Abstract
TP63, a member of the p53 gene family gene, encodes the ΔNp63 protein and is one of the most frequently amplified genes in squamous cell carcinomas (SCC) of the head and neck (HNSCC) and lungs (LUSC). Using an epiallelic series of siRNAs with intrinsically different knockdown abilities, we show that the complete loss of ΔNp63 strongly impaired cell proliferation, whereas partial ΔNp63 depletion rendered cells hypersensitive to cisplatin accompanied by an accumulation of DNA damage. Expression profiling revealed wide-spread transcriptional regulation of DNA repair genes and in particular Fanconi anemia (FA) pathway components such as FANCD2 and RAD18 - known to be crucial for the repair of cisplatin-induced interstrand crosslinks. In SCC patients ΔNp63 levels significantly correlate with FANCD2 and RAD18 expression confirming ΔNp63 as a key activator of the FA pathway in vivo. Mechanistically, ΔNp63 bound an upstream enhancer of FANCD2 inactive in primary keratinocytes but aberrantly activated by ΔNp63 in SCC. Consistently, depletion of FANCD2 sensitized to cisplatin similar to depletion of ΔNp63. Together, our results demonstrate that ΔNp63 directly activates the FA pathway in SCC and limits the efficacy of cisplatin treatment. Targeting ΔNp63 therefore would not only inhibit SCC proliferation but also sensitize tumors to chemotherapy.
INTRODUCTION
Squamous cell carcinoma (SCC) is a common and lethal human cancer, and relatively little progress has been made in improving outcomes for SCC due to a poor understanding of its underlying molecular pathogenesis (1,2). While SCCs typically lack somatic oncogene-activating mutations, they exhibit frequent amplification of the p53 gene family member TP63 (1–6). Recent tumor genome sequencing studies have revealed TP63 amplification in 20–25% of head and neck SCC (HNSCC) and 25–50% of lung SCC (LUSC) (4,6). Overexpression of the major TP63 isoform ΔNp63 is even more common with an upregulation reported in >80% of HNSCC cases (7).
Physiologically, p63 has been shown to be essential for epidermal development during embryogenesis (8). The N-terminally truncated isoform ΔNp63 is mainly expressed in basal epithelia and regulates stem cell maintenance, proliferation and differentiation processes in concert with the full-length p63 protein TAp63 (9,10). ΔNp63 was shown to override oncogenic Ras induced senescence in keratinocytes enabling malignant transformation and to drive squamous transdifferentiation of Kras/p53-induced lung adenocarcinomas into SCC (11,12), together indicating a contribution of aberrant ΔNp63 expression to SCC development. Moreover, a number of studies indicate an essential requirement for p63 in SCC development and maintenance. Heterozygosity of the mouse homolog Trp63 prevented SCC development in a model of ASPP2-haploinsufficiency (13), and deletion of Trp63 in DMBA-induced SCC of the skin resulted in rapid tumor regression (2). Mechanistically, ΔNp63 has been ascribed an essential pro-survival function in SCC preventing the expression of pro-apoptotic bcl-2 members via inhibition of pro-apoptotic TAp73 or the recruitment of repressive histone deacetylases HDAC1 and 2 to pro-apoptotic TAp73 target promoters (14,15). Yet, other studies observed a pro-proliferative effect of ΔNp63 independent of the inhibition of other p53-family members involving chromatin remodeling via H2A.Z deposition (16). In summary, p63 has clear oncogenic functions in SCC development and evidence suggests that it is also required for SCC maintenance making it an interesting target for the development of novel therapies.
To investigate the suitability of ΔNp63 as a therapeutic target in SCC, we revisited its function in a panel of established HNSCC cell lines focusing on a potential synergism with cisplatin as the first-line chemotherapy for SCC. Cisplatin forms covalent adducts with purine bases which include highly toxic DNA interstrand crosslinks (ICL) that in replicating cells progress to deleterious double-strand breaks (DSBs) (3,17,18). Cisplatin resistance is often the result of increased ICL repair that requires the Fanconi anemia (FA) pathway to coordinate three critical DNA repair processes, including nucleolytic incision, translesion DNA synthesis and homologous recombination (17,19). Central to this pathway is FANCD2, which upon monoubiquitination coordinates the multiple DNA repair activities required for the resolution of crosslinks (17,19). The activity status of FANCD2 is fine-tuned by E3 ubiquitin ligases such as FANCL or RAD18 and the deubiquitinase USP1 (17–19).
We observed that, under unstressed conditions, ΔNp63 is essential for proliferation but not survival of HNSCC cells. However, under cisplatin treatment, ΔNp63 strongly promotes DNA repair and cell survival. We identified the FA pathway for DNA ICL repair as a ΔNp63 target. Its central factor FANCD2 contains an enhancer with a p63 response element that is directly bound and aberrantly activated by ΔNp63 in SCC. As FANCD2 is found to be essential for a cytotoxic cisplatin response, p63 targeting could prevent repair of cisplatin-induced ICL via the FA pathway and improve the chemotherapy response of p63-overexpressing SCC.
MATERIALS AND METHODS
Cell culture
HNSCC cell lines have been described elsewhere (20,21). H1299 cells were obtained from the American Tissue Collection Center (ATCC). Cell lines with available reference data were authenticated by short tandem repeat analysis at the Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany. Cells were maintained in high-glucose Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich), 100 U/ml penicillin and 100 μg/ml streptomycin (Life technologies) at 37°C with 5% CO2. Cisplatin (NeoCorp) was used at indicated concentrations.
Normal human epidermal keratinocytes (NHEK) from pooled juvenile foreskins (PromoCell) were cultured in ready-to-use Keratinocyte Growth Medium 2 (C20011, PromoCell) with 0.06 mM CaCl2. NHEK cultures were transduced with pInducer20-ΔNp63α lentivirus produced in 293T cells as previously described (22,23). Transduced cells were selected for 7 days with 150 μg/ml G418 (Geneticin, Gibco) and ΔNp63 expression was induced with 2 μg/ml doxycyclin for 2 days.
siRNA transfections were performed with Lipofectamine RNAiMax (Life Technologies) following manufacturer's instructions with a final concentration of 10 nM. As negative controls served mock- (transfected without siRNA) and nsi-transfected (non-targeting control siRNA) cells. Polo-like kinase 1 (PLK1)-targeting siRNA was used as a positive control for cell death and reduced viability measurements. siRNA sequences are listed in Supplementary data.
Colony formation and viability assays
For colony formation assays, cells were transfected with siRNAs, re-seeded at low density and treated with cisplatin for 24 h. Outgrown colonies were fixed in ice-cold 70% ethanol and stained with Giemsa (Carl Roth). Cell viability was measured with the CellTiter-Glo® Luminescent Cell Viability Assay (Promega). Relative cell viability was calculated as the ratio of average luminescence intensity of treated samples to controls.
Cell cycle and death analysis
Cell cycle and subG1 analysis by propidium-iodide staining was performed as previously described (24). Briefly, cells were fixed in 70% ethanol overnight and stained with 10 μg/ml propidium iodide supplemented with 100 μg/ml RNase A. Cells were analyzed on an Accuri C6 cytometer (BD Biosciences) and cell cycle profiles evaluated by ModFit (Verity Software House). For cell death measurement by DIP analysis, cells were simultaneously fixed and stained in 1 μg/ml DAPI in methanol for 15 min at −20°C. After automated imaging on the BD Pathway 855 microscope (BD Biosciences), cell death was quantified by the DAPI intensity per pixel (DIP) on a single-cell level using AttoVision software (BD Biosciences) (see Supplementary Figure S4).
Luciferase reporter assay
The p63 binding site (p63BS, Figure 5A) of the FANCD2 –10 kb region was cloned upstream of a firefly luciferase reporter (pGL4.23 [luc2/minP], Promega). H1299 were transiently transfected with the reporter in combination with a constitutive Renilla luciferase reporter plasmid (phRL-TK, Promega) for normalization and a ΔNp63α-expressing pcDNA3.1 plasmid. As transfection reagent Lipofectamine 2000 (Life technologies) was used following manufacturer's instructions. Twenty four hours after transfection cells were lysed in passive lysis buffer (Promega) and dual luciferase activity measured with the Beetle and Renilla Juice Luciferase Assay Kits (PJK). Relative reporter activity was calculated by the ratio of firefly to Renilla luminescence intensity and expressed as fold activation of empty vector control.
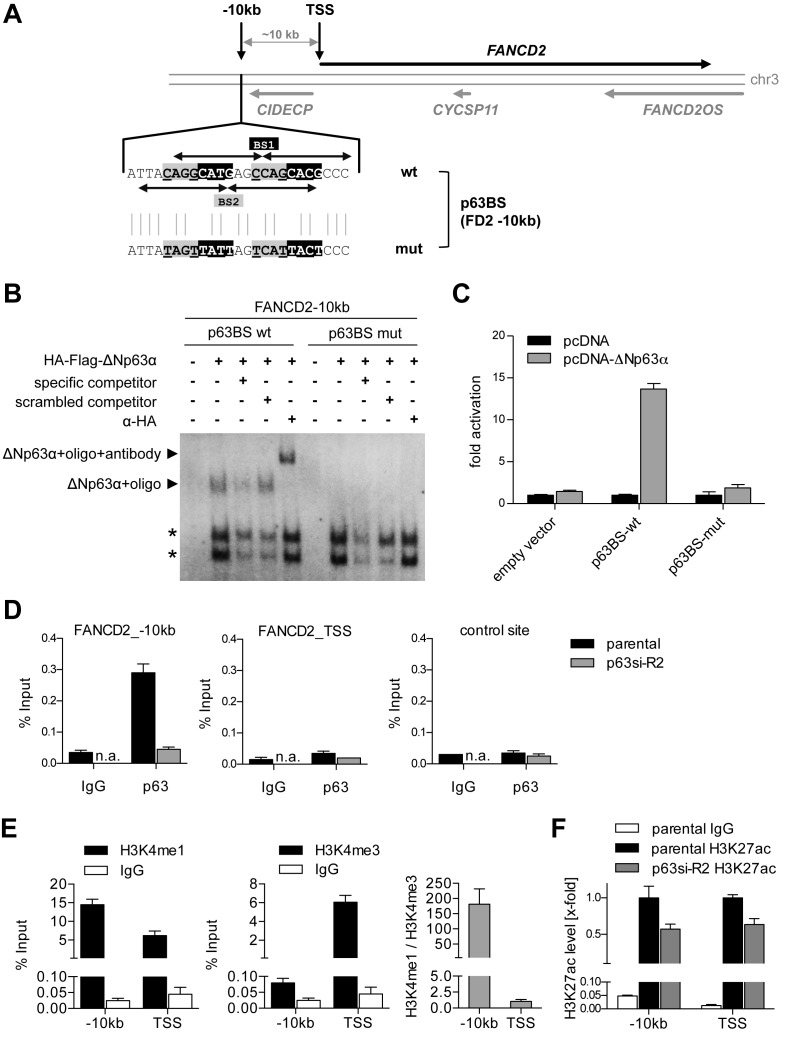
Figure 5.
ΔNp63 binds and regulates an enhancer upstream of FANCD2. (A) Genomic locus of FANCD2 with identified p63 binding site (BS) 10 kb upstream of the FANCD2 transcription start site (TSS). Arrows indicate decameric half-sites of two intertwined p63BS 1 and 2. Mutation of p63BS (p63BS mut) of the central CnnG motif is depicted. (B) EMSA for binding of in vitro translated HA-tagged ΔNp63 to wild-type and mutated FANCD2 −10 kb p63BS. Specificity of the complex was confirmed by competition with unlabeled specific or scrambled competitor and supershift with HA-antibody. Asterisks indicate unspecific complexes. (C) Luciferase reporter assay analyzing ΔNp63-mediated activation of a firefly luciferase via wild-type but not mutated FANCD2 −10 kb p63BS. Bars show relative activation of the luciferase reporter compared to empty vector control (pcDNA) (mean +SD (n = 3)). (D) Chromatin immunoprecipitation (ChIP) demonstrating in vivo binding of endogenous ΔNp63 at the FANCD2 −10 kb but not TSS or negative control region in UT-SCC-74A before (parental) and after depletion of p63 (p63si-R2). Binding is shown as mean % input +SD (n = 2). IgG, negative control ChIP; n.a., not analyzed. (E) Analysis of chromatin state at FANCD2 −10 kb and TSS site by H3K4me1 and H3K4me3 ChIP compared to IgG control. Histone modification is shown as mean % input +SD (n = 2) and H3K4me1/me3 ratio. (F) ChIP analyzing histone H3 lysine 27 acetylation (H3K27ac) as a marker for activity. FANCD2 −10 kb and TSS were evaluated in untreated (parental) and p63-depleted (p63si-R2) UT-SCC-74A. Bars show relative H3K27ac level of % input at −10 kb and TSS region normalized to parental cells (n = 3).
Gene expression analysis
Gene expression analysis was performed as previously described (24). Total RNA was isolated with the RNeasy Mini Kit (Qiagen) and cDNA synthesis performed with the SuperScript VILO cDNA synthesis kit (Life technologies) following manufacturer's instructions. Gene expression was quantified on a LightCycler 480 (Roche) using the ABsolute QPCR SYBR Green Mix (Thermo Scientific). Data were evaluated by the ΔΔCt method, β-actin and 28S-rRNA were used as housekeeping genes.
For gene expression profiling total RNA was reverse transcribed with the two-color (Cy3, Cy5) Quick Amp Labeling Kit (Agilent) following manufacturer's instructions. After purification with the NucleoSpin RNA Clean-up kit (Macherey-Nagel) individual samples (Cy5) each with a mixed reference (Cy3) were hybridized to the Human Gene Expression 4x44K v2 chip (Agilent). Slides were scanned with the DNA microarray scanner and images analyzed with the Feature Extraction software (Agilent). Intensity data for each array were normalized by the LOESS method, expression levels (M-value) calculated by the ratio of Cy5 to Cy3 and filtered for a minimum intensity (A-value) of 5. Differential gene expression was calculated by dividing M-values of p63-siRNAs by the mean of control-siRNAs and expressed as log2 fold change (FC). To assess global pathway changes, gene set enrichment analysis (GSEA) was performed with the Java tool v.2.13 using default parameters with exception for permutation type (gene sets) and a minimum gene set size of 20 genes (25). Visualization of significantly altered pathways was realized with Cytoscape using the Enrichment Map Plugin (26).
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed essentially as previously described (27). Briefly, in vitro translated HA-tagged ΔNp63α (TnT® Quick coupled Transcription/Translation Set, Promega) was incubated with [32P]-end labeled oligonucleotides in 20 mM HEPES (pH 7.8), 0.5 mM EDTA (pH 8.0), 6 mM MgCl2, 60 mM KCl, 1 mM DTT, 120 ng sheared salmon sperm DNA for 30 min at RT. Specificity of the complex was verified by competition with a 200-fold molar excess of specific or scrambled, cold oligonucleotide or a supershift using an HA-tag specific antibody. Complexes were separated on a 3.5% native polyacrylamide gel (37.5:1 acrylamide/bisacrylamide) in 0.5x Tris-borate-EDTA (TBE) buffer and visualized by autoradiography.
Chromatin-Immunoprecipitation
Chromatin-Immunoprecipitation (ChIP) was performed as previously described (24). Briefly, cells were fixed in 1% PFA and lysed in 0.5% SDS, 10 mM EDTA, 50 mM Tris pH 8.1 supplemented with protease inhibitors. Chromatin was sheared by sonication (Bioruptor, Diagenode) and diluted in 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.1, 167 mM NaCl. After pre-clearing, 1% was removed for input measure and the protein of interest was precipitated by incubation with a specific antibody (see Supplementary Data) overnight at 4°C. Complexes were bound to Protein G sepharose beads (GE Healthcare) and washed once with Low Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 150 mM NaCl), once with High Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 500 mM NaCl), once with LiCl Immune Complex Wash Buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA, 10 mM Tris-HCl pH 8.1) and twice with TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Complexes were eluted by boiling in 10% Chelex-100 (Bio-Rad) and de-crosslinked by incubation with proteinase-K (Applichem). Extracted DNA was analyzed by qPCR and binding calculated as % input.
Protein expression analysis
Western Blot analysis was done as described previously (23). Briefly, cells were lysed in NP–40 lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA pH 8.0, 2% NP-40) supplemented with protease inhibitors (complete ULTRA tablets mini, Roche). Protein concentration was measured by Bradford assay (BioRad). Equal amounts were separated on NuPAGE SDS gels (Life technologies) and transferred onto a nitrocellulose membrane. Antigens were detected as previously described (23) with antibodies listed in Supplementary data.
Immunofluorescence
Immunofluorescence stainings were performed in 96-well μClear imaging plates (Greiner bio-one), documented and quantified on a single-cell level with the automated fluorescence microscope BD Pathway 855 using the software Attovision (BD Biosciences). Cells were fixed in 3.7% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) or, for DNA damage detection, in ice-cold methanol/acetone (1:1) for 15 min. PFA-fixed samples were washed two times with 0.1 M glycin in PBS and permeabilized in PBS with 0.1% NP-40 (each 5 min, RT). Blocking was performed in permeabilization buffer supplemented with 5% FBS for 1 h at room temperature (RT). Cells were incubated with primary antibodies (listed in Supplementary data) diluted in blocking buffer for 1 h at 37°C. After washing in blocking buffer, samples were incubated with fluorescent secondary antibodies in blocking buffer supplemented with DAPI (200 nM) for nuclear counterstain and incubated for 1 h at room temperature. After washing in permeabilization buffer, cells were kept in PBS, imaged and quantified.
Immunofluorescence of EdU incorporation
Cells were incubated with 10 μM 5-Ethynyl-2′-deoxyuridine (EdU) for 1 h. Cells were fixed and stained with the Click-iT® EdU Alexa Fluor 488 Kit (Life Technologies) following manufacturer's instruction. The percentage of EdU-positive cells was evaluated by threshold application in comparison to aphidicolin- and non-EdU-treated cells.
Immunofluorescence of Platinum-DNA adducts
Platinum-DNA adducts of cisplatin-treated cells were detected as previously described (28). Briefly, cells were placed on Superfrost Plus Gold slides (Thermo Scientific) and air-dried. After fixation in methanol cells underwent a mild alkali-denaturation treatment followed by a two-step proteolytic digestion with pepsin and proteinase K to remove cellular proteins. After blocking, Platinum-DNA adducts were detected using the Pt-[GG]-specific antibody R-C18 (28) and a Cy3-labeled secondary antibody (Dianova). Nuclei were counterstained by DAPI.
Immunohistochemistry
Paraffin-embedded tissue microarrays (TMAs) for HNSCC and lung SCC were obtained from BioMax, US (HN803a, BC041115a). Tissue sections were incubated in Tris-EDTA pH 9.0 for antigen retrieval and endogenous peroxidase was blocked by H2O2. Primary antibodies were incubated in antibody diluent (Dako) and detected by secondary biotinylated antibodies as listed in Supplementary data. After incubation with streptavidin-coupled peroxidase (Medac Diagnostika), signals were visualized by AEC (3-amino-9-ethylcarbazole) or DAB (3,3′-diaminobenzidine) (Life technologies). Nuclei were counterstained with Haemalaun (Merck) and mounted in Mowiol. Staining intensity was scored on a scale from 0–3, analyzed by contingency tables and statistically evaluated with the X2 test using GraphPad Prism v5.04.
RESULTS
ΔNp63 is essential for proliferation but not survival in SCC
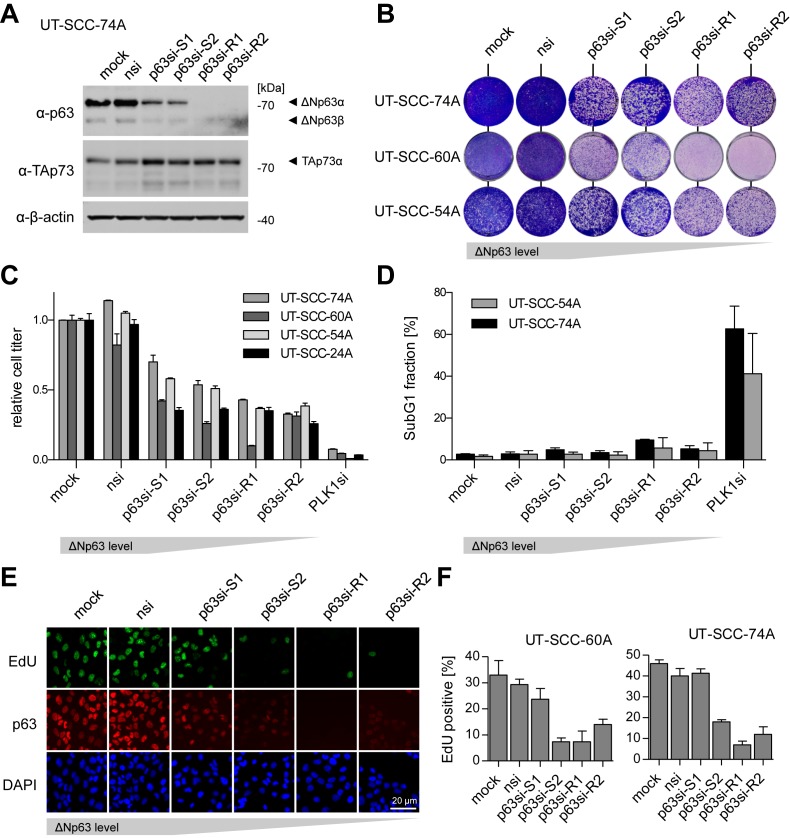
To study ΔNp63 in SCC, we first analyzed p63 protein expression in human HNSCC and LUSC samples (Supplementary Figure S1). More than 60% of HNSCC and >75% of LUSC expressed significantly higher p63 levels than either normal tissues or lung adenocarcinoma. In parallel, we characterized a panel of established HNSCC cell lines for expression of all p53 family members (Supplementary Figure S2). Virtually all of them showed high levels of ΔNp63 and 9 out of 16 a concomitant overexpression of TAp73 consistent with previous studies (Supplementary Figure S2A and B) (29). For functional analysis we depleted p63 with an epiallelic series of siRNAs yielding different degrees of ΔNp63 knockdown (Figure 1A). We included siRNAs targeting both p63 isoforms TAp63 and ΔNp63 (p63si-S1 and p63si-R2) as well as siRNAs specific for ΔNp63 as the major p63 isoform in SCC (p63si-S2 and p63si-R1). The depletion of ΔNp63 led to reduced colony formation and cell titer which was caused by impaired proliferation but not cell death (Figure 1C–F). A possible explanation could be the previously described release of TAp73 from its inactivation by ΔNp63 (14,29). However, the anti-proliferative effect of p63 depletion was independent of TAp73 as co-depletion of p73 did not rescue the phenotype and reduced proliferation was comparable in TAp73-negative cell lines (Supplementary Figure S3A–C). Therefore, we concluded that ΔNp63 is essential for proliferation but not survival in SCC independent of TAp73.
Figure 1.
ΔNp63 is essential for proliferation but not survival in squamous cell carcinoma (SCC). (A) Western Blot evaluating knockdown efficiency and specificity of siRNAs targeting p63 (p63si-S1/2,-R1/2) in the head and neck SCC (HNSCC) cell line UT-SCC-74A compared to controls (mock, nsi); β-actin: loading control. (B) Colony formation assay of HNSCC cell lines after p63 knockdown. (C) Relative cell titer of p63-depleted HNSCC cell lines. Bars show mean relative cell titer +SD normalized to mock treated cells (n = 3). PLK1si: positive control. (D) SubG1 flow cytometry analysis of propidium iodide-stained cells after p63-depletion. Bars show mean % +SEM (n = 2). PLK1si: positive control. (E) Representative immunofluorescence images illustrating EdU incorporation by proliferating UT-SCC-74A after p63-depletion. DAPI: nuclear counterstain. (F) Quantification of EdU-positive cells by immunofluorescence after p63-depletion. Bars show mean % +SD of EdU-positive cells (n = 3).
ΔNp63 modulates cisplatin response
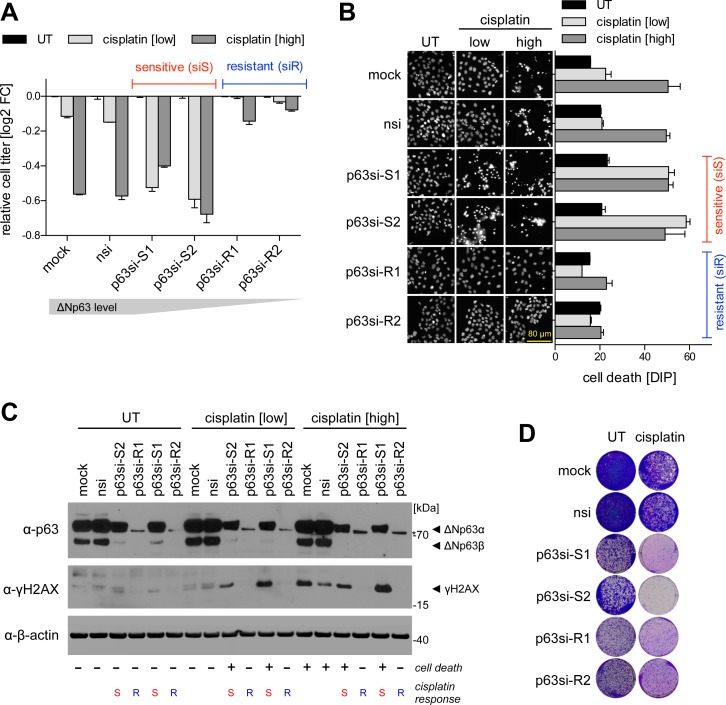
ΔNp63 was described to control the DNA damage response to cisplatin as the first-line chemotherapy for advanced stages of SCC (14,30–34). When we depleted ΔNp63 in HNSCC cells using the epiallelic series of siRNAs we noted completely opposite effects on the cisplatin response depending on knockdown efficiency: partial depletion of ΔNp63 (p63si-S1/S2) sensitized toward cisplatin, whereas the complete knockdown (p63si-R1/R2) rendered cells more resistant (Figure 2A). Sensitivity was associated with induction of cell death (Figure 2B, Supplementary Figure S4), but was again not caused by release and activation of pro-apoptotic TAp73 since co-depletion of TAp73 did not rescue from cell death (Supplementary Figure S3D and E). Sensitized cells showed increased levels of DNA damage at low doses of cisplatin as demonstrated by the DNA DSB marker γH2A.X (Figure 2C). Resistance observed upon complete p63 depletion is most likely caused by the lack of proliferation since cells artificially arrested in G0/G1 phase were equally resistant (Supplementary Figure S5). Moreover, γH2A.X was absent in resistant cells even at high cisplatin doses consistent with DSBs originating from collapsed replication forks at sites of cisplatin-induced DNA crosslinks (18). Importantly, although complete p63 depletion rendered cells largely resistant to cisplatin, clonogenicity was strongly reduced irrespective of cisplatin treatment indicating that the anti-proliferative effect of an efficient p63 knockdown is dominant (Figure 2D). In summary, targeting ΔNp63 has therapeutically beneficial effects in SCC by enhancing cisplatin cytotoxicity at low levels of inhibition or by reducing cell proliferation and clonogenicity at higher levels of depletion.
Figure 2.
ΔNp63 modulates cisplatin response. p63 knockdown results either in increased sensitivity (siS) or resistance (siR) to cisplatin treatment in SCC cells. UT: untreated; cisplatin: low: 8 μM, high: 16 μM. (A) Relative cell titer after cisplatin-treatment of p63-depleted UT-SCC-74A. Cells were transfected with siRNAs for 48 h followed by cisplatin treatment for 24 h. Bars show mean relative cell titer as log2 fold change (FC) +SD of untreated condition (n = 2). (B) Cell death evaluation by DAPI intensity per pixel (DIP) analysis (Supplementary Figure S4). Cells were treated as in A. Left: Representative images of DAPI-stained cells. Right: Quantification. Bars show mean DIP value +SD (n = 2). (C) Western Blot of the DNA DSB marker γH2A.X. Cells were transfected with siRNAs for 48 h followed by cisplatin treatment for 8 h. p63 blot shows knockdown efficiencies of siRNAs. β-actin: loading control. Cisplatin responses (− viable, + dead; S sensitive, R resistant) are indicated below the blots. (D) Colony formation of p63-depleted UT-SCC-74A cells without (same as Figure 1B) or with 24 h cisplatin treatment [1 μM].
ΔNp63 regulates DNA damage repair
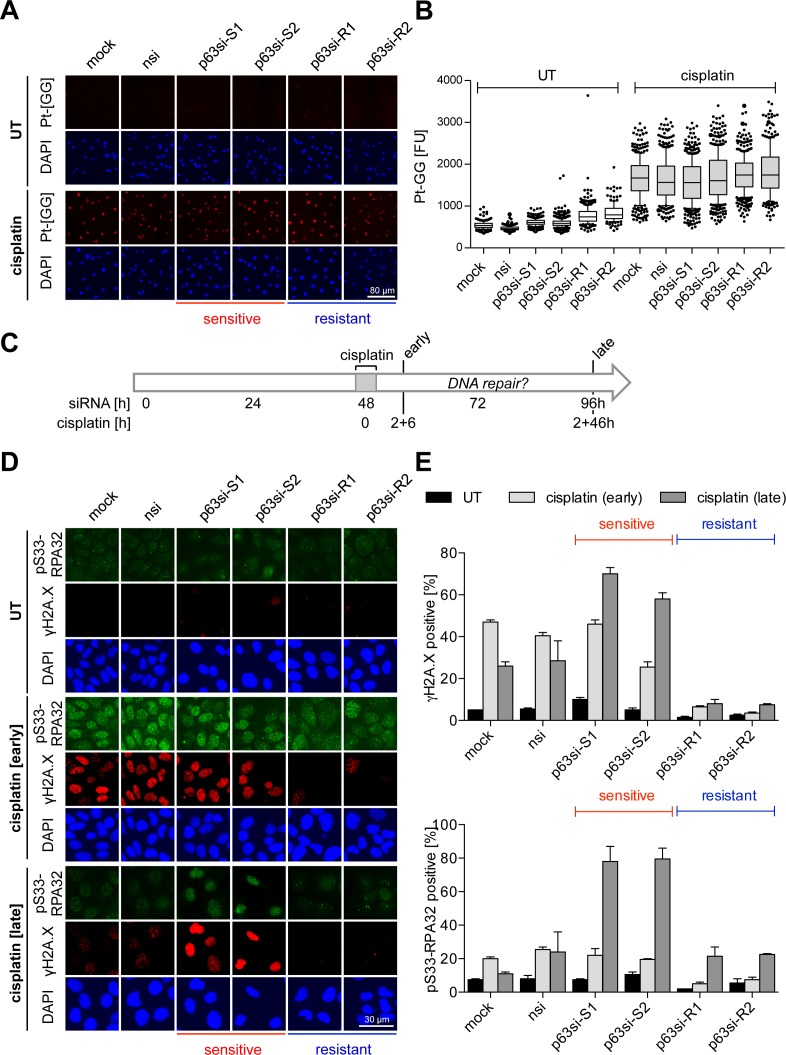
Cisplatin cytotoxicity is predominantly caused by ICLs (18). To exclude that differences in cisplatin response are due to variations in the abundance of these lesions, we measured the most prevalent guanine–guanine intrastrand crosslinks (Pt-[GG]) using a specific antibody (28). Immunofluorescence showed no major changes in the induction of Pt-[GG] shortly after cisplatin treatment irrespective of the degree of ΔNp63 depletion (Figure 3A and B). To explain differences in DNA damage levels, we hypothesized that ΔNp63 levels affect the DNA repair capacity. We therefore measured in a time course experiment γH2A.X and phosphorylated replication protein A (pS33-RPA32) that accumulate during DNA replication as a consequence of unresolved DNA damage (Figure 3C–E). Early after short-term cisplatin exposure, control cells showed strong DNA damage that was resolved later. Resistant cells—despite equal initial levels of cisplatin adducts—showed only weak RPA32 phosphorylation and failed to induce γH2A.X, likely because of their proliferative arrest. Strikingly, in cisplatin-sensitized cells γH2A.X and pS33-RPA32 progressively increased over time indicating defective damage repair when p63 expression is reduced.
Figure 3.
ΔNp63 regulates DNA damage repair. UT-SCC-74A were transfected with siRNAs for 48 h followed by cisplatin treatment [8 μM] for 2 h. Cells were analyzed by immunofluorescence at indicated time points. Cisplatin response is indicated as sensitive or resistant. (A and B) Detection of platinum-DNA adducts with Pt-[GG]-specific antibody (28) 6 h after cisplatin treatment. (A) Representative immunofluorescence images of Pt-[GG] adducts. DAPI: nuclear counterstain. (B) Quantification of platinum-DNA adducts from A by single-cell analysis. Box plots show fluorescence intensities [FU] of single cells, whiskers: 10th and 90th percentiles. (C–E) Time course experiment for analysis of DNA damage induction and repair. (C) Experimental setup: siRNA transfection, cisplatin treatment (for 2 h) and time points of analysis (early, 2 + 6 h and late, 2 + 46 h) are indicated. (D) Representative immunofluorescence images of DNA damage markers γH2A.X and serine 33-phosphorylated (pS33) RPA32. DAPI: nuclear counterstain. (E) Quantification of DNA damage markers from (D). Bars show mean % +SEM (n = 2) of cells positive for γH2A.X (top) and pS33-RPA32 (bottom).
ΔNp63 transactivates the Fanconi anemia pathway for DNA crosslink repair
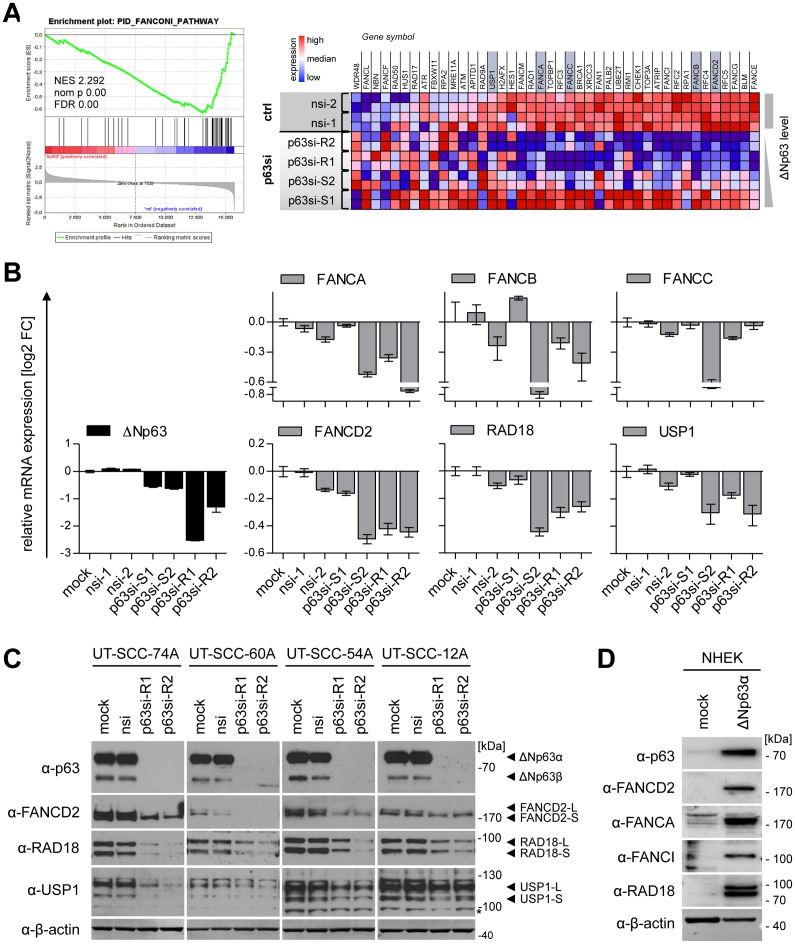
As ΔNp63 is a transcription factor, we analyzed gene expression profiles after p63-knockdown by cDNA microarray analysis (Supplementary Figure S6). Gene set enrichment analysis confirmed established functions of p63 like epidermal development and cell cycle regulation. Moreover, a plethora of DNA repair pathways was altered upon p63 depletion in line with previous studies on mouse embryonic fibroblasts and primary keratinocytes (35–37). Interestingly, we identified the FA pathway (Figure 4A) as a prominent novel p63-target in SCC – a pathway essential for repair of DNA crosslinks induced by agents like cisplatin (17–19). qPCR and Western Blot analysis of major components of this pathway like FANCD2 and its upstream regulators RAD18 and USP1 confirmed a p63-dependent gene expression in several HNSCC cell lines (Figure 4) which cannot be explained solely by a cell cycle-dependent regulation (Supplementary Figure S7). When primary keratinocytes were transduced with ΔNp63α this proved sufficient to induce strong expression of FANCD2 along with the FA pathway genes FANCA, FANCI and RAD18 (Figure 4D). We therefore conclude that ΔNp63 transactivates the FA pathway in SCC.
Figure 4.
ΔNp63 transactivates the Fanconi anemia pathway. UT-SCC-74A were transfected with p63-targeting siRNAs and controls (mock, nsi) in duplicates and harvested 72 h later. The transcriptome was analyzed with a human cDNA microarray and altered pathways evaluated by GSEA (see Supplementary Figure S6). (A) Enrichment plot (left) and corresponding heatmap (right) of the gene set ‘PID_FANCONI_PATHWAY’ illustrates p63-dependent expression (p63si versus control (ctrl) samples). Genes marked in grey were validated in B. NES, normalized enrichment score; nom p, nominal P-value; FDR, false discovery rate. (B) qPCR validation of p63-dependent regulation for selected FA pathway genes. mRNA levels were normalized to mock-treated sample and expressed as relative log2 fold change [log2 FC]. Bars represent mean +SD (n = 2). (C) Western Blot of FA pathway proteins FANCD2, RAD18 and USP1 after ΔNp63 knockdown in indicated HNSCC cell lines. (D) Western Blot for expression of FA pathway genes FANCD2, FANCA, FANCI and RAD18 in primary normal human epidermal keratinocytes transduced with ΔNp63α lentivirus. β-actin: loading control. -L: long (mono-ubiquitinated) isoform, -S: short isoform, * unspecific band.
ΔNp63 binds and regulates an enhancer upstream of FANCD2
The central hub in the FA pathway is FANCD2 (19), which we identified as a novel ΔNp63-regulated gene in SCC. To mechanistically characterize its regulation by ΔNp63, we searched for p63 binding sites in publicly available ChIPseq data sets. p63 ChIPseq data from primary keratinocytes (35) showed binding of p63 ∼10 kb upstream of FANCD2 but not to its promoter region (Figure 5A). By motif search we identified a putative p63 binding site (BS) consisting of two intertwined p63 motifs. In vitro EMSA demonstrated sequence-specific binding of ΔNp63α, the major isoform in SCC, to this site (Figure 5B). Moreover, transactivation of a luciferase-reporter by ΔNp63α through this site provided evidence for its functionality (Figure 5C). Sequence specificity was confirmed by mutating strongly conserved pyrimidines which prevented binding and transactivation (Figure 5B and C).
Importantly, we could confirm in vivo binding of endogenous ΔNp63 to the −10 kb region but not to the transcription start site (TSS) of FANCD2 by ChIP-qPCR analysis in SCC cells (Figure 5D). Analysis of histone modifications from the Encyclopedia of DNA elements (ENCODE) data sets (38) revealed the presence of typical enhancer marks with high mono-methylation and low tri-methylation of histone H3K4 (H3K4me1 high, H3K4me3 low) in multiple cell lines including primary keratinocytes (Supplementary Figure S8) (39). We could confirm the presence of these enhancer marks in SCC at the −10 kb p63 response element but not the TSS (Figure 5E). In contrast to primary NHEK (Supplementary Figure S8), we could detect histone H3K27 acetylation (H3K27ac) as an activation mark at the −10 kb enhancer in SCC cells (Supplementary Figure S8, Figure 5F). Most importantly, depletion of ΔNp63 reduced H3K27 acetylation at the enhancer and TSS (Figure 5F). We conclude that ΔNp63 in SCC binds to and aberrantly activates an enhancer 10 kb upstream of FANCD2.
FANCD2 limits the cytotoxic cisplatin response in SCC
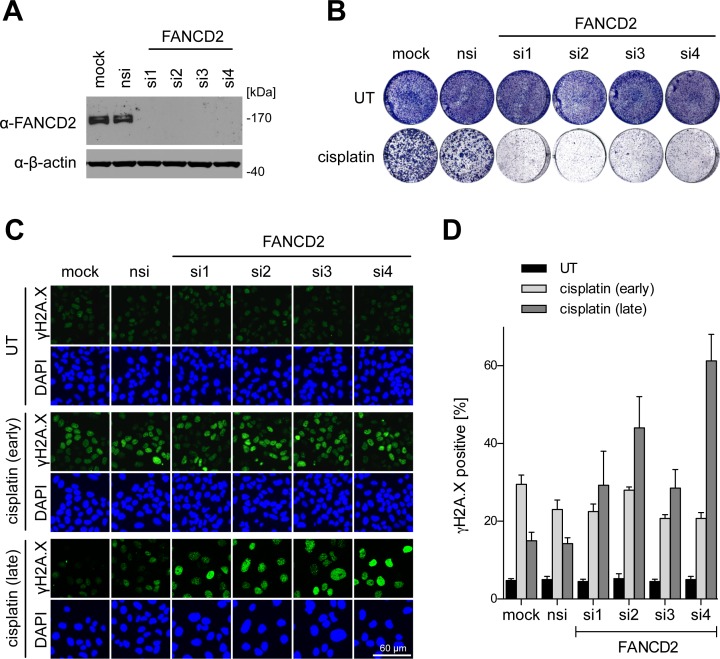
To assess the relevance of FANCD2 for the cisplatin response in SCC, we depleted FANCD2 by siRNAs and analyzed colony formation before and after cisplatin treatment (Figure 6A and B). FANCD2 knockdown had no effect on proliferation of untreated cells but strongly enhanced the cisplatin response leading to reduced colony formation (Figure 6B). Enforced expression of FANCD2 rescued the cisplatin sensitivity induced by FANCD2 knockdown, excluding an off-target effect (Supplementary Figure S9). However, enforced expression of FANCD2 was not sufficient to overcome either the proliferative arrest or the cisplatin sensitivity of ΔNp63-depleted cells (Supplementary Figures S9 and S10). This is consistent with FANCD2 being not the only FA pathway component downregulated by ΔNp63 depletion (Figure 4) and further supported by cisplatin sensitization following knockdown of FANCA, FANCL, FANCI or RAD18 (Supplementary Figure S11). Consistent with its predicted contribution to DNA damage repair, depletion of FANCD2 antagonized the reduction of γH2A.X after cisplatin exposure in SCC cells and led to an accumulation of γH2A.X indicative of deficient DNA repair (Figure 6C and D). High-level expression of FANCD2, which in SCC is maintained by p63, therefore limits the cytotoxic effects of cisplatin.
Figure 6.
FANCD2 limits the cytotoxic cisplatin response in SCC. UT-SCC-74A were transfected with four independent siRNAs targeting FANCD2 compared to mock and nsi controls. (A) Western Blot evaluating knockdown efficiency of FANCD2-targeting siRNAs. β-actin: loading control. (B) Colony formation of FANCD2-depleted cells with or without 24 h cisplatin treatment [2 μM]. (C) Representative immunofluorescence images of FANCD2-depleted cells stained for γH2A.X early (6 h) and late (46 h) after short-term (2 h) cisplatin treatment. Experiment was performed as depicted in Figure 3C. (D) Quantification of (C). Bars show mean % +SD of γH2A.X-positive cells in treated (cisplatin) and untreated (UT) cells (n = 4).
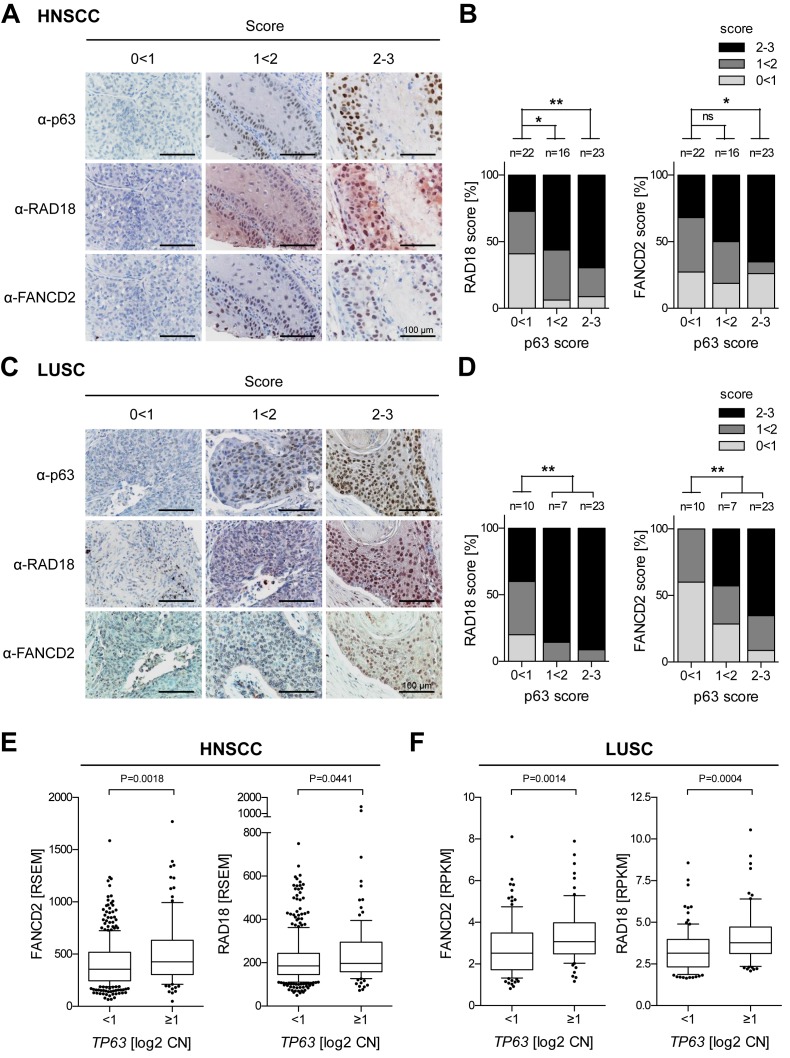
p63 levels correlate with RAD18 and FANCD2 expression in SCC patients
To explore a potential clinical relevance of our results, we evaluated expression levels of p63 in parallel to the FA pathway components RAD18 and FANCD2 in SCC patients by immunostaining TMAs from HNSCC and LUSC patients. Statistical analysis revealed a significant correlation of p63 with RAD18 and FANCD2 protein levels in both tumor types (Figure 7A–D). In addition, we analyzed copy number and RNAseq data from The Cancer Genome Atlas (TCGA) (4,6). Consistent with p63 transactivating FA pathway genes, a copy number gain of TP63 was found to correlate significantly with elevated expression of FANCD2 and RAD18 in both HNSCC and LUSC (Figure 7E and F).
Figure 7.
p63 levels correlate with RAD18 and FANCD2 expression in SCC patients. (A–D) Tissue microarrays of paraffin-embedded SCC samples (A and B) HNSCC; (C and D) LUSC were immunostained for p63, FANCD2 and RAD18. Protein levels were scored as low (0 < 1), intermediate (1 < 2) and high (2–3). (A and C) Representative images. (B and D) Distribution of RAD18 and FANCD2 scores plotted against p63 levels. n = number of samples analyzed; statistical significance tested by χ2 test. P-value ***<0.001, ** 0.001<0.01, * 0.01<0.05, ns >0.05. (E and F) FANCD2 and RAD18 overexpression in HNSCC (E, n = 493) and LUSC (F, n = 178) with TP63 copy number (CN) gain [log2 CN ≥ 1]. RNAseq and CN data were taken from The Cancer Genome Atlas (TCGA). Box plots show RNA expression (RSEM or RPKM), whiskers: 10th and 90th percentiles. Statistical significance tested by Mann–Whitney test.
DISCUSSION
In this study, we extensively characterized the function of ΔNp63 in order to assess its suitability as a therapeutic target in SCC. In line with previous reports we could confirm its essential oncogenic role for proliferation of SCC cells (16,40,41). However, a pro-survival function as described by others could not be observed in the panel of investigated HNSCC cell lines (14,29). Although previous studies have demonstrated the release of tumor-suppressive TAp73 from ΔNp63/TAp73 complexes upon ΔNp63 depletion (14,29), anti-proliferative effects of ΔNp63 were independent of TAp73 in our experiments despite high expression levels of tumor-suppressive TAp73 in some of the cell lines. This has also been observed by others and different mechanisms for ΔNp63's pro-proliferative activity have been reported such as the direct regulation of genes and microRNAs involved in cell cycle control and the modification of the chromatin landscape through p63-dependent recruitment of histone deacetylases and deposition of the histone variant H2A.Z (3,11,15,16,42). ΔNp63 is therefore a key regulator of SCC proliferation by a variety of mechanisms which may differ between cell types.
In our experiments we did not focus further on the underlying mechanism of cell proliferation control, but instead investigated in more detail its interplay with genotoxic stress, in particular DNA damage induced by the chemotherapeutic agent cisplatin which is the most widely used drug for the treatment of recurrent and/or metastatic SCC (1,43). In contrast to unstressed conditions, we observed ΔNp63 to strongly modulate survival of SCC cells exposed to cisplatin. Intriguingly, opposite effects were observed depending on ΔNp63 expression levels: sensitization occurred at intermediate levels – resistance at low levels. We ascribe these conflicting outcomes to the simultaneous regulation of DNA replication and DNA repair by ΔNp63. On the one hand, ΔNp63 drives expression of DNA repair genes explaining the accumulation of cisplatin-induced DNA damage upon downregulation of ΔNp63. On the other hand, complete loss of ΔNp63 triggered a G0/G1 arrest. As cisplatin-induced lesions are particularly toxic during S-phase when replication forks encountering crosslinks collapse and form DNA breaks (18), cisplatin-crosslinks are expected to be less cytotoxic in a ΔNp63-deficient, non-replicating compared to a ΔNp63-expressing, replicating cell. Consistently, we observed similar amounts of cisplatin-induced DNA crosslinks irrespective of ΔNp63 levels (Figure 3A and B) and resistance to cisplatin in cells arrested in G0/G1 by a variety of different mechanisms (Supplementary Figure S5). Importantly, although highly efficient depletion of ΔNp63 strongly reduces the cytotoxicity of cisplatin, tumor cell proliferation and clonogenicity are nevertheless effectively inhibited under these conditions (Figure 2D). Thus, SCC patients would benefit from ΔNp63 inhibition both from impaired tumor cell proliferation, which occurs primarily at maximum ΔNp63 inhibition, and from sensitization to chemotherapy at lower than maximal inhibition levels.
p63-mediated transcriptional control of DNA repair pathways, such as homologous recombination (35,37), poly(ADP-ribose) polymerase 1 signaling (44) and nucleotide excision repair (45,46), has been observed previously. While these studies focused on primary cell types such as mouse embryonic fibroblasts and keratinocytes, we here provide first evidence that ΔNp63 transcriptionally regulates a plethora of genes involved in DNA repair pathways also in human tumor cells with striking consequences for the chemotherapy response. Among these, we identified the FA pathway as a novel ΔNp63 target in SCC, which is crucial for the repair of ICLs induced by crosslinking agents like cisplatin (17–19). In particular, the expression of FANCD2 and its upstream regulators RAD18 and USP1 are dependent on ΔNp63 and their expression correlated significantly with p63 protein levels and TP63 copy number across a large number of human SCC samples of the lung and head and neck.
FANCD2 has been recently described as a p53-repressed target in primary keratinocytes whose expression can be induced by p53-depletion in a p63-dependent manner (35). ChIPseq experiments revealed p63 binding to a region ∼10 kb upstream of FANCD2 in primary human keratinocytes (35). We observed ΔNp63 binding to this region also in SCC and identified a functional ΔNp63 response elements consisting of two intertwined p63 motifs that are sufficient to direct sequence-specific ΔNp63 binding and transactivation. This region bears histone modification marks characteristic of an enhancer (high H3K4me3, low H3K4me1) consistent with preferential binding of ΔNp63 to enhancers (35,47,48). However, H3K27 acetylation as a sign of enhancer activity was only detected in SCC (Figure 5F), not in primary keratinocytes (Supplementary Figure S8), and was dependent on ΔNp63. This suggests that aberrant overexpression of ΔNp63 in SCC drives pathological activation of this FANCD2 enhancer. In support of this, we observed strong FANCD2 induction upon ectopic ΔNp63α expression in primary normal human epidermal keratinocytes, indicating that ΔNp63 is sufficient to trigger FANCD2 upregulation (Figure 4D). In analogy, other tumors frequently amplify Myc and oncogenic Myc expression levels were similarly shown to activate enhancers driving tumor-progression genes which are inactive at physiological Myc levels (49). It will therefore be interesting to compare p63 target genes in non-transformed keratinocytes with physiological p63 expression and malignant SCC cells with amplified p63 to better understand differences in physiological and pathological functions of p63, respectively.
FANCD2 is the central factor of the FA pathway which is activated by ICLs and orchestrates downstream DNA repair through translesion DNA synthesis, nucleotide excision repair and homologous recombination (17,19). Indeed, FANCD2 is essential for repair of cisplatin-induced DNA damage in SCC as its depletion sensitized cells to cisplatin. FANCD2 is therefore on the one hand a mediator of oncogenic functions of ΔNp63 in SCC, on the other it has been classified as a tumor suppressor gene as its deletion predisposes to the development of multiple epithelial tumor types including SCC (50–52). Although confusing at first sight, this is not unexpected for a DNA repair gene. For example, deficiency of the O6-methylguanine-DNA methyltransferase (MGMT), which repairs the mutagenic DNA lesion O6-methylguanine, predisposes mice to cancer while at the same time predicting increased responsiveness of glioblastoma patients to the alkylating drug temozolomide (53). Similarly, as a central component of the genome maintenance machinery FANCD2 functions to prevent tumor development while at the same time counteracting cancer therapies that rely on the induction of DNA damage for tumor cell killing. The FA pathway is therefore contributing to tumor suppression but when overly active renders tumor cells resistant to treatment with DNA crosslinking drugs such as cisplatin.
DNA repair and tumor suppression by FANCD2 requires an activating monoubiquitination that is antagonized by the ubiquitin-specific protease USP1 (54,55). Intriguingly, we identified not only FANCD2 but also USP1 to be upregulated by ΔNp63 (Figure 4). USP1-knockout mice show elevated levels of monoubiquitinated FANCD2 and are resistant to Ras-driven skin carcinogenesis (52). However, USP1-deficiency also results—unexpectedly—in DNA crosslinker sensitivity, indicating that deubiquitination is critical for FANCD2 functions in DNA repair (56,57). USP1 upregulation by ΔNp63 is therefore consistent with improved DNA crosslink repair in SCC cells.
Targeting FANCD2 in combination with ICL-inducing drugs would therefore be an attractive approach for improved chemotherapy. Recently, mTOR has been described as an upstream regulator of FANCD2 in rhabdomyosarcoma offering a rationale for mTOR inhibition together with cisplatin in tumor therapy (58). Indeed, cisplatin combination therapy with the mTOR inhibitor everolimus has demonstrated significantly enhanced efficacy over cisplatin monotherapy in several in vitro and in vivo studies in multiple types of cancer including SCC (59,60). Furthermore, first clinical trials demonstrated a benefit of combining mTOR inhibitors with cisplatin for HNSCC treatment (61). Yet, to which extent FANCD2 inhibition contributes and whether the therapeutic benefit depends on p63 expression levels, remains to be elucidated.
In summary, we characterized ΔNp63 as a key factor promoting both proliferation and DNA repair in SCC with major impact on the therapeutic chemotherapy response. These results make ΔNp63 a promising target for the development of novel therapeutic strategies. First, inhibition of ΔNp63 would be beneficial for tumor therapy by preventing SCC cell proliferation. Second, targeting ΔNp63 would prevent repair of DNA lesions such as interstrand crosslinks via the FA pathway and thereby sensitize SCC cells to the current first-line therapy with cisplatin. Our identification of p63-driven DNA repair highlights the potential for therapeutic targeting of DNA repair enzymes in TP63 amplified tumors, even while targeting p63 as a transcription factor itself remains a challenge.
ACCESSION NUMBERS
Microarray data have been deposited in EBI ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3670.
Supplementary Material
Acknowledgments
We thank Thomas Carey (University of Michigan, USA) and Reidar Grénman (University of Turku, Finland) for providing HNSCC-derived cell lines, Alan D'Andrea for providing FANCD2 expression plasmids, Michael Krause and Angelika Filmer for performing microarray experiments, Sigrid Bischofsberger for excellent technical assistance in immunohistochemistry. The authors are grateful to the members of the Stiewe laboratory for fruitful discussions and support of the project. A.C.B., M.M. and A.N. acknowledge support from Universitätsklinikum Giessen und Marburg (UKGM).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG [TRR81, KFO210, STI 182/7–1 to T.S.]; European Research Council, Deutsche Krebshilfe [111250]; Deutsche José Carreras Leukämie-Stiftung, Von-Behring-Röntgen-Stiftung, Rhön Klinikum AG and Universities of Giessen and Marburg Lung Center [UGMLC, LOEWE]. Funding for open access charge: Publication charges funded by the European Research Council [P73CANCER].
Conflict of interest statement. None declared.
REFERENCES
- 1.Leemans C.R., Braakhuis B.J.M., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Ramsey M.R., Wilson C., Ory B., Rothenberg S.M., Faquin W., Mills A.A., Ellisen L.W. FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J. Clin. Invest. 2013;123:3525–3538. doi: 10.1172/JCI68899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voet D., Cibulskis K., McKenna A., Lander E.S., Gabriel S., Imielinski M., Helman E., Chun H.-J.E., Mungall A.J., Pleasance E., et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hibi K., Trink B., Patturajan M., Westra W.H., Caballero O.L., Hill D.E., Ratovitski E.A., Jen J., Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zangen R., Ratovitski E., Sidransky D. DeltaNp63alpha levels correlate with clinical tumor response to cisplatin. Cell Cycle. 2005;4:1313–1315. doi: 10.4161/cc.4.10.2066. [DOI] [PubMed] [Google Scholar]
- 8.Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 9.Senoo M., Pinto F., Crum C.P., Mckeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 10.King K.E., Ha L., Camilli T., Weinberg W.C. Delineating molecular mechanisms of squamous tissue homeostasis and neoplasia: focus on p63. J. Skin Cancer. 2013:632028. doi: 10.1155/2013/632028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keyes W.M., Pecoraro M., Aranda V., Vernersson-Lindahl E., Li W., Vogel H., Guo X., Garcia E.L., Michurina T.V., Enikolopov G., et al. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8:164–176. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X., Li F., Fang Z., Gao Y., Li F., Fang R., Yao S., Sun Y., Li L., Zhang W., et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat. Commun. 2014;5:1–13. doi: 10.1038/ncomms4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tordella L., Koch S., Salter V., Pagotto A., Doondeea J.B., Feller S.M., Ratnayaka I., Zhong S., Goldin R.D., Lozano G., et al. ASPP2 suppresses squamous cell carcinoma via RelA/p65-mediated repression of p63. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17969–17974. doi: 10.1073/pnas.1309362110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocco J.W., Leong C.-O., Kuperwasser N., DeYoung M.P., Ellisen L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Ramsey M.R., He L., Forster N., Ory B., Ellisen L.W. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. 2011;71:4373–4379. doi: 10.1158/0008-5472.CAN-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallant-Behm C.L., Ramsey M.R., Bensard C.L., Nojek I., Tran J., Liu M., Ellisen L.W., Espinosa J.M. ΔNp63α represses anti-proliferative genes via H2A.Z deposition. Genes Dev. 2012;26:2325–2336. doi: 10.1101/gad.198069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deans A.J., West S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H., D'Andrea A.D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Räsänen K., Virtanen I., Salmenperä P., Grenman R., Vaheri A. Differences in the nemosis response of normal and cancer-associated fibroblasts from patients with oral squamous cell carcinoma. PLoS One. 2009;4:e6879. doi: 10.1371/journal.pone.0006879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lansford C., Grenman R., Bier H., Somers K.D., Kim S.Y., Whiteside T.L., Clayman G., Carey T.E. Head and neck cancers. In: Masters J, editor. Human cell culture, Cancer cell lines part 2. Vol. 2. Dordrecht: Kluwer Academic Press; 1999. pp. 185–255. [Google Scholar]
- 22.Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T., et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charles J.P., Fuchs J., Hefter M., Vischedyk J.B., Kleint M., Vogiatzi F., Schäfer J.A., Nist A., Timofeev O., Wanzel M., et al. Monitoring the dynamics of clonal tumour evolution in vivo using secreted luciferases. Nat. Commun. 2014;5:3981. doi: 10.1038/ncomms4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlereth K., Beinoraviciute-Kellner R., Zeitlinger M.K., Bretz A.C., Sauer M., Charles J.P., Vogiatzi F., Leich E., Samans B., Eilers M., et al. DNA binding cooperativity of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell. 2010;38:356–368. doi: 10.1016/j.molcel.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer M., Bretz A.C., Beinoraviciute-Kellner R., Beitzinger M., Burek C., Rosenwald A., Harms G.S., Stiewe T. C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res. 2008;36:1900–1912. doi: 10.1093/nar/gkn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liedert B. Adduct-specific monoclonal antibodies for the measurement of cisplatin-induced DNA lesions in individual cell nuclei. Nucleic Acids Res. 2006;34:e47. doi: 10.1093/nar/gkl051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeYoung M.P., Johannessen C.M., Leong C.-O., Faquin W., Rocco J.W., Ellisen L.W. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66:9362–9368. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- 30.Sen T., Sen N., Brait M., Begum S., Chatterjee A., Hoque M.O., Ratovitski E., Sidransky D. DeltaNp63alpha confers tumor cell resistance to cisplatin through the AKT1 transcriptional regulation. Cancer Res. 2011;71:1167–1176. doi: 10.1158/0008-5472.CAN-10-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., Jeong J.S., Okamura J., Sook-Kim M., Zhu H., Guerrero-Preston R., Ratovitski E.A. Global tumor protein p53/p63 interactome: making a case for cisplatin chemoresistance. Cell Cycle. 2012;11:2367–2379. doi: 10.4161/cc.20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y., Bell L.N., Okamura J., Kim M.S., Mohney R.P., Guerrero-Preston R., Ratovitski E.A. Phospho-ΔNp63α/SREBF1 protein interactions: bridging cell metabolism and cisplatin chemoresistance. Cell Cycle. 2012;11:3810–3827. doi: 10.4161/cc.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee A., Upadhyay S., Chang X., Nagpal J.K., Trink B., Sidransky D. U-box-type ubiquitin E4 ligase, UFD2a attenuates cisplatin mediated degradation of DeltaNp63alpha. Cell Cycle. 2008;7:1231–1237. doi: 10.4161/cc.7.9.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan M., Luong P., Hudson C., Gudmundsdottir K., Basu S. c-Abl phosphorylation of ΔNp63α is critical for cell viability. Cell Death Dis. 2010;1:e16. doi: 10.1038/cddis.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDade S.S., Patel D., Moran M., Campbell J., Fenwick K., Kozarewa I., Orr N.J., Lord C.J., Ashworth A.A., McCance D.J. Genome-wide characterization reveals complex interplay between TP53 and TP63 in response to genotoxic stress. Nucleic Acids Res. 2014;42:6270–6285. doi: 10.1093/nar/gku299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll D.K., Carroll J.S., Leong C.-O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y.-L., Sengupta S., Gurdziel K., Bell G.W., Jacks T., Flores E.R. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet. 2009;5:e1000680. doi: 10.1371/journal.pgen.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2012;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 40.Yang X., Lu H., Yan B., Romano R.-A., Bian Y., Friedman J., Duggal P., Allen C., Chuang R., Ehsanian R., et al. ΔNp63 versatilely regulates a Broad NF-κB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71:3688–3700. doi: 10.1158/0008-5472.CAN-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hau P.M., Yip Y.L., Huen M.S.Y., Tsao S.W. Loss of ΔNp63α promotes mitotic exit in epithelial cells. FEBS Lett. 2011;585:2720–2726. doi: 10.1016/j.febslet.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 42.Candi E., Amelio I., Agostini M., Melino G. MicroRNAs and p63 in epithelial stemness. Cell Death Differ. 2015;22:12–21. doi: 10.1038/cdd.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salama J.K., Vokes E.E. New radiotherapy and chemoradiotherapy approaches for non-small-cell lung cancer. J. Clin. Oncol. 2013;31:1029–1038. doi: 10.1200/JCO.2012.44.5064. [DOI] [PubMed] [Google Scholar]
- 44.Montariello D., Troiano A., Malanga M., Calabrò V., Quesada P. p63 involvement in poly(ADP-ribose) polymerase 1 signaling of topoisomerase I-dependent DNA damage in carcinoma cells. Biochem. Pharmacol. 2013;85:999–1006. doi: 10.1016/j.bcp.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 45.Ferguson-Yates B.E., Li H., Dong T.K., Hsiao J.L., Oh D.H. Impaired repair of cyclobutane pyrimidine dimers in human keratinocytes deficient in p53 and p63. Carcinogenesis. 2008;29:70–75. doi: 10.1093/carcin/bgm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Lin M., Zhang C., Wang D., Feng Z., Hu W. TAp63γ enhances nucleotide excision repair through transcriptional regulation of DNA repair genes. DNA Repair (Amst) 2012;11:167–176. doi: 10.1016/j.dnarep.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sethi I., Sinha S., Buck M.J. Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes. BMC Genomics. 2014;15:1042. doi: 10.1186/1471-2164-15-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kouwenhoven E.N., Oti M., Niehues H., van Heeringen S.J., Schalkwijk J., Stunnenberg H.G., van Bokhoven H., Zhou H. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 2015;16:863–878. doi: 10.15252/embr.201439941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabò A., Kress T.R., Pelizzola M., de Pretis S., Gorski M.M., Tesi A., Morelli M.J., Bora P., Doni M., Verrecchia A., et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Houghtaling S., Timmers C., Noll M., Finegold M.J., Jones S.N., Meyn M.S., Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houghtaling S., Granville L., Akkari Y., Torimaru Y., Olson S., Finegold M., Grompe M. Heterozygosity for p53 (Trp53+/-) accelerates epithelial tumor formation in fanconi anemia complementation group D2 (Fancd2) knockout mice. Cancer Res. 2005;65:85–91. [PubMed] [Google Scholar]
- 52.Park E., Kim H., Kim J.M., Primack B., Vidal-Cardenas S., Xu Y., Price B.D., Mills A.A., D'Andrea A.D. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol. Cell. 2013;50:908–918. doi: 10.1016/j.molcel.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerson S.L. MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 54.Nijman S.M.B., Huang T.T., Dirac A.M.G., Brummelkamp T.R., Kerkhoven R.M., D'Andrea A.D., Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 55.Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D'Andrea A.D. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 56.Oestergaard V.H., Langevin F., Kuiken H.J., Pace P., Niedzwiedz W., Simpson L.J., Ohzeki M., Takata M., Sale J.E., Patel K.J. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol. Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., D'Andrea A.D. Inactivation of murine Usp1 results in genomic instability and a fanconi anemia phenotype. Dev Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen C., Oswald D., Phelps D., Cam H., Pelloski C.E., Pang Q., Houghton P.J. Regulation of FANCD2 by the mTOR pathway contributes to the resistance of cancer cells to DNA double-strand breaks. Cancer Res. 2013;73:3393–3401. doi: 10.1158/0008-5472.CAN-12-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou G., Zhang Q., Wang L., Liu M., Wang J., Xue L. mTOR inhibitor rapamycin alone or combined with cisplatin inhibits growth of esophageal squamous cell carcinoma in nude mice. Cancer Lett. 2010;290:248–254. doi: 10.1016/j.canlet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 60.Hirashima K., Baba Y., Watanabe M., Karashima R.-I., Sato N., Imamura Y., Nagai Y., Hayashi N., Iyama K.-I., Baba H. Aberrant activation of the mTOR pathway and anti-tumour effect of everolimus on oesophageal squamous cell carcinoma. Br. J. Cancer. 2012;106:876–882. doi: 10.1038/bjc.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pendleton K.P., Grandis J.R. Cisplatin-based chemotherapy options for recurrent and/or metastatic squamous cell cancer of the head and neck. Clin. Med. Insights Ther. 2013;5:103–116. doi: 10.4137/CMT.S10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.