Abstract
Cadicivirus (CDV) is unique amongst picornaviruses in having a dicistronic genome with internal ribosomal entry sites (IRESs) preceding both open reading frames. Here, we investigated initiation on the 5′-terminal IRES. We report that the 982-nt long 5′UTR comprises 12 domains (d1-d12), five of which (d8-d12, nts 341–950) constitute a divergent Type I IRES. It comprises central elements (the apex of d10, d11 and the following polypyrimidine tract) that are homologous to corresponding elements in canonical Type 1 IRESs, and non-canonical flanking domains (d8, d9 and d12). In vitro reconstitution revealed that as with canonical Type I IRESs, 48S complex formation requires eukaryotic initiation factors (eIFs) 1, 1A, 2, 3, 4A, 4B and 4G, and the poly(C) binding protein 2 (PCBP2), and starts with specific binding of eIF4G/eIF4A to d11. However, in contrast to canonical Type I IRESs, subsequent recruitment of 43S ribosomal complexes does not require direct interaction of their eIF3 constituent with the IRES-bound eIF4G. On the other hand, the CDV IRES forms a 40S/eIF3/IRES ternary complex, with multiple points of contact. These additional interactions with translational components could potentially stimulate recruitment of the 43S complex and alleviate the necessity for direct eIF4G/eIF3 interaction.
INTRODUCTION
Translation initiation on most eukaryotic mRNAs occurs by the scanning mechanism (1). First, a 43S preinitiation complex is formed containing a 40S ribosomal subunit, the eIF2·GTP/Met-tRNAiMet ternary complex and eIFs 3, 1 and 1A. The 43S complex attaches to the cap-proximal region of mRNA and scans to the initiation codon, where it forms a 48S initiation complex with established codon-anticodon base-pairing. Attachment of 43S complexes is mediated by eIFs 4F, 4A and 4B. eIF4F comprises three subunits: the cap-binding protein eIF4E, the DEAD-box RNA helicase eIF4A and eIF4G, a scaffold for eIF4E and eIF4A, which also binds eIF3. eIF4G and eIF4B both stimulate eIF4A's helicase activity. Group 4 eIFs bind to the cap-proximal region of mRNA and unwind its secondary structure to allow attachment of 43S complexes, whose recruitment is promoted by the eIF4G–eIF3 interaction (2,3). Group 4 eIFs also assist 43S complexes during scanning. However, scanning on highly structured 5′ untranslated regions (UTRs) additionally requires DHX29, a DExH-box protein that binds directly to the 40S subunit (4–6). eIF1, in cooperation with eIF1A, ensures the fidelity of initiation codon selection, discriminating against initiation at non-AUG codons and AUGs that have poor nucleotide context or are too close to the 5′-end of mRNA. Finally, eIF5 and eIF5B mediate joining of 48S complexes with 60S ribosomal subunits to form elongation-competent 80S ribosomes (7,8).
In contrast, numerous viral mRNAs utilize 5′end-independent initiation mechanisms that are collectively known as ‘internal ribosomal entry’. Internal ribosomal entry sites (IRESs) are structured RNAs that mediate ribosomal recruitment to an internal location in the mRNA, and are classified into a few major groups, based on shared sequence motifs and a common structural core. Different classes of IRES use different mechanisms for initiation, but they are all based on specific non-canonical interactions with canonical components of the translational apparatus (1). Importantly, all IRESs use only a subset of canonical eIFs, and thus evade regulatory mechanisms that inhibit the canonical initiation process.
IRESs were first discovered in two members of the Picornaviridae, poliovirus (PV) and encephalomyocarditis virus (EMCV) (9,10), which exemplify two major IRES Types. Type I IRESs include PV, human rhinovirus (HRV), enterovirus 71 (EV71) and bovine enterovirus (BEV) (11–13). Type II IRESs include EMCV, foot-and-mouth disease virus (FMDV) and Theiler's murine encephalomyelitis virus (TMEV) (14,15). Type I and Type II IRESs are both ∼450-nt long, but they have unrelated sequences and structures, except for a Yn-Xm-AUG motif at their 3′-border, in which a Yn pyrimidine tract (n = 8–10 nt) is separated by a spacer (m = 18–20 nt) from an AUG triplet. Type I IRESs consist of five domains, designated dII-dVI. The AUG of the Yn-Xm-AUG motif is sequestered in dVI and does not serve as an initiation codon. Translation of the viral polyprotein starts at an AUG codon that is located from ∼30 nt (in HRV) to ∼160 nt (in e.g. PV) downstream. Type II IRESs also comprise five domains, designated H-L (16), followed by the Yn-Xm-AUG motif, whose AUG serves as the initiation codon for the viral polyprotein. On FMDV and TMEV IRESs, initiation can also occur at downstream AUG codons.
Type I and II IRESs require almost the full set of canonical eIFs, and for both classes, the mechanism of initiation is based on specific interaction of the central, eIF4A-binding domain of eIF4G (‘eIF4Gm’) with dV and J-K domains of Type I and II IRESs, respectively (17–20). Importantly, these eIF4G-binding sites have unrelated sequences and structures. Binding of eIF4G/eIF4A induces restructuring of downstream regions, which is thought to be essential for subsequent attachment of the 43S complex (19,21). In the case of the Type I PV IRES, attachment of 43S complexes requires direct interaction between eIF4G and eIF3 (13), whereas it is dispensable on the Type II EMCV IRES (20). On Type II IRESs, the 43S complex attaches immediately to the initiation codon of the Yn-Xm-AUG motif without prior scanning, and eIF1 and eIF1A are not essential for the process (17,22), whereas the initiation codon of Type I IRESs is reached by scanning (13,17,22). The nature of this process varies, depending on the stability of dVI. If it is relatively unstructured (e.g. in BEV), 43S complexes scan through it, and eIF1 becomes essential, because 43S complexes have to bypass the AUG triplet in dVI. If dVI is stable (as in PV and EV71), the majority of initiation events occur without inspection of dVI. Notably, initiation at the downstream AUG on the Type II FMDV IRES also involves scanning and is dependent on eIF1 (23).
Thus, initiation on Type I and II IRESs does not require eIF4E and the N-terminal region of eIF4G to which it binds, and picornaviruses exploit this to abrogate cellular cap-dependent translation by encoding viral proteases that cleave eIF4G, yielding a C-terminal fragment that binds eIF4A and eIF3 and is still sufficient to sustain viral translation (e.g. 24).
Importantly, initiation on both classes of picornavirus IRES requires specific IRES trans-acting factors (ITAFs) in addition to canonical eIFs. The common ITAF for Type II IRESs is the pyrimidine tract binding protein (PTB) (17,25), whereas all Type I IRESs require the poly(C) binding protein 2 (PCBP2) (13,26–28). PTB and PCBP2 both have multiple RNA-binding domains, and thus could bind simultaneously to different sites in an IRES, thereby maintaining it in an active conformation (e.g. 29). The requirement for ITAFs is conditional, and may differ even between isolates of a virus (e.g. 30). Moreover, some members of an IRES group may require additional ITAFs: thus, unlike EMCV and TMEV IRESs, the FMDV IRES also requires ITAF45 (25). Cell type-specific differences in ITAF expression levels may determine viral tissue tropism (25).
Interestingly, members of the Kobuvirus, Oscivirus and Salivirus genera of Picornaviridae have hybrid IRESs, with a central domain that is homologous to dIV of Type I IRESs followed by an eIF4G-binding domain that is homologous to domain J of Type II IRESs and the Yn-Xm-AUG motif (31,32). The initiation codon for the viral polyprotein, which is part of this motif, is sequestered in the stable hairpin domain L and initiation on these IRESs requires DHX29 in addition to PTB and a subset of canonical eIFs (31,32).
Taken together, characterization of initiation on picornavirus IRESs revealed that although members of each class of IRES share sequence and structural characteristics that distinguish them from members of other groups and define the general mechanism of their initiation, there is also significant variability within each class that results in variations in the mechanism of initiation, e.g. the requirements for DHX29, the degree of dependence on specific ITAFs, or the mode and factor requirements for ribosomal reaching of the initiation codon. Recent advances in sequencing technology and programs for bioinformatic analysis have resulted in the identification of numerous highly divergent picornaviruses, and characterization of their IRESs would be expected to reveal further variations of existing core mechanisms of initiation.
Cadicivirus (CDV) is a naturally occurring dicistronic (DC) picornavirus (33). It encodes two ORFs encoding structural and non-structural polyproteins, respectively, which are related to the P1 and P2-P3 genomic coding regions in picornaviruses of the genus Rosavirus (34,35) (Figure 1A). Rosaviruses have a Type II IRES in the 5′UTR whereas CDV has two IRESs, in the 5′UTR and in the intergenic region, both with homology to Type I IRESs (33). The CDV 5′UTR and IGR IRESs both contain structures equivalent to PV domain V, but the structures of upstream elements in both IRESs diverge substantially from each other and from those in conventional Type I IRESs (33; see below). Nevertheless, the level of translation promoted by the two CDV IRESs is similar, and interestingly, is comparable to that of the Type II EMCV IRES, at least in transfected MDCK cells (33). The chimeric nature of the CDV genome suggests that it is the product of multiple recombination events, one of which accounts for the presence of unrelated 5′UTRs in CDV and Rosavirus. Here, we have determined the structure of the divergent Type I IRES in the CDV 5′UTR and compared the mechanism of its function with that observed on canonical Type I IRESs.
Figure 1.
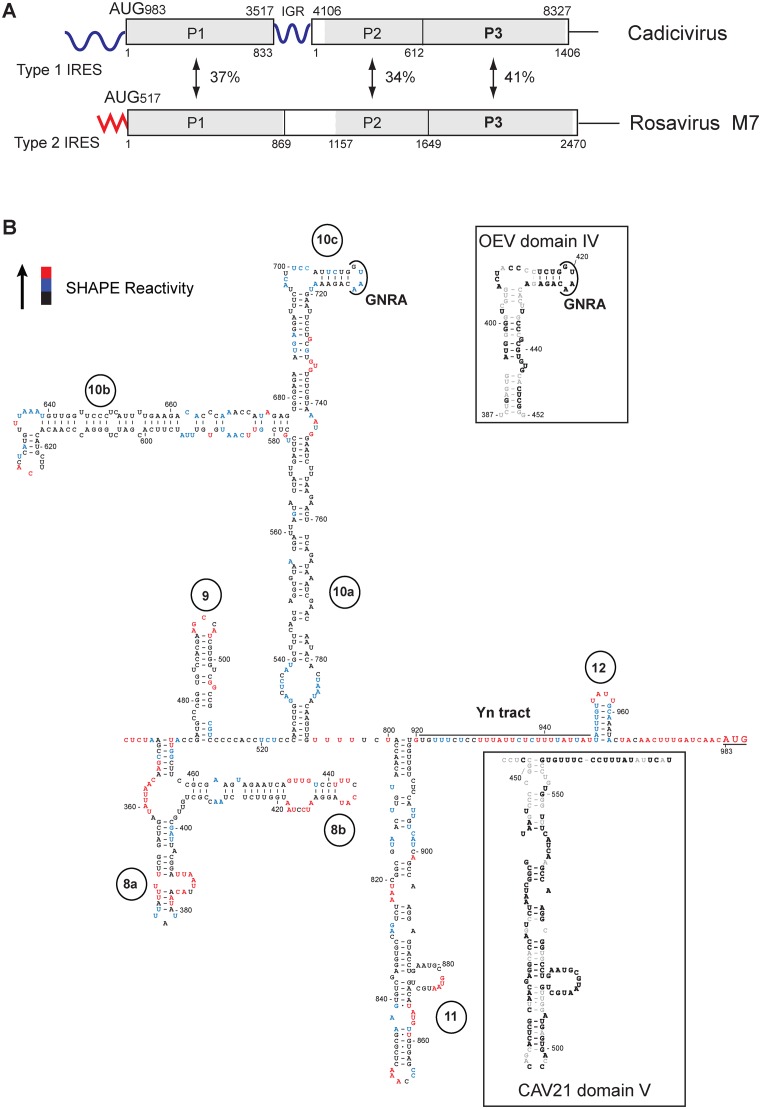
Structure and genomic context of the CDV 5’UTR IRES. (A) Schematic representations of CDV strain 209 and Rosavirus M7 genomes, showing the 5’UTR (including the IRES), P1 structural and P2-P3 non-structural protein coding regions, the 3’UTR and in CDV, the intergenic region (IGR), and labeled to indicate amino acid sequence identity between homologous regions. Numbers shown above and below the borders of domains refer to nucleotides and amino acid residues, respectively. (B) Secondary structure model of the CDV strain 209 IRES, color-coded to show reactivity to NMIA (see key at upper left and data shown in Supplementary Figure S1). The initiation codon (AUG983) is underlined. The upper inset panel shows the apical region of dIV of the Type I Ovine enterovirus IRES, showing the GNRA tetraloop and sequence identity with the corresponding element of CDV subdomain 10c (bold font). The lower inset panel shows the apical region of dV and the Yn tract of the Type I IRES of Coxsackievirus A21, showing sequence identity with CDV d11 and downstream sequences (bold font).
MATERIALS AND METHODS
Plasmids
Expression vectors were for His6-tagged eIF1 and eIF1A (22), eIF4A and eIF4B (17), eIF4AR362Q (36), eIF4AS33C and eIF4AS42C (19), wt, cysteine-less, T829C and D929DC eIF4GI736–1115 (21), eIF4GI653–1115, eIF4GI736–1008 and eIF4GI736–988 (20), DHX29 (37), Escherichia coli methionyl tRNA synthetase (38), ITAF45 (25), 9G8, PCBP1, PCBP2, human glycyl-tRNA synthetase (GARS), hsp27, PTB-1, La, unr-5 and unr+5 (13).
The DC transcription vector Stem-DC-XL-CDV was made (GenScript, Piscataway, NJ, USA) by inserting DNA between Pst1 and BamH1 sites of pUC57 that corresponded to a T3 promoter followed by a hairpin (GGGCCCGACCCGGTGACGGGTCGGGCCC) (ΔG = −32.40 kcal/mol), an SP6 promoter, a variant of nt 4–1168 of Xenopus laevis cyclin B2 mRNA (Genbank J03167) (XL), a 142-nt long linker terminating with Xma1/Sma1 restriction sites, a T7 promoter, a hairpin (GGGCGCGCGAGGGCGTGAGCCCTCGCGCGCCCG)(ΔG = -38.10 kcal/mol) and nt 1–1771 of CDV strain 209 (with substitutions that introduced AUG triplets at codons 170, 189, 200, 206 and 221 of the 228 a.a.-long (24.2 kDa) coding sequence, consecutive UGA and UAA stop codons and a 3′-terminal Msc1 restriction site. The plasmid was modified by replacing the XL ORF1 by an analogous BglII-Sma1 fragment from DC-Aichivirus containing a longer (nt 2–1369) cyclin B2 sequence (31). Transcription yields DC mRNA with or without a 5′-terminal hairpin, or a monocistronic (MC) mRNA with a 5′-terminal hairpin (Figure 2A), depending on the choice of polymerase and of the restriction site used for linearization.
Figure 2.
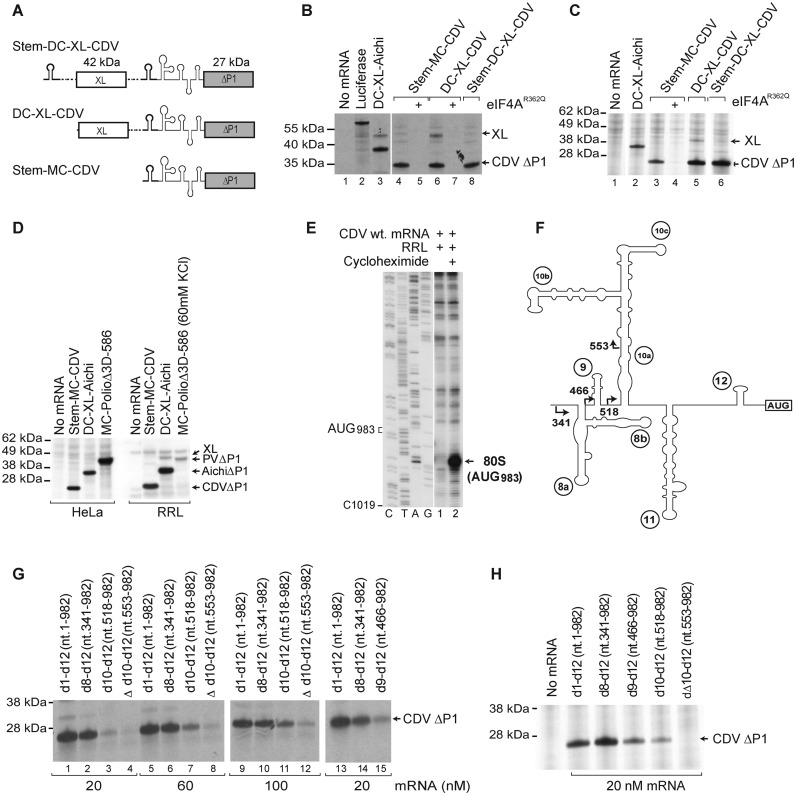
The 5’ border of the CDV 5’UTR IRES. (A) Schematic representation of monocistronic (MC) and dicistronic (DC) CDV mRNAs, with the CDV 5’UTR and a stable hairpin (thick line) placed upstream of the first and/or the second cistron. (B-D) Translational activity of the IRES in (B and D) RRL and (C and D) HeLa cell-free translation extract, and its sensitivity to inhibition by dominant-negative eIF4AR362Q. Luciferase and DC Aichivirus mRNAs (31) were used as controls for the efficiency of translation. (E) Toe-print analysis of 80S ribosomes formed on wt CDV MC mRNA in RRL in the presence of cycloheximide. (F) Model of the IRES, with arrows indicating the borders of 5’-terminal truncations of the 5’UTR. (G and H) Effect of these truncations, introduced into Stem-MC mRNA, on IRES activity in (G) RRL and (H) HeLa cell-free translation extract.
The Stem-DC-XL-CDV vector and derivatives thereof were used to generate variants with deletions and/or substitutions in the CDV 5′UTR. Initially, mutants were made with progressive deletions from the 5′-end of the 5′UTR, including Δnt 1–552 (NorClone Biotech Laboratories, London, Ontario) and Δnt 1–340, Δnt 1–465, Δnt 1–517 (generated by polymerase chain reaction (PCR) using forward primers containing a T7 promoter). Novel AUG triplets in good context (AUG944 and AUG950) were inserted into the 5′UTR via the substitutions TTATTAT941–947 →ACCATGG (DNA Express Inc, Montreal) and TTATTTG9947 -953→ACCATGG (NorClone Biotech), respectively. Domain 12 of the CDV IRES was replaced by dVI of PV Type 1 Mahoney by inserting a synthetic CDV nt 466–980 fragment containing this domain with flanking CDV nucleotides (GenScript) between BstEII and Bcl1 sites of DC Stem-XL-Stem-CDV. The resulting vector was modified further to insert new AUG triplets in good ‘Kozak’ context 26 and 35 nt downstream of dVI by substitutions CTTGATCTTACAGGC989–1003 →ATTATGGTTACAATG989–1003 (NorClone Biotech).
The vector Globin_N1-sc, for transcription of a variant of rabbit β-globin mRNA, was made (GenScript) by inserting DNA between EcoRV and HindIII sites of pUC57 that corresponded to an EcoR1 restriction site, a T7 promoter, the sequence GATGG, nt 2–197 of rabbit β-globin mRNA (Genbank NM_001082260.2) with 10 substitutions between nt 116 and 133 (to allow binding of a primer to a non-native sequence) followed by an A50 tail and an Ssp1 restriction site.
DNA for T7 polymerase-mediated transcription of PV nt 116–632 linked to ∼100-nt long fragment of the PV 3D ORF was generated by PCR using the Polio(AUG586)3D plasmid (39) and the primers: 5′-ATGCATGTAATACGACTCACTATAGGGAGCGTCAAAACCAAGTTCAATAGAAGGG-’3 (in which a T7 polymerase promoter is underlined and PV nt 116–137 are italicized), and 5′-GCACTGGTGGACAAATCAAG-3′ (which is complementary to PV nt 6314–6333).
Sequences
Sequences were analyzed from CDVs 209 (Genbank JN819202), 236 (JN819203) and 244U (JN819204), Rosaviruses M-7 (JF973686) and GA7403 (KJ158169), Norway rat rosavirus NYC-A15 (KJ950906), HRV A7 (DQ473503), Enterovirus C (Human coxsackievirus A21 strain Kuykendall) (AF546702), Enterovirus E1 strains VG-5–27 (D00214) and 82Sh2R (AJ250674), Enterovirus E2 strains PS 42; ATCC VR-758 (DQ092792) and D 3/98 (DQ092790), Enterovirus E3 strains D 14/3/96 (DQ092786) and HY12 (KF748290), Enterovirus E4 PAK-NIH-21E5 (JQ690748) and AK_NIH_48E3 (JQ690749), Enterovirus F1 strains BEV-261 (DQ092770) and alpaca/IL (KC748420), Enterovirus F2 strains PS87 (AY508696) and BHM26 (HQ917060), Enterovirus F3 strains PS87/Belfast (DQ092794) and PS-87 (X79368), Enterovirus F4 Strains W1 (New Zealand) (AY462106) and W6 (AY462107), Enterovirus F strain Wye 8875 (USA)(AY724745), Enterovirus G (Ovine enterovirus strain ovine/TB4-OEV/2009/HUN)(JQ277724) and enterovirus C109 NICA08–4327 (GQ865517).
Modeling of secondary and tertiary RNA structures
Secondary structure elements were modeled essentially as described (40), using probabilistic (Pfold; http://www.daimi.au.dk/_compbio/pfold/) (41) and posterior decoding approaches (CentroidFold; http://www.ncrna.org/centroidfold) (42) and were verified and refined by free energy minimization using Mfold (http://mfold.rna.albany.edu/?q_mfold) (43) and pKiss (http://bibiserv2.cebitec.uni-bielefeld.de/pkiss) (44), in all instances using default parameters. Computational analysis was done using series of overlapping RNA sequences.
SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) analysis
CDV RNA (1 pmol) was denatured, refolded and modified by incubation for 45 min at 37°C in 9 μl buffer (100 mM HEPES, pH 8.0, 6 mM MgCl2, 100 mM NaCl) with 1 μl DMSO or 1 μl 130 mM N-methylisatoic anhydride (NMIA; Sigma-Aldrich) in DMSO, as described (45). RNA was recovered by ethanol precipitation and dissolved in RNAse-free water. Sites of modification were determined by primer extension using avian myeloblastosis virus reverse transcriptase (AMV RT) and [32P]-labeled primers complementary to CDV strain 209 nt 94–114 (primer 1), nt 192–212 (primer 2), nt 297–320 (primer 3), nt 394–415 (primer 4), nt 518–535 (primer 5), nt 659–676 (primer 6), nt 730–751 (primer 7), nt 834–852 (primer 8), nt 914–934 (primer 9), nt 1016–1037 (primer 10) or nt 1086–1108 (primer 11).
In vitro translation
mRNAs (0.4 pmol unless stated) were translated in the Flexi Rabbit reticulocyte lysate (RRL) System (Promega) or the HeLa cell-free Protein Expression System (Takara Bio) (20 μl reaction volume) supplemented with 0.5 mCi/ml [35S]methionine (43.5 TBq/mmol) in the absence or in the presence of 4 μg of eIF4AR362Q for 60 min at 37°C. Translation products were analyzed by electrophoresis using NuPAGE 4–12% Bis-Tris-Gel (Invitrogen), followed by autoradiography and quantification on a phosphoimager.
Purification of initiation factors, ribosomal subunits and aminoacylation of tRNA
40S ribosomal subunits, eIF2, eIF3 and eIF4F were purified from RRL (46). Recombinant eIF1, eIF1A, eIF4A, eIF4B, eIF4Gm, DHX29 and E. coli methionyl tRNA synthetase were expressed and purified from E. coli (37,38,46). Recombinant Hsp27, La and His6-tagged unr−5 and unr+5 isoforms, 9G8, GARS, ITAF45, PCBP1, PCBP2 and PTB1 were expressed and purified from E. coli (13,47). Native unfractionated tRNA (Promega) and partially purified native tRNAiMet (48) were aminoacylated using E. coli methionyl tRNA synthetase, as described (46).
Assembly and analysis of ribosomal complexes
48S complexes were assembled by incubating 1 pmol CDV mRNA with 1.5 pmol 40S subunits, 3.5 pmol Met-tRNAiMet (unless otherwise stated), 2.5 pmol eIF2, 2 pmol eIF3, 7 pmol eIF1, 7 pmol eIF1A, 5 pmol eIF4A, 2 pmol eIF4B, 3 pmol eIF4Gm or its derivatives, 1 pmol eIF4F, 1 pmol DHX29 and 1.4 pmol PCBP1 or PCBP2, and 5 pmol PTB-1, 5 pmol ITAF45, 2 pmol 9G8, 2 pmol hsp27, 2 pmol GARS, 2 pmol unr + 5, 2 pmol unr-5 or 5 pmol La, as indicated, for 15 min at 37°C in 20 μl buffer A (20 mM Tris pH 7.5, 100 mM KCl, 1 mM DTT, 2.5 mM MgCl2, 0.25 mM spermidine) supplemented with 1 mM adenosine triphosphate (ATP) and 0.4 mM Guanosine triphosphate (GTP). Assembled 48S complexes were analyzed by toe-printing using AMV RT and 32P-labeled primer 11 as described (46). cDNA products were resolved in a 6% polyacrylamide sequencing gel.
For toe-printing analysis of 80S complexes assembled in RRL, 1 pmol CDV mRNA was incubated for 10 min at 37°C in the Flexi RRL System (Promega) (20 μl reaction volume) supplemented with 20 μg cycloheximide. The reaction mixture was then diluted with buffer A to 100 μl final volume, and assembled 80S complexes were analyzed by toe-printing using AMV RT and 4 pmol 32P-labeled primer 11 as described (46). cDNA products were resolved in a 6% polyacrylamide sequencing gel.
Analysis of the interaction of the CDV IRES with 40S subunits, eIF3, eIF4Gm and eIF4A
A total of 2 pmol of CDV mRNA was incubated with the indicated (Figures 6E and 7D) combinations of eIF4Gm (8 pmol), eIF4A (20 pmol), 40S subunits (4 pmol) and eIF3 (4 pmol) for 15 min at 37°C in 40 μl buffer A supplemented with 1 mM ATP, and then analyzed by primer extension inhibition for 2 h at 20°C, using AMV RT and [32P]-labeled primer 11. cDNA products were resolved in a 6% sequencing gel.
Figure 6.
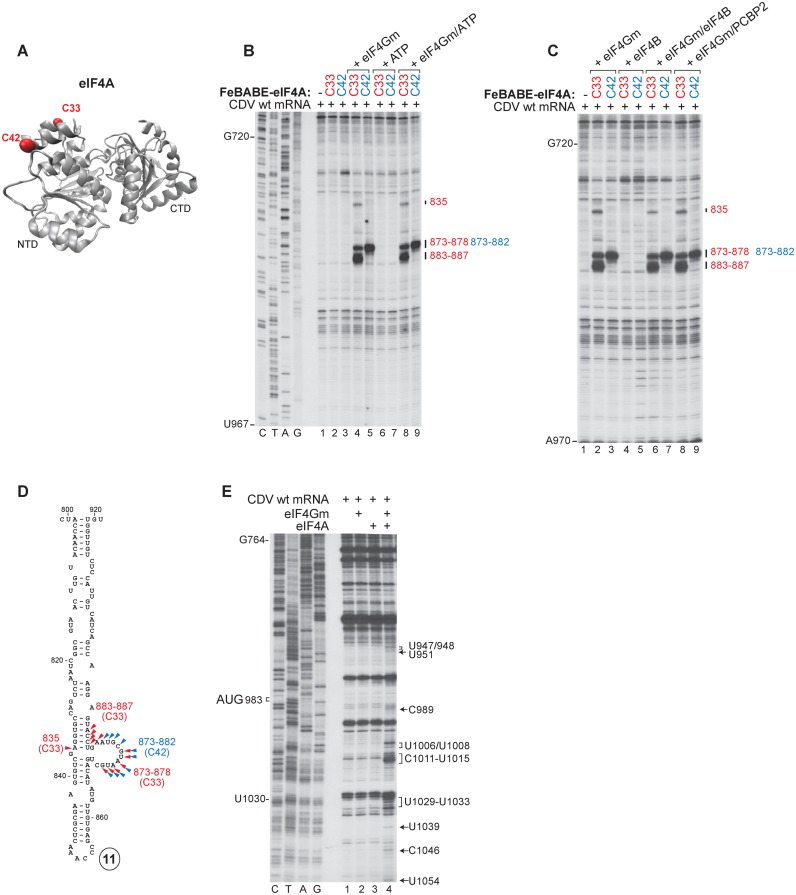
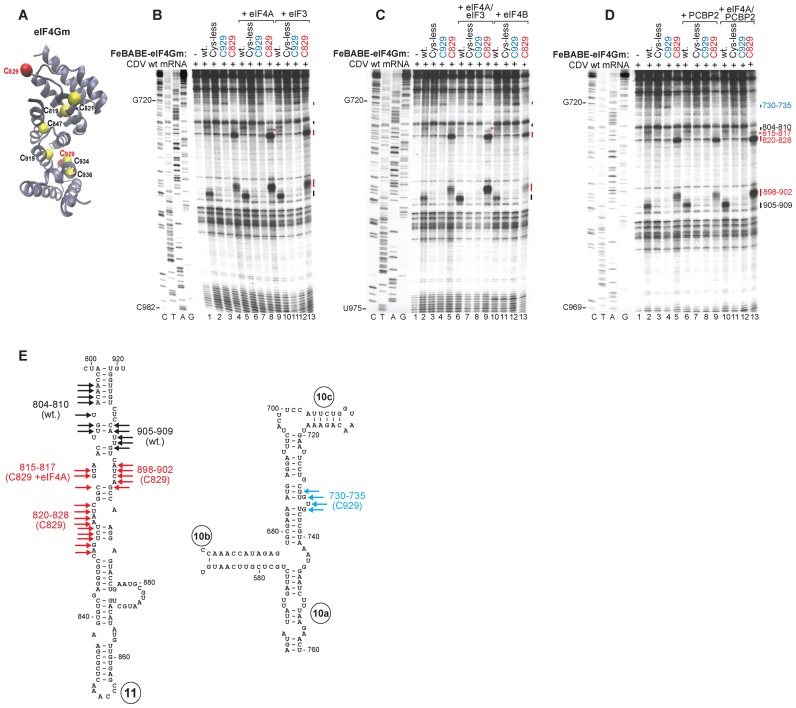
Interaction of the CDV IRES with eIF4A. (A) Ribbon diagram of eIF4A in the ATP/RNA-bound conformation (PDB: 3HX7) with red spheres indicating introduced (C33, C42) cysteines. (B and C) Primer extension analysis of directed hydroxyl radical cleavage of wt CDV mRNA from Fe(II)-tethered eIF4AS33C and eIF4AS42C (B) alone or in the presence of eIF4Gm, ATP or eIF4Gm and ATP and (C) in the presence of eIF4Gm, eIF4B, eIF4Gm/eIF4B or eIF4Gm/PCBP2. (D) Sites of hydroxyl radical cleavage from eIF4AS33C (red arrowheads) and eIF4AS42C (blue arrowheads) mapped onto a model of IRES d11. (E) Toe-prints induced on the IRES by interaction with combinations of eIF4A and eIF4Gm.
Figure 7.
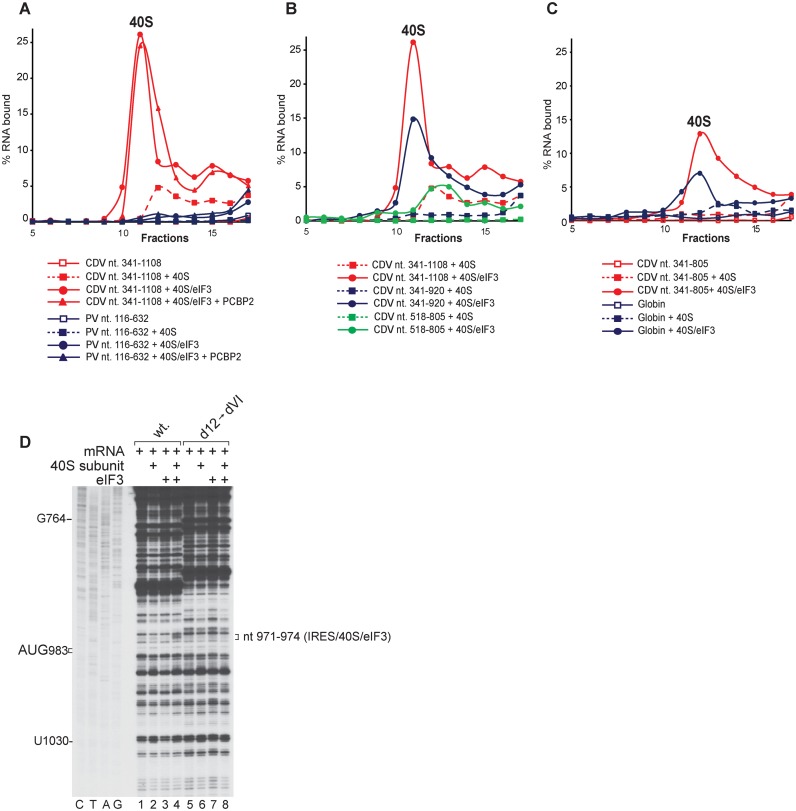
Binding of the CDV IRES to 40S subunits depending on the presence of eIF3. (A–C) Binding of 40S subunits and 40S/eIF3 complexes to the IRES, assayed by SDG centrifugation. Complexes were assembled by incubating (A) [32P]UTP-labeled CDV nt 341–1108, or PV mRNA comprising nt 116–632 and 96 nt of the 3D ORF, (B) [32P]UTP-labeled CDV nt 341–1108, nt 341–920 or nt 518–805 mRNAs (as indicated) and (C) [32P]UTP-labeled CDV nt 341–805 or nt 2–197 of β-globin mRNA with 40S subunits in the absence and in the presence of eIF3 and PCBP2, as indicated. Sedimentation was from right to left. Upper gradient fractions have been omitted for clarity. (D) Toe-printing analysis of binding of 40S subunits and eIF3 to wt and [d12→dVI] mutant Stem-MC CDV mRNAs. Toe-prints are indicated on the right.
Directed hydroxyl radical cleavage
Wt and cysteine-less eIF4Gm, eIF4GmT829C, eIF4GmD929DC, eIF4AS33C and eIF4AS42C were derivatized with Fe(II)-BABE as described (21). Derivatized proteins were separated from unincorporated reagent by buffer exchange on Microcon YM-30 filter units. To investigate hydroxyl radical cleavage, 10 pmol of Fe(II)-BABE-derivatized wt and cys-less eIF4Gm, eIF4GmT829C, eIF4GmD929DC, eIF4AS33C or eIF4AS42C were incubated at 37°C for 10 min in 40 μl buffer A with 4 pmol wt CDV mRNA in the presence/absence of 20 pmol of eIFs and/or PCBP2 and 1 mM ATP, as indicated in Figures 5 and 6. Cleavage reactions were initiated with 0.05% H2O2 and 5 mM ascorbic acid, incubated on ice for 10 min, and quenched using 20 mM thiourea (21). Sites of hydroxyl radical cleavage were determined by primer extension using AMV RT and 32P-labeled primer 10. cDNA products were resolved in a 6% sequencing gel.
Figure 5.
Interaction of the CDV IRES with eIF4G. (A) Ribbon diagram of eIF4Gm (PDB: 1HU3) with yellow spheres indicating native (C819, C821, C847, C919, C934 and C936) and red spheres showing introduced (C829, C929) cysteines. (B–D) Primer extension analysis of directed hydroxyl radical cleavage of wt CDV mRNA from Fe(II)-tethered wt eIF4Gm, eIF4GmT829C and eIF4GmD929DC in the presence and absence of (B) eIF4A or eIF3, (C) eIF4A/eIF3 or eIF4B and (D) PCBP2 or eIF4A/PCBP2. (E) Sites of hydroxyl radical cleavage from eIF4GmT929C (blue arrows), eIF4GmD829DC (red arrows) and wt eIF4Gm (black arrows) mapped onto models of IRES d11 and the apex of d10.
Analysis of ribosomal complexes by sucrose density gradient centrifugation
40S/IRES and 40S/eIF3/IRES complexes were assembled by incubating 40S subunits (6 pmol), eIF3 (6 pmol) and 2 pmol [32P]UTP-labeled RNA corresponding to CDV nt 341–1108, 341–920, 341–805 or 518–805, PV nt 116–632 linked to 96 nts of the 3D ORF, or β-globin nt 2–197, as indicated in Figure 7, and analyzed by centrifugation through 10–30% sucrose density gradients (SDGs) prepared in buffer A (46). The optical density of fractionated gradients was measured at 260 nm, and the presence of [32P]-labeled mRNA was monitored by Cherenkov counting.
RESULTS
The structure of the CDV 5′-untranslated region
The initial model of the CDV 5′UTR suggested that even some closely related sequences in it and in Type I IRESs (e.g dV in the PV IRES) form significantly different structures (33). We re-examined the CDV 5′UTR using complementary bioinformatic approaches. Application of an explicit evolutionary model to aligned sequences (41) yielded probabilistic models of domains 1, 2, 5, most of domain 10 and helical elements in domains 3, 4, 6, 7, 8, 9 and 11. A posterior decoding approach (42) confirmed these models and identified domain 12 and elements in domains 3, 4, 6, 8b and 9. The resulting model was refined by free energy minimization (43,44) and by assessing its compatibility with the results of chemical modification of the IRES by NMIA (Supplementary Figure S1). This analysis identified a series of single or branched hairpin domains, extending from domain 1 near the 5′-terminus to domain 12, 17-nt upstream of the initiation codon (Supplementary Figure S2).
In the resulting model (Figure 1B), structural homology is apparent between elements of the CDV 5′UTR and of canonical Type I IRESs, such as the apex of CDV d10c and Ovine enterovirus dIV (53% nt identity) (upper inset panel), apical and central regions of CDV d11 and human coxsackievirus A21 dV (79% identity) and their Yn tracts (lower inset panel). There are also major differences between the CDV 5′UTR and canonical Type I IRESs: the CDV 5′UTR lacks equivalents of dII and dIII, its d10 has a central three-way helical junction instead of the four-way junction present in dIV, and the base of d11 is extended relative to dV (lower inset panel). Domain 12 is shorter than dVI, and lacks its characteristic CUUAUGG motif (49).
Mapping the borders of the 5′UTR CDV IRES
Picornavirus 5′UTRs are modular, and the IRES commonly follows elements required for RNA replication. To determine if the CDV 5′UTR functions as an IRES in RRL and HeLa cell-free extract, we investigated its activity in MC CDV mRNA (comprising the 5′UTR and adjacent coding region) containing a stable 5′-terminal stem (ΔG = −38.10 kcal/mol) and in DC mRNAs, in which the stem, CDV 5′UTR and adjacent coding region were inserted downstream of a cyclin B2 cistron (XL), with or without a second 5′-terminal stem (ΔG = −32.40 kcal/mol) (Figure 2A). While translation of ORF1 from DC mRNA was strongly reduced by the 5′-terminal stem, the CDV 5′UTR ensured similarly robust levels of translation in all constructs, which were comparable to the level of translation driven by the Aichivirus IRES (Figure 2B–D). Thus, the CDV 5′UTR possesses an IRES that is fully functional in RRL. In contrast to the PV Type 1 IRES, translation promoted by the CDV IRES was as robust in RRL as in a HeLa cell-free extract, indicating that its activity did not depend on factors that are present in limiting amounts in RRL (Figure 2C and D). Initiation on the CDV IRES was readily inhibited by the dominant negative eIF4AR362Q mutant (50) (Figure 2B and C), indicating that the IRES function requires eIF4G and eIF4A.
To verify that translation initiates at AUG983, at the beginning of ORF1, the position of 80S ribosomes assembled on the IRES and arrested by inclusion of cycloheximide in translation reactions was mapped by toe-printing, which involves extension by reverse transcriptase of a primer annealed to the ribosome-bound mRNA. Eukaryotic ribosomes yield toe-prints +16–18-nt downstream of the P-site codon (e.g. 37). Following ribosomal arrest on the CDV IRES, prominent toe-prints appeared +17–18-nt downstream of AUG983 (Figure 2E), confirming that initiation in vitro took place at the authentic initiation codon.
To map the 5′-border of the IRES, the effect of progressive truncations (Figure 2F) was assayed using Stem-MC CDV mRNAs at different concentrations in in vitro translation in RRL (Figure 2G). Deletion of domains 1–7 (nt 1–340) had only a small effect on IRES function, reducing translation by ∼20% (Figure 2E, lane 2). In contrast, further deletion of d8 (nt 1–465) reduced IRES activity by >80% (Figure 2E, lane 15), and this inhibition was only slightly exacerbated by additional removal of d9 (nt 1–517) (Figure 2E, lane 3). Further truncation of part of d10 (nt 1–552) almost abrogated IRES function (Figure 2E, lane 4). The effects of these deletions were reduced by elevation of mRNA concentrations (Figure 2E, lanes 5–12), which likely compensated for the reduced affinity of truncated IRESs for components of the translational apparatus. Similarly, translation of this panel of mRNAs in a HeLa cell-free translation extract showed that nt 341–982 (domains 8–12) were sufficient for full IRES activity, that it was progressively reduced by removal of domains 8 and 9, and that it was abrogated by further deletion of part of domain 10 (Figure 2H).
Factor requirements for initiation on the CDV IRES
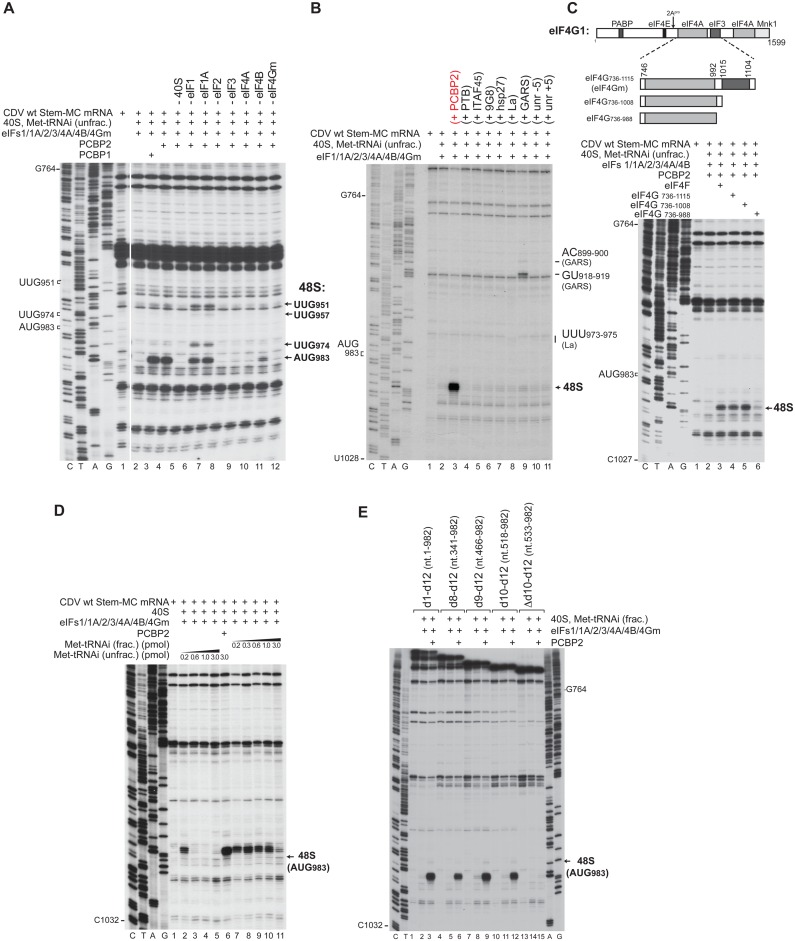
To investigate the factor requirements for initiation on the CDV IRES, we employed an in vitro reconstitution approach, in which 48S complexes were assembled on the Stem-MC CDV mRNA using individual purified translational components. 48S complex formation was monitored by toe-printing. 40S subunits, Met-tRNAiMet (obtained by aminoacylation of total native mammalian tRNA, referred to here as ‘unfractionated’ Met-tRNAiMet) and canonical eIFs (1, 1A, 2, 3, 4A, 4B and the central eIF4G736–1115 domain, termed ‘eIF4Gm’) were not sufficient for 48S complex formation (Figure 3A, lane 2). However, inclusion into reaction mixtures of PCBP2, which is required for initiation on all Type I IRESs tested to date (13,26–28), resulted in efficient 48S complex formation at CDV AUG983 (Figure 3A, lane 4; Figure 3B, lane 3). PCBP exists in four isoforms, each containing three K-homology (KH) domains. PCBP1 and PCBP2 are closely related, differing principally in the linker between KH2 and KH3 domains. PCBP1 was able to substitute fully for PCBP2 in promoting 48S complex formation (Figure 3A, lane 3). In contrast to PCBP1 and PCBP2, hsp27 (which binds to eIF4Gm), 9G8 (which binds to murine PCBP2) and several ITAFs that have been implicated in picornavirus IRES function, including La, PTB, ITAF45 and both isoforms of upstream of N-Ras (unr) (51) did not complement the activity of canonical eIFs in supporting 48S complex formation on the CDV IRES (Figure 3B). GARS bound specifically and stably to the CDV IRES, yielding toeprints at nt 899–900 and nt 918–919 in domain 11, whereas binding of La yielded weak toeprints at nt 973–975, a short distance upstream of the initiation codon AUG983 (Figure 3B, lanes 8, 9). These two proteins also interact with the Type I PV IRES (52,53). Inclusion of GARS led to a marginal level of 48S complex formation on the CDV IRES, whereas La had no effect.
Figure 3.
Factor requirements for initiation on the CDV IRES. Toe-printing analysis of 48S complex formation on CDV MC mRNAs containing (A–D) full-length or (E) 5’-terminally truncated derivatives of the 5’UTR, in the in vitro reconstituted system in the presence of 40S subunits, unfractionated or partially purified (‘fractionated’) Met-tRNAiMet, eIFs, PCBP1, PCBP2, PTB1, ITAF45, 9G8, hsp27, La, GARS, unr-5, unr+5, eIF4F, eIF4G736–1115, eIF4G736–1108 or eIF4G736–988, as indicated. Toe-prints caused by 48S complexes assembled on AUG983 or at near-cognate initiation codons (UUG951, UUG957, UUG974) are indicated on the right. (C, upper panel) Schematic representation of full-length and truncated fragments of eIF4G1, showing sites of factor binding and cleavage by 2Apro.
Systematic omission revealed that eIF2, eIF3, eIF4A and eIF4Gm were essential for 48S complex formation at AUG983 (Figure 3A, lanes 8–10, 12), whereas eIF4B had a strong stimulatory effect (Figure 3A, lane 11). Omission of eIF1 and eIF1A individually only moderately reduced initiation at AUG983, but importantly, it affected the fidelity of initiation, leading to assembly of 48S complexes at the upstream near-cognate codons UUG951, UUG957 and UUG974 (Figure 3A, lanes 6 and 7).
Direct eIF3/eIF4G interaction is essential for initiation on the Type I PV IRES but not on the Type II EMCV IRES (13,20). To investigate whether eIF3/eIF4G interaction is required for 48S complex formation on the CDV IRES, eIF4Gm truncation mutants lacking the C-terminal eIF3-binding domain (Figure 3C, upper panel) were employed. In contrast to initiation on the canonical Type I PV IRES, deletion of the eIF3-binding region (eIF4G736–1008) did not affect eIF4G's ability to promote 48S complex formation on the CDV IRES (Figure 3C, lane 5). Further truncation of the linker between eIF4A- and eIF3-binding regions (eIF4G736–988) very strongly reduced 48S complex formation likely due to weakening of the eIF4G/eIF4A interaction (Figure 3C, lane 6).
PCBP2 and domains 8 and 9 of the IRES enhance its translational competitiveness
Commercially available preparations of mammalian tRNA contain RNA contaminants (both short and long) that could compete with the CDV IRES in the in vitro reconstituted system (54). To investigate 48S complex formation on the CDV IRES in conditions of minimized potential competition, total tRNA was partially purified by size exclusion chromatography (48). Met-tRNAiMet obtained by aminoacylation of partially purified total native mammalian tRNA will be referred to as ‘fractionated’ Met-tRNAiMet.
To assay the influence of competitors on initiation on the CDV IRES, we compared 48S complex formation at different concentrations of fractionated or unfractionated Met-tRNAiMet. Surprisingly, at the lowest concentration of Met-tRNAiMet (either fractionated or unfractionated), substantial 48S complex formation occurred even in the absence of PCBP2 (Figure 3D, lanes 2 and 7). However, even a small elevation of the concentration of unfractionated Met-tRNAiMet abrogated PCBP2-independent 48S complex formation (Figure 3D, lanes 3–5), whereas fractionated Met-tRNAiMet showed an inhibitory effect only at a much higher amount (Figure 3D, lanes 7–11). Importantly, inclusion of PCBP2 with the highest concentration of unfractionated tRNA led to efficient 48S complex formation at AUG983 (Figure 3D, lane 6), indicating that PCBP2 enables the IRES to compete effectively for access to the translational apparatus.
In vitro reconstitution of 48S complexes on 5′-terminally truncated CDV IRESs done using fractionated Met-tRNAiMet showed that IRES function was maintained after progressive deletion of domains 1–7, 1–8 and 1–9, and was abrogated only after partial deletion of d10 (Figure 3E). Domains 8 and 9 are therefore required for IRES function in RRL but not in the in vitro reconstituted system, likely because the latter contain lower levels of competitor RNA. Thus, d8 and d9 enhance the ability of the IRES to compete effectively in the cytoplasmic milieu.
The ribosomal landing site on the CDV IRES and the influence of mutations in d12 on initiation at AUG983
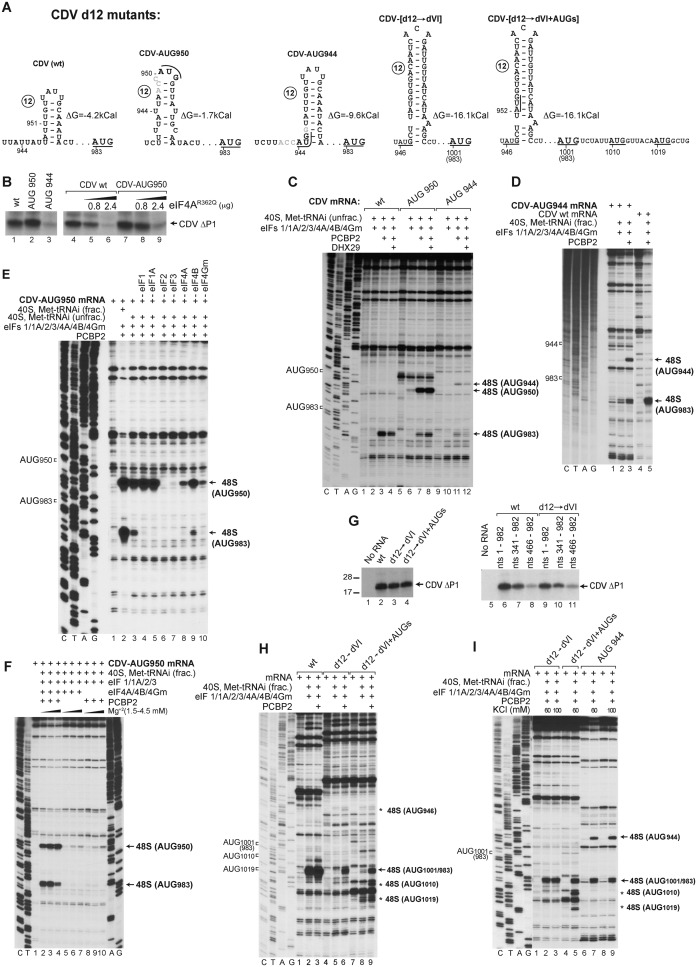
Formation of 48S complexes at the near-cognate initiation codons UUG951 and UUG974 in the absence of eIF1 suggests that 43S complexes can ‘enter’ the IRES at or upstream of nt 951 and most likely reach the authentic initiation codon AUG983 by subsequent scanning. Therefore to investigate the influence of the presence of upstream AUGs and the stability of d12 on the ability of 43S complexes to reach AUG983, we generated CDV mutants containing novel upstream AUG triplets and a reconfigured d12 with altered stability (Figure 4A). In the first mRNA (designated CDV-AUG950), a new AUG in good context (AccAUG950G) was introduced at nt 950 in frame with AUG983, which also reduced the stability of d12 (ΔG = −1.7 kcal/mol) compared to the wt d12 (ΔG = −4.2 kcal/mol). In the second mRNA (designated CDV-AUG944), an additional in-frame AUG in good context (AccAUG944G) was placed at nt 944, which substantially stabilized d12 (ΔG = −9.6 kcal/mol). In the third mRNA (designated CDV-[d12→dVI]), CDV d12 was replaced by the even more stable PV dVI (ΔG = −16.1 kcal/mol), which also created two additional upstream AUG triplets in suboptimal contexts at nts 946 and 952 (AuuAUG946C and CuuAUG952G), which are not in frame with the authentic initiation codon. In this mRNA, the naturally occurring AUG983 +18-nt downstream of dVI is referred to as AUG1001 because dVI is larger than d12. In the fourth mRNA (designated CDV-[d12→dVI + AUGs]), two additional in-frame AUGs in good contexts were introduced at nts 1010 and 1019 (AuuAUG1010G and AcaAUG1019G) downstream of AUG1001.
Figure 4.
Ribosomal attachment to the CDV IRES is followed by scanning to the initiation codon. (A) Details of the sequence and structure of IRES d12 and flanking nucleotides in wt and d12 mutant CDV mRNAs. (B and G) Translation of wt and d12 mutant Stem-MC CDV mRNAs in RRL in the absence and in the presence of eIF4AR362Q (μg, as indicated). (C–F, H and I) Toe-printing analysis of 48S complex formation on wt and d12 mutant Stem-MC CDV mRNAs in the presence of 40S subunits, eIFs, PCBP2, DHX29 and unfractionated Met-tRNAiMet (unless indicated otherwise), (F and I) at various Mg2+ and KCl concentrations, respectively. Toe-prints caused by 48S complexes assembled on the wt initiation codon (AUG983) or at the novel initiation codons AUG944 or AUG950, AUG1010 or AUG1019 are indicated on the right. The wt initiation codon is designated AUG1001/983 in (H and I).
Translation of Stem-MC CDV-AUG950 mRNA in RRL showed that AUG950 was efficiently used as an initiation codon (evident by the slight upward shift of the translation product) (Figure 4B, lane 2). In contrast, translation of CDV-AUG944 mRNA, from either new or authentic AUGs, was very weak (Figure 4B, lane 3). As in the case of the wt IRES (Figure 4B, lanes 4–6), translation of CDV-AUG950 mRNA was also inhibited by the eIF4AR362Q mutant (Figure 4B, lanes 7–9). The results of in vitro reconstitution in the presence of unfractionated Met-tRNAiMet were entirely consistent with translation in RRL. Thus, on CDV-AUG950 mRNA, efficient 48S complex formation occurred at AUG950, at the expense of initiation at AUG983 (Figure 4C, lanes 7), whereas on CDV-AUG944 mRNA, initiation on both AUG944 and AUG983 was very weak (Figure 4C, lane 11). The fact that AUG944 could still be recognized as the initiation codon (albeit with low efficiency) indicates that 43S complexes can ‘enter’ the IRES at or upstream of nt 944. The low level of initiation at AUG944 and AUG983 on CDV-AUG944 mRNA was likely due to the greater stability of d12 in the mutant mRNA, which could impair attachment of 43S complexes and/or subsequent scanning. Unlike initiation on the Aichivirus IRES that shares features of both Type I and II IRESs (31), but similarly to that on the canonical Type I PV IRES (13), DHX29 did not enhance 48S complex formation on mutant or wt CDV mRNAs (Figure 4C, lanes 4, 8 and 12).
48S complex formation on CDV-AUG944 mRNA was moderately enhanced by using fractionated tRNA (Figure 4D). The use of fractionated tRNA also strongly enhanced relative initiation at AUG983 on CDV-AUG950 mRNA (Figure 4E, compare lanes 2 and 3). The RNA contaminants present in unfractionated tRNA could compete with the IRES for RNA-binding factors that are required for scanning, e.g. eIF4B. Sequestration of such factors would have a stronger effect on initiation at AUG983 than at AUG950 because of its higher dependence on scanning. Consistently, the effect of elevated Mg2+ concentrations, which impair scanning, also favored initiation at AUG950 over initiation at AUG983 (Figure 4F, lanes 2–4). Analogous experiments have shown that initiation by direct ribosomal loading onto the initiation codon e.g. on the Type II EMCV IRES is more resistant to elevated Mg2+ concentrations than initiation by mechanisms that also involve substantial scanning e.g. on the Type I PV IRES (13,55). Systematic factor omission revealed that eIF2 and eIF3 were essential, whereas eIF1 and eIF1A were dispensable for initiation at AUG950 (Figure 4E, lanes 4–7). Interestingly, even with unfractionated tRNA, low-level 48S complex formation at AUG950 occurred in the absence of eIF4A or eIF4Gm (Figure 4E, lanes 8 and 10). It also occurred in the absence of all group 4 eIFs, with an efficiency that was similar to that observed in the absence of PCBP2 (Figure 4F, lanes 5–10). eIF4A/eIF4Gm-independent 48S complex formation at AUG950 was not due to contamination of eIF3 by eIF4F, because 48S complexes did not form on the EMCV IRES in the absence of eIF4A/eIF4Gm with the same factor preparations (data not shown).
Surprisingly, replacement of d12 by the significantly more stable PV dVI had only a minor effect on translation of CDV IRES mRNA in RRL (Figure 4G, compare lanes 2 and 3). Translation of CDV-[d12→dVI] mRNA was also not significantly altered by the presence of downstream in-frame AUG1010 and AUG1019 (Figure 4G, lane 4). Progressive 5′-terminal truncation of wt and CDV-[d12→dVI] mutant mRNAs also had very similar effect on their translation (Figure 4G, lanes 6–11). However, in the in vitro reconstituted reactions done using even fractionated Met-tRNAiMet, 48S complex formation at AUG1001 on CDV-[d12→dVI] mRNA was much weaker than at AUG983 on the wt IRES and was strongly dependent on PCBP2 (Figure 4H, lanes 1–6). Almost no 48S complexes formed at the upstream AUG946 and AUG952 on CDV-[d12→dVI] mRNA. On CDV-[d12→dVI + AUGs] mRNA, substantial leaky scanning past AUG1001 and 48S complex formation at the downstream AUG1010 and AUG1019 were observed (Figure 4H, lanes 7–9). However, the fact that the presence of AUG1010 and AUG1019 did not increase translation of CDV-[d12→dVI + AUGs] mRNA compared to CDV-[d12→dVI] mRNA indicates that AUG1001 is efficiently selected as the initiation codon in RRL. Initiation on canonical Type I IRESs has a low [K+] optimum (13,55), and in in vitro reconstituted reactions, was significantly more efficient at 60 mM K+ than at 100 mM K+ (8). However, reduction of [K+] from 100 to 60 mM did not enhance initiation on CDV-[d12→dVI] or CDV-AUG944 mRNA (Figure 4I). This observation suggests that RRL contains an additional factor that complements the activities of PCBP2 and eIFs 1, 1A, 2, 3, 4A, 4B and 4Gm in promoting initiation on the CDV-PV hybrid IRES.
Specific interactions of eIF4G/eIF4A with the CDV IRES
Initiation on canonical Type I IRESs relies on the specific interaction of its domain V with eIF4Gm, which is enhanced by eIF4A (13,19). In light of the strong homology between dV of Type I IRESs and CDV d11 (Figure 1B, lower inset panel), we investigated whether eIF4Gm/eIF4A also bind directly to the CDV IRES and whether their modes of binding are compatible with that observed on canonical Type I IRESs, using the directed hydroxyl radical cleavage (DHRC) technique. In this approach locally generated hydroxyl radicals cleave mRNA in the vicinity of Fe(II) tethered to a cysteine residue on the surface of mRNA-bound protein via the linker 1-(p-bromoacetamidobenzyl)-EDTA (BABE), after which cleavage sites are mapped by primer extension.
eIF4Gm comprises five α-helical ‘HEAT repeats’ that contain six native Cys residues (C819, C821, C847, C919, C934 and C936) (56) (Figure 5A). We employed wt eIF4Gm, a cysteine-less variant (‘cys-less’) and C829 and C929 single-cysteine eIF4Gm mutants, which are fully active and have previously been used to characterize eIF4Gm's interaction with Type I and II IRESs (13,19,21,29,31,32). As in the case of canonical Type I IRESs, eIF4Gm alone bound specifically to the CDV IRES, inducing prominent cleavage in d11 at nt 804–810 and nt 905–909 from wt eIF4Gm, and at nts 820–828 and 898–902 from eIF4Gm-[C829], and weak cleavage in d10 at nt 730–735 from eIF4Gm-[C929] (Figure 5B, lanes 2–5; Figure 5E). Cleavage induced by eIF4GM-[C829] on the CDV IRES and on the Type I PV and EV71 IRESs (19) occurs at analogous positions; cleavage at nt 730–735 in d10 by eIF4Gm-[C929] parallels weak cleavage from this residue in the apical region of EV71 IRES dIV (19).
Inclusion of eIF4A enhanced cleavage at nt 820–828 and 898–902 and led to the appearance of cleavage at nt 815–817 (Figure 5B, lanes 6–9), which are all located in the central region of d11 (Figure 5E). No significant changes in cleavage were observed on inclusion of eIF3, eIF4B or PCBP2 (Figure 5B, lanes 10–13; Figure 5C, lanes 10–13; Figure 5D, lanes 6–9), or of eIF3 or PCBP2 with eIF4A (Figure 5C, lanes 6–9; Figure 5D, lanes 10–13). Taken together, these data indicate that eIF4Gm binds to the CDV IRES in the same way as to canonical Type I IRESs, and that its binding is similarly enhanced by eIF4A.
We next investigated eIF4Gm-mediated recruitment of eIF4A to the CDV IRES and eIF4A's orientation in IRES/eIF4Gm/eIF4A complexes, also using the DHRC technique. eIF4A comprises two RecA domains (Figure 6A) and binds via its N-terminal domain to the C-terminal helix of eIF4Gm. For DHRC experiments, we employed cys-less and C33 and C42 single-cysteine eIF4A mutants (Figure 6A), which have previously been used to characterize the position of eIF4A on Type I and Type II IRESs (19,29,31). eIF4A alone, with or without ATP, did not induce cleavage of the CDV IRES, but in the presence of eIF4Gm, strong cleavage at nt 873–878 and nt 883–887, and weak cleavage at nt 835 in d11 occurred from eIF4A-[C33] (Figure 6B, lane 4; Figure 6D). Strong eIF4Gm-dependent cleavage was also induced by eIF4A-[C42] at nt 873–882 (Figure 6B, lane 5; Figure 6D). Inclusion of eIF4B in the absence of eIF4Gm did not lead to IRES cleavage, and neither eIF4B nor PCBP2 influenced the sites or the intensity of cleavage in its presence (Figure 6C, lanes 2–9). The locations of cleavage sites in d11 (Figure 6D) are directly comparable to eIF4A-induced sites of cleavage in canonical Type I IRESs (19).
Binding of eIF4G/eIF4A to Type I and II IRESs results in the appearance of toe-prints at the 3′ border of the IRES, which were attributed to induced conformational changes (19,21,31). Incubation of the CDV IRES with eIF4A and eIF4Gm together (but not individually) also led to the appearance of specific toe-prints upstream and downstream of AUG983 at nts 947–948, 951, 989, 1006–1008, 1011–1015, 1029–1033, 1039, 1046 and 1054 (Figure 6E; data not shown). Thus, the mechanism of action of eIF4G/eIF4A on the CDV IRES appears to be similar to that on canonical Type I and Type II IRESs.
Direct binding of the CDV IRES to 40S/eIF3 complexes
In the in vitro reconstituted system, 48S complex formation on the CDV IRES was not dependent on the direct interaction of eIF3 with IRES-bound eIF4G (Figure 3B). Recruitment of 43S complexes must therefore involve additional contacts between the IRES and components of this complex. We therefore investigated the potential interaction of the IRES with 40S subunits and eIF3. SDG centrifugation, which can be used to assay formation of stable complexes between mRNA and 40S subunits, showed that d8-d12 of the CDV IRES (nt 341–1108) could only weakly bind to individual 40S subunits, but associated with them very efficiently in the presence of eIF3 (Figure 7A, red). The binding was not influenced by PCBP2. In contrast, the PV IRES did not bind appreciably to 40S subunits alone or in the presence of eIF3 (Figure 7A, blue). Truncated forms of the CDV IRES comprising d8-d11 (nt 341–920) (Figure 7B, blue) and d8-d10 (nt 341–805) (Figure 7C, red) did not bind directly to 40S subunits, but formed stable complexes with them in the presence of eIF3, albeit with lower efficiency than d8-d12. Binding of CDV d10 (nt 518–805) to 40S subunits in the presence of eIF3 was very inefficient (Figure 7B, green) and occurred only at a level comparable to that of β-globin mRNA (Figure 7C, blue). In primer extension experiments, 40S subunits and eIF3 together induced toe-prints at nt 971–974 in the vicinity of the initiation codon (Figure 7D; data not shown). Thus, specific interactions between the CDV IRES and eIF3 and/or 40S subunits could potentially stimulate attachment of 43S complexes and alleviate the requirement for direct binding of eIF3 to eIF4G.
DISCUSSION
Structure of the CDV 5′UTR IRES
The ∼450-nt long canonical Type I IRESs comprise five domains (dII–dVI). Domains dII, dIV and dV are essential, whereas domains dIII and dVI are not (57,58). dV and dVI are connected by a conserved Yn-Xm-AUG motif, in which AUG constitutes a part of dVI. Consistent with the lack of an absolute requirement for the accessory domains III and VI adjacent to the dIV/dV core for Type I IRES function, numerous picornaviruses have dIII and/or dVI that are smaller than in the archetypal PV IRES (e.g. Falcovirus A1 and Enteroviruses E1, E2, E3, E4, F1, F2, F3 and F4), or an enlarged dVI (e.g. enterovirus 109 and HRV A7) (59–61).
The CDV IRES encompasses nts 341–950 (domains 8–12) and is thus larger than canonical Type I IRESs. The proposed structure of many helical elements in the IRES is supported by the pattern of compensatory sequence variation in strains 236, 209 and 244U. Although the CDV IRES contains the equivalent of the dIV/dV core of canonical Type I IRESs (domains d10/d11 in CDV), its 5′-flanking d8 and d9 are highly divergent and not homologous to dII and dIII, and the 3′-flanking d12 is smaller and less structured than the Type I dVI and does not contain an AUG triplet to complete the Yn-Xm-AUG motif. The strongest sequence conservation occurs in CDV d11 and dV of canonical Type I IRESs, which contain the site for specific binding of eIF4G. Even though the very apex of d10 is strongly homologous to the corresponding element in dIV of canonical Type I IRESs, d10 has a three-way helical junction instead of the conventional four-way junction observed in dIV, and its branching ‘10b’ subdomain is very elongated. The 5′-terminal region of the CDV IRES consists of the large Y-shaped d8, which has no apparent equivalent in any picornavirus IRES and the following hairpin d9 is shorter than the Type I dIII and lacks its characteristic E-loop motif (62).
Factor requirements for initiation on the CDV IRES and their functions in this process
The combination of canonical (d11) and divergent (d8, d9, d10 and the weakly structured region encompassing d12) elements of the CDV IRES determines common and specific aspects of its mechanism of function. In vitro reconstitution revealed that as in the case of canonical Type I IRESs, efficient initiation at the native initiation codon AUG983 of the CDV IRES requires eIFs 1, 1A, 2, 3, 4A, eIF4B and 4Gm, and a single ITAF, PCBP2. eIFs 2, 3, 4A and eIF4Gm are essential for the process. eIF4Gm specifically interacts with the region of d11, which is highly homologous to the eIF4Gm-binding site in canonical Type I IRESs (13,19). Association of the CDV IRES with eIF4Gm is stimulated by eIF4A, whose orientation in the CDV IRES/eIF4Gm/eIF4A complexes is also similar to that of canonical Type I IRESs (19). Binding of eIF4Gm/eIF4A induces RT stops in the area upstream and downstream of the initiation codon. Similar stops caused by eIF4Gm/eIF4A in the vicinity of the initiation codon have also been observed on Type I and II IRESs (19,21,31), and were attributed to local restructuring of the mRNA, potentially facilitating attachment of the 43S complex. Thus, the initial steps of binding of eIF4Gm, recruitment of eIF4A and possible restructuring of a downstream region are a common characteristic of initiation on the CDV and canonical Type I and II IRESs.
In addition to recognition of the IRES and recruitment of eIF4A, eIF4Gm plays a key role in promoting entry of 43S complexes onto canonical Type I IRESs via its interaction with eIF3 (13). However, initiation on the CDV IRES does not depend on this function of eIF4Gm, implying that in its absence, other interactions between the IRES and component(s) of the 43S complex could be instrumental in mediating ribosomal attachment to the IRES. SDG centrifugation experiments showed that the CDV IRES forms a stable ternary complex with the 40S subunit and eIF3. Analysis of the 40S/eIF3/IRES ternary complex by toe-printing revealed stops at nt 971–974, a short distance upstream of the native initiation codon AUG983, that could potentially correspond to the leading edge of the 40S subunit. The interaction of the CDV IRES with 40S subunits and eIF3 likely involves multiple points of contact, because 3′-terminally truncated forms of the CDV IRES comprising d8-d11 and d8-d10 also form stable ternary complexes with the 40S subunit and eIF3, albeit with reduced efficiency compared to the complete IRES comprising d8-d12. The interaction of the CDV IRES with 40S/eIF3 has striking parallels with the observation that 40S subunits also form dispersed contacts with the Type II EMCV IRES (63), initiation on which similarly does not depend on the direct eIF3/eIF4G interaction (20). Although the direct interaction of the CDV IRES with the 40S/eIF3 complex is intriguing, there is currently no reason to assume that eIF3 and eIF4G do not interact during viral translation in virus-infected cells, and thus that their interaction would not have a role in recruitment of 43S complexes to these IRESs. However, even in circumstances in which the eIF4G/eIF3 interaction can be established, specific binding of the IRES with the components of the 43S complex could increase the affinity of the eIF4G-bound IRES to the 43S complex and/or facilitate docking of viral mRNA into the mRNA-binding channel of the 40S subunit after initial capture of the 43S complex. The affinity of the CDV IRES for the components of the 43S complex could also be responsible for the observed low-level 48S complex formation in the absence of group 4 eIFs (Figure 4E and F).
Experiments done with CDV IRES mutants containing additional AUGs revealed that 43S complexes ‘enter’ the IRES upstream of AUG983 (at or before nt 944) and then reach it by scanning. This mechanism of initiation codon selection is similar to that observed on the canonical BEV IRES, in which dVI is also not very stable (13). The fidelity of AUG983 recognition is compromised in the absence of eIF1 or eIF1A, allowing 48S complexes to form at near-cognate upstream UUGs. However, in contrast to initiation on the BEV IRES, eIF1 and eIF1A are not completely essential, because in the CDV IRES, d12 does not contain an AUG to complete the Yn-Xm-AUG motif, whereas the efficiency of initiation at upstream UUGs is relatively low. Interestingly, as in the case of canonical Type I IRESs, 48S complex formation was strongly enhanced by eIF4B, even though the region preceding the initiation codon is not highly structured. Consistent with the scanning mechanism of initiation codon selection, stabilization of d12 strongly reduced initiation at AUG983, and similarly to canonical Type I IRESs (13), DHX29 was not able to relieve this inhibition. Interestingly, replacement of d12 by the even more stable PV dVI inhibited initiation on the CDV IRES only in an in vitro reconsituted system, but not in RRL. This suggests that RRL contains an additional factor that promotes initiation on the CDV-PV hybrid IRES by facilitating specific unwinding of the PV dVI or its efficient bypassing by 43S complexes.
As in the case of canonical Type I IRESs, initiation on the CDV IRES required PCBP2. 48S complex formation on the CDV IRES showed a lower dependence on PCBP2 when purified, rather than crude, Met-tRNAiMet was used in an in vitro reconstituted system, which indicates that PCBP2 enhanced the IRES's ability to compete with other mRNAs for the canonical components of the translation apparatus. There is no evidence that PCBP2 binds directly to any initiation factor, and our data indicate that it does not enhance binding of 40S subunits or 40S/eIF3 complexes to the IRES in SDG assays. Whereas direct stable interactions of PCBP2 with factors or the 40S subunit are thus unlikely to account for PCBP2's role in promoting initiation, binding of PCBP2 with the canonical Type I IRESs at multiple sites (13,64–66) suggests that it could constrain these IRESs in the active conformation. On canonical Type I IRESs, PCBP2 binds to the upper part of dIV, with the strongest interactions involving subdomains IVc and IVb (13,64–66). The corresponding region of the CDV IRES has very low sequence homology with canonical Type I IRESs, and it will be very important to determine the exact binding site of PCBP2 on the CDV IRES. Another interesting question is the role of the 5′-terminal flanking domains d8 and d9 of the CDV IRES, which becomes apparent only in conditions of competition with other mRNAs. Future studies will focus on determination of the influence of these domains on the affinity of the CDV IRES to eIF4Gm/eIF4A, PCBP2 and 40S/eIF3.
Thus, elucidation of the structure and mechanism of initiation on the CDV IRES here, and previous characterization of initiation on Aichivirus and related IRESs, which share structural characteristics with both Type I and II IRESs (31,32), together show that alteration of IRES structure by substitution and recombination results in the diversification of initiation mechanisms that remain based on the core characteristics of initiation on canonical Type I IRESs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [AI51340 to C.U.T.H., GM59660 to T.V.P.] Funding for open access charge: NIH [AI51340].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeFebvre A.K., Korneeva N.L., Trutschl M., Cvek U., Duzan R.D., Bradley C.A., Hershey J.W., Rhoads R.E. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villa N., Do A., Hershey J.W., Fraser C.S. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 2013;288:32932–32940. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abaeva I.S., Marintchev A., Pisareva V.P., Hellen C.U., Pestova T.V. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhote V., Sweeney T.R., Kim N., Hellen C.U., Pestova T.V. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3150–E3159. doi: 10.1073/pnas.1208014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisareva V.P., Pisarev A.V., Komar A.A., Hellen C.U., Pestova T.V. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unbehaun A., Borukhov S.I., Hellen C.U., Pestova T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 9.Jang S.K., Kräusslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 11.Borman A., Jackson R.J. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology. 1992;188:685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- 12.Thompson S.R., Sarnow P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology. 2003;315:259–266. doi: 10.1016/s0042-6822(03)00544-0. [DOI] [PubMed] [Google Scholar]
- 13.Sweeney T.R., Abaeva I.S., Pestova T.V., Hellen C.U. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014;33:76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kühn R., Luz N., Beck E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1990;64:4625–4631. doi: 10.1128/jvi.64.10.4625-4631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bandyopadhyay P.K., Wang C., Lipton H.L. Cap-independent translation by the 5′ untranslated region of Theiler's murine encephalomyelitis virus. J. Virol. 1992;66:6249–6256. doi: 10.1128/jvi.66.11.6249-6256.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duke G.M., Hoffman M.A., Palmenberg A.C. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pestova T.V., Hellen C.U., Shatsky I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova T.V., Shatsky I.N., Hellen C.U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Breyne S., Yu Y., Unbehaun A., Pestova T.V., Hellen C.U. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomakin I.B., Hellen C.U., Pestova T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolupaeva V.G., Lomakin I.B., Pestova T.V., Hellen C.U. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pestova T.V., Borukhov S.I., Hellen C.U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 23.Andreev D.E., Fernandez-Miragall O., Ramajo J., Dmitriev S.E., Terenin I.M., Martinez-Salas E., Shatsky I.N. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gradi A., Svitkin Y.V., Imataka H., Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilipenko E.V., Pestova T.V., Kolupaeva V.G., Khitrina E.V., Poperechnaya A.N., Agol V.I., Hellen C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 26.Blyn L.B., Towner J.S., Semler B.L., Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter B.L., Nguyen J.H., Ehrenfeld E., Semler B.L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamarnik A.V., Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y., Abaeva I.S., Marintchev A., Pestova T.V., Hellen C.U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaminski A., Jackson R.J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y., Sweeney T.R., Kafasla P., Jackson R.J., Pestova T.V., Hellen C.U. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011;30:4423–4436. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweeney T.R., Dhote V., Yu Y., Hellen C.U. A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J. Virol. 2012;86:1468–1486. doi: 10.1128/JVI.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo P.C., et al. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J. Virol. 2012;86:2797–2808. doi: 10.1128/JVI.05481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phan T.G., Vo N.P., Simmonds P., Samayoa E., Naccache S., Chiu C.Y., Delwart E. Rosavirus: the prototype of a proposed new genus of the Picornaviridae family. Virus Genes. 2013;47:556–558. doi: 10.1007/s11262-013-0968-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim E.S., Cao S., Holtz L.R., Antonio M., Stine O.C., Wang D. Discovery of rosavirus 2, a novel variant of a rodent-associated picornavirus, in children from The Gambia. Virology. 2014;454–455:25–33. doi: 10.1016/j.virol.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skabkin M.A., Skabkina O.V., Dhote V., Komar A.A., Hellen C.U., Pestova T.V. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lomakin I.B., Shirokikh N.E., Yusupov M.M., Hellen C.U., Pestova T.V. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pestova T.V., Hellen C.U., Wimmer E. A conserved AUG triplet in the 5’ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology. 1994;204:729–737. doi: 10.1006/viro.1994.1588. [DOI] [PubMed] [Google Scholar]
- 40.Asnani M., Kumar P., Hellen C.U. Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology. 2015;478:61–74. doi: 10.1016/j.virol.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knudsen B., Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato K., Hamada M., Asai K., Mituyama T. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janssen S., Giegerich R. The RNA shapes studio. Bioinformatics. 2015;31:423–425. doi: 10.1093/bioinformatics/btu649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson K.A., Merino E.J., Weeks K.M. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 46.Pisarev A.V., Unbehaun A., Hellen C.U., Pestova T.V. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 47.Kolupaeva V.G., de Breyne S., Pestova T.V., Hellen C.U. In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 2007;430:409–439. doi: 10.1016/S0076-6879(07)30016-5. [DOI] [PubMed] [Google Scholar]
- 48.Pestova T.V., Hellen C.U. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of transl.ation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wimmer E., Hellen C.U., Cao X. Genetics of poliovirus. Annu. Rev. Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 50.Pause A., Méthot N., Svitkin Y., Merrick W.C., Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flather D., Semler B.L. Picornaviruses and nuclear functions: targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015;6:594. doi: 10.3389/fmicb.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andreev D.E., Hirnet J., Terenin I.M., Dmitriev S.E., Niepmann M., Shatsky I.N. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res. 2012;40:5602–5614. doi: 10.1093/nar/gks182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meerovitch K., Svitkin Y.V., Lee H.S., Lejbkowicz F., Kenan D.J., Chan E.K., Agol V.I., Keene J.D., Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borman A.M., Bailly J.L., Girard M., Kean K.M. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcotrigiano J., Lomakin I.B., Sonenberg N., Pestova T.V., Hellen C.U., Burley S.K. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 57.Dildine S.L., Semler B.L. The deletion of 41 proximal nucleotides reverts a poliovirus mutant containing a temperature-sensitive lesion in the 5’ noncoding region of the genomic RNA. J. Virol. 1989;63:847–862. doi: 10.1128/jvi.63.2.847-862.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuge S., Nomoto A. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5’ noncoding sequence in viral replication. J. Virol. 1987;61:1478–1487. doi: 10.1128/jvi.61.5.1478-1487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zell R., Sidigi K., Henke A., Schmidt-Brauns J., Hoey E., Martin S., Stelzner A. Functional features of the bovine enterovirus 5’-non-translated region. J. Gen. Virol. 1999;80:2299–2309. doi: 10.1099/0022-1317-80-9-2299. [DOI] [PubMed] [Google Scholar]
- 60.Yozwiak N.L., Skewes-Cox P., Gordon A., Saborio S., Kuan G., Balmaseda A., Ganem D., Harris E., DeRisi J.L. Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J. Virol. 2010;84:9047–9058. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boros Á., Pankovics P., Simmonds P., Pollák E., Mátics R., Phan T.G., Delwart E., Reuter G. Genome analysis of a novel, highly divergent picornavirus from common kestrel (Falco tinnunculus): the first non-enteroviral picornavirus with type-I-like IRES. Infect. Genet. Evol. 2015;32:425–431. doi: 10.1016/j.meegid.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Bailey J.M., Tapprich W.E. Structure of the 5’ nontranslated region of the coxsackievirus b3 genome: Chemical modification and comparative sequence analysis. J. Virol. 2007;81:650–668. doi: 10.1128/JVI.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chamond N., Deforges J., Ulryck N., Sargueil B. 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs. Nucleic Acids Res. 2014;42:10373–10384. doi: 10.1093/nar/gku720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gamarnik A.V., Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5’ untranslated region of the poliovirus genome. J. Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sean P., Nguyen J.H., Semler B.L. Altered interactions between stem-loop IV within the 5’ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: effects on IRES-mediated translation and viral infectivity. Virology. 2009;389:45–58. doi: 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zell R., Ihle Y., Effenberger M., Seitz S., Wutzler P., Görlach M. Interaction of poly(rC)-binding protein 2 domains KH1 and KH3 with coxsackievirus RNA. Biochem. Biophys. Res. Commun. 2008;377:500–503. doi: 10.1016/j.bbrc.2008.09.156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.