Abstract
Sequencing small quantities of DNA is important for applications ranging from the assembly of uncultivable microbial genomes to the identification of cancer-associated mutations. To obtain sufficient quantities of DNA for sequencing, the small amount of starting material must be amplified significantly. However, existing methods often yield errors or non-uniform coverage, reducing sequencing data quality. Here, we describe digital droplet multiple displacement amplification, a method that enables massive amplification of low-input material while maintaining sequence accuracy and uniformity. The low-input material is compartmentalized as single molecules in millions of picoliter droplets. Because the molecules are isolated in compartments, they amplify to saturation without competing for resources; this yields uniform representation of all sequences in the final product and, in turn, enhances the quality of the sequence data. We demonstrate the ability to uniformly amplify the genomes of single Escherichia coli cells, comprising just 4.7 fg of starting DNA, and obtain sequencing coverage distributions that rival that of unamplified material. Digital droplet multiple displacement amplification provides a simple and effective method for amplifying minute amounts of DNA for accurate and uniform sequencing.
INTRODUCTION
Single cell sequencing is an invaluable tool in microbial ecology and has enhanced the analysis of communities ranging from the ocean (1) to the human mouth (2). Because the majority of microorganisms cannot be cultured (3), obtaining sufficient quantities of DNA for sequencing requires significant amplification of single-cell genomes. However, existing methods for accomplishing this are prone to amplification bias, making sequencing inefficient and costly. Consequently, there has been a sustained effort to develop new methods to uniformly amplify small quantities of DNA.
A powerful method is to modify the polymerase chain reaction (PCR) to enable non-specific amplification. For example, Primer Extension Preamplification (PEP) and Degenerate Oligonucleotide-Primed PCR (DOP-PCR) use modified primers and thermal cycling conditions to enable non-specific annealing and amplification of most DNA sequences (4,5). These methods are simple and accessible, but amplification bias remains a major challenge: the products typically do not fully cover the original template and possess significant variation in coverage (6,7). Multiple Annealing and Looping Based Amplification Cycles (MALBAC) reduces this bias with primers that cause amplicons to self-anneal in a loop; this suppresses exponential amplification of dominant products and equalizes amplification across the templates (8). Nevertheless, the specialized polymerase required for this reaction is prone to copy errors that propagate through cycling, resulting in increased error rates (8).
Multiple displacement amplification (MDA) enables non-specific amplification with minimal error through the use of the highly accurate enzyme Φ29 DNA polymerase (9). In addition, Φ29 DNA polymerase displaces Watson–Crick base-paired strands, enabling exponential amplification of template molecules without thermally-induced denaturation (7). Nevertheless, two major problems persist with MDA: amplification of contaminating DNA (10) and highly uneven amplification of single-cell genomes (11,12). These problems yield numerous challenges when sequencing MDA-amplified material, including incomplete genome assembly, gaps in genome coverage and biased counts of replicated sequences, which are of biological relevance in a variety of applications such as assessing copy number variants in cancer. Due to its simplicity and accuracy, several strategies have been employed to reduce MDA amplification bias, including augmenting reactions with trehalose (13), reducing reaction volumes (14) and using nanoliter-scale microfluidic chambers to reduce the diversity in isolated pools (15,16). While these methods mitigate the problems associated with MDA, robust and uniform amplification of low-input material remains a challenge.
In this paper, we describe digital droplet MDA (ddMDA), an alteration to the MDA reaction in which single template molecules are compartmentalized in millions of picoliter droplets. Compartmentalizing and amplifying single molecules affords a number of benefits for obtaining accurate sequence data with uniform coverage. Because the molecules are isolated, each amplifies to saturation irrespective of when the reaction initiates – a stochastic process that, in bulk, is the primary source of bias (17). As we show, this greatly reduces bias. The ‘compartmentalization’ is analogous to that in digital droplet PCR (ddPCR), in which PCR reagents are isolated in millions of droplets, enabling accurate quantification of nucleic acids (18). In this work, we describe microfluidic and accessible non-microfluidic methods for generating the droplet compartments, and demonstrate uniform amplification and high-coverage sequencing of single Escherichia coli cells comprising just 4.7 femtograms of starting DNA. Our method combines the accuracy of Φ29 DNA polymerase with the uniformity of compartmentalized amplification, and should be valuable whenever low-input samples must be sequenced accurately, such as in forensics and single-cell analysis.
MATERIALS AND METHODS
Generating shaken emulsion droplets
Shaken emulsions are generated by adding 30 μl of HFE-7500 fluorinated oil (3M, catalog no. 98-0212-2928-5) and 2% (w/w) PEG-PFPE amphiphilic block copolymer surfactant (RAN Technologies, catalog no. 008- FluoroSurfactant-1G) to 30 μl of MDA reaction mixture. Alternatively, HFE-7500 fluorinated oil with 2% PicoSurf1 (Dolomite Microfluidics) can be used. The combined mixture is vortexed at 3000 rpm for 10 s using a VWR vortexer, creating droplets ranging in diameter from 15 μm to 250 μm (Supplementary Figure S1). At the conclusion of incubation, 10 μl of perfluoro-1-octanol (Sigma Aldrich) is added, the mixture vortexed to coalesce the droplets and the aqueous layer extracted with a pipette. A detailed protocol for shaken emulsion formation can be found in Supplementary Protocol S1.
Generating monodisperse microfluidic emulsion droplets
The poly(dimethylsiloxane) (PDMS) microfluidic device used to generate monodisperse emulsions is fabricated by pouring uncured PDMS (10.5:1 polymer-to-crosslinker ratio) over a photolithographically-patterned layer of photoresist (SU-8 3025, MicroChem) on a silicon wafer (19). The device is cured in an 80°C oven for 1 h, extracted with a scalpel and inlet ports added using a 0.75 mm biopsy core (World Precision Instruments, catalog no. 504529). The device is bonded to a glass slide using O2 plasma treatment and channels are treated with Aquapel (PPG Industries) to render them hydrophobic. Finally, the device is baked at 80°C for 10 min. Commercial microfluidic droplet makers and pumps may also be used to generate monodisperse emulsions for ddMDA.
The MDA reaction mixture and HFE-7500 fluorinated oil with 2% (w/w) PEG-PFPE amphiphilic block copolymer surfactant (RAN Biotechnologies) are loaded into separate 1 ml syringes and injected at 300 and 500 μl/h, respectively, into a flow-focusing droplet maker using syringe pumps (New Era, catalog no. NE-501) controlled with a custom Python script (https://github.com/AbateLab/Pump-Control-Program). Alternatively, HFE-7500 fluorinated oil with 2% PicoSurf1 (Dolomite Microfluidics) may also be usable and is available for purchase. The droplet maker generates monodisperse droplets ∼26 μm in diameter (Supplementary Figure S1), collected into a PCR tube. Droplets in this size range are stable during the ddMDA reaction. At the conclusion of incubation, 10 μl of perfluoro-1-octanol is added, the emulsion vortexed to coalesce the droplets and the aqueous layer extracted with a pipette. A detailed protocol for microfluidic device fabrication and emulsification can be found in Supplementary Protocol S2. Furthermore, a schematic of the droplet maker used can be found in Supplementary Figure S2.
Biological reactions
Extraction, fragmentation and amplification of genomic DNA
Purified E. coli K12(DH10B) cells are obtained from New England BioLabs (catalog no. C3019H), lysed and purified using PureLink Genomic DNA Mini Kit (Life Technologies, catalog no. K1820-00). Ten kilobase fragments are gel-extracted following a 10-min digestion with NEBNext dsDNA Fragmentase (NEB, catalog no. M0348S) of 800 ng DNA and quantified using a NanoDrop (Thermo Scientific). MDA reactions are performed using REPLI-g single cell kit (Qiagen, catalog no. 150343). Purified DNA (0.05 pg, 0.5 pg and 5 pg) is incubated with 3 μl Buffer D2 and 3 μl H2O for 10 min at 65°C. After stopping by adding 3 μl stop solution, the reaction is divided in two and a master mix comprising nuclease-free H2O, REPLI-g Reaction Buffer and REPLI-g DNA Polymerase is added to each partition. The MDA reactions are either incubated at 30°C for 16 h in bulk or as an emulsion.
Single E. coli cell sorting and whole genome amplification
OneShot TOP10 chemically competent E. coli cells (Life Technologies, catalog no. C4040–10) are cultured in LB media for 12 h, diluted in water and stained with 0.25x SYBR Green I (Life Technologies, catalog no. S-7563). Following cell stain, the cell solution is imported into a BD FACS Aria II. Single positive events are sorted into 10 separate wells of a 96-well plate. Added to each well are 3 μl Buffer D2 and 3 μl H2O, after which the plate is heated at 98°C for 4 min. This heat step lyses the cells and fragments the genomic DNA to adequate lengths for ddMDA (5–15 kilobases). After heating, the reaction is stopped by adding 3 μl stop solution to each well. Next, master mix comprising nuclease-free H2O, REPLI-g Reaction Buffer and REPLI-g DNA Polymerase is added to each well. The MDA reactions are either incubated at 30°C for 16 h in bulk or as an emulsion.
Digital droplet PCR and MDA
The digital PCR and MDA experiments are performed with phage lambda genomic DNA as template (NEB, catalog no. N3011S). For digital PCR, the template is mixed in bulk with primers (IDT), TaqMan probe (IDT) and 2X Platinum Multiplex PCR Master Mix (Life Technologies, catalog no. 4464268) in a total volume of 100 μl. The sequences of the primers and probes are – Lambda Fwd: 5′-GCC CTT CTT CAG GGC TTA AT-3′; Lambda Rev: 5′CTC TGG CGG TGT TGA CAT AA-3′; Lambda Probe: 5′/6-FAM/AT ACT GAG C/ZEN/A CAT CAG CAG GAC GC/3IABkFQ/-3′. Primers and probe are used at concentrations of 1 μM and 250 nM, respectively, and target a 110-basepair region in the lambda phage genome. Reaction mixture and HFE-7500 fluorinated oil with 2% (w/w) PEG-PFPE amphiphilic block copolymer surfactant are loaded into separate 1 ml syringes and injected at 300 and 600 μl/h, respectively, into the flow-focusing device. After collecting the emulsion in PCR tubes, the oil underneath the emulsion is removed using a pipette and replaced with FC-40 fluorinated oil (Sigma-Aldrich, catalog no. 51142-49-5) with 5% (w/w) PEG-PFPE amphiphilic block copolymer surfactant. This oil/surfactant combination yields greater stability during the cycled ddPCR reaction than the HFE oil combination. The emulsion is transferred to a T100 thermocycler (BioRad) and cycled with the following program: 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 60°C for 90 s and 72°C for 20 s, followed by a final hold at 12°C.
For digital MDA, the template is mixed with reagents from the REPLI-g single cell kit as described previously, and combined with a DNA dye (EvaGreen, Biotium). The reaction mixture is emulsified through a flow-focusing device connected to syringes containing the reaction mixture and HFE-7500 fluorinated oil with 2% (w/w) PEG-PFPE amphiphilic block copolymer surfactant. The collected emulsion is incubated at 30°C for 16 h. Since thermocycling is not required, FC-40 replacement is not necessary for digital MDA.
Sequencing and bioinformatics
Library prep and sequencing parameters
Bacterial libraries are prepared from 1 ng genomic DNA from each sample using the Nextera XT sample preparation kit (Illumina). The resulting libraries are quantified using a high sensitivity Bioanalyzer chip (Agilent), a Qubit Assay Kit (Invitrogen) and qPCR (Kapa Biosystems). Bacterial libraries vary between 800–1000 bp in fragment size. All libraries are pooled in equimolar proportions and sequenced using an Illumina MiSeq with 150 bp paired-end reads (Figure 3), an Illumina HiSeq with 100 bp paired-end reads (Figure 4) and additional Illumina MiSeq with 150 bp paired-end reads (Figure 5).
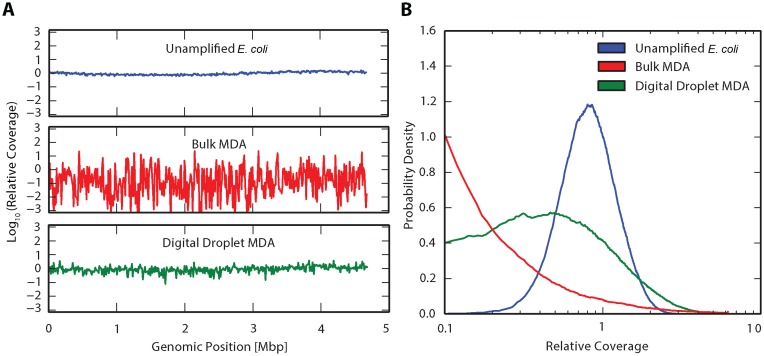
Figure 3.
Impact of compartmentalized amplification on coverage uniformity. (A) Relative coverage, defined as the number of reads for each base divided by the mean number of reads for the whole genome (29), plotted versus genome position. Relative coverage was measured for three scenarios: unamplified E. coli (top), standard bulk MDA (middle), and digital droplet MDA (bottom) and consolidated into 10 kbp bins. (B) Probability density as a function of relative coverage for Unamplified E. coli, Bulk MDA and digital droplet MDA. While coverage distribution has negligible undercovered reads for Unamplified E. coli, Bulk MDA shows a significant fraction of bases with very low coverage, a known property of MDA. Digital droplet MDA appears as a mixture of these distributions, indicating that coverage is enhanced.
Figure 4.
Comparison of bias for three different MDA methods for three input DNA concentrations. The mathematical metric definitions are provided in the supplemental information. Plots on the right show each metric normalized to the bulk MDA measurements averaged over all three input DNA concentrations. (A) Dropout rate, defined as the fraction of bases covered at less than 10% the mean coverage, plotted against input DNA concentration. (B) Coverage spread, measured as the root mean square of the relative coverage. (C) Informational entropy, defined as ∫p log(1/p), where p is the probability of observing reads within defined windows of the genome.
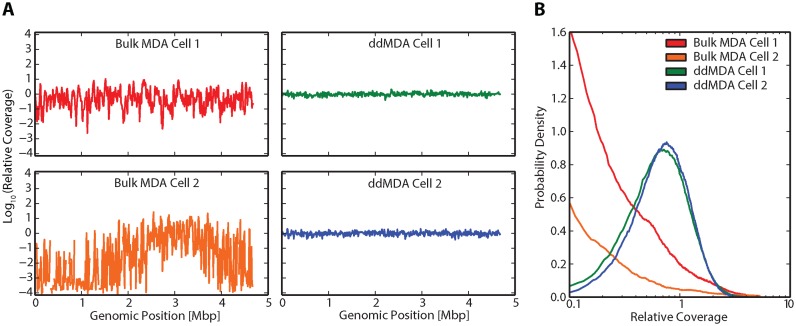
Figure 5.
ddMDA of single E. coli cells significantly enhances coverage uniformity. (A) Relative coverage, defined as the number of reads for each base divided by the mean number of reads for the whole genome, plotted versus genome position. Relative coverage is measured for two cells amplified via bulk MDA (first panel) and two cells amplified via ddMDA (second panel) consolidated into 10 kbp bins. Gaps in coverage plots represent complete dropout of a given 10 kbp bin. (B) Probability density as a function of relative coverage for two cells amplified via bulk MDA and two cells amplified via ddMDA. The two cells amplified by bulk MDA show a significant fraction of bases with very low coverage, while the cells amplified by ddMDA show much more uniform coverage.
Sequencing analysis
Sequencing data are mapped to the E. coli K12 DH10B reference genome using the BWA Whole Genome Sequencing program available on BaseSpace (Illumina). Mapped data are converted to SAM files and pileup files are generated using SAMtools. Genomic coverage as a function of genome position is determined by parsing the number of aligned reads from the pileup file, dividing each read number by the average read number, and consolidating the normalized data into 10 000 bp bins.
RESULTS
Digital droplet MDA workflow
Single cell sequencing necessitates the unbiased amplification of tiny quantities of DNA. Because of its low error rate and ability to amplify long genomic regions, MDA is a powerful method for amplifying low-input DNA. However, traditional bulk MDA does not constrain the exponential character of the reaction: sequences that begin amplifying early tend to comprise a disproportionately large fraction of the final DNA library, resulting in these regions being sequenced with high coverage while others are sequenced with low coverage (Figure 1A). Uneven coverage creates major challenges when sequencing, including inefficient use of reads, difficulty confidently assembling genomes with low-covered regions and un-sequenced gaps in the genome (Figure 1A, right panel).
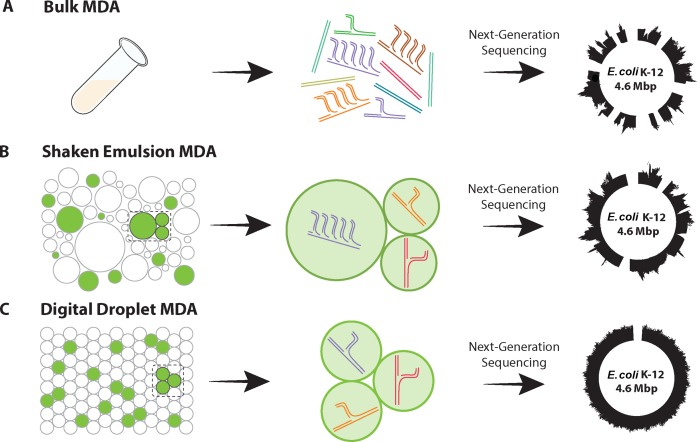
Figure 1.
Illustration of how compartmentalized Multiple displacement amplification (MDA) enhances sequencing coverage (A) Uncompartmentalized amplification does not constrain the exponential activity of Φ29 DNA Polymerase, leading to sequencing bias. (B) Compartmentalization of reaction in a shaken emulsion enhances sequencing coverage; however, the polydispersity of the emulsion leads to some sequencing bias. (C) Compartmentalization of reaction generated using a microfluidic device yields even greater sequencing coverage due to the high uniformity of the reaction.
One strategy for increasing amplification uniformity is to compartmentalize the MDA reaction in millions of isolated reactors. Just as nanoliter-based MDA yields more uniform single-cell sequences (15,16), compartmentalized digital MDA constrains the reaction to single molecules, resulting in better coverage of each molecule and more uniform representation of all molecules in the final library. A simple method for compartmentalizing reactions is to emulsify the solution containing the DNA to be amplified in oil by vigorously shaking the mixture. If a surfactant is present, stable aqueous droplets suspended in oil are produced, each of which amplifies a template molecule (Figure 1B). Because the isolated reactors are not physically connected, the reactions occur independently and in parallel, allowing each compartment to amplify to saturation. Consequently, the representation of each template in the amplified product is more uniform. Nevertheless, ‘shaken’ emulsions consist of polydisperse droplets in which volume can vary by thousands of times; because the number of product molecules at saturation scales with the volume of the reactor, polydispersity produces bias.
A simple way to remove bias due to droplet polydispersity is to compartmentalize the molecules in droplets of equal volume, which can be achieved using microfluidic emulsification (Figure 1C). Just as in the shaken emulsion case, ensuring that single molecules are amplified requires that the template concentration be set such that a percentage of droplets, typically <10%, contain a molecule, in accordance with Poisson statistics (20). This reduces the number of product molecules generated, but provides better uniformity (Figure 1C, right panel). Moreover, since MDA is an efficient reaction yielding copious DNA (7,9), the small number of productive droplets provides more than enough material for sequencing.
Non-specific quantification of DNA with digital droplet MDA (ddMDA)
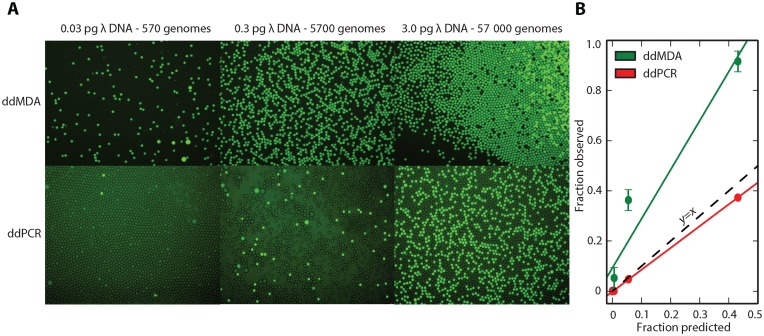
ddMDA enables uniform amplification of DNA by compartmentalizing and amplifying single template molecules in isolated droplet reactors. If a fluorescent reporter is included that indicates when a given droplet undergoes amplification, and thus contains a template molecule, the method can also be used to quantify nucleic acids in solution by counting the fraction of fluorescent droplets. This process is similar to ddPCR, a more accurate alternative to qPCR for measuring DNA concentration, except that whereas ddPCR counts known templates, ddMDA quantitates any template amplifiable with the reaction, including templates of unknown sequence (21). To illustrate this, we apply ddMDA to quantify the concentrations of Lambda phage genomic fragments in solution, comparing the results with ddPCR (Figure 2). The small Lambda phage genome offers a convenient source of DNA for quantification of amplification and contamination. Because ddPCR uses specific primers and probes, fewer fluorescent droplets are observed for the same concentration compared to ddMDA (Figure 2A). In addition, the TaqMan probe required by ddPCR leads to higher background fluorescence than the non-specific dye used in ddMDA (Figure 2A). Moreover, whereas the prediction based on Poisson encapsulation of single molecules is close to the ddPCR data (Figure 2B, red line), digital MDA systematically overestimates concentration (Figure 2B, green line). This can be rationalized by the specific nature of PCR versus the non-specific nature of MDA: ddPCR yields approximately one fluorescent droplet for each target genome in the sample, but ddMDA does so for every genomic fragment amplifiable with the reaction. As the DNA concentration increases, the probability of multiple template molecules encapsulated in the droplets increases too, leading to a larger fraction of droplets with two, three or more molecules. Nevertheless, since this is accounted for by the Poisson distribution, the method can still be used at these concentrations, although precision is reduced. Thus, fragmented or highly contaminated DNA will yield higher concentrations using ddMDA compared to ddPCR. This is key to the effectiveness of ddMDA non-specific DNA quantitation and for amplifying low-input DNA without regards to sequence.
Figure 2.
Demonstration of digital droplet MDA and its utility for nonspecific DNA quantification (A) Fluorescence microscopy images of droplets subjected to digital droplet MDA (ddMDA-upper row) and digital droplet PCR (ddPCR-lower row) for three concentrations of input material. Fluorescence was obtained using Eva Green (ddMDA) and Taqman probe (ddPCR). The disparity between digital MDA and PCR quantification corresponds to the nonspecific nature of MDA compared to specific PCR amplification (B) Fraction of observed versus predicted droplets. Fraction of fluorescent droplets is predicted assuming Poisson encapsulation of whole genomes. While ddPCR encapsulates one positive droplet per genome, ddMDA encapsulates one positive droplet per DNA segment. This enables nonspecific quantitation of nucleic acids and allows for the calculation of contamination and fragmentation of the sample.
Next generation sequencing of ddMDA-amplified DNA
To investigate the effectiveness of ddMDA for amplifying low-input DNA for sequence analysis, we sequence samples prepared in different ways and compare the results: unamplified E. coli DNA (no amplification bias), E. coli DNA amplified using bulk MDA (the current standard) and E. coli DNA amplified using monodisperse ddMDA (the best-case scenario of compartmentalized reactions). We use E. coli genomes rather than Lambda phage genomes due to their greater size and complexity, thus offering greater applicability to next generation sequencing techniques. The starting concentration for the MDA reactions is 0.5 pg, corresponding to the genomes of ∼100 E. coli cells. The unamplified sample, not surprisingly, exhibits extremely uniform coverage with the exception of long-ranged systematic variation that may be representative of the bacteria's natural DNA replication cycle (Figure 3A, top row). When the sample is subjected to bulk MDA, we observe substantial amplification bias causing significant over- and under-coverage of regions (Figure 3A, middle row). In contrast, when the MDA amplification is constrained in monodisperse droplets, no subset of templates dominates the final product, resulting in uniform coverage across the genome (Figure 3A, bottom row).
To further quantify the differences in sequencing bias for these preparation methods, we plot the probability density of coverage levels for the three samples (Figure 3B). As expected, unamplified E. coli DNA has a narrow distribution, with little variation in coverage. In contrast, the coverage of DNA amplified by bulk MDA is extremely broad, with many regions exhibiting very low or very high coverage. This variation causes a number of challenges. The limited data for under-covered regions make it challenging to assemble long sequences spanning these regions, since the low-coverage junctions cannot be determined with high confidence. Additionally, the high-coverage regions are wasteful of sequencing, since these regions are already covered adequately; they comprise a large fraction of sequencing data but offer little additional information. DNA amplified by ddMDA has a coverage distribution similar to the unamplified best-case scenario, but with larger bias. ddMDA thus yields amplified DNA that approaches the uniformity of unamplified material. To further validate the utility of ddMDA as a reliable whole genome amplification method, we compare sequenced DNA from a commercially available PCR-based single cell WGA kit (PicoPLEX WGA, NEB) to that of ddMDA. The PicoPLEX WGA kit gives rise to less bias than that seen in bulk MDA. However, the uniformity in coverage observed in ddMDA still outperforms that seen in PicoPLEX (Supplementary Figure S3). This demonstrates the unique ability of ddMDA to provide minimal amplification bias.
To compare differences in sequence bias obtained with the different methods of preparation, we prepare fresh samples using the three amplification methods described in Figure 1 (bulk MDA, shaken emulsion MDA and ddMDA) at three different input concentrations: 5 pg (∼1000 genomes), 0.5 pg (∼100 genomes) and 0.05 pg (∼10 genomes). Next-generation sequencing of these samples reveals that, indeed, bulk MDA yields poor uniformity in sequencing coverage, while ddMDA and shaken emulsion MDA exhibit significantly improved uniformity (Supplementary Figure S4).
Next-generation sequencing of unknown genomes necessitates near-complete coverage of all regions. However, amplification can result in biased genomic representation, in which low-abundance regions may not be sufficiently covered during sequencing. To quantify the frequency of this occurrence for the different preparation methods and concentrations, we use a dropout metric that represents the number of genomic regions that are significantly under-covered (Figure 4A). Specifically, we analyze the fraction of bases covered at less than 10% of the mean coverage for each sample (the equation is in Supplementary Figure S5). In the bulk MDA samples, a significant fraction of the genome is not detected for low and moderate input concentrations (Figure 4A, red curve). For higher input concentrations, the fraction of under-coverage is lower, but still significant. In the shaken emulsion MDA samples, compartmentalization results in a marked reduction of dropout for all three concentrations (Figure 4A, blue curve); however, substantial dropout is still observed. The ddMDA samples further reduce the number of dropout regions and maintain low dropout even down to 10 genome equivalents of E. coli DNA (Figure 4A, green curve). This trend in which bulk MDA results in the worst data and ddMDA the best is evident when all three concentrations are normalized to the bulk preparation and averaged (Figure 4A, right panel).
Another important factor in sequencing low-input DNA is the efficiency of sequencing – specifically, ensuring that each additional read that is sequenced provides maximum new information content. If significant coverage spread exists, small, highly covered regions can comprise a large fraction of the sequenced reads, thus requiring increased sequencing expenditure to observe the low-covered regions. To quantify this disparity in coverage, we use a metric that estimates coverage spread, calculated as the root mean square of the relative coverage (Figure 4B). The mathematical definition of the spread is in Supplementary Figure S5. The trend between samples is similar to the trend in the dropout metric, since regions that are under-covered also tend to drop out, and is also evident when the points are normalized to the bulk results and averaged (Figure 4B, right panel). This shows that compartmentalized MDA significantly reduces coverage disparity, maximizing the useful information content in the reads that are obtained and, consequently, allowing an equal amount of new information to be obtained with less total sequence expenditure compared to bulk MDA.
Another valuable metric for estimating uniformity of coverage and the likelihood of being able to generate an accurate assembly is the informational entropy, a measurement used to estimate the randomness of a signal, such as the coverage signals obtained from Figure 3A. When sequencing unknown genomes, high entropy representing a coverage distribution that is maximally randomized over the entire sequence is ideal. The informational entropies are similar for ddMDA and shaken emulsion MDA, and both perform better than bulk MDA (Figure 4C). As before, the trend is present when normalizing and averaging over input concentrations (Figure 4C, right panel). The definition for informational entropy is in Supplementary Figure S5. These data demonstrate that compartmentalized ddMDA is an effective means to maximally cover the genome with minimal sequencing expenditure.
Next generation sequencing of ddMDA-amplified DNA from single cells
To illustrate the utility of ddMDA for single cell whole genome amplification, we use it to sequence single E. coli cells. Amplifying single cells is of enormous importance for single cell analysis, such as for studying uncultivable microbes and individual cancer cells. Though valuable, the procedure is more complex than when working with purified DNA, because the single cell must be reliably lysed and its genome fragmented. Furthermore, a number of precautions must be taken to minimize contamination and DNA loss, such as UV exposure and sterile procedures. To sequence single cells, we FACS-sort E. coli cells individually into wells, lyse the cells, heat-fragment the genomes and emulsify the resultant solution with ddMDA reagents, in accordance with the protocol. We compare two cells amplified with ddMDA to two different cells amplified with standard bulk MDA. As expected, we observe significant amplification bias in the bulk-amplified samples (Figure 5A, first panel). Bulk MDA Cell 2, in particular, possesses a massive amount of under-amplification, yielding complete dropout of several regions (denoted by gaps in the coverage plot). The two cells amplified by ddMDA, on the other hand, have uniform coverage (Figure 5A, second panel). These results are further illustrated by analyzing the probability densities of the four samples (Figure 5B). The dramatic difference in coverage uniformity between bulk and ddMDA is further illustrated by looking at the probability mass function and Lorenz curve of each sample (Supplementary Figure S6), generated using htSeqTools (22). Though contamination and DNA loss are a concern, the dramatic difference in coverage between bulk MDA and ddMDA demonstrate the adaptability of this technique to single bacterial cells.
DISCUSSION
ddMDA is a simple method for amplifying small amounts of DNA that significantly enhances sequencing coverage compared to available methods. It is also useful for non-specifically detecting and quantifying nucleic acids in solution. Quantification via ddMDA is similar to ddPCR quantification in that it has a low level of detection and allows absolute counting of molecules without a standard curve (21). In contrast to ddPCR, however, ddMDA does not require specific probes, enabling the quantification of unknown sequences. These advantages make ddMDA valuable for quantitating DNA in low-abundance settings, such as clean rooms and extra-terrestrial habitats. When used with ddPCR, ddMDA is also effective for detecting fragmentation and contamination during DNA amplification (23).
In addition to its utility for nonspecific DNA quantification, ddMDA is valuable for whole genome amplification of limited DNA samples. Efficient amplification of these samples necessitates the development of new techniques, including modifications of existing PCR techniques (PEP, DOP-PCR, MALBAC) (4,5,8) and isothermal amplification techniques (MDA). MDA performed on single-cell genomes in nanoliter chambers yields enhanced sequencing coverage (15,16), but reducing reaction volume further and increasing the number of compartments is difficult with this approach. Droplet microfluidics represents a powerful alternative for compartmentalizing samples in millions of monodisperse droplets, enabling, as we show, digital MDA on single molecules and yielding extremely uniform sequencing coverage. Furthermore, by applying this approach to single cell genomes, ddMDA provides accurate and uniformly covered sequencing data.
A barrier to implementing ddMDA is the requirement of microfluidic emulsification. While a number of available commercial instruments (BioRad, RainDance, Dolomite) can be used to generate monodisperse emulsions for ddMDA, many may prefer a simpler and more accessible protocol that requires no microfluidics or specialized equipment. Shaken emulsion MDA, in which polydisperse droplets are generated by vortexing the sample with an emulsifier, is simple and accessible. Even though polydispersity of the resultant droplets yields in some bias, the method is a significant improvement over bulk MDA. Thus, the simplicity and accessibility of shaken emulsion MDA may make it the preferred method for labs lacking microfluidic expertise.
An additional barrier to implementing ddMDA is the need for reliable and chemically inert oils and surfactants. In this work, the fluorinated oil HFE-7500 is used with 2% (w/w) PEG-PFPE amphiphilic block copolymer as the stabilizing surfactant, due to the reliability of this formulation for producing thermostable, chemically inert droplets. However, if cost and ease of access are a concern, a number of other non-ionic surfactants may be used, such as PicoSurf1 (Dolomite Microfluidics) or mineral oil formulations commonly used in emulsion PCR.
Many important sequencing applications involve mammalian cells, which have much larger genomes than the E. coli cells to which we have applied the method. The E. coli genome is ∼4.7 million base pairs and relatively simple to amplify and sequence. The diploid human genome, on the other hand, is complex and possesses over 6 billion base pairs. Due to the larger genome size, more fragments must be generated for a fixed fragment length, which in turn necessitates more droplets to ensure single-molecule Poisson encapsulation. For example, for a 10-kb fragment size, there will be 600 000 fragments, requiring ∼6 million droplets to ensure 1 in 10 loading rates during the ddMDA reaction. There is, however, immense flexibility in the workflow and this is well within the comfort zone of ddMDA: using ∼30 μm droplets, for example, a 6 million droplet emulsion consumes ∼140 μl of ddMDA reagent and takes ∼30 min to generate with microfluidic flow focusing, both of which are reasonable. In addition, droplet volume, fragment length and the emulsification method can all be altered to optimize for the experiment. For example, higher-throughput droplet generation method such as parallel droplet generation (24), hierarchical droplet splitting (25) and bubble triggered droplet generation (26), each provide >10X throughput in droplet generation, and can be used in combination.
The ability to uniformly amplify minute quantities of DNA is valuable for a variety of biological applications. Forensic investigation, for instance, requires amplification and sequencing of samples well below the sensitivity limits of routine DNA analysis (27). Incorporating ddMDA into these samples should yield enhanced uniformity of whole genome amplification, thus improving draft genomes and follow-on analyses of the data. In addition, single cell analysis is becoming an increasingly valuable tool for identifying tumor growth, evolution and potentially effective therapies (28). ddMDA on individual tumor cells should provide more accurate sequences of cancer-associated mutations, especially copy-number variants, for tracking the progression and evolution of the disease.
Supplementary Material
Acknowledgments
We thank Phil Romero for reading the manuscript and providing input on sequencing analysis. We also thank Tristan Tao and Sunay Rajbhandari for technical assistance. Data for this study were acquired from the Center for Advanced Technology at UCSF.
Footnotes
Present address: Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, 1700 4th Street, San Francisco, CA 94158, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation through a CAREER Award [DBI-1253293]; National Institutes of Health (NIH) [HG007233-01, R01-EB019453-01, DP2-AR068129-01]; Defence Advanced Research Projects Agency Living Foundries Program [contract numbers HR0011-12-C-0065, N66001-12-C-4211, HR0011-12-C-0066]. Funding for open access charge: NIH [DP2-AR068129-01].
Conflict of interest statement. None declared.
REFERENCES
- 1.Yoon H.S., Price D.C., Stepanauskas R., Rajah V.D., Sieracki M.E., Wilson W.H., Yang E.C., Duffy S., Bhattacharya D. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- 2.Marcy Y., Ouverney C., Bik E.M., Lösekann T., Ivanova N., Martin H.G., Szeto E., Platt D., Hugenholtz P., Relman D.A., et al. Dissecting biological ‘dark matter’ with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutchison C.A. III, Venter J.C. Single-cell genomics. Nat. Biotechnol. 2006;24:657–658. doi: 10.1038/nbt0606-657. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Cui X., Schmitt K., Hubert R., Navidi W., Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Telenius H., Carter N.P., Bebb C.E., Nordenskjöld M., Ponder B.A., Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 6.Cheung V.G., Nelson S.F. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14676–14679. doi: 10.1073/pnas.93.25.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J., et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong C., Lu S., Chapman A.R., Xie X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteban J.A., Salas M., Blanco L. Fidelity of phi29 DNA Polymerase. J. Biol. Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 10.Raghunathan A., Ferguson H.R., Jr, Bornarth C.J., Song W., Driscoll M., Lasken R.S. Genomic DNA amplification from a single bacterium genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean F.B., Nelson J.R., Giesler T.L., Lasken R.S. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosono S., Faruqi A.F., Dean F.B., Du Y., Sun Z., Wu X., Du J., Kingsmore S.F., Egholm M., Lasken R.S. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan X., Urban A.E., Palejev D., Schulz V., Grubert F., Hu Y., Snyder M., Weissman S.M. A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15499–15504. doi: 10.1073/pnas.0808028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchison C.A III, Smith H.O., Pfannkoch C., Venter J.C. Cell-free cloning using phi29 DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcy Y., Ishoey T., Lasken R.S., Stockwell T.B., Walenz B.P., Halpern A.L., Beeson K.Y., Goldberg S.M.D., Quake S.R. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gole J., Gore A., Richards A., Chiu Y.-J., Fung H.-L., Bushman D., Chiang H.-I., Chun J., Lo Y.-H., Zhang K. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat. Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigue S., Malmstrom R.R., Berlin A.M., Birren B.W., Henn M.R., Chisholm S.W. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J., Bright I.J., Lucero M.Y., Hiddessen A.L., Legler T.C., et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia Y., Whitesides G.M. Soft Lithography. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 20.Köster S., Angilè F.E., Duan H., Agresti J.J., Wintner A., Schmitz C., Rowat A.C., Merten C.A., Pisignano D., Griffiths A.D., et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 21.White R.A., Blainey P.C., Fan H.C., Quake S.R. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Planet E, Attolini C.S.-O, Reina O., Flores O., Rossell D. htSeqTools: high-throughput sequencing quality control, processing and visualization in R. Bioinformatics. 2015;28:589–590. doi: 10.1093/bioinformatics/btr700. [DOI] [PubMed] [Google Scholar]
- 23.Blainey P.C., Quake S.R. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romanowsky M.B., Abate A.R., Rotem A., Holtze C., Weitz D.A. High throughput production of single core double emulsions in a parallelized microfluidic device. Lab Chip. 2012;12:802–807. doi: 10.1039/c2lc21033a. [DOI] [PubMed] [Google Scholar]
- 25.Abate A.R., Weitz D.A. Faster multiple emulsification with drop splitting. Lab Chip. 2011;11:1911–1915. doi: 10.1039/c0lc00706d. [DOI] [PubMed] [Google Scholar]
- 26.Abate A.R., Weitz D.A. Air-bubble-triggered drop formation in microfluidics. Lab Chip. 2011;11:1713–1716. doi: 10.1039/c1lc20108e. [DOI] [PubMed] [Google Scholar]
- 27.Hanson E.K., Ballantyne J. Whole genome amplification strategy for forensic genetic analysis using single or few cell equivalents of genomic DNA. Anal. Biochem. 2005;346:246–257. doi: 10.1016/j.ab.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D., et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross M.G., Russ C., Costello M., Hollinger A., Lennon N.J., Hegarty R., Nusbaum C., Jaffe D.B. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.