to (persistent) epigenetic defects that may eventually alter cell function (5).
While the essential role of DSB-proximal chromatin reorganization in regulating repair factor access and function is well established (6,7), its potential impact on transcriptome integrity is only beginning to be understood. Many DSB-induced chromatin changes, such as the phosphorylation of histone H2AX (γ-H2AX), are restored to the pre-damage state once the break is repaired, which has led to an access (or prime)-repair-restore model of chromatin-directed DSB repair (8,9). Consistent with this, recent findings demonstrate transcriptional repression in cis to DSBs, which depends on DNA damage signaling and ceases after DSB repair is complete (10–13). This work is, however, limited to highly proliferative tumor cell lines, and its implications for animal tissues, which often exhibit little or no proliferation, remain unclear. Moreover, contrasting these transient effects, persisting DNA damage-induced chromatin changes have been observed in cells that undergo irreversible proliferation arrest, or senescence. The latter are thought to depend on the continued presence of DNA lesions, suggesting that unrepaired DNA damage may result in chronic epigenetic deregulation, particularly in the context of age-related cellular decline (14–17). Remarkably, persistent H2AX phosphorylation was recently observed in the absence of ongoing DNA damage in quiescent old hematopoietic stem cells (HSCs) (18). The initial formation of γ-H2AX was attributed to replication stress triggered by HSC expansion and its continued presence had a significant impact on rDNA expression and ribosome biogenesis. Beyond break-proximal chromatin reorganization, DNA damage has been linked to a redistribution of DNA repair-relevant chromatin modifiers to sites of damage, which can result in the epigenetic deregulation of ostensibly undamaged genomic loci normally controlled by these proteins (19). Together, these findings highlight the potential impact of DSB formation on the control of gene expression programs, which has in turn been proposed to contribute to the aging process (5).
It is of note that epigenetic dysfunction in a small subset of cells may be sufficient to affect entire tissues, and possibly organismal aging, as evidenced by the systemic impact of aberrantly expressed inflammatory cytokines derived from senescent cells (senescence-associated secretory phenotype) (20). Indeed, DNA damage-driven inflammatory responses have been linked to aging in several organisms (4,21). The distinction between cell-intrinsic and systemic consequences of DSB induction is, thus, critical to advance our understanding of the role of DSBs in age-associated functional decline. However, despite numerous cell-based reporter systems for DSB induction, there is a scarcity of tools to follow the consequences of DSBs for cell and tissue function in higher organisms. Here, we generated a transgenic mouse model, which allows for both temporally and spatially controlled DSB induction at a defined number of genomic loci, as well as the fluorescence-based detection and isolation of cells exposed to DSBs. We interrogate the impact of DSB formation and repair on transcriptional integrity at and beyond DSB sites in vivo and reveal an unexpected capacity of primary cells to maintain transcriptome integrity in response to DSBs.
MATERIALS AND METHODS
Generation of PpoSTOP knock-in mice and mouse analyses
An improved estrogen receptor nuclear translocation domain (ERT2) with N-terminal HA tag was cloned upstream of the I-PpoI cDNA (kind gift from M. Kastan). The resulting HA-ERT2-I-Ppo-I cDNA was inserted into the STOP-eGFP-Rosa26 targeting vector (Addgene plasmid 11739, (22)) upstream of the IRES-eGFP cassette. Gene targeting was performed in C57BL/6 Bruce4 ES cells as described previously (22). Neomycin-resistant ES cells were analyzed for correct transgene integration by Southern Blot analysis of EcoRI digested genomic DNA using a 5′ Rosa26 probe. The resulting knock-in allele is referred to as PpoSTOP. Targeted ES cells were injected into C57BL/6 albino (cBRD/cBRD) blastocysts and chimeric males were crossed to C57BL/6 females. PpoSTOP/+ mice were bred to lck-Cre transgenic mice for T lineage-specific ERT2-I-PpoI expression. To induce nuclear translocation of ERT2-I-PpoI, PpoSTOP/+; lck-Cre mice were subjected to 2–4 intraperitoneal injections of 1 mg TAM (Sigma, resuspended in corn oil) at 24 h intervals. Animals were analyzed 4 h after the final TAM injection.
In vitro T cell culture
Naïve splenocytes were enriched for CD4+ T cells by negative selection using the EasySep Mouse CD4+ T-cell isolation Kit (Stem Cell Technologies) according to the manufacturer's instructions. CD4+ T cells were then maintained in phenol-free RPMI 1640 medium supplemented with 10% charcoal stripped-FBS (Gemini Bioproducts), 1x Non-Essential Amino Acids, 1x Vitamin solution, 1 mM sodium pyruvate, 10 mM HEPES, penicillin/streptomycin (all Invitrogen) and 50 μM β-mercaptoethanol (Sigma) for 24 h before DSB induction. Medium was supplemented with 2 ng/ml IL-7 (R&D Systems). For DSB induction, cultures were treated with 1 μM 4-OH-TAM for 3 h, washed and maintained in medium for the indicated time frames. For DDR inhibition, T cells were treated with 20 μM ATMi (Ku-55933, Calbiochem) and 10 μM DNA-PKi (NU7026, R&D Systems) or DMSO starting 1 h before DSB induction. For BrdU labeling, cultured CD4+ T cells were treated for 6 h with 10 μM BrdU.
Immunophenotyping and cell sorting
Single cell suspensions of nucleated cells from thymus or spleen were stained using antibodies against CD4, CD8, CD5, CD19 or IgM (eBiosciences) and analyzed for GFP expression in the respective subsets, dead cells were identified using 7-AAD and excluded from the analysis. To detect apoptosis, cells were stained with Annexin V and 7AAD (BD Pharmingen). To detect DNA damage, cells were stained for cell surface markers, fixed and permeabilized using Cytofix/Cytoperm solution (BD Pharmingen), followed by intra-cellular staining for γ-H2AX (Cell Signaling, Novus Biologicals), dead cells were excluded using the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Life technologies). For BrdU analyses, cells were fixed, DNase-treated and stained with anti-BrdU-FITC and 7AAD (BD Pharmingen). FACS acquisition was performed on FACS Calibur or LSRII flow cytometers (Becton Dickinson) and cell sorting on the FACS Aria II (Becton Dickinson). FACS data were analyzed using Flowjo software (Tree Star, Inc.).
Immunofluorescence
Cells were plated onto poly-L-lysine coated coverslips and fixed in 4% paraformaldehyde (PFA) in PBS. Fixed cells were permeabilized with 0.5% TritonX-100 and blocked in 3% BSA. Two-step immunostaining was performed in 1% BSA in PBS using α-phospho-S139-H2AX (γ-H2AX, Millipore) and α-mouse Alexa Fluor 568 antibodies (Life Technologies). Nuclei were counter-stained with 5 μg/ml Hoechst 33342 (Life Technologies). Confocal z-stacks (0.3 μm z-resolution) were taken with a Zeiss LSM780 microscope. Mean nuclear intensities from maximum intensity projections were measured using ImageJ.
Western blotting
Total thymocytes were lysed in RIPA buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) in the presence of 1x complete mini protease inhibitor mix (Roche) on ice. Ten micrograms of total protein from each sample was resolved on a 10% SDS-PAGE and transferred to PVDF membrane. The blots were then probed with antibodies against HA or GAPDH (Santa Cruz).
Analysis of I-PpoI cutting efficiency
Genomic DNA from the indicated cell subsets was analyzed by qPCR using I-PpoI site spanning primers (see Supplementary Table S1). Data were normalized to the relative DNA content based the average Ct value (Ctcontrol) of three non-I-PpoI-associated genomic DNA segments (Chr2-Ppp1r1c, Chr6-Hoxa9, Chr7-H19-ICR, see Supplementary Table S1). The relative amount of intact I-PpoI spanning DNA was calculated as x = 2−ΔCt, where ΔCt = CtI-PpoI – Ctcontrol. Values from PpoSTOP/+, lck-Cre mice were then normalized to lck-Cre controls.
RNA analysis
Total RNA was extracted using the RNeasy Mini Kit according to the manufacturer's instructions (Qiagen). cDNA was synthesized from 0.2–1 μg of total RNA using the ThermoScript RT-PCR system (Invitrogen) and expression of the indicated genes was analyzed by qRT-PCR using a LightCycler 480 II (Roche), see Supplementary Table S1 for primer sequences.
Mutation analysis
Genomic DNA from FACS-sorted CD4+ T cells or CD19+ B cell was extracted using DNeasy Blood & Tissue kit (Qiagen). PCR amplification of a 1 kb fragment flanking the chromosome 17 I-PpoI site was performed using primers listed in Supplementary Table S1. PCR products were sub-cloned using the TOPO-TA Cloning Kit (Invitrogen), followed by sequencing of plasmid DNA.
RNA sequencing and analysis
Total RNA from FACS-sorted CD4+ splenic or thymic T cells and IgM+ splenic B cells from female littermates was extracted using the RNeasy Mini Kit (Qiagen). For library construction, 0.1–1 μg of total RNA was subjected to mRNA capture with oligo-dT coated magnetic beads. The mRNA was fragmented, followed by random-primed cDNA synthesis, standard Illumina library prep with end-repair, adapter ligation and PCR amplification. Libraries were sequenced on a HiSeq2000 instrument with TruSeq v3.0 chemistry (Illumina, San Diego, CA, USA) for 101 cycles in paired-end mode. Reads were mapped to the mouse reference genome (mm10) using TopHat v.2.0.11 (23) with Bowtie2 v.2.2.1 (24). The UCSC transcripts for mm10 from iGenomes (Illumina, San Diego, CA) were used for all analyses. TopHat alignments were performed supplying the gene models (–G option), and reporting one alignment per read (–g 1 option). A summary of experiments and read depth is provided in Supplementary Table S2. Gene level expression estimates in units of expected Fragments per Kilobase per Million Mapped Reads (FPKM) were calculated with Cuffdiff v.2.1.1 (25) using default parameters. Cuffdiff tests the hypothesis of a significant difference in the number of fragments originating from one gene compared to another, in excess of the estimated fragment count variance in terms of dispersion, P-values < 0.01 as reported by Cuffdiff were used to define differential expression. All samples were treated as replicates of a single global condition to estimate dispersion. Low abundance transcripts (FPKM < 1 in both samples), genes annotated on chrY or unplaced assembly genes (chromosome unknown) were excluded from the analysis, Complete gene level abundance estimates and differential expression results are provided in Supplementary Table S3. Functional enrichments within lists of differentially expressed genes were identified with the DAVID tool (26) (Supplementary Table S4).
I-PpoI cut site identification
The canonical I-PpoI endonuclease cut site is CTCTCTTAAGGTAGC. As set of 25 degenerate cut sites was identified based on variants with only modest differences in cutting efficiency compared to the wild-type cut site (27,28). Genomic locations of cut sites were mapped using Fuzznuc (EMBOSS, (29)). In-house perl scripts were used to convert Fuzznuc output to browser extensible data (BED) format (Supplementary Table S5). Chromosome ideograms were drawn with the Idiographica tool (30). BEDTools (31) was used for coordinate intersections between cut site locations and gene annotation.
RESULTS
A mouse model for endonuclease-mediated DSB induction at endogenous genomic loci
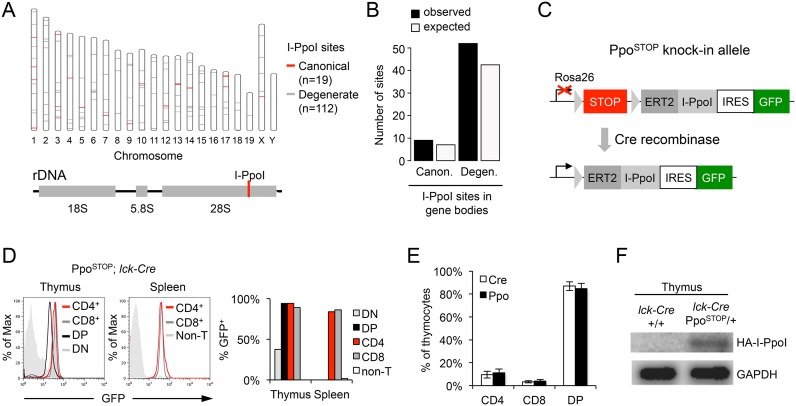
To induce locally defined DSBs in mice, we took advantage of the fungal homing endonuclease I-PpoI. I-PpoI targets a 15 bp DNA motif and has been used successfully to promote DSB formation in human cell lines (12,32). The mouse genome contains 19 bona fide I-PpoI sites and at least 112 degenerate sites that exhibit comparable I-PpoI-mediated cutting in vitro (27,28). I-PpoI sites are distributed randomly, occurring with expected frequencies in genic and intergenic regions. One canonical site is located within the 28S rDNA coding region within the repetitive 45S rDNA locus (Figure 1A and B, Supplementary Table S5). To temporally control I-PpoI activity in vivo, the I-PpoI cDNA was fused to a 4-hydroxy-tamoxifen (4OH-TAM)-responsive, modified estrogen-receptor nuclear translocation domain (ERT2, (33)). The ERT2-I-PpoI cDNA was placed under the control of a loxP-flanked transcriptional STOP cassette to allow for Cre recombinase-dependent, tissue-specific ERT2-I-PpoI expression (22). An internal ribosomal entry site (IRES) followed by an eGFP coding region downstream of the ERT2-I-PpoI cDNA allows for the identification and purification of I-PpoI expressing cells based on GFP signal. A single copy of the resulting PpoSTOP transgene was inserted into the mouse genome under the control of the ubiquitously expressed Rosa26 promoter by gene targeting (Figure 1C).
Figure 1.
Mouse model for I-PpoI-mediated DSB induction. (A) Map of canonical (red) and degenerate I-PpoI sites (gray) across the mouse genome. (B) Observed and expected distribution of I-PpoI sites in gene bodies. (C) Schematic of the inducible PpoSTOP knock-in allele targeted downstream of the mouse Rosa26 locus promoter. Triangles depict loxP sites, STOP represents a transcription terminator cassette, which is removed in the presence of Cre recombinase. (D) GFP expression in T cells from PpoSTOP, lck-Cre mice. Histograms depict GFP intensity in the indicated T cell subsets, DP: CD4+CD8+ double-positive, DN: CD4−CD8− double-negative, Non-T: non-T cells. The fraction of GFP positive cells in each subset is shown in the bar graph. (E) Frequency of thymic T cell subsets in PpoSTOP, lck-Cre mice (n = 4, gated on GFP+ cells) and lck-Cre controls (n = 7). (F) Western blot for HA-I-PpoI expression in the thymus of the indicated mouse strains. GAPDH served as loading control.
To investigate the consequences of DSB induction in a cell population that can be readily isolated and manipulated, heterozygous PpoSTOP/+ mice were crossed to the T lineage-specific lck-Cre transgene (34). Lck-Cre transgenic mice initiate Cre expression during early T cell development within the CD4/CD8 double-negative (DN) stage. Consistent with this, GFP expression was detected in ∼40% of PpoSTOP/+; lck-Cre DN thymocytes and reached > 90% in the more differentiated CD4/CD8 double-positive (DP) and CD4+ or CD8+ T cell subsets. No GFP+ cells were detected in non-T lineage cells or lck-Cre control mice (Figure 1D and data not shown). The distribution of GFP+ thymic T cell subsets was comparable to lck-Cre controls, indicating that expression of the PpoSTOP transgene does not significantly alter T cell development (Figure 1E). Thymic expression of the ERT2-I-PpoI protein was confirmed by Western blot (Figure 1F).
PpoSTOP mice allow for spatially and temporally controlled DSB formation in vivo
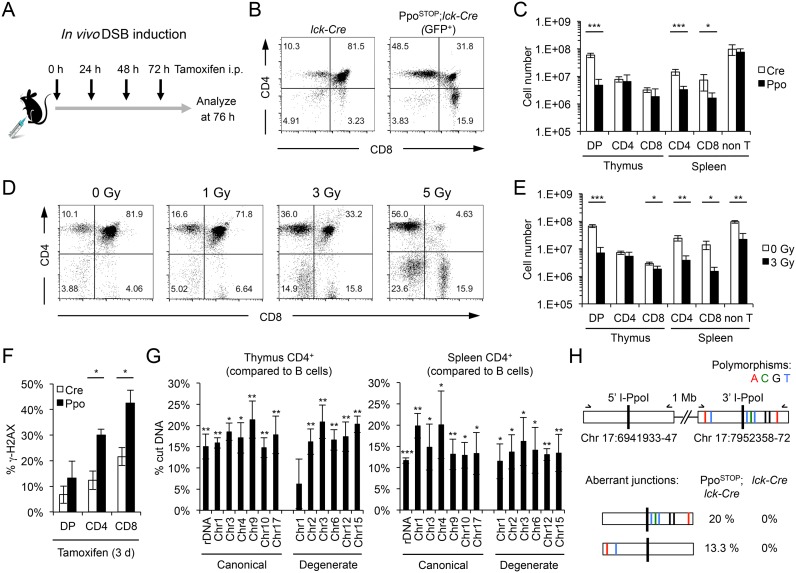
To determine the efficiency of DSB induction, PpoSTOP; lck-Cre mice and lck-Cre controls were treated with TAM for three consecutive days (Figure 2A). The same feeding regimen resulted in robust activation of an ERT2-containing reporter transgene (ERT2-Cre) in mouse thymocytes (Supplementary Figure S1, (35)). In agreement with previous reports demonstrating reduced viability of immature DP as well as mature CD4+ or CD8+ T cells in response to genotoxic stress (36–38), I-PpoI activation resulted in a loss of GFP+ cells in these subsets that was comparable to ∼3 Gy of ionizing radiation (IR). No reduction in non-T cell numbers was detected in PpoSTOP; lck-Cre mice, consistent with T cell-specific DNA damage induction (Figure 2B–E). Moreover, I-PpoI induction caused a pronounced increase in γ-H2AX+ T cells compared to lck-Cre controls (Figure 2F, Supplementary Figure S2A). Notably, DSB levels were compatible with cell survival in a sizeable fraction of T cells, particularly within the more mature CD4+ and CD8+ subsets. To investigate the extent of DSB induction in these cells, DNA from PpoSTOP; lck-Cre or lck-Cre control T cell subsets was subjected to DSB-spanning qPCR analysis. Live T cells were isolated by flow cytometry based on CD4/CD8 surface marker expression and, in the case of PpoSTOP; lck-Cre mice, expression of GFP. Unique I-PpoI spanning primer sets were generated for 12 randomly selected canonical or degenerate I-PpoI sites, as well as for the I-PpoI site within the 28S rDNA. All tested I-PpoI sites were subject to DSB formation, with comparable cutting efficiencies for canonical and degenerate sites ranging from 10% to 20% (Figure 2G, Supplementary Figure S2B, C). Notably, I-PpoI-mediated DSB induction was similar after two or three days of TAM treatment (Supplementary Figure S2D), suggesting a constant rate of DSB formation and repair.
Figure 2.
Efficiency of I-PpoI-induced DSB formation in vivo. (A) Schematic of TAM-mediated I-PpoI induction. Black arrows indicate time points of TAM injection. (B) Thymic CD4/CD8 T cell profiles in GFP+ PpoSTOP, lck-Cre mice and lck-Cre controls after 3 days of TAM treatment. A representative pair is shown. (C) Total numbers of indicated cell subsets from mice in (B), n = 4 mice per group. (D) Thymic CD4/CD8 T cell profiles 24 h after increasing doses of ionizing radiation (IR). (E) Total numbers of indicated cell subsets from mice in (D, 3 Gy), n = 3 mice per group. Unless noted otherwise, P-values are based on Student's two-tailed t-test, *P < 0.05, **P < 0.01, ***P < 0.001. (F) Fraction of γ-H2AX+ cells in the indicated thymic T cell subsets. TAM was injected as in (A), Mean and SEM are from three independent experiments. (G) Cutting efficiency of canonical and degenerate I-PpoI sites in FACS-sorted thymic and splenic CD4+ T cells were calculated based on the reduction in I-PpoI site abundance compared to three non-I-PpoI associated genomic control loci (see Supplementary Figure S2B for details). The DSB-induced increase in cut I-PpoI sites in T cells relative to B cells is shown. Mean and S.E.M. are from at least three independent experiments. Average cutting efficiencies were 17.2 ± 2.3% for canonical versus 16.2 ± 5.3% for degenerate sites in the thymus, and 15.0 ± 3.4% versus 13.6 ± 1.5% in the spleen. P-values are in relation to splenic B cells. (H) Mutation analysis of I-PpoI flanking DNA from splenic CD4+ T cells. A pair of neighboring and highly homologous I-PpoI sites on chromosome 17 was amplified and sequenced. Annotated polymorphisms in the downstream I-PpoI site are indicated. The frequency of aberrant 5′-3′ hybrid junctions is shown for the PpoSTOP, lck-Cre and lck-Cre genotypes (n = 15 clones per sample).
Based on previous work revealing a half life of ∼2–4 h for the majority of IR-induced DSBs in mouse lymphocytes (39), we estimate that, at a DSB frequency of 10–20%, most I-PpoI sites will be cut at least once per cell over the course of the experiment. As a proxy for previous DSB formation and repair, we sequenced genomic DNA derived from an I-PpoI site on chromosome 17 to assess break/repair-associated mutagenesis following three days of I-PpoI induction. In agreement with efficient, error-free religation of clean I-PpoI breaks, no point mutations were detected at the DSB site (40). However, 33% of break-spanning DNA segments yielded a chimeric DNA sequence, in which one end of the I-PpoI-flanking DNA was joined to that of a second, polymorphic I-PpoI site located ∼1 Mb downstream. No evidence for aberrant junctions was observed in break-flanking DNA from lck-Cre controls, demonstrating I-PpoI-dependent formation of these distal fusions (Figure 2H). Given that the joining of proximal DSB ends is considerably more efficient than that of more distant ones (41), we expect the fraction of alleles that have undergone DSB formation and repair to be significantly greater than 33%. Together, these findings demonstrate that the PpoSTOP allele allows for efficient, temporally controlled and tissue-specific DSB induction in mice.
DSB induction reveals transcriptome stability with few DDR-related gene expression changes
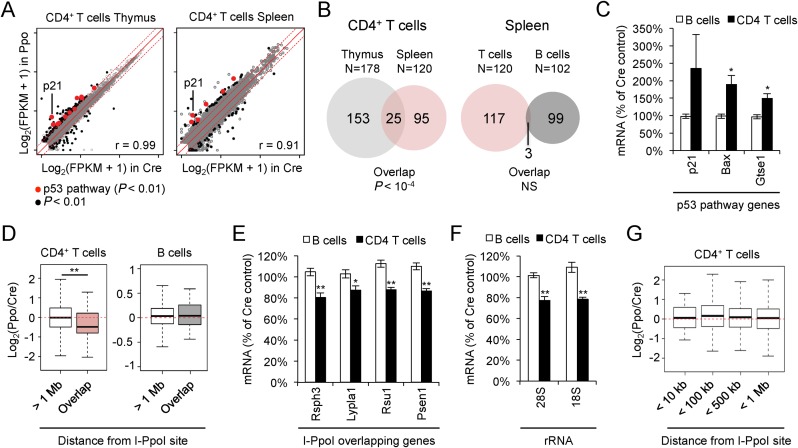
We next used this animal model to investigate the consequences of DSBs for transcriptional integrity. Following three days of TAM treatment, CD4+ T cells from PpoSTOP; lck-Cre mice and lck-Cre controls were FACS-purified (>98% CD4+, Supplementary Figure S3A) and subjected to RNA sequencing (RNA-Seq). Analyses were limited to cells surviving I-PpoI-mediated DSB formation (live cells), and splenic B cells isolated from the same animals served as a negative control. Notably, RNA-seq profiles showed a strong correlation between control and I-PpoI-expressing T cells (Figure 3A), and the number of genes that were significantly altered upon I-PpoI induction was only moderately larger than the non-DSB-related variation observed between B cells from PpoSTOP; lck-Cre mice and lck-Cre controls (Figure 3B, Supplementary Table S3). Remarkably, however, 21% of genes altered in I-PpoI-expressing T cells isolated from the spleen were also deregulated in the thymus (P < 10−4, Figure 3B). Pathway analysis of this subset revealed ‘p53 signaling’ (mmu04115) as the top overrepresented KEGG pathway (P = 8.5 × 10−5), and ‘response to DNA damage stimulus’ (GO:0006974) as well as ‘induction of apoptosis by intracellular signals’ (GO:0008629) as significantly enriched gene ontology terms (Figure 3A, Supplementary Table S4). I-PpoI-induced gene expression changes were validated for a subset of p53 target genes by quantitative RT-PCR (qRT-PCR). No activation of DNA damage response genes was observed in splenic B cells from PpoSTOP; lck-Cre mice and, accordingly, no significant overlap was detected between genes altered in B and T cells (Figure 3B, C, Supplementary Figure S3B). These findings demonstrate that DSB induction results in relatively few gene expression changes in vivo, many of which are DDR-related.
Figure 3.
Impact of I-PpoI induction on gene expression. (A) Correlation of gene expression patterns in PpoSTOP, lck-Cre (Ppo) and lck-Cre control (Cre) CD4+ thymic T cells (left) and CD4+ splenic T cells (right). Significantly altered genes are shown in black (P < 0.01, see Methods), genes in the KEGG pathway ‘p53 signaling’ are shown in red (P < 0.01). Dashed lines depict 2-fold changes. (B) Venn diagram depicting overlap between differentially expressed genes in thymic and splenic CD4+ T cells (left), or splenic CD4+ T cells and B cells (right). (C) Validation of I-PpoI induced changes in the expression of select p53 target genes in sorted splenic B or T cells. mRNA was normalized to GAPDH, β-actin and Rpl13a. Shown are I-PpoI-induced changes in mRNA levels relative to Cre controls. Mean and SEM are from four independent experiments. (D) DSB-associated gene expression changes in I-PpoI-expressing cells compared to Cre control cells. I-PpoI-overlapping genes and I-PpoI-distal genes (>1 Mb) are shown for the indicated lymphocyte subsets. Boxplots show median, 25th and 75th percentile, whiskers extend to 1.5 times the interquartile range. **P < 0.01, two-sample Wilcoxon test. (E) I-PpoI-induced gene expression changes at I-PpoI-overlapping genes were analyzed by qRT-PCR and normalized as in (C). Rsph3a overlaps a Chr. 17 canonical I-PpoI site, Lypla1, Rsu1 and Psen1 overlap degenerate I-PpoI sites on Chr. 1, Chr. 2 and Chr. 12, respectively, see Figure 2G for cutting efficiencies. Mean and SEM are from the same four independent experiments used in (C). (F) I-PpoI-induced changes in rRNA expression were analyzed by qRT-PCR as in (E) for 18S and 28S rRNAs, both derived from a single 45S precursor. (G) Gene expression changes in I-PpoI-flanking genes, binned by distance from the break site. I-PpoI-expressing CD4+ T cells were compared to Cre control cells. Boxplots are as in (D).
DSBs induce cell-autonomous repression of DSB-bearing but not DSB-flanking genes in vivo
In addition to the activation of bona fide DDR targets, DSBs were previously shown to interfere with the expression of break-proximal genes in tumor cell lines (11–13,42). We, thus, sought to assess the impact of DSB induction on the expression of genes with intragenic I-PpoI sites in vivo. While the extent of I-PpoI-induced gene deregulation did not reach significance in individual genes, the expression of all I-PpoI-bearing genes combined showed a significant decrease when compared to genes >1 Mb away from DSBs (∼28% median reduction, Figure 3D, Supplementary Figure S3C). No DSB-associated reduction in gene expression was observed for I-PpoI-containing genes in splenic B cells from the same animals (Figure 3D, Supplementary Figure S3C). I-PpoI-induced gene expression changes were independently validated using qRT-PCR, focusing on four abundantly expressed I-PpoI-containing genes. In agreement with the RNA-Seq results, upon DSB induction, all four genes exhibited a 10–20% reduction in mRNA levels in I-PpoI transgenic T cells compared to lck-Cre controls (Figure 3E). Similar results were observed for I-PpoI-induced DSBs at the rDNA, extending earlier in vitro findings (11) (Figure 3F). The I-PpoI-induced decrease in gene expression correlated well with the fraction of DSB-containing alleles (Figure 2G), suggesting that DSB-induced gene deregulation does not persist following repair of the lesion. Notably, and in contrast to I-PpoI-containing genes, I-PpoI flanking genes within up to 1 Mb of the break site showed expression patterns comparable to non DSB-associated genes (Figure 3G, Supplementary Figure S3C). Together, these findings reveal that, although moderate transcriptional changes can be detected in DSB-bearing genes, persistent DSB formation and repair is associated with a surprisingly stable transcriptome in vivo.
DSB-induced gene repression in primary cells is reversible and depends on DNA damage signaling
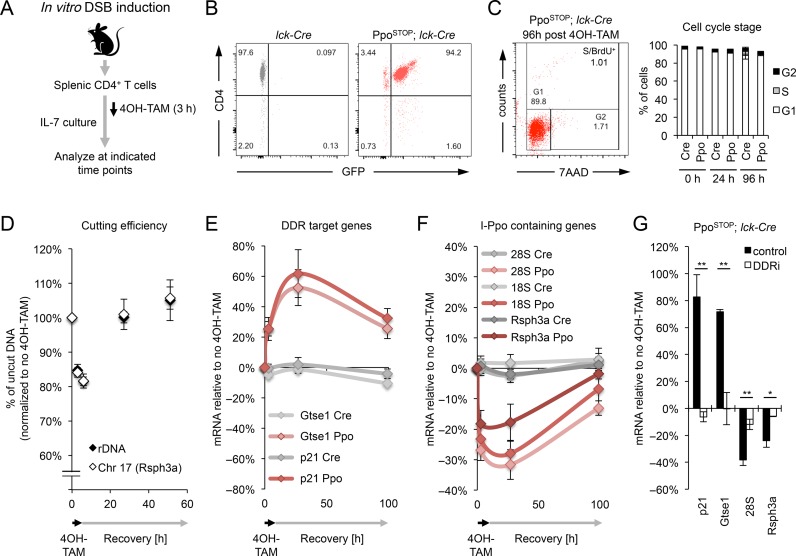
Our in vivo data collectively suggest a dynamic restoration of I-PpoI-associated gene expression changes in mature, resting T cells. However, DSB repair kinetics and its consequences for transcriptome integrity have yet to be investigated in non-dividing cells. Taking advantage of our mouse model, we, therefore, established an in vitro system, which allows for the transient activation of I-PpoI in ex vivo isolated primary CD4+ T cells, employing culture conditions that promote homeostatic T cell survival without inducing proliferation (Figure 4A–C) (43). To monitor DSB repair over time, cultured T cells from PpoSTOP; lck-Cre mice or lck-Cre controls were treated with 4OH-TAM for 3 h, followed by a recovery period of up to 4 days (see Figure 4A). DNA and RNA were collected at several time points both during and after I-PpoI induction. The efficiency of DSB induction in vitro was similar to that observed in vivo as measured by both I-PpoI site cutting (15–20% of a given site) and γ-H2AX induction (∼60% of cells), and break repair was completed within 24–48 h, in agreement with previous observations (39) (Figure 4D, Supplementary Figure S4). While 12 of 13 genomic I-PpoI sites analyzed were restored with largely comparable kinetics, one DSB site showed a reproducible delay in repair (Supplementary Figure S4A), consistent with a role for (epi)genomic context in modulating DSB repair efficiency (44–47). DNA damage-induced activation of the p53 targets p21 and Gtse1 peaked at 24 h, indicating prolonged activation of downstream DDR effectors (Figure 4E). In line with our in vivo findings, we observed a concomitant loss of I-PpoI-proximal gene expression. Notably, expression of the latter returned to near pre-damage levels coincident with subsiding DDR target gene activation (Figure 4E, F). DSB-induced, dynamic gene deregulation did not require cell division (Figure 4C), underlining its physiological relevance beyond highly proliferative tumor cell lines (10–13). Importantly, while we detect an increase in apoptosis coincident with maximal DDR activation 24 h after I-PpoI induction, the number of live cells only modestly decreased between 24 and 96 h after 4OH-TAM, excluding continued cell loss as a predominant cause for restored gene expression (Supplementary Figure S5A, B). To determine if DSB-associated gene deregulation is directly affected by DNA damage signaling, we performed 4OH-TAM treatment in the presence or absence of inhibition of the DDR mediators ATM and DNA-PK. Consistent with impaired DDR activation, ATM/DNA-PK inhibition abrogated the activation of p21 and Gtse1 despite comparable I-PpoI mediated DSB induction (Figure 4G, Supplementary Figure S5C). Importantly, inhibition of DDR signaling also rescued the reduction in DSB-proximal gene expression (Figure 4G). Together, these findings demonstrate that DSB-induced transcriptional changes in primary cells are transient/reversible and depend on DDR activation.
Figure 4.
I-PpoI induction in vitro reveals transient, DDR-dependent gene repression in primary cells. (A) Schematic for DSB induction in purified splenic CD4+ T cells in vitro. 4OH-TAM was added for 3 h and removed thereafter. (B) FACS analysis of cultured T cells indicating CD4+ T cell purity and GFP expression in cells from lck-Cre (grey) and PpoSTOP; lck-Cre mice (red). (C) Cell cycle profiles of cultured T cells based on DNA content (7AAD) and BrdU incorporation. A representative experiment is shown, mean and S.D. are from three (Cre) or two (Ppo) independent in vitro cultures. (D) Cutting efficiency of I-PpoI sites analyzed in (E, F). Genomic DNA was isolated at the indicated time points during and after I-PpoI induction. Shown is the fraction of uncut DNA compared to lck-Cre controls. Mean and SEM are from at least 4 independent in vitro cultures. (E, F) qRT-PCR analysis of I-PpoI-induced changes in the expression of DDR target genes (E) and I-PpoI-containing genes (F) at the indicated time points after I-PpoI induction in PpoSTOP; lck-Cre (Ppo) and lck-Cre (Cre) control T cells. Samples were normalized to GAPDH, β-actin and Rpl13a. Expression changes are relative to no 4OH-TAM. Mean and S.E.M. are from five independent in vitro cultures. (G) qRT-PCR analysis of I-PpoI-associated gene expression changes in PpoSTOP; lck-Cre T cells in the presence or absence of ATM/DNA-PK inhibition (DDRi) at time of maximal I-PpoI effects (based on E, F). Mean and S.E.M. are from three independent in vitro cultures.
DISCUSSION
DSBs are known to promote a dynamic reorganization of break-proximal chromatin, with the potential to affect the expression of both break-proximal and ostensibly undamaged genes (10,13,19,45,48–50). However, the contribution of DSBs to epigenetic dysfunction in vivo remains unclear to date. Using a mouse model for both spatially and temporally controlled DSB induction, we show here that, at DNA damage levels compatible with cell survival, the mammalian transcriptome is sufficiently robust to accommodate continued DSB exposure, even in the absence of cell division. Our findings further suggest that DSB repair is necessary and sufficient to ensure the maintenance and/or restoration of break-proximal gene expression profiles and, by extension, epigenetic integrity in vivo.
Among the relatively small number of genes we found to be significantly deregulated in response to DSBs, a significant fraction could be attributed to DNA damage signaling (Figure 3A–C). Notably, the proximity of genes to DSBs did not predictably alter their expression, with the exception of DSB-bearing genes (Figure 3D, E) (12). This observation is consistent with the finding that γ-H2AX does not propagate on active genes and that transcription can be accurately maintained within γ-H2AX domains (49). However, a contribution of DSB-induced epigenetic reorganization of undamaged loci, perhaps due to the DSB-induced redistribution of chromatin modifiers, cannot be excluded at this time (19). Consistent with cell-intrinsic epigenetic deregulation being a minor consequence of continued DSB exposure in vivo, a recent study shows that DNA damage-induced, age-associated functional decline can be attributed in large part to systemic consequences of DSBs, including cell death, tissue atrophy and the ensuing, non-cell-autonomous inflammatory response (4).
Despite remarkably stable gene expression at the population level, we provide evidence for DDR-dependent, transient transcriptional repression of break-proximal genes in DSB-bearing primary cells (Figures 3D and 4F). The latter is in agreement with several reports implicating the DDR mediators ATM and DNA-PK in DSB-proximal repressive chromatin formation as well as transcriptional silencing (9–13,50). Importantly, we show here that the restoration of DSB-induced gene expression changes can occur in the absence of proliferation, ruling out a specific requirement for S phase progression and, by extension, homology-directed repair in this process. Together with previous work in cycling cells (10–13), our findings, thus, suggest that most tissues have the potential to restore DSB-induced gene deregulation, although the kinetics of DSB repair and DNA damage-associated gene silencing may differ depending on cell type and/or cell cycle state.
If DSB repair is impaired, as has been reported in aged cells (51,52), DNA damage may result in prolonged gene repression, with potentially harmful consequences for cell function. The latter is of particular relevance for rDNA-associated DSBs and the ensuing reduction in rRNAs, since the regulation of mRNA translation represents a conserved mechanism of longevity control (53). Depletion of ribosome subunits or interference with the target of rapamycin/ribosomal protein S6 kinase pathway, which functions to activate protein translation, were found to alter the lifespan of a variety of species including mammals (54–56). Notably, while I-PpoI-induced DSBs are artificial in nature, the rDNA is naturally prone to DSB formation, as it presents a unique challenge to DNA replication due to frequent non-canonical palindrome structures as well as canonical tandem repeats (57,58). Consistent with this, it was recently reported that replication stress can result in the persistent formation of rDNA-associated γ-H2AX foci and a concomitant decrease in rRNA/ribosome biogenesis in quiescent old HSCs (18). Together with our work, these findings raise the intriguing possibility that a DNA damage-associated reduction in rRNA availability may, over time, interfere with efficient mRNA translation and thereby affect cell function and aging.
It is of note that stochastic variations in gene expression in aged tissues are thought to be a contributing factor to age-associated organ decline even if gene expression across the tissue remains unaltered (59). The transient repression of DSB-proximal genes observed here may at least in part account for this phenomenon. Moreover, although our results do not point to epigenetic deregulation as a major consequence of short-term DSB-induction (days) in vivo, we cannot rule out that chronic exposure to DNA damage as observed during aging (years) may result in DSB-associated chromatin changes over time. Finally, DNA damage-induced gene deregulation may be an important contributor in cell subsets with persisting DNA lesions, such as senescent cells (4,14–16,60). We anticipate that the mouse model presented here will greatly facilitate future studies of both cell-autonomous and cell-non-autonomous cellular changes and organ pathologies in the context of DNA damage and aging.
ACCESSION NUMBER
Sequencing data are publicly available in the Gene Expression Ominbus (GEO) under accession GSE74855.
Supplementary Material
Acknowledgments
We thank Tom Misteli and Andre Nussenzweig for critical reading of the manuscript, Kathy McKinnon for help with flow cytometry and cell sorting, Bao Tran and the CCR Sequencing Core for RNA Seq, Elena Kuznetsova for animal care and Michael Kastan for the I-PpoI construct. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH); NCI; Center for Cancer Research. Funding for open access charge: Intramural Research Program of the National Institutes of Health (NIH); NCI; Center for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombard D.B., Chua K.F., Mostoslavsky R., Franco S., Gostissa M., Alt F.W. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 4.White R.R., Milholland B., de Bruin A., Curran S., Laberge R.M., van Steeg H., Campisi J., Maslov A.Y., Vijg J. Controlled induction of DNA double-strand breaks in the mouse liver induces features of tissue ageing. Nat. Commun. 2015;6:6790. doi: 10.1038/ncomms7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberdoerffer P., Sinclair D.A. The role of nuclear architecture in genomic instability and ageing. Nat. Rev. Mol. Cell Biol. 2007;8:692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- 6.Shi L., Oberdoerffer P. Chromatin dynamics in DNA double-strand break repair. Biochim. Biophys. Acta. 2012;1819:811–819. doi: 10.1016/j.bbagrm.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smeenk G., van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu. Rev. Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- 8.Smerdon M.J. DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 9.Soria G., Polo S.E., Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruhlak M., Crouch E.E., Orlov M., Montano C., Gorski S.A., Nussenzweig A., Misteli T., Phair R.D., Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 12.Pankotai T., Bonhomme C., Chen D., Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 13.Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbig U., Ferreira M., Condel L., Carey D., Sedivy J.M. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 15.Rodier F., Munoz D.P., Teachenor R., Chu V., Le O., Bhaumik D., Coppe J.P., Campeau E., Beausejour C.M., Kim S.H., et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sedelnikova O.A., Horikawa I., Zimonjic D.B., Popescu N.C., Bonner W.M., Barrett J.C. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 17.Di Micco R., Sulli G., Dobreva M., Liontos M., Botrugno O.A., Gargiulo G., dal Zuffo R., Matti V., d'Ario G., Montani E., et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat. Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C., et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S.K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coppe J.P., Patil C.K., Rodier F., Sun Y., Munoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ermolaeva M.A., Segref A., Dakhovnik A., Ou H.L., Schneider J.I., Utermohlen O., Hoppe T., Schumacher B. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501:416–420. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki Y., Derudder E., Hobeika E., Pelanda R., Reth M., Rajewsky K., Schmidt-Supprian M. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Argast G.M., Stephens K.M., Emond M.J., Monnat R.J., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- 28.Wittmayer P.K., McKenzie J.L., Raines R.T. Degenerate DNA recognition by I-PpoI endonuclease. Gene. 1998;206:11–21. doi: 10.1016/s0378-1119(97)00563-5. [DOI] [PubMed] [Google Scholar]
- 29.Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 30.Kin T., Ono Y. Idiographica: a general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics. 2007;23:2945–2946. doi: 10.1093/bioinformatics/btm455. [DOI] [PubMed] [Google Scholar]
- 31.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berkovich E., Monnat R.J., Jr, Kastan M.B. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat. Protoc. 2008;3:915–922. doi: 10.1038/nprot.2008.54. [DOI] [PubMed] [Google Scholar]
- 33.Feil R., Wagner J., Metzger D., Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 34.Hennet T., Hagen F.K., Tabak L.A., Marth J.D. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. U.S.A. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh S.K., Williams C.A., Klarmann K., Burkett S.S., Keller J.R., Oberdoerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J. Exp. Med. 2013;210:987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke A.R., Purdie C.A., Harrison D.J., Morris R.G., Bird C.C., Hooper M.L., Wyllie A.H. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 37.Lowe S.W., Schmitt E.M., Smith S.W., Osborne B.A., Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 38.Zheng L., Asprodites N., Keene A.H., Rodriguez P., Brown K.D., Davila E. TLR9 engagement on CD4 T lymphocytes represses gamma-radiation-induced apoptosis through activation of checkpoint kinase response elements. Blood. 2008;111:2704–2713. doi: 10.1182/blood-2007-07-104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rube C.E., Grudzenski S., Kuhne M., Dong X., Rief N., Lobrich M., Rube C. DNA double-strand break repair of blood lymphocytes and normal tissues analysed in a preclinical mouse model: implications for radiosensitivity testing. Clin. Cancer Res. 2008;14:6546–6555. doi: 10.1158/1078-0432.CCR-07-5147. [DOI] [PubMed] [Google Scholar]
- 40.Odersky A., Panyutin I.V., Panyutin I.G., Schunck C., Feldmann E., Goedecke W., Neumann R.D., Obe G., Pfeiffer P. Repair of sequence-specific 125I-induced double-strand breaks by nonhomologous DNA end joining in mammalian cell-free extracts. J. Biol. Chem. 2002;277:11756–11764. doi: 10.1074/jbc.M111304200. [DOI] [PubMed] [Google Scholar]
- 41.Bothmer A., Robbiani D.F., Di Virgilio M., Bunting S.F., Klein I.A., Feldhahn N., Barlow J., Chen H.T., Bosque D., Callen E., et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., Cammarata M., Perez M., Agarwal P., Brodbelt J.S., et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rathmell J.C., Farkash E.A., Gao W., Thompson C.B. IL-7 enhances the survival and maintains the size of naive T cells. J. Immunol. 2001;167:6869–6876. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 44.Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;4:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgess R.C., Burman B., Kruhlak M.J., Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9:1703–1717. doi: 10.1016/j.celrep.2014.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodarzi A.A., Jeggo P., Lobrich M. The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Rep. 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 47.Lemaitre C., Grabarz A., Tsouroula K., Andronov L., Furst A., Pankotai T., Heyer V., Rogier M., Attwood K.M., Kessler P., et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014;28:2450–2463. doi: 10.1101/gad.248369.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayrapetov M.K., Gursoy-Yuzugullu O., Xu C., Xu Y., Price B.D. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khurana S., Kruhlak M.J., Kim J., Tran A.D., Liu J., Nyswaner K., Shi L., Jailwala P., Sung M.H., Hakim O., et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014;8:1049–1062. doi: 10.1016/j.celrep.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garm C., Moreno-Villanueva M., Burkle A., Petersen I., Bohr V.A., Christensen K., Stevnsner T. Age and gender effects on DNA strand break repair in peripheral blood mononuclear cells. Aging Cell. 2013;12:58–66. doi: 10.1111/acel.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ju Y.J., Lee K.H., Park J.E., Yi Y.S., Yun M.Y., Ham Y.H., Kim T.J., Choi H.M., Han G.J., Lee J.H., et al. Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence. Exp. Mol. Med. 2006;38:686–693. doi: 10.1038/emm.2006.81. [DOI] [PubMed] [Google Scholar]
- 53.Mehta R., Chandler-Brown D., Ramos F.J., Shamieh L.S., Kaeberlein M. Regulation of mRNA translation as a conserved mechanism of longevity control. Adv. Exp. Med. Biol. 2010;694:14–29. doi: 10.1007/978-1-4419-7002-2_2. [DOI] [PubMed] [Google Scholar]
- 54.Harrison D.E., Strong R., Sharp Z.D., Nelson J.F., Astle C.M., Flurkey K., Nadon N.L., Wilkinson J.E., Frenkel K., Carter C.S., et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selman C., Tullet J.M., Wieser D., Irvine E., Lingard S.J., Choudhury A.I., Claret M., Al-Qassab H., Carmignac D., Ramadani F., et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steffen K.K., MacKay V.L., Kerr E.O., Tsuchiya M., Hu D., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lebofsky R., Bensimon A. DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol. Cell Biol. 2005;25:6789–6797. doi: 10.1128/MCB.25.15.6789-6797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voineagu I., Narayanan V., Lobachev K.S., Mirkin S.M. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bahar R., Hartmann C.H., Rodriguez K.A., Denny A.D., Busuttil R.A., Dolle M.E., Calder R.B., Chisholm G.B., Pollock B.H., Klein C.A., et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 60.Rodier F., Coppe J.P., Patil C.K., Hoeijmakers W.A., Munoz D.P., Raza S.R., Freund A., Campeau E., Davalos A.R., Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.