Abstract
DNA-based antisense oligonucleotides (ASOs) elicit cleavage of the targeted RNA by the endoribonuclease RNase H1, whereas siRNAs mediate cleavage through the RNAi pathway. To determine the fates of the cleaved RNA in cells, we lowered the levels of the factors involved in RNA surveillance prior to treating cells with ASOs or siRNA and analyzed cleavage products by RACE. The cytoplasmic 5′ to 3′ exoribonuclease XRN1 was responsible for the degradation of the downstream cleavage products generated by ASOs or siRNA targeting mRNAs. In contrast, downstream cleavage products generated by ASOs targeting nuclear long non-coding RNA Malat 1 and pre-mRNA were degraded by nuclear XRN2. The downstream cleavage products did not appear to be degraded in the 3′ to 5′ direction as the majority of these products contained intact poly(A) tails and were bound by the poly(A) binding protein. The upstream cleavage products of Malat1 were degraded in the 3′ to 5′ direction by the exosome complex containing the nuclear exoribonuclease Dis3. The exosome complex containing Dis3 or cytoplasmic Dis3L1 degraded mRNA upstream cleavage products, which were not bound by the 5′-cap binding complex and, consequently, were susceptible to degradation in the 5′ to 3′ direction by the XRN exoribonucleases.
INTRODUCTION
mRNA surveillance mechanisms are utilized by organisms to ensure fidelity and quality of RNA molecules. A number of surveillance mechanisms function at various steps of RNA biogenesis to mark aberrant RNA molecules for degradation. In eukaryotes, these mechanisms are known to function in both the nucleus and cytoplasm. Fidelity checks of mRNA molecules in the nucleus result in the degradation of improperly processed transcripts both before and after export into the cytoplasm.
Eukaryotic mRNAs contain stabilizing elements, a 5′ 7-methylguanosine cap and a 3′ poly(A) tail that bind to the protein complex containing the 5′-cap binding protein eIF4E and to the poly(A) binding protein (PABP), respectively, which protect the mRNA from exonuclease degradation. Degradation of mRNAs may be deadenylation-dependent or deadenylation-independent or may involve endonucleolytic mechanisms. The deadenylation-independent mechanism is associated with nonsense mediated decay (NMD), which involves decapping followed by 5′ to 3′ degradation by the XRN exoribonucleases (1,2). The deadenylation-dependent mechanism begins with the shortening of the 3′-poly(A) tail by either the CCR4-NOT or PARN deadenylases (3–5). Removal of the poly(A) tail results in the release of PABP and the 5′-cap binding complex (6–11). Once deadenylation is complete, the LSM1–7 complex binds and recruits the decapping enzymes DCP1 and 2 (12–14). Once deadenylated and decapped, the mRNA is degraded in the 5′ to 3′ direction by either the cytoplasmic XRN1 exoribonuclease or by the nuclear XRN2 exoribonuclease and in the 3′ to 5′ direction by the multi-protein exosome complex (4,15–18).
The exosome is a multi-protein complex containing the six structural subunits Mtr3, Rrp41, rRp42, Rrp43, Rrp45 and Rrp46/EXOSC5 arranged in a ring configuration (18–21). The structural core is capped with the Csl4, Rrp4 and Rrp40 proteins (22). Depending on its subcellular localization, the structural core recruits the various catalytically active exonuclease subunits Rrp6/Exosc10, the nuclear exonuclease Rrp44/Dis3 or cytoplasmic exonuclease Dis3L1 (18–21). These exonuclease subunits bind to the core ring structure at the surface opposite the exosome cap structure (22).
A highly efficient means of destroying mRNA begins with endonucleolytic cleavage of the mRNA. Several endogenous endoribonucleases have been shown to target mRNAs including PMR1, IRE1 and the RNase MRP (23–28). These endoribonucleases are both highly specific and tightly regulated. RNase MRP, for example, is restricted to the nucleolus and mitochondria (24,25,29). Following endonucleolytic cleavage of the mRNA, the 5′ fragment (upstream cleavage product) and 3′ fragment (downstream cleavage product) is presumably degraded by the RNA surveillance machinery, although the specific mechanism is unclear.
Short interfering RNAs (siRNAs) may initiate endonucleolytic cleavage of mRNAs mediated by endoribonuclease Argonaute-2 (Ago2), which is followed by degradation of the downstream cleavage products by either XRN1 or XRN4 (30–33). This finding is consistent with the cytoplasmic localization of XRN1 and Ago2. In addition, some studies have claimed that siRNAs function in the nucleus (34,35). However, the activity of siRNAs in the nucleus has been controversial. In our laboratory, we have not observed reproducible Ago2 mediated siRNA activity in the nucleus (36). Micro-RNAs (miRNA) have also been shown to degrade mRNA, but in contrast to siRNAs, miRNA mediated degradation does not involve endonucleolytic cleavage of the mRNA. Instead, miRNA mediated degradation of the mRNA occurs in P-bodies requiring the RNA-binding protein GW182 and the DCP1:2 decapping complex (37). DNA-based antisense oligonucleotides (ASOs) have been shown to degrade mRNAs and nuclear-retained RNAs via RNase H1-mediated endoribonucleolytic cleavage (38). Surprisingly, despite decades of use, the mechanisms by which these mRNA cleavage products are degraded remain poorly understood.
To better understand the mechanisms involved in degrading the cleavage products generated by RNase H1-activating ASOs and siRNAs, the levels of the key factors involved in RNA surveillance were reduced in cells using siRNAs or an shRNA. The cells were then treated with ASOs or siRNA targeting a nuclear-retained non-coding RNA, pre-mRNA or mRNAs. The stabilities of the cleavage products were determined by PCR, 5′-RACE or 3′-RACE. The binding interactions between the target RNA cleavage products and the 5′-cap and poly(A) protein complex were determined by immunoprecipitation with PABPC1 or eIF4E. In this study, we identified both the factors involved in the degradation of the upstream and downstream cleavage products as well as the proteins bound to the cleavage products.
MATERIALS AND METHODS
Preparation of oligonucleotides
Synthesis and purification of modified ASOs were performed using an Applied Biosystems 380B automated DNA synthesizer as described previously (38). DNA and RNA oligonucleotide probes used for 5′- or 3′-RACE and quantitative real-time PCR (RT-qPCR) are listed in Supplementary Figure S1 and Supplementary Materials. The sequences or catalog numbers of siRNAs used in this study are listed in Supplementary Materials.
Transfection of HeLa cells with siRNA, shRNA and ASOs
HeLa cells were maintained in Dulbecco's Modified Eagle Media (DMEM, Life Technologies), supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in an atmosphere of 7.5% CO2/92.5% air. Cells were grown to ≈70–80% confluency in 10-cm dishes and transfected with a final concentration of 10 nM siRNA using RNAiMAX (Life Technologies) for 4 h using the protocol supplied by the manufacturer. After 48 h, cells were transfected with either a final concentration of 50 nM target ASO or 10 nM siRNA using Lipofectamine 2000 (Life Technologies) or RNAiMAX for 4 h, respectively. As negative controls, cells were mock-transfected using the same transfection reagent but without ASO or siRNA. The mock-transfected cells are referred to as untreated controls (UTC). For shRNA treated cells, the shRNAs (Vigene Biosciences) are delivered via lentivirus transduction to cells overlayed with DMEM at a multiplicity of infection (MOI) of 100–500. Transfection with the target ASO or siRNA was performed 144 h post shRNA adenovirus treatment as described previously.
Western blot
At 24 h post-transfection, cells were lysed in RIPA buffer (Sigma or Santa Cruz Biotechnology) for 15–30 min on ice. The lysate was cleared by top speed centrifugation before the addition of 4X LDS sample buffer (Life Technologies) and heated at 95°C for 3 min. Samples were separated in a 4–12% Bis-Tris polyacrylamide gel (Life Technologies) according to the manufacturer's instructions. The proteins were transferred to a nitrocellulose membrane via the iBlot system (Life Technologies). The nitrocellulose membrane was blocked in a solution of PBST (PBS containing 0.1% Tween) and 5% non-fat milk for 1 h. The blot was then incubated in a solution containing primary antibody diluted 1:500–1:1000 in PBST overnight (Santa Cruz Biotechnology for XRN1: sc-98459, XRN2: sc-99237, GAPDH: sc-365062. Abcam for EXOSC5 : ab168804, Histone H3: ab1791. Sigma for Tubulin: T6199). The blot was washed four times in PBST followed by incubation with secondary antibody conjugated to horse-radish peroxidase (Bio-Rad Laboratories) diluted 1:2500 in PBST for 1 h. Following three washes in PBST and a final wash in PBS, ECL reagent (GE Healthcare or Millipore) was applied to the membrane for 5 min followed by exposure to Kodak film from 30 sec to 20 min.
RNA isolation and RT-qPCR
At 24 h post-transfection, total RNA was isolated using TRIzol (Life Technologies) per the manufacturer's instructions. Briefly, growth medium was removed from cells, and cells were washed once with PBS. TRIzol was added directly to the plate, and cells scraped and harvested. The aqueous phase of a phenol-chloroform extraction was mixed with isopropanol to precipitate the total RNA. The RNA was further treated with DNase I (Life Technologies) to avoid false-positives caused by DNA contamination and then re-precipitated. For RT-qPCR, 1–2ug total RNA was used to synthesize cDNA using StepOne Real-Time PCR System at 48°C for 30 mins, followed by qPCR using Express One–Step Superscript RT-qPCR Kit (Life Technologies). Primer probe sets include 2 primers (a forward and reverse) as well as an internal probe and were selected to be either upstream or downstream of the amplicon site (Supplementary Figure S1). For all RT-qPCR reactions, we report means and standards deviations from 3 independent experiments with triplicate samples in each experiment and the data are normalized to Ribogreen (Life Technologies). The primer probe sets used in RT-qPCR are listed in Supplementary Figure S1 and Supplementary Materials.
Nuclear/cytoplasmic fractionation and PCR
Untreated or treated cells were washed briefly in PBS to remove any residual media and pelleted. The cell pellet was resuspended in NP-40 lysis buffer (150 mM NaCl, 0.6% NP-40, 10 mM Tris HCl, pH 8.0 and 0.1 mM EDTA) and incubated on ice for 10 min. The lysate was spun at 4400 RPM for 1.5 min, and the supernatant (cytoplasmic fraction) was collected. The pellet (nuclear fraction) was washed in DOC wash buffer (150 mM NaCl, 0.6% NP-40, 10 mM Tris HCl, pH 8.0, 0.1 mM EDTA and 0.5% deoxycholate), spun at 7500 RPM for 1.5 min and lysed in RIPA buffer. Cytoplasmic and nuclear RNAs were prepared using TRIzol. Purities of the fractions were determined by western blot analysis or RT-qPCR of known nuclear and cytoplasmic proteins and RNAs, respectively. To assess target reduction in each fractionated compartment, equal amounts of RNAs (≈50 ng) from each preparation were used in RT-qPCR reaction with gene-specific primer probe sets and AgPath-ID Onestep RT-PCR kit (Life Technologies), based on the manufacturer's procedures. RT-qPCR was performed in Step-one qPCR system, in following programs: 48°C, 30 min, 94°C, 10 min, following by 40 cycles of 94°C, 20” and 60°C, 1’. The primer probe sets used in RT-qPCR are listed in Supplementary Figure S1 and Supplementary Materials.
5′- and 3′-RACE
RACE was performed using Life Technologies RACE system according to the manufacturer's instructions with the following modifications: The CIP and TAP procedures were not performed due to the fact that RNase H- and Ago2-mediated cleavages generate 5′-phosphorylated RNA required for ligation of the 5′-RACE adapter. Total RNA input was increased to 2–5 μg and was ligated to the specific adapter using RNA ligase (Life Technologies). Reverse transcription and amplification was performed as described in the materials and methods for PCR with the cycling extended to 37X. Following outer PCR, nested PCR was performed. For 3′-RACE, the upstream fragment contains a 3′-OH after RNase H and Ago2 cleavage so the 3′-adapter contains both a 5′- and a 3′-Phosphate (this is to prevent ligation of the adapter to itself to try to maximize ligation to the target RNA).
The sequences of the primers are listed in Supplementary Figure S1. The PCR products were gel purified in a 2% agarose TBE gel and extracted and purified using QIAquick gel extraction kit (QIAGEN) following the manufacturer's instructions. The samples were ligated to pCR 4-TOPO vector by TA cloning (Life Technologies). After transformation into Mach1 cells, plasmid DNA was isolated from several colonies. The correct inserts were verified by EcoRI digestion and further verified by sequencing (Retrogen or Genewiz).
Poly(A) isolation and immunoprecipitation
Poly(A) isolations were performed using 20 μg of total RNA and the Oligotex mini kit (QIAGEN) following the manufacturer's instructions. Poly(A) RNA was quantified by RT-qPCR using StepOne Real-Time PCR System (Life Technologies). The primers and probes used for RT-qPCR are listed in Supplementary Figure S1.
PABPC1 and EIF4E were immunoprecipitated from lysates following the Magna RIP Protocol (Millipore). Briefly, magnetic Protein A/G beads (Millipore) containing PABPC1 antibody (Millipore 03–101), EIF4E antibody (Abcam76256) or Rabbit IgG (Millipore) were incubated with equal amounts of the treated lysate for 2 h at 4°C. The beads were washed five times in wash buffer provided in the kit followed by a final wash in PBS. Following the final wash, samples were treated with TRIzol and the RNA was isolated as described previously. RNA in the supernatant was analyzed using RT-qPCR, 5′-RACE or 3′-RACE.
RESULTS
Downstream cleavage products are degraded by the XRN exoribonucleases
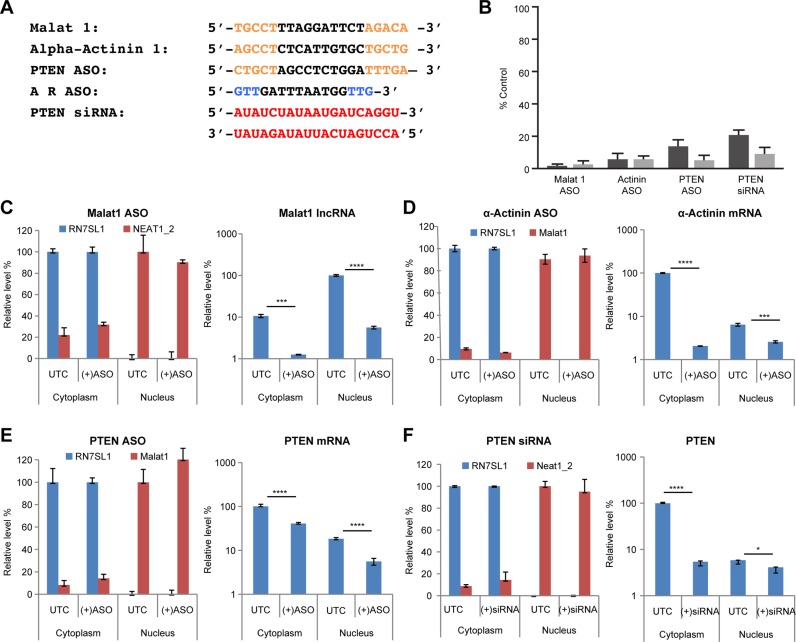
We designed ASOs to target α-Actinin and PTEN mRNAs and Malat 1, a nuclear retained long non-coding RNA, and an siRNA to target PTEN mRNA (Figure 1A). The activities of the ASOs and siRNA were determined by RT-qPCR using primer/probe sets (PPS) designed to bind either the upstream or downstream cleavage products (Supplementary Figure S1A). The ASOs and siRNAs used in this manuscript are the products of extensive screens to identify the most potent and selective reagents and typically several siRNAs and ASOs were studied and these served as controls (data not shown). The ASOs and siRNA reduced complementary target RNAs by 80–97% compared to the levels in control cells treated with transfection reagent alone (Figure 1B). The upstream and downstream cleavage products appeared to be degraded with similar efficiencies (Figure 1B). Nuclear and cytoplasmic fractions were prepared and the purity of the fractions was determined by RT-qPCR using PPS designed to bind Malat1 or NEAT1 nuclear lncRNAs and the cytoplasmic RN7SL1 RNA (Figure 1C–F, left panels, and Supplementary Figure S1A; 39,40). Note that RN7SL1 RNA was barely detected in the nuclear fractions, indication that there was little cytoplasmic contamination in the nuclear fractions. The targeted RNAs were quantified in nuclear and cytoplasmic fractions using primers designed to bind downstream of the ASO or siRNA binding sites. Malat1 RNA was observed to be enriched in the nuclear fraction, and treatment with the Malat1 ASO resulted in the loss of the Malat1 RNA in the nuclear fraction (Figure 1C, right panel). The α-Actinin and PTEN mRNAs were detected in both the cytoplasmic and nuclear fractions, though the majority of the mRNAs were observed in the cytoplasmic fraction, as expected (Figure 1D,E, right panels). Treatment with their respective ASOs resulted in loss of the mRNA from both fractions (Figure 1D,E, right panels). Consistent with the expected Ago2-mediated cleavage upon treatment of cells with siRNA, reduction of PTEN mRNA was observed in the cytoplasmic fraction, but not obviously in the nuclear fraction, of the cells treated with PTEN siRNA (Figure 1F, right panel). Similar results were obtained using primers targeting the regions upstream of the ASO and siRNA hybridization sites (data not shown).
Figure 1.
ASO- or siRNA-mediated reduction of target RNA and subcellular localization of targeted RNAs. (A) In sequences of the ASOs and siRNA, the orange, blue, red and black letters represent, respectively, 2′-methoxyethylribonucleotides, 2′-constrained ethylribonucleotides, 2′-ribonucleotides and 2′-deoxyribonucleotides. The siRNA was prepared by annealing the guide strand (top sequence) with complementary length matched RNA (bottom sequence). ASOs had phosphorothioate linkages; linkages in the siRNA were phosphodiester. (B) Target RNA levels were determined 24 h post ASO or siRNA treatment by RT-qPCR and are shown as a percentage of untreated controls. Dark and light gray bars correspond to, respectively, the upstream and downstream target RNA cleavage products. (C–F) Cells treated with ASOs targeting Malat1 (C), α-Actinin (D), PTEN (E) or with a siRNA targeting PTEN (F) were collected and cytoplasmic and nuclear fractions were separated. The purity of the nuclear and cytoplasmic fractions (left panels) was determined by RT-qPCR using primers targeting, respectively, the cytoplasmic RNA RN7SL1 and nuclear NEAT1 or Malat1 RNA. The levels of the RN7SL1 RNA are shown as a percentage of the level in the cytoplasmic fraction in control cells; whereas the levels of NEAT1 or Malat1 RNA are shown as a percentage of the level in the nuclear fraction in control cells. RT-qPCR analyses of the target RNA downstream cleavage products were performed on the cytoplasmic and nuclear fractions from control cells treated with transfection reagent only (UTC) or ASO/siRNA treated cells (right panels, Y-axis is shown as logarithmic scale). The qPCR primers are located downstream to the cleavage sites on the target RNAs. The PPS used for the RT-qPCR are listed in Supplementary Figure S1A. The predicted sizes (base pairs) of the PCR products are: Malat1 495; α-Actinin 432; PTEN ASO 435; PTEN siRNA 435. The error bars represent standard deviation from three independent experiments. Statistics analysis was performed using unpaired t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
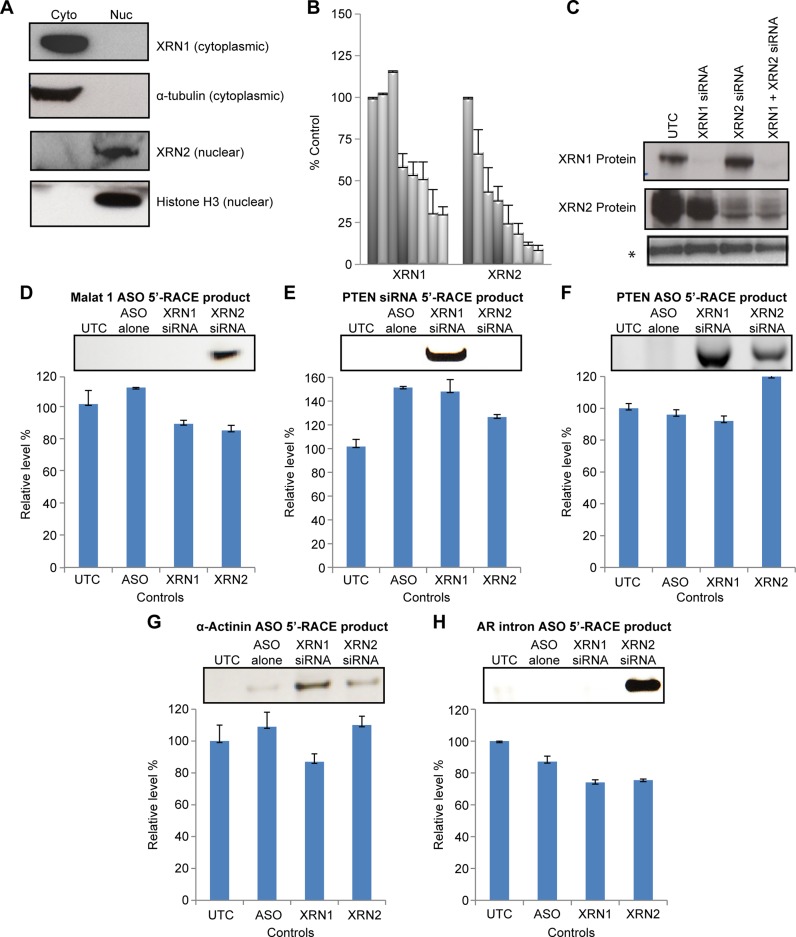
Consistent with previous reports, western blot analysis of the XRN proteins following cell fractionation showed that XRN1 was predominantly localized in the cytoplasm and XRN2 was predominantly in the nucleus (Figure 2A). Thus, if XRNs are involved in the degradation of the downstream cleavage products, XRN1 and XRN2 would degrade, respectively, cytoplasmic and nuclear RNA fragments. To evaluate this, siRNAs targeting the XRN mRNAs were used to reduce XRN levels in cells. The activities of the siRNAs targeting the XRN mRNAs were determined by qRT-PCR (Figure 2B). Greater than 90% reductions in XRN proteins were observed 48-h post siRNA treatment (Figure 2C). After treating cells with an XRN-targeted siRNA, cells were treated with ASO or siRNA targeting α-Actinin, PTEN, or Malat1, and the downstream cleavage products were detected by 5′-RACE. Ligation of the 5′-RACE adaptor requires 5′-phosphorylated RNA and RNase H and Ago2 generate downstream cleavage products with a 5′-phosphate; therefore, only the downstream cleavage products and not intact RNA or upstream cleavage products were amplified. As a control, RN7SL1 RNA was detected by RT-qPCR from 10% of input RNAs used in the 5′-RACE, and the relative levels were comparable, as shown below each panel of the 5′-RACE products (Figure 2D–H).
Figure 2.
XRN exonucleases degrade downstream RNA cleavage products. (A) Subcellular localization of XRNs was determined western blots performed on cytoplasmic (Cyto) and nuclear (Nuc) fractions of HeLa cells. Purity of the fractions was determined using antibodies to histone H3 and α-tubulin. (B) siRNA-mediated reduction of XRN mRNAs were quantified 48 h post treatment. Cells were transfected with 3-fold serial dilutions of the siRNAs ranging in concentration from 14 pM to 10 nM. The mRNA levels were determined by RT-qPCR and are shown as percentages of control cells (UTC) treated with transfection reagent only. The mean and standard errors reported are based on three trials. The PPS used for the RT-qPCR are listed in Supplementary Figure S1A. Note that the XRN1 siRNA had no effect on XRN2 or vice versa. (C) Western blot analyses of XRN proteins from untreated (UTC) and siRNA-treated cells 48 h post siRNA treatment. A non-specific band (*) serves as a control for loading. (D–H) Cells were either treated or not with siRNA targeting XRN1 or XRN2 mRNA for 4 h, or were treated with transfection reagent only (UTC). Cells were then treated with Malat1 ASO (panel D), PTEN siRNA (Panel E), PTEN ASO (Panel F), α-Actinin ASO (Panel G) or AR ASO (Panel H), and 5′-RACE was performed 48 h later to detect downstream cleavage products. The adapter and primers used for the 5′-RACE are listed in Supplementary Figure S1B. RN7SL1 RNA was detected by RT-qPCR from 10% of input RNA for RACE and used as loading controls, as listed below each panel of the 5′ RACE results. The error bars represent standard deviation from three independent experiments. The predicted sizes (base pairs) of the 5′-RACE products are: Malat 1 504; α-Actinin 470; PTEN ASO 331; PTEN siRNA 266.
5′-RACE products corresponding to the Malat1 RNA downstream cleavage product following the reduction of XRN2 but not after reduction of XRN1 were detected (Figure 2D). The stabilization of the cleavage product in the absence of XRN2 suggests that XRN2 degrades the downstream cleavage products of Malat1. The degradation of the Malat1 RNA downstream cleavage products by XRN2 is consistent with the nuclear localization of both the enzyme and Malat1 RNA (Figure 1D). A second nuclear target was then analyzed. An ASO was designed to target an intronic region of the androgen receptor (AR) pre-mRNA (Figure 2H). 5′-RACE products were again observed in extracts of cells pretreated with siRNA targeting XRN2 but not those pretreated with XRN1 siRNA (Figure 2H). Sequencing of the 5′-RACE products corresponding to the Malat1 RNA and the androgen receptor pre-mRNA confirmed the identity of the RACE products and showed that the sites of cleavage were within the regions complementary to the ASOs, consistent with an RNase H1-mediated cleavage (Supplementary Figures S2A and S2B).
In contrast, XRN1 appears to be involved in the degradation of the PTEN mRNA downstream cleavage product that resulted from treatment with the PTEN siRNA. 5′-RACE products were observed in cells deficient in XRN1, and no 5′-RACE products were observed when XRN2 levels were significantly reduced (Figure 2E). siRNA-mediated Ago2 cleavage activity occurs in the cytoplasm, and the results reported here are consistent with the cytoplasmic localization of XRN1 (Figure 1E). Sequencing of the 5′-RACE product confirmed the specificity and showed that the sites of cleavage were at mRNA nucleotides expected to be opposite the tenth and eleventh nucleotides from the 5′-terminus of the siRNA guide strand in the mRNA:siRNA duplex (Supplementary Figure S2C).
Consistent with the observation, that ASO-mediated degradation of PTEN and α-Actinin RNAs occurred in both the nucleus and the cytoplasm (Figure 1D), both XRN1 and XRN2 appear to be involved in the degradation of downstream cleavage products resulting from treatment of cells with PTEN and α-Actinin ASOs (Figure 2F,G). Sequencing of 5′-RACE products were consistent with an RNase H1-induced cleavage mechanism (Supplementary Figures S2D and S2E).
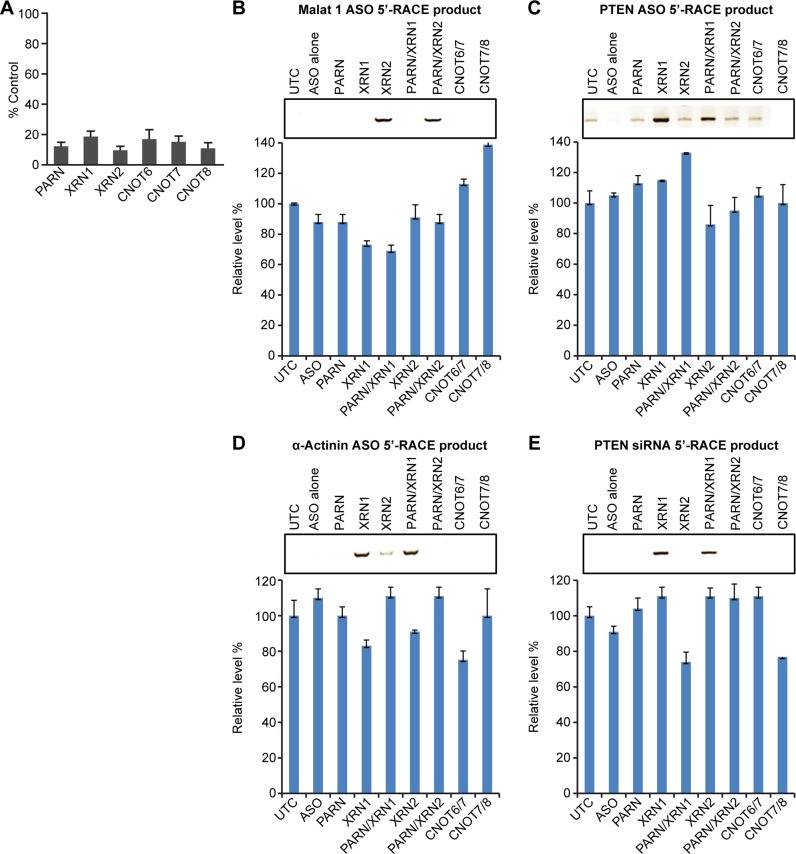
Next we determined whether degradation of the downstream RNA cleavage products involved deadenylation of the RNA by reducing the levels of the deadenylases PARN and CNOT proteins in the cells prior to treatment with target ASOs or siRNA. The siRNA treatments reduced the levels of the targeted RNAs by at least 80% (Figure 3A). In cells depleted of PARN or CNOT RNAs, no detectable 5′-RACE products were observed for targeted RNAs in cells treated with Malat1 ASO (Figure 3B), PTEN ASO (Figure 3C), α-Actinin ASO (Figure 3D) or PTEN siRNA (Figure 3E). A faint 5′-RACE band was observed in cells treated with PTEN ASO; however, sequence analysis of the product showed no sequence homology with PTEN mRNA (data not shown). The failure to detect enrichment of 3′ fragment in PARN reduced cells appears not to be due to insufficient reduction of the protein, since similar treatment with the PARN siRNA led to accumulation of polyadenylated snoRNA ACA45 (Supplementary Figure S3), a class of snoRNAs known to be targets of PARN (41). 5′-RACE products were only observed with the reduction of XRN exoribonucleases suggesting that the deadenylase does not contribute significantly to degradation of RNA downstream cleavage products (Figure 3B–E). As a control, the relative levels of RN7SL1 RNA detected by RT-qPCR from 10% of input RNAs for 5′-RACE were comparable, as presented below each panel.
Figure 3.
Deadenylases do not contribute significantly to degradation of downstream RNA cleavage products. (A) Cellular levels of XRN1, XRN2, and indicated deadenylase mRNAs 48 h post siRNA treatment. The mRNA levels were determined by RT-qPCR and are shown as a percentage of untreated control. The mean and standard errors reported are based on three trials. The PPS used for the RT-qPCR are listed in Supplementary Figure S1A. (B–E) Cells were treated with individual or combined XRN or deadenylase siRNAs. After 4 h, cells were treated with Malat1 ASO (Panel B), PTEN ASO (Panel C), α-Actinin ASO (Panel D) or PTEN siRNA (Panel E). 5′-RACE of the RNA downstream cleavage products was performed after 48 h. RN7SL1 RNA was detected by RT-qPCR from 10% of input RNA for RACE and used as loading controls, as shown below each panel of the RACE products. The error bars represent standard deviation from three independent experiments. The adapter and primers used for the 5′-RACE are listed in Supplementary Figure S1B.
Given that deadenylases did not appear to be involved in the degradation of the downstream cleavage products, we evaluated the polyadenylation states of the downstream cleavage products. Specifically, poly(A) isolations were performed from cells treated with the XRN siRNAs prior to treatment with Malat1 ASO, α-Actinin ASO, or PTEN ASO or siRNA (Figure 4A). qRT-PCR amplification was performed using PPS that hybridize to regions up or downstream of the ASO/siRNA hybridization sites (Supplementary Figure S1A). A previous report showed that Malat1 RNA does not have a poly(A) tail, and little to no poly(A) RNA was detected with either Malat1 RNA primer set (Figure 4A; 42). The levels of poly(A) mRNA isolated from cells treated with the target specific ASOs or siRNA ranged from 7% to 27% of the levels in control cells treated with transfection reagent alone when analysis was performed using the downstream PPS (Figure 4A). Similar results were obtained when the upstream PPS was used to amplify mRNA, suggesting that these co-immunoprecipitated poly(A) mRNAs likely correspond to the full-length mRNAs (Figure 4A).
Figure 4.
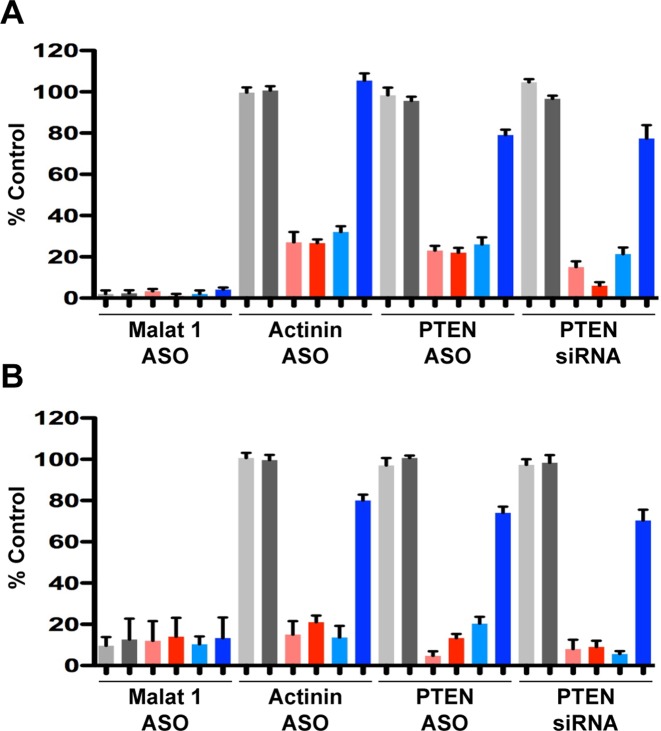
Downstream target RNA cleavage products are polyadenylated and bound to PABPC1. (A) Poly(A) RNA was isolated from control cells treated with transfection reagent alone (gray bars), cells treated with target ASOs or siRNA alone (red bars), or from cells treated with both target ASOs/siRNA and XRN1 and XRN2 siRNAs (blue bars). (B) Levels of the target RNA cleavage products associated with immunoprecipitated PABPC1 from untreated cells (gray bars), cells treated with target ASOs or siRNA alone (red bars) or cells treated with both target ASOs/siRNA and XRN1 and XRN2 siRNAs (blue bars). Levels of the target RNA cleavage products were determined 48 h post treatment by RT-qPCR and are shown as a percentage of untreated controls. The mean and standard errors reported are based on three trials. Light and dark hues bars represent, respectively, the target RNA upstream and downstream cleavage products. The PPS used for the RT-qPCR are listed in Supplementary Figure S1A.
We then depleted XRN proteins from cells prior to treatment with ASO or siRNA and analyzed poly(A)-isolated RNA using upstream and downstream PPS (Supplementary Figure S1A). Target mRNA levels detected with the upstream PPS were similar when cells were treated with both the XRN siRNAs and target-specific ASOs or siRNA or only target-specific ASOs or siRNA (Figure 4A). The upstream PPS is likely detecting full-length mRNA. In contrast, when the targeted mRNA was detected using the downstream PPS, levels of poly(A) isolated mRNA ranged from 60% to 120% for cells treated with both the XRN siRNAs and target ASOs or siRNAs compared to the values for the control cells treated with transfection reagent alone (Figure 4A). This indicated that a significant fraction of the downstream cleavage products was polyadenylated supporting our hypothesis that deadenylases do not contribute significantly to the degradation of downstream cleavage products.
Next we determined whether the polyadenylated downstream cleavage products were bound to PABP proteins by immunoprecipitating PABPC1 and associated RNAs from cells depleted of XRN proteins and treated with ASOs or siRNA. Although eukaryotic cells contain one nuclear and four cytoplasmic PABP proteins, PABPC1 has been shown to be the most abundant isoform (43). Approximately 10% of the Malat1 RNA, which lacks a poly(A) tail, co-immunoprecipitated with PABPC1 (Figure 4B; 42). Absent XRN reduction but with ASO or siRNA treatment, the levels of α-Actinin and PTEN mRNAs that co-immunoprecipitated with PABPC1 ranged from 12% to 22% of the levels in control cells treated with transfection reagent alone (Figure 4B). In contrast, the amounts of α-Actinin and PTEN mRNA co-immunoprecipitated with PABPC1 from cells treated with both the XRN siRNAs and the target ASOs or siRNA were 4–5-fold higher than those in cells treated with the target ASOs or siRNA alone (Figure 4B). As a negative control, immunoprecipitation using IgG only co-selected less than 2% of input Malat1, PTEN or Actinin RNAs (data not shown). These data suggest that a significant fraction of the polyadenylated downstream cleavage products bind PABPC1.
Upstream cleavage products are degraded by the exosome and XRNs
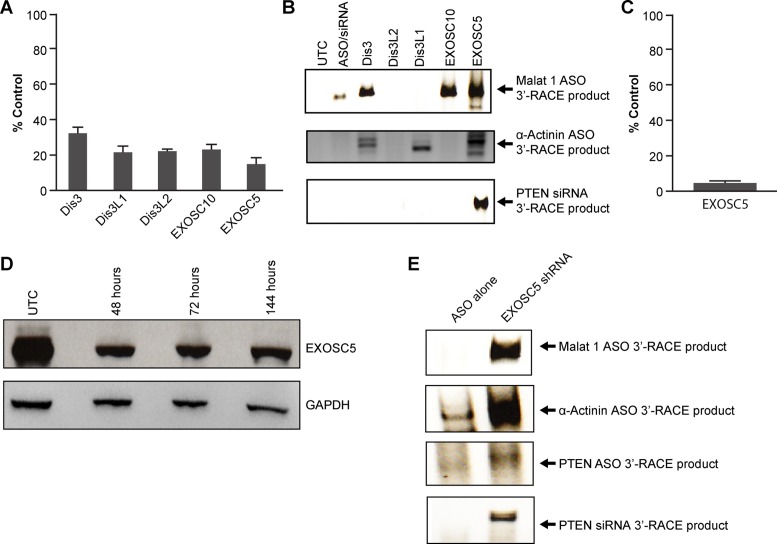
The levels of the catalytic components Dis3 and EXOSC10 and the key structural component EXOSC5 of the exosome were reduced in cells using siRNAs in order to determine whether the exosome was involved in the degradation of the target RNA upstream cleavage products. siRNAs targeting the 3′ to 5′ exoribonucleases Dis3L2 and Dis3L1 were also tested. The siRNAs used reduced levels of the mRNAs encoding the exosome subunits to less than 30% of levels in untreated cells (Figure 5A). To evaluate the stabilities of upstream cleavage products in conditions in which exosome components were depleted, we used 3′-RACE with 5′-phosphorylated RACE adaptors since the upstream cleavage products contain a 3′ hydoxyl. Under these conditions only the upstream cleavage products but not the intact mRNA or the downstream cleavage products were amplified. 3′-RACE products were observed for the nuclear-retained Malat1 RNA when nuclear Dis3, EXOSC10 or EXOSC5 were reduced prior to treating the cells with the Malat1 ASO (Figure 5B). 3′-RACE products of the α-Actinin mRNA in cells treated with α-Actinin ASO were observed only when cells were pretreated with nuclear Dis3, cytoplasmic Dis3L1 or EXOSC5 siRNAs (Figure 5B). Finally, cells treated with the PTEN siRNA exhibited PTEN 3′-RACE products only after reduction of EXOSC5 (Figure 5B). EXOSC5 is required for the formation of the exosome core, and reducing the levels of this key structural subunit in the cell likely inhibits exosome activity by preventing assembly of the core structure (18,19). Taken together, these data suggest that the exosome participates in the degradation of the upstream cleavage products in the nucleus and cytoplasm.
Figure 5.
Exosome complex degrades upstream cleavage products. (A) Cellular levels of indicated mRNAs 48 h post siRNA treatment. The mRNA levels were determined by RT-qPCR and are shown as a percentage of untreated controls. The mean and standard errors reported are based on three trials. (B) 3′-RACE of the target RNA upstream cleavage products from ASO or siRNA treated and control cells (UTC) treated with transfection reagent only. 3′-RACE was performed 48 h post treatment with Malat 1, α-Actinin or PTEN ASOs or siRNA alone or with both target-specific ASO/siRNA and exosome/exonuclease siRNAs. The predicted sizes of the 3′-RACE products are: Malat1 378; α-Actinin 423; PTEN ASO 439; PTEN siRNA 386. (C) Cellular level of EXOSC5 mRNA 48 h post shRNA treatment. (D) Western blot analysis of EXOSC5 protein from untreated (UTC) and siRNA treated cells 48 h to 144 h post siRNA treatment. (E) 3′-RACE of upstream cleavage products was performed on extracts of cells treated with Malat1, α-Actinin, or PTEN ASOs or siRNA alone or first treated 144 h with EXOSC5 shRNA and then with target-specific ASO/siRNA. The PPS used for the RT-qPCR are listed in Supplementary Figure S1A. The adapter and primers used for the 3′-RACE are listed in Supplementary Figure S1C.
Given the importance of EXOSC5 protein in the activity of the exosome and to further validate the role of the exosome in the degradation of the target RNA upstream cleavage products, we treated cells with an shRNA targeting EXOSC5. The shRNA reduced EXOSC5 mRNA levels by greater than 95% compared to those in control cells treated with transfection reagent alone (Figure 5C). In addition, shRNA-mediated reduction of EXOSC5 protein was observed for up to 144 h (Figure 5D). Cells treated with the target ASOs or siRNA alone resulted in little to no 3′-RACE products (Figure 5E). Conversely, 3′-RACE products were observed in cells treated with both the EXOSC5 shRNA and target ASOs or siRNA (Figure 5E), indicating that the exosome is involved in degradation of upstream cleavage fragments. The 3′-RACE products were confirmed by sequencing and were consistent with expected cleavage mechanisms (Supplementary Figure S2F–I). We also reduced a number of other nucleases such as ERI1 and CNOT6L and reduction of these did not stabilize either 5′ or 3′ fragment (Supplementary Figure S4).
To determine whether upstream cleavage products associate with PABP, cells were treated with the shRNA targeting EXOSC5. To ensure that the downstream cleavage product/PABP complex remained intact, cells were also treated with siRNAs targeting XRNs. Finally, cells were treated with either ASOs or siRNAs targeting α-Actinin or PTEN mRNAs, and the levels of upstream cleavage products co-immunoprecipitated with PABPC1 were determined using the upstream PPS (Supplementary Figure S1). Unlike the target RNA downstream cleavage products, the upstream cleavage products did not co-immunoprecipitate with PABPC1 (Figure 6A).
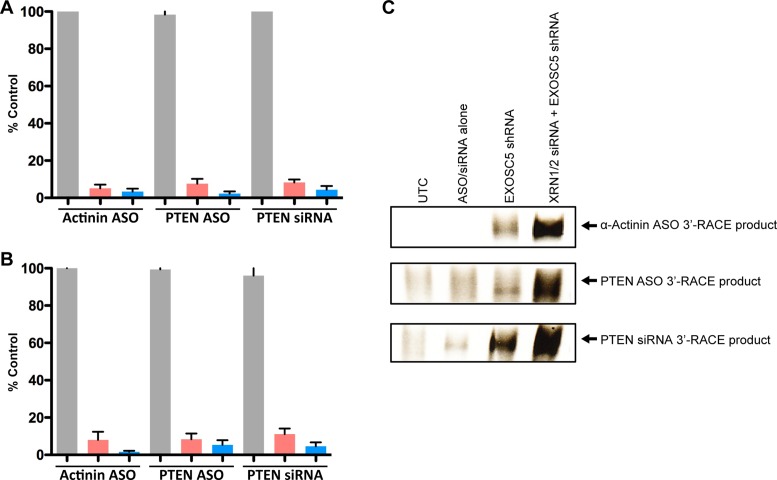
Figure 6.
Upstream cleavage products are not bound by PABPC1 or eIF4E and are degraded by both XRNs and the exosome complex. (A and B) Levels of the target RNA upstream cleavage products associated with (A) PABPC1 or (B) eIF4E from untreated cells (gray bars), cells treated with target ASOs or siRNA alone (red bars), or cells treated first with XRN siRNAs and EXOSC5 shRNA and then with ASO or siRNA (blue bars). Levels of the target RNA cleavage products were determined 48 h post treatment by RT-qPCR and are shown as a percentage of their respective untreated controls (transfection reagent only). (C) Cells were treated with EXOSC5 shRNA or XRN siRNAs and EXOSC5 shRNA and then with ASO or siRNA. 3′-RACE of the target RNA upstream cleavage products was performed 48 h post treatment. The PPS used for the RT-qPCR are listed in Supplementary Figure S1A. The adapter and primers used for the 3′-RACE are listed in Supplementary Figure S1C.
Next, we immunoprecipitated the 5′-cap binding protein eIF4E and associated RNA from cells treated with the EXOSC5 shRNA, XRN siRNAs, and/or target ASOs or siRNA. eIF4E was chosen since it binds cytoplasmic mRNAs which represent majority of mature mRNAs, even though a small portion of mRNAs may associate with CBC20/80 complex in the nucleus (44). The upstream cleavage products of α-Actinin and PTEN mRNAs did not co-immunoprecipitate with eIF4E (Figure 6B). The lack of binding between the upstream cleavage products and either PABPC1 or eIF4E suggests that the 5′ terminus of the upstream cleavage products may be susceptible to 5′ to 3′ exonuclease degradation. To determine whether the XRNs participate in the degradation of the upstream cleavage products, we performed 3′-RACE from cells in which the levels of the EXOSC5 only or both EXOSC5 and XRNs were reduced prior to treatment with the target ASOs or siRNA. The number of amplification cycles was reduced in order to better differentiate the 3′-RACE signals. 3′-RACE products were observed in extracts of cells depleted of EXOSC5 (Figure 6C). Importantly, the signal from 3′-RACE products appeared to be enhanced when both EXOSC5 and XRN protein levels were reduced compared to EXOSC5 alone suggesting that both the exosome and XRNs participate in the degradation of upstream cleavage products resulting from ASO or siRNA treatment (Figure 6C).
DISCUSSION
The RNA surveillance mechanism relies predominately on exoribonuclease activity to degrade unnecessary or aberrant RNA. Both antisense oligonucleotides and siRNAs have significant clinical potential. These agents induce cleavage of a targeted RNA by recruiting RNase H1 and the RNAi machinery, respectively. We sought to determine the fates of the products of ASO- and siRNA-mediate cleavage.
mRNAs are protected from exonucleases with a 5′ 7-methylguanosine cap and 3′-poly(A) tail, whereas non-coding RNAs often have stable structures that protect against exoribonuclease degradation. For example, the nuclear-retained long non-coding RNA Malat1 has a 7-methylguanosine cap and a triple helical structure, instead of a poly(A) tail, protects the 3′ terminus (42). Given the complex nature of mRNA biogenesis, these RNAs contain additional stabilizing factors. Some mRNAs are further protected in the cytoplasm against exoribonuclease degradation by a protein complex containing 5′-cap and poly(A) binding proteins, which effectively sequester the termini of the mRNA. As a result, degradation of these mRNAs begins with dissociation of the cap/poly(A) protein complex from the mRNA by shortening of the poly(A) tail, decapping the mRNA or initiation of translation (4).
An effective RNA degradation mechanism that circumvents the prerequisite for disruption of the 5′-cap/poly(A) complex involves production of unprotected termini through endoribonucleolytic cleavage mediated by endoribonucleases RNase H1 and Ago2. These unprotected termini are ideal substrates for exoribonucleases since cleavage by RNase H1 and Ago2 produces a 5′-phosphorylated terminus, the preferred substrate for XRN exoribonucleases (15,16). The substrate preferences of the XRN enzymes are likely the result of the synergistic activities between the XRNs and the decapping enzymes DCP1 and 2, which also produce unprotected 5′-phosphorylated RNA (45). Whether the cleavage products generated by RNase H1 and Ago2 evolved to function synergistically with the XRN enzymes is unclear, but the structure of the RNase H1 and Ago2 substrates suggest that it is unlikely. RNase H1 and Ago2 cleave the RNA within a double-stranded structure and the position of the scissile phosphate within the double-stranded structure is such that the only possible orientation for an inline nucleophilic attack of the scissile phosphate results in cleavage products with a 3′ hydroxyl and 5′ phosphate (46).
Given their prominent role in the RNA surveillance machinery, it was not surprising that the XRN exoribonucleases were involved in the degradation of upstream and downstream products resulting from ASO- and siRNA-mediated cleavage. The XRN enzymes degraded the target RNA upstream cleavage products, presumably following decapping of the target RNA; our analysis indicated that these products did not contain the 5′-cap binding protein eIF4E and likely were decapped (Figure 6B). Nuclear XRN2 was responsible for degrading the unprotected 5′ terminus of the downstream cleavage products generated by ASOs targeting the nuclear-retained Malat1 and androgen receptor pre-mRNA. Consistent with the observed cytoplasmic activity of the siRNA targeting the PTEN mRNA, the downstream cleavage of the PTEN mRNA was degraded by cytoplasmic XRN1 (Figure 2D). Conversely, the downstream cleavage products of the α-Actinin and PTEN mRNAs generated by ASO treatment were degraded by both XRN1 and XRN2 (Figure 2D). Consistent with these observations and the fact that RNase H1 is found in both the nucleus and cytoplasm, we observed ASO-mediated cleavage of mRNAs in both the nucleus and cytoplasm (Figure 1D). As ASOs are capable of targeting regions of pre-mRNAs, XRN2 may degrade downstream cleavage products resulting from these events.
The exosome complex containing the nuclear Dis3 and/or EXOSC10 catalytic subunits was responsible for the 3′ to 5′ degradation of the nuclear-retained RNA upstream cleavage products (Figure 5B,E). Both the nuclear Dis3 exosome complex and cytoplasmic Dis3L1 exoribonuclease were responsible for the 3′ to 5′ degradation of the mRNA upstream cleavage products suggesting that degradation likely occurs in both cellular compartments (Figure 5B,E). Interestingly, cytoplasmic Dis3L1 did not appear to play a role in the degradation of the upstream cleavage products generated by the siRNA suggesting that different pathways maybe involved in the degradation of siRNA and ASO cleavage products (Figure 5B). For example, RISC components are known to localize in P-bodies where siRNA mediated mRNA cleavage and degradation may occur (47), whereas RNase H1 was not detected in P-bodies (our unpublished data). Perhaps due to the redundant nature of activities, reduction of either of the catalytic subunits of the exosome alone did not significantly enhance the stability of the target RNA upstream cleavage products (Figure 5B). The most consistent enhancement in the stability of the upstream cleavage products were observed following the reduction of EXOSC5. Reductions in levels of this scaffolding protein presumably inhibited assembly of an active exosome complex (5B and 5E). Surprisingly, the exosome did not play a significant role in the degradation of downstream cleavage products (Figure 4).
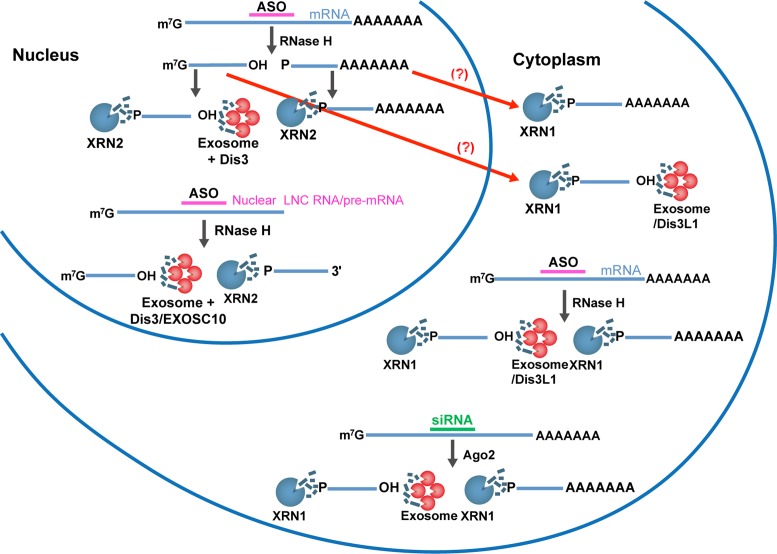
Taken together, our analysis reveals the major degradation pathways for cleavage products generated by treatment of cells with ASOs and siRNAs (additional minor pathways may exist, as we have not studied all possible nucleases; Figure 7). Following siRNA-mediated Ago2 cleavage of mRNA in the cytoplasm, the downstream cleavage products, containing a poly(A) tail and bound by PABPC1, are degraded predominantly in the 5′ to 3′ direction by XRN1. The upstream cleavage products, which were not bound by the 5′-cap binding protein eIF4E, are presumably decapped and then degraded in the 5′ to 3′ direction by XRN1 and in the 3′ to 5′ direction by the exosome complex. A similar mechanism was observed for products of the ASO-mediated RNase H1 cleavage of the target mRNAs which mainly localize in the cytoplasm. Consistent with these observations, the 3′ to 5′ cytoplasmic exoribonuclease Dis3L1 also appeared to be involved in the degradation of the mRNA upstream cleavage products generated by ASOs (Figure 5B). In the case of the nuclear-retained target RNAs, degradation of the downstream cleavage products involved XRN2, whereas the upstream cleavage products were degraded by the exosome complex containing nuclear Dis3 and/or EXOSC10 and exoribonuclease XRN2. Our data do not support exclusion of the possibility that for some RNAs, RNase H1 could cleave the target in both the nucleus and cytoplasm and some fragments generated in the nucleus might escape nuclear degradation and be exported to the cytoplasm and processed there.
Figure 7.
Degradation pathway for RNA cleavage products generated by treatment with ASO or siRNA. In the nucleus, XRN2 and either the exosome containing Dis3 and /or EXOSC10 degrades the nuclear-retained target RNA upstream cleavage products and XRN2 degrade the downstream cleavage products. The same pathway may also be involved in the degradation of mRNA cleavage products generated by ASOs in the nucleus and/or these cleavage products may be exported to the cytoplasm for processing by XRN1, the exosome and/or Dis3L1. In the cytoplasm, mRNA cleavage products resulting from treatment with ASOs or siRNAs are degraded by XRN1, the exosome and/or Dis3L1.
Supplementary Material
Acknowledgments
We thank Donna Parrett for excellent administrative support. We thank Wen Shen for helpful discussions and Tracy Reigle for figure preparation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ionis Pharmaceuticals has funded the work for this manuscript. Funding for open access charge: Ionis Pharmaceuticals, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Badis G., Saveanu C., Fromont-Racine M., Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Conti E., Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Boil. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 4.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell. Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 5.Goldstrohm A.C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell. Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 6.Hershey J.W.B., Merrick W.C. In: Translational Control of Gene Expression. Mathews MB, editor. Vol. 39. NY: Cold Spring harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- 7.Tarun S.Z., Jr, Sachs A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 8.Imataka H., Gradi A., Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs A. In: Translational Control of Gene Expression. Mathews MB, editor. Vol. 39. NY: Cold Spring harbor Laboratory Press; 2000. pp. 447–465. [Google Scholar]
- 10.Kahvejian A., Roy G., Sonenberg N. The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:293–300. doi: 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- 11.Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissan T., Rajyaguru P., She M., Song H., Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tharun S., He W., Mayes A.E., Lennertz P., Beggs J.D., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 15.Chang J.H., X S., Tong L. In: Ribonucleases. Nicholson AW, editor. Vol. 26. Heidelberg: Springer; 2011. pp. 167–192. [Google Scholar]
- 16.Geisler S.C., J. In: Eukaryotic RNases and Their Partners in RNA Degradation and Biogenesis. Chanfreau GF, Tamanoi F, editors. Vol. 31. Academic Press; 2012. pp. 97–114. [Google Scholar]
- 17.Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 18.Schmid M., Jensen T.H. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Lykke-Andersen S., Brodersen D.E., Jensen T.H. Origins and activities of the eukaryotic exosome. J. Cell Sci. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- 20.Staals R.H., Bronkhorst A.W., Schilders G., Slomovic S., Schuster G., Heck A.J., Raijmakers R., Pruijn G.J. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 22.Tomecki R., Kristiansen M.S., Lykke-Andersen S., Chlebowski A., Larsen K.M., Szczesny R.J., Drazkowska K., Pastula A., Andersen J.S., Stepien P.P., et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernokalskaya E., Dubell A.N., Cunningham K.S., Hanson M.N., Dompenciel R.E., Schoenberg D.R. A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA. 1998;4:1537–1548. doi: 10.1017/s1355838298980451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F., Peng Y., Schoenberg D.R. Endonuclease-mediated mRNA decay requires tyrosine phosphorylation of polysomal ribonuclease 1 (PMR1) for the targeting and degradation of polyribosome-bound substrate mRNA. J. Biol. Chem. 2004;279:48993–49002. doi: 10.1074/jbc.M409776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollien J., Weissman J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 27.Xiao S., Scott F., Fierke C.A., Engelke D.R. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Ann. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill T., Cai T., Aulds J., Wierzbicki S., Schmitt M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gill T., Aulds J., Schmitt M.E. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J. Cell Biol. 2006;173:35–45. doi: 10.1083/jcb.200512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 31.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 32.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Gen. Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 33.Orban T.I., Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robb G.B., Brown K.M., Khurana J., Rana T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 36.Liang X.H., Vickers T.A., Guo S., Crooke S.T. Efficient and specific knockdown of small non-coding RNAs in mammalian cells and in mice. Nucleic Acids Res. 2011;39:e13. doi: 10.1093/nar/gkq1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 39.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Englert M., Felis M., Junker V., Beier H. Novel upstream and intragenic control elements for the RNA polymerase III-dependent transcription of human 7SL RNA genes. Biochimie. 2004;86:867–874. doi: 10.1016/j.biochi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Berndt H., Harnisch C., Rammelt C., Stohr N., Zirkel A., Dohm J.C., Himmelbauer H., Tavanez J.P., Huttelmaier S., Wahle E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.D., Joshua-Tor L., Sharp P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Gen. Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kini H.K., Kong J., Liebhaber S.A. Cytoplasmic poly(A) binding protein C4 serves a critical role in erythroid differentiation. Mol. Cell. Biol. 2014;34:1300–1309. doi: 10.1128/MCB.01683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonatopoulos-Pournatzis T., Cowling V.H. Cap-binding complex (CBC) Biochem. J. 2014;457:231–242. doi: 10.1042/BJ20131214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 46.Berger I., Tereshko V., Ikeda H., Marquez V.E., Egli M. Crystal structures of B-DNA with incorporated 2′-deoxy-2′-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability. Nucleic Acids Res. 1998;26:2473–2480. doi: 10.1093/nar/26.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jagannath A., Wood M.J. Localization of double-stranded small interfering RNA to cytoplasmic processing bodies is Ago2 dependent and results in up-regulation of GW182 and Argonaute-2. Mol. Biol. Cell. 2009;20:521–529. doi: 10.1091/mbc.E08-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.