Abstract
Successful and accurate completion of the replication of damage-containing DNA requires mainly recombination and RAD18-dependent DNA damage tolerance pathways. RAD18 governs at least two distinct mechanisms: translesion synthesis (TLS) and template switching (TS)-dependent pathways. Whereas TS is mainly error-free, TLS can work in an error-prone manner and, as such, the regulation of these pathways requires tight control to prevent DNA errors and potentially oncogenic transformation and tumorigenesis. In humans, the PCNA-associated recombination inhibitor (PARI) protein has recently been shown to inhibit homologous recombination (HR) events. Here, we describe a biochemical mechanism in which PARI functions as an HR regulator after replication fork stalling and during double-strand break repair. In our reconstituted biochemical system, we show that PARI inhibits DNA repair synthesis during recombination events in a PCNA interaction-dependent way but independently of its UvrD-like helicase domain. In accordance, we demonstrate that PARI inhibits HR in vivo, and its knockdown suppresses the UV sensitivity of RAD18-depleted cells. Our data reveal a novel human regulatory mechanism that limits the extent of HR and represents a new potential target for anticancer therapy.
INTRODUCTION
DNA is continuously damaged by intrinsic and extrinsic factors, which may lead to the stalling of the replication fork during the S-phase of the cell cycle and, eventually, to its collapse, resulting in the formation of a single-stranded gap (ssDNA gap) behind the replication fork (1). DNA damage tolerance (DDT) pathways have evolved to ensure successful completion of DNA replication. DDT can act either coupled with DNA replication or post-replicatively at the ssDNA gaps (2–4) and is controlled by RAD6/RAD18 (5,6). DDT consists of two sub-branches: translesion (TLS) DNA synthesis involving specialized DNA polymerases that directly bypass the lesion (7) and template switching, involving repair from the newly synthesized sister chromatid to fill in the ssDNA gap (8,9). As an alternative to the RAD6/RAD18-dependent pathway, stalled replication forks can also be rescued by recombination (10,11). Homologous recombination (HR)-mediated repair can act either on ssDNA gaps left behind the replication fork or on double-strand breaks emanating from the collapsed replication fork (12,13). However, unrestrained HR can lead to aberrant recombination between repetitive sequences leading to chromosome aberrations and loss of heterozygosity, which are associated with carcinogenesis (14–16).
The yeast Srs2 protein is a well characterized anti-recombinase, which restrains unscheduled and excessive HR, and its deletion suppresses the UV-sensitive phenotype of yeast Δrad18 cells (17–20). Srs2 is recruited to replication forks by SUMOylated PCNA (S-PCNA) where it displaces Rad51 recombinase from the ssDNA via its 3′-5′ helicase activity (21–24). In addition, we have recently described that Srs2 regulates recombination through interference with the DNA extension step of the HR pathway, which limits extension length and crossover (CO) formation (25). This activity is mediated by specific interaction between Srs2 and SUMO-PCNA. PCNA is a homotrimer, ring-like molecule, which increases the processivity of the replicative DNA polymerases (26–28) and recruits a broad range of DNA repair factors to the site of action on the DNA (29,30). In response to DNA damage, PCNA becomes post-translationally modified via the ubiquitylation of the K164 residue (5) creating a second binding domain that recruits TLS polymerases to the site of their action (31,32) to promote direct bypass of the lesion (33–35). PCNA can be further polyubiquitylated promoting another, error-free branch of DDT (36–38). Alternatively, PCNA can be SUMOylated on the very same residue (5,39–40). This modification suppresses HR presumably by recruiting Srs2 to the site of action, thus, promotes DDT only indirectly (25,41–42). Recently, we and others have shown that SUMOylation of human PCNA suppresses HR, similarly to its yeast counterpart (39,40).
Intensive research has been carried out for a long time on how HR is regulated in humans, but no direct ortholog of Srs2 has been identified so far. Up-to-date, several functional homologs of Srs2 have been described in humans. These include RTEL1, which can dissociate the D-loop structure (43); FBH1 and RECQL5β, which dismantle the RAD51 filaments (44–46), and the recently described PCNA-associated recombination inhibitor protein (PARI) (40). Although these anti-recombinases are capable of dismantling RAD51 filaments via their strong adenosine triphosphatase (ATPase) activity, PARI seems to only slightly attenuate ssDNA binding by RAD51, because it is associated with an inherently low ATPase activity. PARI, which is mainly present in vertebrates, is unique among the functional Srs2 homologs in human as it is the only one that interacts with SUMOylated PCNA through a functional SUMO-interacting motif (SIM) and a PIP box. Although the SIM and PIP motifs are important for the function of PARI, the molecular mechanism of its action has not been explored yet (40).
Here, we investigate the molecular mechanism by which PARI may regulate the frequency of HR events in humans. We found that PARI, similarly to Srs2, acts along with RAD18 to promote replication completion by promoting DDT mechanisms in humans. The biochemical mechanism of its function is based on the inhibition of D-loop extension by DNA polymerase δ. A C-terminal part of PARI and intact PIP and SIM motifs are sufficient and necessary for the inhibition of HR events, while the UvrD-like helicase domain is dispensable for the inhibition. Our results suggest that the regulation of DNA polymerase δ by PARI and PCNA at the D-loop structure is an important strategy to down-regulate HR events, representing an active contribution to genome stability in human cells.
MATERIALS AND METHODS
Protein purification
GST-Sae2/Sae1 (pIL1055), GST-Ubc9 (pIL1054), GST-Flag-SUMO1 (pIL2576), PCNA (pIL2179), RPA, Rad51, Rad54, RFC and DNA polymerase δ were purified as described previously (39,47–48). The purification of Ub- and polyUb-PCNA has been published previously (49).
C12ORF48/PARI was cloned from a human cDNA library into the pCR8 gateway entry vector (Life Technologies) and verified by sequencing. For recombinant protein production, PARI was subcloned into His-Flag-tagged baculovirus transfer vector pVL1392. Baculovirus was produced by co-transfection of transfer vector with Bsu36I linearized BakPAK6 baculovirus DNA (Clontech) into Sf9 insect cells. High Five insect cells were infected with the baculovirus and incubated at 28°C for 40 h. Cells were washed twice in phosphate buffered saline (PBS), resuspended in 40 ml of lysis buffer (25 mM HEPES-NaOH pH 7.5, 300 mM NaCl, 1.5 mM MgCl2, 10% Glycerol, 0.2% Triton X-100, Leupeptin, Aprotinin), sonicated and cleared by centrifugation at 20 000 g for 30 min at 4°C. The lysate was filtered through a 0.45 μM filter and loaded onto a 10 ml poly-prep chromatography column (Bio-Rad) packed with 2 ml TALON resin (Clontech). The column was washed 4× with 10 ml lysis buffer supplemented with 10 mM imidazole before elution with 200 mM imidazole. Eluted proteins were dialyzed against lysis buffer before loading onto a 10 ml poly-prep chromatography column packed with 1 ml Anti-Flag M2 affinity gel (Sigma-Aldrich). The column was washed 3× with 10 ml of lysis buffer before elution with 6 × 0.5 ml lysis buffer supplemented with 200 μg/ml 3× FLAG Peptide. The eluted fractions were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and stored at −80°C.
Polymerase chain reaction fragments were cloned into EcoRI and SalI sites of expression vector pMAL2cX resulting in wild-type pMAL2cX-PARI333–579, SIM mutant pMAL2cX-PARI368–579, PIP mutant pMAL2cX-PARI333–569 and PIP SIM double mutant pMAL2cX-PARI368–569. All of the construct were confirmed by DNA sequencing. The wild-type and mutant forms of PARI333–579 were expressed as MBP fusion proteins in Escherichia coli BL21 RIPL cells (induction: 11°C, 0.2 mM IPTG, 24 h). All purification steps were performed at 4°C. Ten grams of E. coli cell paste was sonicated in 50 ml of lysis buffer C (50 mM Tris–HCl, 10% sucrose (w/v), protease inhibitors, 10 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 0.01% (v/v) Nonidet-P40, and 500 mM KCl, pH7.5). The crude lysate was clarified by centrifugation (100 000 x g for 60 min). The supernatant was batched for 2 h with 2 ml of amylose resin (NEB) equilibrated with buffer T (25 mM Tris–Cl, 10% (v/v) glycerol, 5 mM EDTA, pH 7.5) containing 500 mM KCl. MBP-PARI fragments were eluted with 6 × 2 ml of 10 mM maltose in buffer T containing 150 mM KCl. After elution, fractions containing MBP-PARI were pooled, and they were diluted with 5 ml of buffer T and loaded onto a 1 ml Mono S column (GE Healthcare) equilibrated in buffer T containing 150 mM KCl. The MBP-PARI fragments were eluted with a 20 ml gradient of 100–500 mM KCl in buffer T. Fractions containing homogenous MBP-PARI were concentrated in a Vivaspin concentrator (Sartorius Stedim Biotech) and stored in 10 μl aliquots at −80°C.
RAD51-K133R - Expression plasmid pET11c-RAD51 (kind gift from P. Sung) was introduced into E. coli BLR(DE3)pLysS cells (Novagen) and the culture was grown to A600 ∼0.7 in 2 × TY media supplemented with ampicillin (100 mg/l) and chloramphenicol (33 mg/l). Expression of the RAD51 protein was induced by the addition of IPTG (1 mM) at 37°C for 3–4 h and cells were harvested (5000 x g). Cells were then resuspended in cell breakage (CBB) buffer (0.05 M Tris–HCl pH 7.5, 10% sucrose, 0.5 mM EDTA, 1 M KCl, 1 mM DTT and 0.01% NP-40, cocktail of protease inhibitors, PMSF), sonicated and centrifuged at 100 000 × g for 60 min. Clarified supernatant was mixed with ammonium sulfate (0.242 mg/ml) to precipitate the RAD51 protein. After centrifugation of the mixture at 9000 x g, the pellet was resuspended in K buffer (20 mM K2HPO4 pH 7.5, 10% glycerol, 0.5 mM EDTA, 1 mM DTT and 0.01% NP-40) and loaded onto a Q Sepharose Fast Flow column (GE Healthcare) pre-equilibrated in K buffer supplemented with 175 mM KCl. The proteins from the column were subsequently eluted with a gradient of 0.2–0.6 M KCl in K buffer. RAD51 peak protein fractions were pooled and loaded onto a hydroxyapatite (Sigma-Aldrich) column equilibrated with T buffer (25 mM Tris–HCl pH 7.5, 10% glycerol, 0.5 mM EDTA, 1 mM DTT and 0.01% NP-40) supplemented with KCl to 100 mM concentration. RAD51 was eluted by 60–260 mM KH2PO4 gradient in T buffer. Pooled peak fractions were loaded on MonoQ column equilibrated with T buffer with 50 mM KCl. Protein was eluted with 200–450 mM KCl gradient in T buffer. Peak fractions were pooled and concentrated using Vivaspin Centrifugal Concentrator (30 000 MWCO PES). Aliquots were stored at −80°C.
HOP2-MND1 and RECQL5β were purified as described previously (50,51).
PCNA SUMOylation
In vitro SUMOylation reaction of PCNA was carried out in 1.2 ml of P0 buffer (40 mM Tris–HCl pH 7.5, 8 mM MgCl2, 100 μg/ml bovine serum albumin (BSA), 10% glycerol, 100 μM adenosine triphosphate (ATP)) in the presence of GST-PCNA (bound to glutathione sepharose beads), Sae1/Sae2 (10 μg), Ubc9 (30 μg) and Flag-SUMO1-GG (600 μg), RFC (1 μg), DNA (50 μg) for 1 h at 37°C.
After the reaction on glutathione beads, untagged enzymes were washed out with T + 100 buffer. Unmodified and SUMOylated PCNA were eluted by 3 h incubation with PreScission protease in T + 100 buffer. The eluted PCNA/SUMO-PCNA fraction was loaded onto anti-Flag beads. After extensive washing with T + 100 buffer, the Flag-SUMO-PCNA fraction was eluted using Flag peptide-containing T + 100 buffer. The elution fraction was collected and used directly in the subsequent assays.
D-loop and primer extension assays
The reaction was performed essentially as described previously (47). Radioactively labeled or unlabeled 90-mer (3 μM) nucleotides were incubated for 5 min at 37°C with RAD51 (1 μM) in 10 μl of buffer R (35 mM Tris-Cl pH 7.4, 1 mM ATP, 1.25 mM MgCl2, 50 mM KCl, 1 mM DTT and an ATP-regenerating system consisting of 20 mM creatine phosphate and 20 μg/ml creatine kinase). After the completion of Rad51 filament formation, Rad54 protein was added (150 nM) and incubated for 3 min at 25°C. The reaction was initiated by the addition of pBluescript replicative form I (50 μM in base pairs). After 5 min incubation at 25°C, the reactions were stopped on ice. Next, RPA (660 nM), PCNA (30 nM), RFC (10 nM) and Polδ (10 nM) were added in buffer O (20 mM Tris-Cl pH 7.5, 5 mM DTT, 0.1 mM EDTA, 150 mM KCl, 40 μg/ml BSA, 8 mM MgCl2, 5% (v/v) glycerol, 0.5 mM ATP and 100 μM each of dGTP, dCTP) to a 30 μl final volume. The mixture was incubated for 5 min at 30°C. The reaction was stopped by cooling on ice followed by addition of PARI (as indicated in the figures). After 2 min incubation at 37°C, the DNA synthesis reaction was started by adding dTTP (100 μM) and dATP to the reaction mixture. If the reaction was monitored using D1 oligonucleotide, 100 μM unlabeled dATP was used. When dATP incorporation was monitored, 0.375 μCi [α-32P]dATP was used followed by 4 min incubation at 37°C. Reactions were stopped by incubating at 37°C for 10 min with sodium dodecyl sulphate (0.5% final) and proteinase K (0.5 mg/ml) and separated on agarose gel (0.8% w/v). The electrophoresis the gel was dried on DE81 paper and exposed on a phosphorimager screen. Scanning and quantification of the results were done using Fuji FLA 9000 imager followed by analysis with the Multi Gauge software (Fuji).
ΦX-based extension assay was carried out similarly to the D-loop extension (47). The reaction (20 μl final volume) was assembled on singly primed ФX174 virion ssDNA (5 nM) in buffer O (20 mM Tris-Cl pH 7.5, 5 mM DTT, 0.1 mM EDTA, 70 mM KCl, 0.5 mM ATP, 40 μg/ml BSA, 8 mM MgCl2, 5% glycerol and 60 μM each of dGTP and dCTP) in the presence of RPA (1 μM), PCNA (1 nM), RFC (1 nM) and Polδ (5 nM) followed by 5 min incubation at 30°C to allow for loading of PCNA on the substrate. After the loading step, inhibition with PARI and extension were carried out as described above for D-loop extension.
Anti-recombinase assay
Fluorescently labeled 90-mer oligonucleotide (3.6 μM) was incubated for 5 min with RAD51-K133R (1 μM) followed by the addition of 1 μl RPA (100 nM) and various amounts of RECQL5β or PARI. Followed by a 4-min incubation, 1 μl HOP2-MND1 (400 nM) was added to a total volume of 10 μl and the mixture was incubated for an additional 1 min. The reaction was initiated by adding 2 μl pBluescript SK(-) replicative form I (50 μM base pairs), incubated for an additional 7 min and the reactions were further processed as stated above.
Cell cultures and cellular protein localization studies
HEK293FT cells were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% Fetal Calf Serum (FCS) (Sigma) at 37°C. Transfections were carried out using Lipofectamine 2000 transfection reagent (Invitrogen) according to the instruction of the manufacturer. After treatment with UV (as indicated), cells were incubated for 6 h and immunostained where indicated. For cellular localizations of Flag-PARI and PCNA, cells were treated with a detergent solution (10 mM TRIS–HCl, 2.5 mM MgCl2, 1% NP-40, 1 mM PMSF) and immunostained using anti-Flag antibody (Sigma M2 A8592) diluted 1:300 and Alexa Fluor 488 (Invitrogen, A11011) diluted 1:500, anti-PCNA antibody (Santa Cruz, sc-56) diluted 1:200. Samples were mounted in 25% glycerol in PBS containing 1 mg/ml DAPI followed by microscopy using an Olympus FV1000 confocal laser scanning microscope and a Leica confocal Laser Scanning Microscope.
PARI PCNA co-localization was quantified by counting all PCNA PARI co-localizations in 50 randomly selected cells. All countings were repeated in at least three independent experiments.
Measurement of the frequency of homologous recombination
HEK293 cells were cultured in DMEM supplemented with 10% FCS and antibiotics. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. A Green Fluorescent Protein (GFP)-based recombination reporter assay was used in HeLa cells. The assay measures the recombination frequency between an integrated DNA fragment encoding a C-terminally truncated GFP and a transiently transfected DNA fragment encoding N-terminally truncated GFP proteins. Briefly, reporter cells were co-transfected with vectors encoding N-terminally truncated GFP proteins and expression plasmids or gene-specific siRNAs and I-SceI cDNA-coding plasmid. After 48 h of transfection, cells were transfected again with the I-SceI expression vector. One day after the second transfection, cells were selected for puromycin resistance, and the frequency of GFP-positive recombinants was analyzed.
Sensitivity assay
Cells were transfected with 100 pmol siRNA using Lipofectamine 2000 (Invitrogen) in 6-well plates. Cell competition-based sensitivity assay was performed as described earlier. After 24 h, the cells were mixed briefly with stable GFP-expressing HeLa cells in a ratio of 1:1 followed by treatment with UV, MMC or Cisplatin as indicated. After 7 days of culturing, the ratio of GFP-negative and GFP-positive cells (surviving cells) was determined by FACS (Guava Easy site System).
Chromosomal aberration assay
HEK293 cells were cultured in 0.05 μg/ml demecolcine (Sigma, D1925)-containing medium for 2 h, followed by a washing step with PBS. Next, cells were re-suspended in 75mM KCl-containing buffer and incubated for 10 min. The samples were fixed by methanol for 2 min, then in methanol:acetic acid (3:1) for 10 min. The fixed cell suspension was dropped onto glass slides and air dried. Staining was carried out with 0.1 μg/ml acridine orange solution (Sigma, A8097).
RESULTS
PARI depletion suppresses the UV sensitivity of RAD18-depleted cells
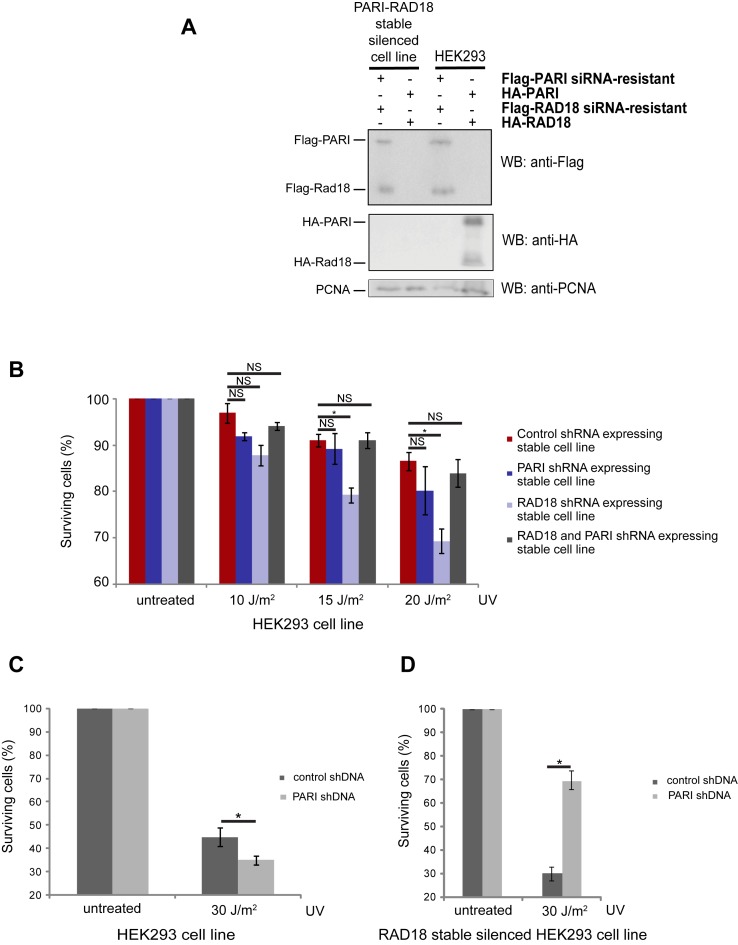
HR is potentially dangerous because it can easily result in genomic rearrangements that eventually lead to cancer (52); thus, it needs to be regulated very tightly (12). In yeasts, the Srs2 protein was identified as a potent regulator of recombination-mediated repair. This anti-recombination function of Srs2 was revealed by the fact that the deletion of SRS2 suppresses the UV hypersensitivity of Δrad18 cells (19,53). To investigate whether PARI also has a suppressing effect after prolonged fork stalling in humans, we analyzed the effect it has on the UV sensitivity of RAD18-depleted human cell lines. First, we created plasmids that allowed us to specifically downregulate PARI and RAD18 using shRNA. Next, we generated HEK293 cell lines that harbored the corresponding plasmids integrated into the genome, expressing shRNA specific to either RAD18 or PARI or both. The efficiency of the depletion was tested by western blotting after the transfection of wild-type or silencing-resistant forms of PARI or RAD18 (Figure 1A). When comparing the UV sensitivity of these different cell lines, we observed that under these circumstances the downregulation of PARI suppressed the sensitivity of the RAD18-silenced cell line (Figure 1B), which supports the notion that PARI represents a functional human Srs2 homolog. Furthermore, transient depletion of PARI resulted in a moderate increase in the UV sensitivity (Figure 1C), which is in agreement with previously published observations (40,49). We also examined the effect of shRNA-mediated silencing of PARI in the RAD18-depleted cell line. The silencing of PARI in this RAD18-deficient cell line suppressed its UV sensitivity (Figure 1D), indicating that PARI phenocopies Srs2 in human cells.
Figure 1.
PARI silencing suppresses the UV sensitivity of RAD18-depleted cells. (A) The efficiency of individual depletions tested by western blotting. (B) UV sensitivity of the RAD18-, PARI- and RAD18/PARI co-depleted HEK 293 cells. (C) HEK293 cells were transfected with control or PARI-specific shRNA. (D) RAD18-depleted HEK293 cells were transfected with control or PARI-specific shRNA. All cell lines were treated with UV; the cells were cultivated for 7 days, and the surviving cells were analyzed by FACS. Mean values of triplicates are shown with SD (error bars). Significant difference is indicated by asterisk.
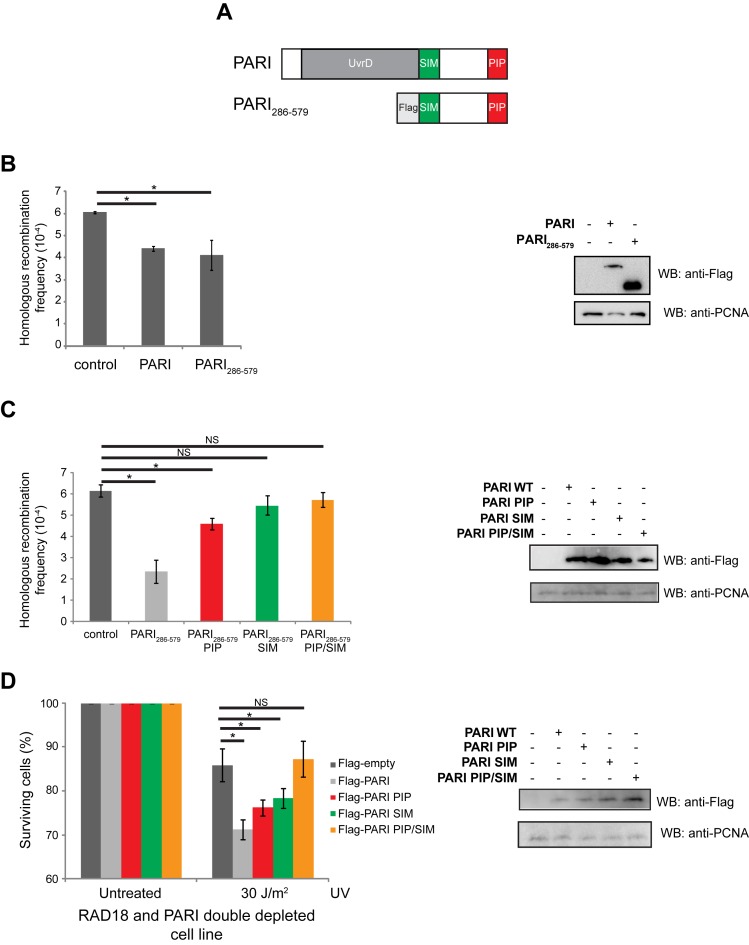
A C-terminal fragment of PARI is sufficient to inhibit the homologous recombination in vivo
In order to understand the in vivo function of PARI as a negative regulator of HR, we tested its effect on the frequency of HR. For this purpose, we used a GFP-based reporter system described previously (47). It has been published that depletion of PARI results in an increase in the frequency of HR (40). In our experiments, overexpression of the full-length PARI protein in this reporter system resulted in the decrease of the frequency of HR, demonstrating its inhibitory function (Figure 2B). To determine how the UvrD domain contributes to PARI's functions in vivo, we expressed the C-terminal PARI286–579 fragment (Figure 2A) in this reporter system. Expression of this fragment resulted in a significant reduction in recombination frequencies (Figure 2B) demonstrating that the C-terminal part of PARI is responsible for the major HR inhibitory activity. To further dissect the role of the PCNA- and SUMO-binding motifs in the suppression of recombination, we used corresponding mutants of the Flag-PARI286–579 fragment described above. Loss of either of the interacting motif alone resulted in a decrease in the suppression of recombination as compared to the Flag-PARI286–579 fragment (Figure 2C). Notably, overexpression of Flag-PARI286–579SIM had a reproducibly milder effect on the frequency of recombination than that of Flag-PARI286–579PIP (Figure 2C), indicating that interaction with the SUMO-moiety is more prominent in the suppression of HR. Importantly, expression of Flag-PARI286–579PIP/SIM had no effect on recombination frequencies, confirming that both PCNA and SUMO interactions are required and sufficient to suppress HR (Figure 2C). To confirm that PARI negatively regulates HR, we tested how the ectopic expression of this functional fragment of PARI (Flag-PARI286–579) affects the UV sensitivity of RAD18-depleted cells. The higher amount of PARI in this cell line, resulted in increased UV sensitivity (Supplementary Figure S1), indicating effective inhibition of HR.
Figure 2.
The C-terminus of PARI has a strong negative regulatory function in vivo. (A) Schematic representation of the wild-type and the truncated PARI fragment. (B) Full-length Flag-PARI and truncated Flag-PARI 286–579 were expressed in a GFP reporter system-containing HT1080 cell line, and the frequency of HR was measured. (C) The PARI286–579 fragment inhibits DSB repair via homologous recombination. The HT1080 cell line containing the I-SceI-induced GFP-based reporter system was transfected with plasmids expressing Flag-PARI286–579 (used in B), and PIP, SIM or PIP/SIM mutant fragments. After 2 days, DSB was induced by the transfection of the I-SceI-expressing plasmid and GFP-positive recombinants were measured after 7 days of cultivation. Mean values of triplicates are shown with SD (error bars). The expression of the individual fragments was analyzed by western blotting. (D) The PARI286–579 fragment sensitizes RAD18-depleted cells to UV. RAD18 and PARI double depleted HEK293 cells were transfected with PARI286–579, PIP, SIM or PIP/SIM double mutants. The cells were treated with UV, cultivated for 7 days and analyzed by FACS. Significant difference is indicated by asterisk.
To further analyze the contribution of the C-terminal part of PARI to its function, we tested the effect of the C-terminal PARI286–579 fragment on the UV sensitivity of RAD18-depleted cells. Expression of shRNA-resistant Flag-PARI286–579 sensitized the cells to UV irradiation, indicating that the C-terminal domain of PARI is, indeed, required for PARI activity in vivo (Figure 2D). To dissect the role of PCNA as well as of SUMO-mediated interactions in the suppression of the UV sensitivity of RAD18-depleted cells, we used mutant versions of Flag- PARI286–579 lacking the PIP box, the SIM or both motifs (Figure 2A). As seen in Figure 2D, expression of either Flag-PARI286–579PIP or Flag-PARI286–579SIM led to a partial increase in the UV sensitivity, indicating their direct and parallel roles in the PARI-mediated suppression of the UV-sensitivity of RAD18 PARI double depleted cells. On the other hand, expression of Flag-PARI286–579PIP/SIM did not sensitize the cells, demonstrating that PCNA- as well as SUMO-binding are required for the in vivo activity of PARI (Figure 2D).
Taken together, our results suggest that the expression of various PARI fragments leads to a decrease in the frequency of recombination and indicate that the UvrD domain is dispensable while both the PCNA- and the SUMO-binding domains are required for its function.
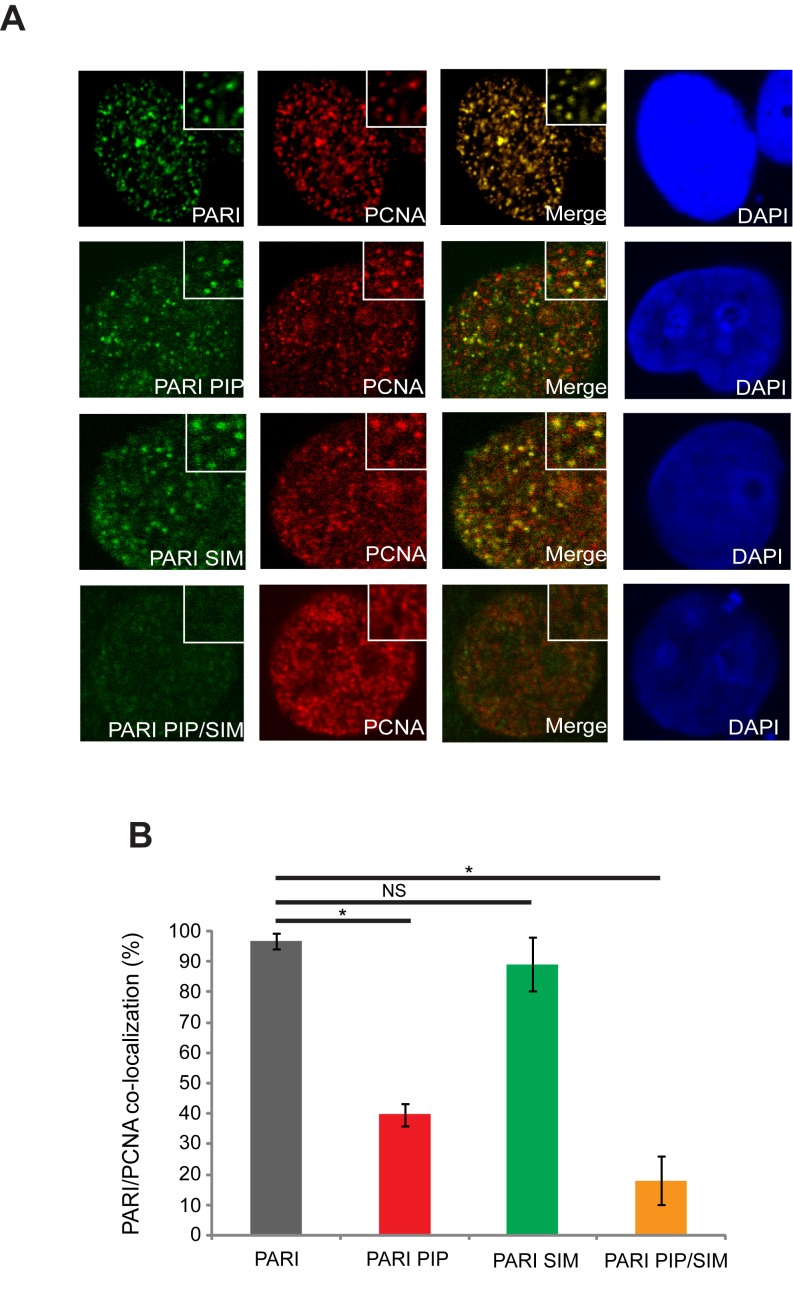
PARI localization depends mainly on the PIP motif
To further examine how the PIP and SIM motifs of PARI contribute to its function, we analyzed the intracellular distribution of PARI. Although it is known that the PIP motif is necessary for the chromatin fraction binding of PARI, the function of the SIM motif has not been revealed yet. Therefore, we used fluorescence microscopy and determined the sub-cellular localization of PARI and its mutants using antibodies against the Flag tag. Whereas wild-type PARI formed discrete nuclear foci that perfectly co-localized with PCNA (Figure 3A), only a subset of PIP box mutant foci co-localized with PCNA. The PARI SIM mutant was also able to form foci co-localizing with PCNA, indicating that the SIM motif is not the major determinant of PARI localization (Figure 3B). Surprisingly, the PARI mutant lacking both the PIP and the SIM motifs were much more defective in foci formation, indicating that PCNA- and SUMO-binding represent two parallel ways of PARI nuclear foci formation (Figure 3B). We also wished to test whether PARI localization would be affected by UV treatment, however we were not able to observe any changes in the localization (data not shown).
Figure 3.
Localization of PARI is dependent on both the SIM and the PIP motifs. (A) Flag-PARI and corresponding PIP, SIM or PIP/SIM mutants were expressed in HEK293 cells. After fixation, cells were stained using anti-Flag and anti-PCNA antibodies. (B) Graphical representation of PARI and PCNA co-localizing foci. Significant difference is indicated by asterisk.
To further corroborate the presence of S-phase-specific PARI foci, we labeled the cells that undergo DNA replication with BrdU and analyzed BrdU and PARI co-localization. As seen in Supplementary Figure S2A, we observed co-localization of PARI and BrdU, indicating that the observed PARI foci are, indeed, specific to the sites of DNA synthesis. While there was significant co-localization with BrdU in mutants lacking either PIP or SIM, mutant lacking both domains resulted in loss of PARI foci, supporting PCNA-independent targeting (Supplementary Figure S2B).
The complementary effect of PIP and SIM domains on PARI foci formation prompted us to test the role of PCNA SUMOylation. To this end, we depleted endogenous PCNA by shRNA and simultaneously expressed the shRNA-resistant PCNA-K164R mutant, which is partially defective in post-translational modification. As can be seen in Supplementary Figure S2D, the expression of mutant PCNA in the cell resulted in a very slight decrease in PARI's ability to form discrete foci, suggesting that the SUMO moiety on PCNA K164 is not the major reason for the additive effect of the PARI PIP and SIM motif mutations on the foci forming ability and pointing to other SUMO-mediated interactions.
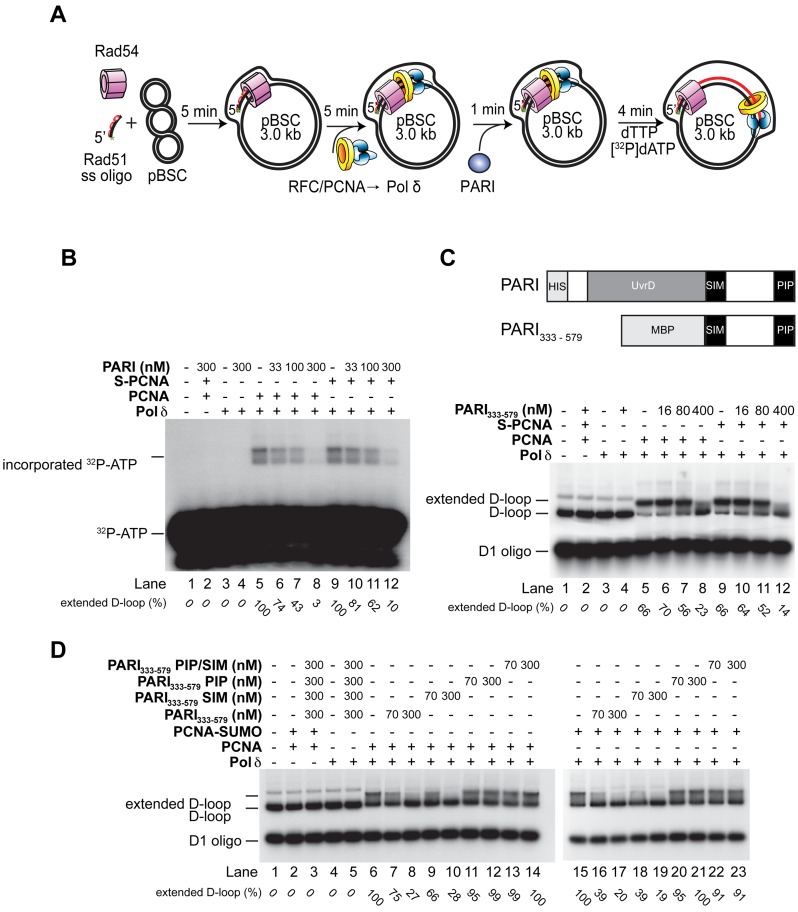
PARI inhibits D-loop extension in vitro
We have recently described the molecular mechanism of the Srs2-mediated regulation of recombination-associated DNA repair synthesis by preventing the formation of toxic recombination intermediates (25). Moreover, the previously described interaction between PARI and PCNA (40,54) and our cell biological findings prompted us to test the effect of PARI on DNA repair synthesis using our D-loop-based in vitro reconstitution assay (Figure 4A). We found that full-length PARI strongly inhibits the D-loop extension reaction (Figure 4B). Since Srs2-mediated inhibition of the extension is dependent on SUMO-PCNA, and we found that the PIP and SIM motifs contribute additively to the function of PARI as a negative regulator of HR, we substituted PCNA with SUMO-PCNA (Figure 4B). However, we did not observe any further increase in the PARI-mediated inhibition in the presence of SUMO-PCNA, indicating that the in vitro inhibition is only dependent on the PARI–PCNA interaction. Next, we used singly primed circular ssDNA instead of the D-loop substrate as a model of the replication fork. Under the same reaction conditions, full-length PARI protein inhibited DNA synthesis with comparable efficiency to the one observed with the D-loop substrate, indicating that this inhibition is independent of the type of DNA substrate (Supplementary Figure S3B). Since PARI-mediated inhibition depends exclusively on the interaction with PCNA, it could be possible that the observed inhibitory effect of PARI on the enzymatic activity of Polδ is a result of a non-specific effect of the interaction between the PIP box and PCNA. To exclude this possibility, we tested other Srs2 ortholog that also bind PCNA (55,56). RTEL1 was not able to inhibit D-loop extension by Polδ (Supplementary Figure S4), proving that the inhibitory effect is specific for PARI.
Figure 4.
PARI inhibits DNA polymerase ä-mediated D-loop extension. (A) Reaction scheme of the D-loop extension assay. (B) PARI inhibits DNA extension in a PCNA-dependent manner. After incubation of the pre-loaded replication complex with PARI, the extension was started with the addition of nucleotides including á-32P labeled dATP. (C) The UvrD domain is not required for the inhibition of D-loop extension. The reaction was carried out using radioactively labeled D1 oligonucleotides in the presence of PCNA or SUMO-PCNA (S-PCNA) as indicated. The pre-loaded replication complex was incubated with increasing concentrations of the PARI333–579 fragment, and the extension was started with the addition of nucleotides to the reactions.(D) PARI-mediated inhibition of D-loop extension depends on its PIP box. Reactions were carried out using radioactively labeled D1 oligonucleotides in the presence of wild-type, PIP box, SIM motif or double mutant PARI333–579 fragments as indicated. Reactions were supplemented either with PCNA or PCNA-SUMO fusion proteins as indicated. All reactions were resolved on 0.8% native agarose gel.
Based on the exclusively PCNA-dependent inhibition of Polδ action by PARI, we presumed that the uncontrolled overexpression of PARI should result in fork stalling and induction of chromosomal aberrations such as chromatid breaks or chromosomal fusions. Indeed, overexpression of Flag-PARI resulted in an increased number of chromosomal aberrations (Supplementary Figure S5A and B). Additionally, we tested how PARI depletion affects the viability of the cells after mild HU treatment, which induces the stalling of the replication fork. As expected from our previous data, the silencing of PARI resulted in decreased survival indistinguishable from RAD18-depleted cells (Supplementary Figure S5C), supporting the importance of PARI in DNA synthesis. Noteworthy, at higher HU concentrations PARI overexpression shows higher sensitivity compare to RAD18-depleted cells, indicating additional role for PARI.
Next, we also analyzed the contribution of the UvrD-helicase domain within PARI to gain further insights into the molecular mechanism of PARI-mediated inhibition. Using a small, C-terminal fragment (PARI333–579), we observed the same inhibitory effect (Figure 4C) as with the full-length protein, indicating that the UvrD domain is not required for the inhibition. Additionally, we also monitored the inhibition reaction using radioactively labeled oligonucleotides to verify the amount of the DNA substrate during the reaction. As shown in Supplementary Figure S3B, while the extension of the D-loop was inhibited the amount of the D-loop template remained constant, indicating that the inhibition does not stem from unwinding the extension substrate.
To further corroborate the hypothesis that the binding of PARI to PCNA represents the molecular mechanism of its inhibition, we tested mutant versions of PARI333–579 lacking the PIP motif (PARI333–579PIP), the SIM motif (PARI333–579SIM) or both (PARI333–579PIP/SIM) in the extension assay. We found that only the PARI fragments harboring an intact PIP motif were able to inhibit D-loop extension (Figure 4D). Since PCNA also undergoes ubiquitylation on the K164, we tested the effect of PCNA (poly)ubiquitylation on the PARI-mediated inhibition of DNA repair synthesis. As seen in Supplementary Figure S3C, the PARI333–579 fragment inhibited DNA repair synthesis in the presence of Ub-PCNA and polyUb-PCNA to the same extent as unmodified PCNA, suggesting that (poly)ubiquitylation of PCNA does not influence PARI activity.
Additionally, we tested how PARI affects the strand invasion mechanism mediated by the RAD51 protein. Although it has been previously described that PARI binds the RAD51 filament and this interaction is able to destabilize the filament (40), we were not able to observed any effect on D-loop formation using an authentic anti-recombinase RECQL5β as positive control (Supplementary Figure S6A). Moreover, the addition of PARI to RECQL5β did not further increase the anti-recombinase activity of RECQL5β (Supplementary Figure S6B), underlining our finding that PARI acts as a negative regulator at another step of HR.
In sum, our data point to the conclusion that it is the PCNA–PARI interaction and not the UvrD-like helicase domain that plays a role in the regulation of D-loop extension. The mechanism suggests a direct competition between PARI and Pol δ for PCNA binding and that the inhibition is independent of either the post-translational status of PCNA or the type of DNA substrate.
DISCUSSION
Replication past damaged templates or naturally occurring replication fork barriers induces replication fork stalling and/or collapse, which can have serious impact on genome maintenance (57). Over the course of evolution, conserved DDT pathways promoting either replication in the presence of damaged templates or filling-in of gaps left behind replication forks have evolved (58). However, very limited information is available regarding the choice of a particular DDT pathway and its regulation. In particular, how HR utilizes single-strand gaps arising after fork collapse remains to be determined. In yeast, Srs2 and its interplay with SUMO-PCNA is critical for the suppression of HR and promotion of DDT (25,42). Recently, the PARI protein was suggested to be the human functional homologue of Srs2 suppressing inappropriate recombination events (40).
In this work, we addressed the mechanism of PARI's regulatory function on the frequency of HR. We overexpressed full-length and truncated versions of PARI as well as its mutant forms lacking either the PIP box, the SIM motif, or both and assessed their effect on recombination frequency using a reporter system containing an I-SceI recognition site. Overexpression of the full-length and the PARI286–579 fragments resulted in equally decreased levels of recombination, which is in agreement with a previous study showing increased recombination frequency after the silencing of PARI (40). Furthermore, it reveals that the UvrD domain of PARI is dispensable for its activity in vivo. Further analysis of the PARI mutants suggested that both PCNA and SUMO binding are independently required for effective suppression of recombination.
The RAD18-dependent damage tolerance pathway represents the most prominent and alternative way of HR. The loss-of-function of a RAD18-independent inhibitor of HR thus results in the suppression of the UV sensitivity of RAD18-deficient cells as described in yeast (53,59–60). Therefore, we directly assessed the effect of PARI and its various fragments on the UV sensitivity of RAD18-depleted cells. Depletion of PARI in a RAD18-deficient cell line resulted in suppression, clearly demonstrating that PARI is a negative regulator of HR. In addition, overexpression of PARI286–579 decreased the viability of RAD18 and PARI double depleted cells, corroborating even further the fact that the UvrD domain of PARI is dispensable for its function. Moreover, using mutant versions of PARI, we determined that both PCNA and SUMO binding are required for its function in vivo, suggesting that perhaps SUMO-PCNA may become a toxic factor in this situation. This is supported by the fact that yeast Srs2 is already known to be targeted to replication forks in a SIM-dependent manner (24). To clarify the contribution of PCNA SUMOylation to the regulation of the PARI function, we analyzed its nuclear distribution. PARI formed discrete replication foci, marked by PCNA or BrdU staining, which were PCNA interaction-dependent. Surprisingly, while the SIM mutant still co-localized with PCNA the double PIP/SIM mutant failed to form any PARI foci. These findings suggest that PARI localization, as well as its in vivo function, require both PCNA- and SUMO-mediated interactions. To underline these observations, we used the K164R mutant of PCNA, which resulted only in a slight reduction of PARI's co-localization with PCNA, indicating that SUMO-PCNA is not a major determinant of the PARI recruitment to the site of action. However, PCNA gets SUMOylated on another, yet unidentified residue as well (39), therefore, we cannot exclude the possibility of a more significant contribution of PCNA SUMOylation to PARI targeting. The possible requirement of SUMO-PCNA in the targeting of PARI can be explained by the presence of a large number of PCNA-interacting proteins that may act as potential competitors for PCNA binding. At present, more than 100 nuclear proteins, involved in diverse aspects of DNA metabolism, have been described to interact with PCNA (61,62). Existence of an additional interaction interface (in this case for SUMOylated PCNA) may increase the affinity as well as the specificity of PARI toward PCNA. However, it is also very likely that PARI targeting depends on other SUMO-modified proteins, which could represent key components of HR regulation in humans. Finally, we analyzed the molecular mechanism of PARI function by taking advantage of our previous work in which we successfully reconstituted recombination-associated DNA synthesis using purified human proteins (47,48). Similarly to Srs2, PARI inhibits DNA synthesis by displacing Pol δ from the complex formed with PCNA but, contrarily to Srs2, this mechanism is biochemically independent of PCNA SUMOylation. Furthermore, the UvrD-like catalytic domain of PARI was not necessary for the regulation of the modeled extension step of HR, and the inhibition depended solely on the PARI–PCNA interaction.
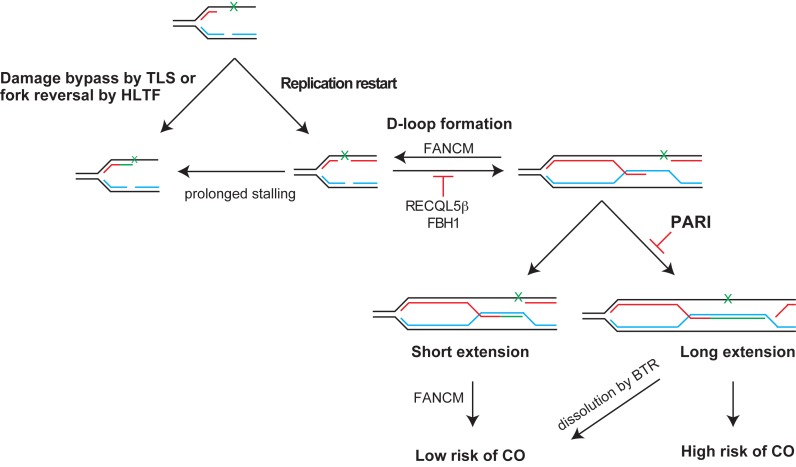
While Srs2 uses two mechanisms to suppress recombination, one that relies on the dismantling of Rad51 filaments and the other on dissociating the complex between SUMO-PCNA and Pol δ, PARI seems to promote exclusively the latter one. We hypothesize that over the course of evolution the dismantling of human RAD51-nucleoprotein filaments has been taken up by other helicases. There are at least four examples of factors (RTEL, BLM, RECQL5β, FBH1 and FANCM) that are able to act as anti-recombinases at the level of either dismantling the RAD51 filament or the D-loop substrate (43–45,63–64). This suggests that PARI may have a relatively low affinity to RAD51 (40) when compared to its affinity toward human PCNA. Therefore, in the context of recombination-associated DNA synthesis in humans, the interaction between PARI and PCNA alone may be sufficient to limit the length of the extension over its global inhibitory effect on HR (Figure 5). PARI thus may be an upstream factor of the FANCM/Mph1 mediated D-loop resolution mechanism (64–66) or the BLM/TOP3/RMI1 (BTR) complex which resolves recombination intermediates (67). Likewise, PARI could suppress the deleterious effects HR-mediated fork restart can cause when activated in the context of repetitive DNA sequences, as recently suggested (68,69). D-loops or short extension products are disassembled by FANCM/Mph1 promoting SDSA and limiting formation of CO. Long extension products can be stabilized by second-end capture resulting in the formation of Holliday junctions that can be dissolved by the BTR (BLM/TOP3/RMI1) complex or resolved by nucleases with a higher risk of CO. Future research in this direction will be required to establish a direct role of PARI in this process.
Figure 5.
PARI regulates the repair of single-strand gaps left behind the replication fork. DNA lesions induce stalling of the replication fork, which can be repaired by the DDT pathway. Prolonged fork stalling will result in fork collapse. After re-priming, single-strand gaps are left behind the replication fork, which can be filled by TLS polymerases as part of the DDT pathway or repaired by TS utilizing components of the HR machinery. RECQL5β - or FBH1 are able to disrupt RAD51 filament thus inhibiting HR and allowing DDT pathway to bypass the lesion and fill the gap. In addition, PARI can also regulate HR by blocking re-assembly of the DNA polymerase complex required for the extension of the invading strand or limiting the length of the extension and resulting in short extension products. D-loops or short extension products are disassembled by FANCM promoting SDSA and limiting formation of crossovers (CO). Long extension products can be stabilized by second-end capture resulting in the formation of Holliday junctions that can be dissolved by the BTR (BLM/TOP3/RMI1) complex or resolved by nucleases with a higher risk of CO.
In summary, we provide evidence that PARI is an important player in the promotion of DDT during DNA replication, by inhibiting unscheduled recombination events. Moreover, the PARI-mediated binding of SUMOylated proteins via its SIM is an important event in PARI's function as a negative regulator of HR. Association with SUMOylated factors may further strengthen binding of PARI to PCNA-containing foci an/or promote its PCNA-independent role. Furthermore, mechanistic analyses indicate that PARI does not suppresses recombination by direct anti-recombinase activity as compare to RECQL5β, but through regulating the extent of DNA repair synthesis thus limiting the consequent formation of potentially deleterious CO products. In the light of the yeast Srs2 protein that carries both activities (21,25), this clearly indicates the evolutionary need for separation of these functions. A better comprehension of pathways maintaining replication forks as well as genome stability is essential not only for the understanding of processes linked to increased risks of oncogenic transformation and cancer development but also for the development of the possible therapeutic potentials they hold.
Supplementary Material
Acknowledgments
We thank Simon Boulton for RTEL1 protein, Mario Spirek for RECQL5b a RAD51-K133R proteins, and Katalin Illesne Kovacs and Anita Nemeth for technical assistance. Author contribution: L.K., P.B. and L.H. designed the research; M.S., M.P., M.L. and K.F. purified proteins; P.B. and V.A. carried out biochemical experiments; L.D. and S.J. carried out in vivo experiments; P.B., L.K. and M.S. wrote the manuscript.
Footnotes
Present address:
Marek Sebesta, Sir William Dunn School of Pathology, University of Oxford, South Parks Road, OX1 3RE Oxford, UK.
Martin Pacesa, Institute of Molecular Life Sciences, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.
Kasper Fugger, The Francis Crick Institute, Clare Hall Laboratory, Blanche Lane, South Mimms EN63 LD, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [GACR13-26629S, GACR207/12/2323]; European Regional Development Fund—(Project FNUSA-ICRC) [CZ.1.05/1.1.00/02.0123]; Research Support Programme [GAMU] - MUNI/M/1894/2014; Hungarian Science Foundation [OTKA PD108611, OTKA NK101225]; Federation of European Biochemical Societies (FEBS) Short Term Fellowship (to P.B.); Faculty of Medicine MU to junior researcher (to V.A.). Funding for open access charge: Czech Science Foundation [GACR13-26629S]; Hungarian Science Foundation [OTKA PD108611, OTKA NK101225].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lopes M., Foiani M., Sogo J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Daigaku Y., Davies A.A., Ulrich H.D. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karras G.I., Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Adar S., Izhar L., Hendel A., Geacintov N., Livneh Z. Repair of gaps opposite lesions by homologous recombination in mammalian cells. Nucleic Acids Res. 2009;37:5737–5748. doi: 10.1093/nar/gkp632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 6.Sale J.E., Lehmann A.R., Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nature Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 8.Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Ramos C.A., Prakash S., Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branzei D., Vanoli F., Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 11.Gangavarapu V., Prakash S., Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krejci L., Altmannova V., Spirek M., Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Lawrence C.W. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abeysinghe S.S., Chuzhanova N., Cooper D.N. Gross deletions and translocations in human genetic disease. Genome Dyn. 2006;1:17–34. doi: 10.1159/000092498. [DOI] [PubMed] [Google Scholar]
- 15.Lisby M., Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Rep. 2009;8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smirnova M., Klein H.L. Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat. Res. 2003;532:117–135. doi: 10.1016/j.mrfmmm.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Marini V., Krejci L. Srs2: the ‘Odd-Job Man’ in DNA repair. DNA Rep. 2010;9:268–275. doi: 10.1016/j.dnarep.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rong L., Klein H.L. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- 19.Schiestl R.H., Prakash S., Prakash L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics. 1990;124:817–831. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Komen S., Reddy M.S., Krejci L., Klein H., Sung P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 2003;278:44331–44337. doi: 10.1074/jbc.M307256200. [DOI] [PubMed] [Google Scholar]
- 21.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 22.Antony E., Tomko E.J., Xiao Q., Krejci L., Lohman T.M., Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 24.Burgess R.C., Lisby M., Altmannova V., Krejci L., Sung P., Rothstein R. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell Biol. 2009;185:969–981. doi: 10.1083/jcb.200810055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkovics P., Sebesta M., Sisakova A., Plault N., Szukacsov V., Robert T., Pinter L., Marini V., Kolesar P., Haracska L., et al. Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 2013;32:742–755. doi: 10.1038/emboj.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gulbis J.M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 27.Yao N., Turner J., Kelman Z., Stukenberg P.T., Dean F., Shechter D., Pan Z.Q., Hurwitz J., O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 28.Prelich G., Tan C.K., Kostura M., Mathews M.B., So A.G., Downey K.M., Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 29.Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 30.Waga S., Hannon G.J., Beach D., Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 31.Acharya N., Yoon J.H., Gali H., Unk I., Haracska L., Johnson R.E., Hurwitz J., Prakash L., Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bienko M., Green C.M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A.R., et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 33.Haracska L., Prakash L., Prakash S. Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc Natl. Acad. Sci. U.S.A. 2002;99:16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson R.E., Haracska L., Prakash S., Prakash L. Role of DNA polymerase eta in the bypass of a (6–4) TT photoproduct. Mol. Cell. Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson R.E., Washington M.T., Haracska L., Prakash S., Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 36.Blastyak A., Hajdu I., Unk I., Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unk I., Hajdu I., Blastyak A., Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Rep. 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Unk I., Hajdu I., Fatyol K., Hurwitz J., Yoon J.H., Prakash L., Prakash S., Haracska L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gali H., Juhasz S., Morocz M., Hajdu I., Fatyol K., Szukacsov V., Burkovics P., Haracska L. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2012;40:6049–6059. doi: 10.1093/nar/gks256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moldovan G.L., Dejsuphong D., Petalcorin M.I., Hofmann K., Takeda S., Boulton S.J., D'Andrea A.D. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haracska L., Torres-Ramos C.A., Johnson R.E., Prakash S., Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 43.Barber L.J., Youds J.L., Ward J.D., McIlwraith M.J., O'Neil N.J., Petalcorin M.I., Martin J.S., Collis S.J., Cantor S.B., Auclair M., et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paliwal S., Kanagaraj R., Sturzenegger A., Burdova K., Janscak P. Human RECQ5 helicase promotes repair of DNA double-strand breaks by synthesis-dependent strand annealing. Nucleic Acids Res. 2014;42:2380–2390. doi: 10.1093/nar/gkt1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simandlova J., Zagelbaum J., Payne M.J., Chu W.K., Shevelev I., Hanada K., Chatterjee S., Reid D.A., Liu Y., Janscak P., et al. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 2013;288:34168–34180. doi: 10.1074/jbc.M113.484493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fugger K., Mistrik M., Danielsen J.R., Dinant C., Falck J., Bartek J., Lukas J., Mailand N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 2009;186:655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebesta M., Burkovics P., Juhasz S., Zhang S., Szabo J.E., Lee M.Y., Haracska L., Krejci L. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Rep. 2013;12:691–698. doi: 10.1016/j.dnarep.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebesta M., Burkovics P., Haracska L., Krejci L. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Rep. 2011;10:567–576. doi: 10.1016/j.dnarep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juhasz S., Balogh D., Hajdu I., Burkovics P., Villamil M.A., Zhuang Z., Haracska L. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012;40:10795–10808. doi: 10.1093/nar/gks850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. The EMBO Journal. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pezza R.J., Petukhova G.V., Ghirlando R., Camerini-Otero R.D. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. The Journal of biological chemistry. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- 52.Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 53.Rong L., Palladino F., Aguilera A., Klein H.L. The hyper-gene conversion hpr5–1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Connor K.W., Dejsuphong D., Park E., Nicolae C.M., Kimmelman A.C., D'Andrea A.D., Moldovan G.L. PARI overexpression promotes genomic instability and pancreatic tumorigenesis. Cancer Res. 2013;73:2529–2539. doi: 10.1158/0008-5472.CAN-12-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bacquin A., Pouvelle C., Siaud N., Perderiset M., Salome-Desnoulez S., Tellier-Lebegue C., Lopez B., Charbonnier J.B., Kannouche P.L. The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells. Nucleic Acids Res. 2013;41:6501–6513. doi: 10.1093/nar/gkt397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vannier J.B., Sandhu S., Petalcorin M.I., Wu X., Nabi Z., Ding H., Boulton S.J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 57.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fumasoni M., Zwicky K., Vanoli F., Lopes M., Branzei D. Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Polalpha/Primase/Ctf4 Complex. Mol. Cell. 2015;57:812–823. doi: 10.1016/j.molcel.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aboussekhra A., Chanet R., Zgaga Z., Cassier-Chauvat C., Heude M., Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence C.W., Christensen R.B. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 1979;139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooper S.E., Hodimont E., Green C.M. A fluorescent bimolecular complementation screen reveals MAF1, RNF7 and SETD3 as PCNA-associated proteins in human cells. Cell Cycle. 2015;14:2509–2519. doi: 10.1080/15384101.2015.1053667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naryzhny S.N. Proliferating cell nuclear antigen: a proteomics view. Cell. Mol. Life Sci. 2008;65:3789–3808. doi: 10.1007/s00018-008-8305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bugreev D.V., Yu X., Egelman E.H., Mazin A.V. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J.E., Sung P., et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitby M.C. The FANCM family of DNA helicases/translocases. DNA Rep. 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Zheng X.F., Prakash R., Saro D., Longerich S., Niu H., Sung P. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Rep. 2011;10:1034–1043. doi: 10.1016/j.dnarep.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 68.Carr A.M., Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol. 2013;425:4733–4744. doi: 10.1016/j.jmb.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 69.Hu L., Kim T.M., Son M.Y., Kim S.A., Holland C.L., Tateishi S., Kim D.H., Yew P.R., Montagna C., Dumitrache L.C., et al. Two replication fork maintenance pathways fuse inverted repeats to rearrange chromosomes. Nature. 2013;501:569–572. doi: 10.1038/nature12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.