Abstract
Topoisomerase V (Topo-V) is the only topoisomerase with both topoisomerase and DNA repair activities. The topoisomerase activity is conferred by a small alpha-helical domain, whereas the AP lyase activity is found in a region formed by 12 tandem helix-hairpin-helix ((HhH)2) domains. Although it was known that Topo-V has multiple repair sites, only one had been mapped. Here, we show that Topo-V has three AP lyase sites. The atomic structure and Small Angle X-ray Scattering studies of a 97 kDa fragment spanning the topoisomerase and 10 (HhH)2 domains reveal that the (HhH)2 domains extend away from the topoisomerase domain. A combination of biochemical and structural observations allow the mapping of the second repair site to the junction of the 9th and 10th (HhH)2 domains. The second site is structurally similar to the first one and to the sites found in other AP lyases. The 3rd AP lyase site is located in the 12th (HhH)2 domain. The results show that Topo-V is an unusual protein: it is the only known protein with more than one (HhH)2 domain, the only known topoisomerase with dual activities and is also unique by having three AP lyase repair sites in the same polypeptide.
INTRODUCTION
Topoisomerases are proteins found in all three domains of life that change the topology of DNA via transient breaks made on either one or both strands of a DNA molecule to allow passage of either a single or a double DNA strand through the break or swiveling of one strand around the other. They are involved in several cellular processes, such as transcription, replication and recombination (1–3). Topoisomerases are the target of several anti-cancer drugs and antibiotics as interfering with topoisomerase activity leads to cell death (4). Topoisomerases have been classified into two types: type I enzymes cleave one DNA strand and pass either a single or a double stranded (ds) DNA through the break before resealing it, whereas type II molecules cleave both DNA strands in concert and pass another dsDNA through the break followed by resealing of the break.
Type I enzymes are classified into three different sub-types: types IA, IB and IC (5). Members of one sub-type show no sequence or structural similarities to members of the other sub-types. Type IC topoisomerases comprise the most recently identified family of topoisomerases (6,7). Although they have many characteristics in common with type IB enzymes (8–10), the structure elucidation of a 61 kDa fragment of Methanopyrus kandleri Topoisomerase-V (Topo-V) (7) revealed that they form a distinct sub-type. In addition to relaxation activity, type IC enzymes show DNA repair activity (11,12), suggesting a close relationship between these two essential functions in hyperthermophiles. M. kandleri Topo-V has been identified only in the archaeal Methanopyrus genus, which is found in deep, hot vents in the ocean (13). Topo-V is a large molecule, formed by 984 amino acids (∼112 kDa) and has optimal activity at 122°C and high salt concentrations (14). Topo-V shows a modular structure with a 30 kDa topoisomerase domain followed by 24 helix-hairpin-helix (HhH) motifs arranged as 12 tandem (HhH)2 domains (7,11). The topoisomerase domain has no sequence or structural similarity to any other topoisomerase (7), the active site is well mapped and biochemical studies have shown that the cleavage and religation reactions are similar to the ones employed by type IB enzymes (8). In addition, single molecule studies have shown that Topo-V relaxes DNA by controlled rotation or swiveling, again in a similar manner to type IB topoisomerases (10). Overall, despite their differences, type IB and IC enzymes relax DNA using the same general mechanism, but using a completely different structural scaffold.
The DNA repair activity of Topo-V is not as well characterized as the topoisomerase activity. Previous studies showed that Topo-V has apurinic/apyrimidinic (AP) lyase and deoxyribose-5-phosphate (dRP) lyase activities and that there are at least two repair active sites in the protein, both located in the (HhH)2 domains (11). Recently, the location of the first repair site was identified and it was established that Topo-V is a Class I AP endonuclease that employs a catalytic lysine to cleave 3′ to the AP site (12). The catalytic lysine of the first repair site was identified as lysine 571. There is at least one more AP lyase site in Topo-V, but its exact location is not known, although it has been mapped to the last 34 kDa of the protein (11).
Here, we present studies that identified two additional AP lyase active sites in Topo-V. The location of a second AP lyase site was mapped to the junction of the 9th and 10th (HhH)2 domains. The structure of a 97 kDa fragment containing the topoisomerase domain as well as the first two repair domains was solved to 2.4 Å resolution. Site directed mutagenesis studies combined with structural analysis suggest that the catalytic lysine in the second AP lyase site is lysine 809. Once the first and second repair sites were removed by mutagenesis, a third repair active site was identified and mapped to (HhH)2 domain 12, at the C-terminus of the protein. Further analysis shows no additional repair sites. These studies establish that Topo-V is an unusual enzyme with four active sites within a single polypeptide, where the first site is involved in DNA relaxation using the topoisomerase active site and the other three sites are involved in DNA repair. The combined topoisomerase and DNA repair functions might be needed to survive the extreme living conditions of M. kandleri. The presence of both DNA repair and relaxation activities in the same polypeptide also suggests a possible synergy between the two activities. Although the way the four active sites may interact is still not understood, it could be mediated by DNA/protein interactions.
MATERIALS AND METHODS
Protein purification
DNA fragments coding for the Topo-V regions corresponding to residues 1–751 (Topo-86) and residues 1–907 (Topo-103) were PCR amplified from a plasmid containing the entire Topo-V sequence (15), with added 5′ NcoI and 3′ NdeI restriction sites, and cloned into the pET15b vector. Fragments corresponding to residues 1–802 (Topo-91) and 1–854 (Topo-97) were obtained by introducing stop codons at the desired position in the full length Topo-V gene. Topo-78 (residues 1–685), Topo-78(ΔRS1) (K566A/K570A/K571A, first repair site inactive) and full length Topo-V were made as previously described (11,12,15). Topo-V(ΔRS1) has K566, K570 and K571 mutated to alanine in the full length backbone. Topo-97(ΔRS2) was made from Topo-97 with K809, K820, K831, K835, K846 and K851 mutated to alanine to remove the second AP lyase site. Topo-V(ΔRS1ΔRS2) was made from Topo-V(ΔRS1) with K809, K820, K831, K835, K846 and K851 mutated to alanine to remove the first two repair sites. Topo-103(ΔRS1ΔRS2) was made by introducing a stop codon after residue 908 in Topo-V(ΔRS1ΔRS2). All the lysine mutants were made by site directed mutagenesis (Quikchange, Stratagene). For protein production, Topo-V was transformed into Escherichia coli BL21(DE3) pLysS cells, Topo-78 was transformed into E. coli BL21(DE3) cells and all other fragments were transformed into E. coli BL21 Rosetta (DE3) cells. Protein induction and purification was done according to previously described protocols (12,16). Pure proteins were concentrated and stored in 50 mM Tris pH 8, 250 mM NaCl and 1 mM DTT, except for Topo-97 and Topo-103 where 500 mM NaCl was used and Topo-V where 1M NaCl was used in the storage buffer for enhanced protein solubility. To ensure that the proteins were well folded, the CD spectra of all the mutants were measured (Supplementary Figure S1). The spectra show no major changes, suggesting that the proteins are well folded and that the changes in activity are not due to folding defects in the mutants.
AP lyase assay
AP lyase assays were done using the same 49-mer DNA and a similar procedure as described before (12,17). The DNA strand containing uracil was synthesized by integrated DNA technologies (IDT, Coralville, IA, USA) with a 5′ 6-FAM (fluorescein) label. The labeled DNA strand was annealed to equimolar amounts of the complementary strand. To produce the abasic site, the oligonucleotide was treated with Uracil DNA Glycosylase (UDG) (NEB) as previously described (17). The standard AP lyase assay contained 50 mM HEPES pH 7.5, 20 mM KCl, 0.5 mM EDTA, 2 mM DTT, 500 nM UDG-treated abasic DNA and 2.5 μM protein, except for the full-length protein where the protein concentration was reduced by 10-fold. Incubation was done at 65°C for 30 min. For the Topo-97 experiments, incubation was followed by treatment with 340 mM sodium borohydride on ice for 30 min. For the Topo-V wild-type, full length mutants and Topo-103 experiments, the sodium borohydride step was not used. In both cases, before gel loading the samples were treated by the addition of formamide dye (96% formamide, 10 mM EDTA) and incubated at 75°C for 2 min. The reaction products were separated on a 16% denaturing polyacrylamide gel (8 M urea). The gel was scanned using a Typhoon 9400 scanner (GE Lifesciences) and bands were quantified using ImageQuantTL version 5.2 (GE Healthcare). Percentage of cleavage was calculated as follows: (intensity of product band / (intensity of substrate band + intensity of product band)) × 100. For Topo-V full-length mutants, the above procedure was modified to include a proteinase K treatment step before gel loading due to the tight binding of the full length protein to DNA that prevented the DNA from entering the gel.
Crystallization, data collection, and structure determination
The Topo-97 fragment was crystallized by vapor diffusion against 0.1 M sodium acetate pH 5.5, 0.2 M NaCl and 7% PEG 8000. The crystals were frozen by supplementing the mother liquor with 20% sucrose as cryoprotectant. Diffraction data were collected at the Life Science Collaborative Access Team station (LS CAT) at the Advanced Photon Source (APS) in Argonne National Laboratory. Data collection and refinement statistics are shown in Supplementary Table I. All data were processed and integrated using XDS (18) and scaled with SCALA (19). The 2.4 Å resolution structure of Topo-97 was initially solved by molecular replacement (MR) with Phaser (20) using the Topo-61 fragment (PDB ID 2CSB (7)) as the search model. The MR map showed poor density for the missing portions in the model. To improve the initial map, a platinum derivatized crystal was used together with the Topo-61 model MR solution using the program Sharp (21). The phases obtained from the heavy atom derivative combined with the partial model improved the electron density, showing clear electron density for almost the entire protein. The program Buccaneer (19) was used to trace the model. Manual model building was done using Coot (22,23) and refinement using Refmac5 (24) and Phenix (25). Electron density for almost the entire molecule was observed with a break in the electron density between residues 597–689. The clear side chain density in the region towards the C-terminal part of the protein helped in the accurate placement of the amino acids after the break in the electron density map. The final Rwork and Rfree for the model are 19.9% and 23.8%, respectively. The final model contains residues 2–596, 690–852 and 184 water molecules. Analysis with Molprobity (26) shows that the model is of excellent quality, with 97.75% of the residues in the Ramachandran favored regions and no disallowed residues, 98.01% of the residues have favored rotamers and there were no poor rotamers. The model has a root mean square deviation (RMSD) of 0.002 Å for bond lengths and 0.57° for bond angles.
Small angle X-ray scattering
A small angle X-ray scattering (SAXS) experiment was performed using Topo-97(ΔRS2) at three different protein dilutions: 1.25 mg/ml, 2.5 mg/ml and 5 mg/ml. All data were collected at the Dupont Northwestern Dow Collaborative Access Team (DND CAT) beamline at the APS using a SAXS, wide angle X-ray scattering (WAXS), medium angle X-ray Scattering (MAXS) triple CCD detector system (27) at room temperature (23°C) using a flow cell to minimize radiation damage. Data processing details are shown in Supplementary Table II. Data for an empty flow cell as well as buffer were collected for each experiment. Data were collected at 12 keV (1.0332 Å) and covered the momentum transfer range of 0.0026 < q < 2.46 Å−1 (q = 4πsinθ/λ, where 2θ is the scattering angle). The scattering data from the detectors were azimuthally integrated and merged resulting in a one-dimensional scattering profile with respect to q and normalized to the incident beam intensity. Scattering due to the buffer was subtracted using local software (27) and taking into account the sample concentration, which was estimated from a 280 nm absorbance measurement of the sample. Further processing was done with the ATSAS SAXS software package (28). Data from the three concentrations were scaled together and merged to produce a final scattering curve in the 0.010649–0.61699 q range (Supplementary Figure S2). Analysis showed a linear Guinier plot resulting in an Rg of 45.65 Å with Fidelity of 0.91. A Dmax for the particle of 181 Å was selected based on the particle distance distribution function P(r) (Supplementary Figure S2). Twenty ab initio models were calculated using DAMMIF (29) to a q value of 0.45 Å−1 and all models were accepted and averaged using DAMAVER (30). The normalized standard deviation (NSD) for each of the models used was under 0.92 Å. An envelope was generated from the bead model and the Topo-97 crystal structure model was positioned in the envelope using Coot by manually fitting the first 596 residues, followed by manual placing of residues 690–852 into the map. The manually placed model captures the general features of the scattering profile and the particle distance distribution function despite the fact that no fitting to the data was performed (Supplementary Figure S2).
Figures
Figures for the atomic models were created using Pymol (31).
RESULTS AND DISCUSSION
Identification of the second AP lyase active site of Topo-V
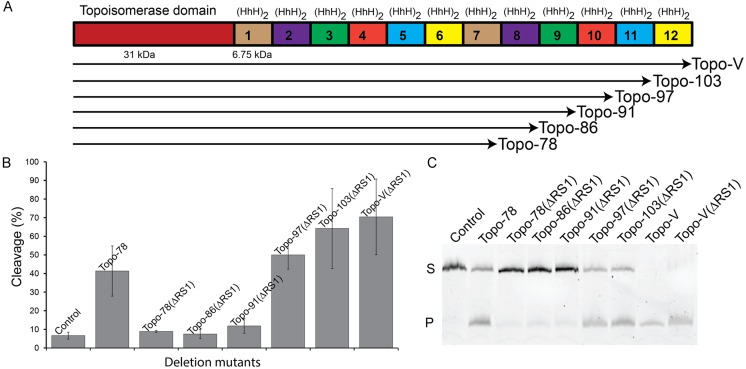
Topo-V is known to contain more than one DNA repair active site (11,12). Previously, the location of the first repair active site was mapped to the junction between the 5th and the 6th (HhH)2 domains and a combination of structural and biochemical studies suggested that lysine 571 is the active site lysine in the first repair site (12). To map the second active site, a series of deletion mutants of Topo-V in the context of a triple mutant (ΔRS1: K566A/K570A/K571A) that removes the repair activity from the first site were constructed and tested for AP lyase activity. The deletion mutants were constructed by removing sequentially one (HhH)2 domain from the C-terminus of the protein based on the location predicted by sequence analysis (15). The deletion mutants constructed included Topo-103(ΔRS1) (residues 1–907), Topo-97(ΔRS1) (residues 1–854), Topo-91(ΔRS1) (residues 1–802) and Topo-86(ΔRS1) (residues 1–751) (Figure 1A). All proteins were purified to homogeneity and the AP-lyase activity of these mutants as well as of the full length protein and the Topo-78 fragment were compared. AP lyase activity using a fluorescently labeled oligonucleotide containing a single abasic site shows activity for the full length protein and for the Topo-103(ΔRS1, (HhH)2 domain 12 deleted) and Topo-97(ΔRS1, (HhH)2 domains 11 and 12 deleted) Figure 1B and C). The Topo-91(ΔRS1) fragment, which contains (HhH)2 domains 1–9, as well as Topo-86(ΔRS1) which contains (HhH)2 domains 1–8 show no AP lyase activity (Figure 1B and C). These results conclusively show that the second repair active site is located in the vicinity of the 10th (HhH)2 domain. The location of the second active site is consistent with previous observations that showed that a 34 kDa C-terminal fragment of Topo-V has repair activity (15).
Figure 1.
Identification of a second AP lyase active site in Topo-V. (A) Schematic representation of the Topo-V protein and the various fragments studied. The N-terminus of the protein corresponds to the topoisomerase domain (red) and is followed by 12 tandem (HhH)2 domains (small boxes). The fragments studied were made by removing (HhH)2 domains from the C-terminus of the protein. (B) Quantification of the AP lyase activity for full-length Topo-V and different Topo-V fragments (3 replications). All fragments were made in a backbone where the first AP lyase active site was inactivated (ΔRS1). All fragments larger than 91 kDa exhibit AP lyase activity, showing that the second repair site is located in the Topo-97 fragment. The wild-type Topo-78 fragment was used as a positive control for AP lyase activity as it contains only one well-characterized repair active site. The partial cleavage of control DNA is due to the inherent instability of abasic DNA at the temperature used for the experiments. In all cases the level of activity reported is based on three replications and the error bars correspond to the standard error. (C) Gel illustrating experiments to locate the second AP lyase active site. The upper band corresponds to uncleaved DNA (S) while the lower band on the gel shows the product after cleavage of the DNA at the abasic site (P). Topo-78: wild-type 78 kDa fragment; Topo-78(ΔRS1): 78 kDa fragment with K566A/K570A/K571A mutations to inactivate the first repair site; Topo-V(ΔRS1): full-length Topo-V with the first repair site inactivated.
Previously it was shown that Topo-V uses a lysine as the nucleophile for the AP lyase activity (12), similar to many other AP lyase repair enzymes. In the case of the first repair site, the active site lysine was identified as K571 (12). To narrow down the position of the active site lysine for the second AP lyase active site, several lysine mutants in the 10th (HhH)2 domain were made in the Topo-97(ΔRS1) backbone and tested for activity. Sequence analysis and structural comparisons did not allow unambiguous identification of a lysine candidate as there are several lysines in the repeat and none of them exactly match the catalytic lysine of the first Topo-V repair site or that in other AP lyases, such as Geobacillus stearothermophilus Endonuclease III (GS Endo III) (32) or human 8-Oxoguanine DNA glycosylase 1 (Hs Ogg1) (33). For this reason, six lysines in this region (K809, K820, K831, K835, K846 and K851) were individually mutated to alanine. In addition, three double mutants (K809A/K820A, K831A/K835A and K846A/K851A) were made and tested for activity. All the single and double mutants show drastic reduction in activity, but not complete loss of AP lyase activity (Supplementary Figure S3). This is very similar to what was observed before when the first repair active site was identified, where single and double mutants retained partial activity and only a triple mutant showed complete loss of activity (12). A Topo-97(ΔRS1) mutant with all six lysines mutated (Topo-97(ΔRS1ΔRS2) did show complete loss of activity (Supplementary Figure S4), confirming that a lysine is the catalytic residue and that the repair active site maps to the 10th (HhH)2 domain.
Topo-V contains three AP lyase active sites
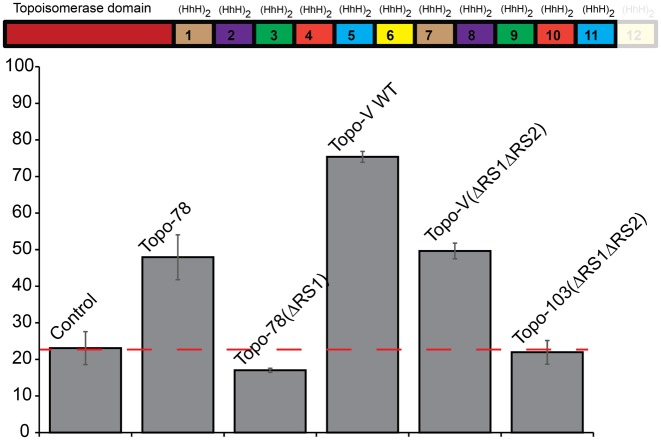
To ensure that there are no additional AP lyase sites in Topo-V, the full length protein was mutated to remove the first two active sites (three lysines in site 1 and six lysines in site 2 were mutated to alanines) and tested for AP lyase activity. This mutant protein (Topo-V(ΔRS1ΔRS2)) showed AP lyase activity, suggesting the presence of additional AP lyase active sites in Topo-V (Figure 2). Given the location of the second active site, additional active site(s) can only be located in either domain 11 or 12 or in both. To test for the presence of the AP lyase site in the last repeat, a deletion of the last (HhH)2 domain that will produce a variant that spans the first 11 (HhH)2 repeats (Topo-103(ΔRS1ΔRS2)) was made using the Topo-V(ΔRS1ΔRS2) backbone. The truncated protein showed no AP lyase activity (Figure 2), confirming the presence of only one more AP lyase site (third AP lyase site) at the extreme C-terminus of the protein and most likely in the last (HhH)2 domain.
Figure 2.
Identification of a third AP lyase active site in Topo-V. (Top) Schematic representation of the Topo-V fragment used to identify the third AP lyase active site. In the fragment, Topo-103, the last (HhH)2 domain has been removed (shown shaded). (Bottom) Quantification of the AP lyase activity for full length Topo-V and the Topo-103 fragment. The wild-type protein with the first two active sites inactivated (Topo-V(ΔRS1ΔRS2)) retained partial activity, indicating the presence of a third repair site. The Topo-103(ΔRS1ΔRS2) fragment, corresponding to the Topo-103 fragment with the first two AP lyase active site mutated, showed no AP lyase activity, consistent with the existence of a third active site in the last (HhH)2 domain. The wild-type Topo-78 fragment was used as a positive control for AP lyase activity as it contains only one well-characterized repair active site. Topo-78: wild-type 78 kDa fragment; Topo-78(ΔRS1): 78 kDa fragment with three mutations to inactivate the first repair site. The partial cleavage of control DNA is due to the inherent instability of abasic DNA at the temperature used for the experiments. In all cases the level of activity reported is based on three replications and the error bars correspond to the standard error. The red dashed line corresponds to the level of cleavage of the control DNA and serves to indicate baseline activity.
Sequence analysis of the full length protein shows the presence of several lysines in both domains 11 and 12, but only (HhH)2 domain 12 has lysines in similar positions to the first two AP lyase sites (Figure 4C). Of these lysines, K912 and K918 appear to be the most likely candidates for the active site residue as they are found in the putative first helix of the (HhH)2 domain. In the case of K918, its predicted location in the (HhH)2 domain matches the location of the putative catalytic lysine in the second repair site. Residues K912 and K918 were mutated to alanine in the Topo-V(ΔRS1ΔRS2) backbone. Similar to what was observed for the first and second AP lyase active sites, single and double mutants (K912A, K918A, K912A/K918A) showed reduced, but not complete loss of activity (Supplementary Figure S5). Two additional lysines, K901 and K904, were selected as possible residues that could contribute to activity, as they are in the predicted helix immediately before the region harboring K908 and K912. K904 is in a similar position to K566, which is one of the lysines that was mutated to abolish activity in the first repair site of Topo-V (12). A mutant with four lysines mutated to alanine (K901A/K904A/K912A/K918A) also showed reduced, but not complete loss of activity (Supplementary Figure S5). For this reason, two more lysines were considered, K942 and K943, which are in a similar position to K831 and K835. A mutant with all six lysines mutated to alanine (K901A/K904A/K912A/K918AK942A/K943A) show no AP lyase activity (Supplementary Figure S6), confirming the presence of a third AP lyase site in the protein. Given that it has only been possible to identify the active site lysines in the first and second active sites by a combination of structural and biochemical studies ((12) and below), the exact identity of the active site lysine in the third repair site remains unknown and only the general location of the repeat(s) harboring the active site is unambiguous.
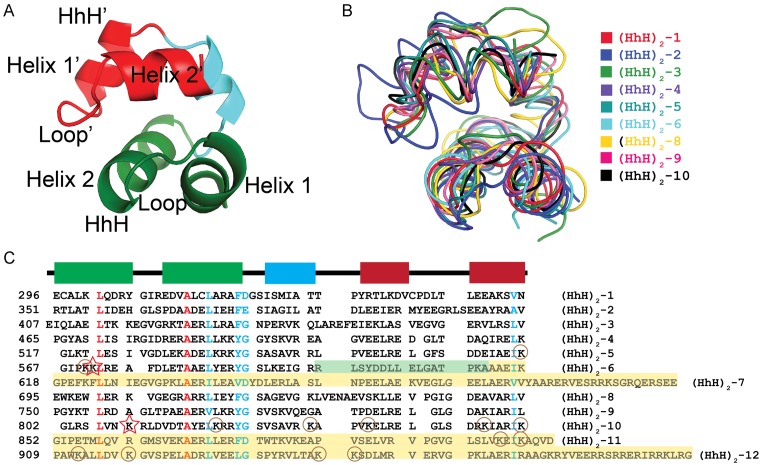
Figure 4.
Similarity amongst the (HhH)2 domains in Topo-V. (A) Schematic diagram illustrating the general architecture of an (HhH)2 domain. Each domain is formed by two HhH motifs (green and red) linked by a short helix (light blue). Topo-V is unique as it has 12 tandem (HhH)2 domains. (B) Superposition of the nine (HhH)2 domains observed in the Topo-97 structure. The (HhH)2 domains were superposed using an SSM alignment. The domains superpose well and in general have a similar structure. In general, adjacent (HhH)2 domains are connected by a short loop, except for domains 2 and 3, where the last helix of domain 2 extends to form the first helix of domain 3. (C) Structure-based sequence alignment of the (HhH)2 domains. The sequences of the 9 ordered (HhH)2 domains were aligned based on the SSM superposition whereas domains 7, 11 and 12 were manually aligned based only on sequence similarity. The alignment shows the presence of a few strictly conserved residues (red) in the first HhH motif and some highly conserved residues (cyan) in both motifs. The catalytic lysines for the first and second AP lyase sites are marked by stars, lysines that were mutated to quantitate activity are shown by circles. Yellow shaded areas correspond to regions whose structure is unknown. In the case of domain 6, the green shaded region represents an area that is ordered in the Topo-78 (green), but disordered in the Topo-97 structure. Secondary structure elements are shown above as a reference and using the same coloring as in panel A.
Structure of Topo-97
The structure of Topo-97 was solved to 2.4 Å resolution by a combination of MR and heavy atom phasing (Supplementary Table I). The structure can be described as formed by three main parts: a topoisomerase domain comprising residues 1–268, followed by the first six tandem (HhH)2 domains extending up to residue 596, and finally a small, rod-like domain, formed by three (HhH)2 domains arranged in tandem and spanning residues 690–852 and corresponding to (HhH)2 domains 8–10 (Figure 3). The 94 amino acids between (HhH)2 domains 6 and 8, which correspond to (HhH)2 domain 7 and part of domain 6, are disordered and could not be modeled. Interestingly, in the crystal structure of Topo-78 (residues 1–685), the model extends to amino acid 612, almost at the end of (HhH)2 domain 6. The Topo-78 crystal structure is almost identical to the corresponding region in the Topo-97 structure (RMSD of 1.1 Å for the main chain atoms). The absence of (HhH)2 domain 7 in both the Topo-78 and Topo-97 crystal structures suggests that this region may be unstructured or highly mobile. Very weak electron density in the experimental map suggests that the latter may be the case. A structure-based sequence alignment of the different (HhH)2 domains of Topo-V shows that each (HhH)2 domain comprises around 55 amino acids, and has key highly conserved amino acids (Figure 4) in the first HhH motif. The disordered (HhH)2 domain 7 contains signature amino acids for the first HhH motif, suggesting that this disordered repeat could form an (HhH)2 domain followed by a short, highly charged region that is present only in (HhH)2 domain 7. It is also possible that this repeat represents a hinge point that allows some flexibility in the molecule by dividing the 12 tandem (HhH)2 domains into two separate groups and that this mobility is needed to accommodate DNA binding.
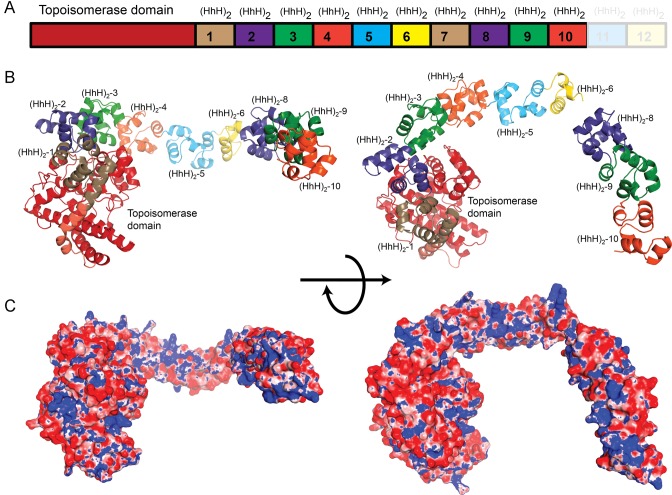
Figure 3.
Crystal structure of the Topo-97 fragment. (A) Schematic diagram of the Topo-V protein. The Topo-97 fragment consists of the topoisomerase and 10 tandem (HhH)2 domains. The last two (HhH)2 domains in the full length protein are not part of the fragment and are shown for reference only. (B) The diagram shows a model of the Topo-97 fragment in two orthogonal views. The seventh putative (HhH)2 domain is not visible in the structure and may represent a hinge point. For reference, the coloring of the (HhH)2 domains is similar to the one in A. (C) Electrostatic surface representations of the Topo-97 fragment of topoisomerase V show that there is a positively charged region along one edge of the (HhH)2 domains, which may represent binding areas for DNA. The electrostatic potential was calculated with the program APBS (37). The surface is colored with a blue to red gradient from +8 to −8 KbT/ec.
As expected, the topoisomerase domain of the Topo-97 structure does not show any major changes from the structures observed previously, both for the isolated topoisomerase domain (16) or the different Topo-V fragments consisting of different number of (HhH)2 domains (Topo-44 (16), Topo-61 (7) and Topo-78 (12)). The RMSD between the isolated topoisomerase domain (16) and the same region in Topo-97 is 1.0 Å for all main chain atoms. Comparison of the common main chain atoms of Topo-78 and Topo-97 gives an RMSD of 1.1 Å, whereas when only the topoisomerase domains in these two structures are compared, the RMSD is 1.2 Å, showing that the two fragments have almost identical structure with very little change in the relative arrangement of the topoisomerase and (HhH)2 domains. It is not surprising that the first six (HhH)2 domains of Topo-97 are observed in almost the same conformation as in Topo-78; in all known structures of Topo-V fragments the first four (HhH)2 domains are always found in the same general conformation, suggesting structural rigidity in the way the topoisomerase domain and the first four to six (HhH)2 domains are arranged in the absence of DNA. (HhH)2 domains 8–10, which have not been observed before, fold in a very similar manner to (HhH)2 domains 3–5 in the other structures of fragments of Topo-V (Figure 4B). (HhH)2 domains 8–10 form a short rod-like structure that extends away from (HhH)2 domain 6, with not much space to accommodate another (HhH)2 domain between domains 6 and 8, suggesting that domain 7 is not structured or oriented in the same manner as the other tandem (HhH)2 domains. Overall, each (HhH)2 domain is well folded and does not share any secondary structure element, except for (HhH)2 domains 2 and 3, which share a short helix.
Calculation of the electrostatic potential on the structure shows a spine of positive density running along one side of the (HhH)2 domains (Figure 3 and Supplementary Figure S7) suggesting an extended DNA binding surface but, as observed before (7), all the (HhH)2 domains are unlikely to be able to engage the DNA simultaneously without local conformational changes to align the domains on the DNA or without large distortions in the DNA. Alternatively, it is plausible that not all the (HhH)2 domains bind DNA and only the ones directly involved in DNA repair bind DNA. This last possibility is not supported by the observation that both the topoisomerase and AP lyase activities of the protein are enhanced by the presence of increasing number of (HhH)2 domains and that the DNA binding affinity of the different fragments is also directly dependent on the number of (HhH)2 domains present, suggesting that the (HhH)2 domains contribute significantly to DNA binding (16). In the absence of a structure of Topo-V in complex with DNA, it is difficult to picture how the tandem (HhH)2 domains interact with DNA.
Comparison of the structures of the (HhH)2 domains
Topo-V is unusual given the number of (HhH)2 domains present in the protein. It is the only known protein with more than one (HhH)2 domain in the same polypeptide. The (HhH)2 domains of Topo-97 all conform to the folding that has been observed previously for this type of domains (34): two HhH motifs linked by a short helix (Figure 4A). To compare the (HhH)2 domains in Topo-V, a secondary structure match (SSM) alignment of all the nine structured (HhH)2 domains in Topo-97 was performed and the superposed structures and the structure-based sequence alignment were compared (Figure 4B). All domains align well, with an RMSD of the Cα atoms varying between 1 Å and 2.2 Å. The alignment confirms that all the (HhH)2 domains have the same overall fold, but the lengths of the helices and loops are variable. In the case of (HhH)2 domains 2 and 3, the linker between the two domains is not a loop, but instead the last helix of domain 2 continues to form the first helix of domain 3. For this reason, domain 2 is structurally distinct from all the other (HhH)2 domains in the protein. (HhH)2 domain 6 is part of the first repair site and is not fully ordered in either the Topo-97 or the Topo-78 structures. In both structures, the first HhH motif in the (HhH)2 domain is well folded whereas the second one is only partially folded.
Structure-based sequence alignment of the (HhH)2 domains shows strict conservation of only two amino acids, a leucine and an alanine, in the first and second helices, respectively, of the first HhH motif (Figure 4C). In addition, the second helix of the first HhH motif also shows a highly conserved hydrophobic residue and a tyrosine or phenylalanine at the C-terminal end. The same conservation is not observed in the helices belonging to the second HhH motif, although a hydrophobic residue is present at the end of the second helix of the second HhH motif. The position of the highly conserved residues in the (HhH)2 domains of Topo-V are consistent with the pattern of sequence conservation that has been observed in (HhH)2 domains present in other proteins (34). In many instances, (HhH)2 domains present in other proteins show the presence of a GxG pattern in the loop linking the helices in the HhH motif (34). The same pattern is not present in most of the HhH motifs in Topo-V, although some motifs show the presence of one glycine. Overall, the sequence analysis shows that for Topo-V there is a stronger amino acid conservation in the first HhH motif compared to the second, and this conservation is consistent with what has been observed in HhH motifs belonging to other proteins (34). This analysis also shows that despite minimal sequence conservation, the (HhH)2 domains are able to maintain the same overall secondary structure.
It is interesting to note that there does not appear to be a sequence signature in the (HhH)2 domains of Topo-V that can be associated with AP lyase activity. The first repair site showed some sequence similarity to the active site of AP lyases harboring an HhH motif, such as Endo III and rat polymerase β (Pol β), and led to the identification of K571 as the nucleophile for the reaction (12). The catalytic lysine of the first AP lyase site of Topo-V is found in the first helix of the 6th (HhH)2 domain, in a similar spatial position to the catalytic lysine in Gs Endo III and Hs Ogg1 (12). Based on the structure-based sequence alignment, no other (HhH)2 domain in Topo-V has a lysine at a position corresponding to K571 (Figure 4C). Domain 10, which harbors the second repair site, and domain 12, which contains the third repair site, do not show strong sequence similarity to (HhH)2 domain 6 (Figure 4C). Several of the (HhH)2 domains of Topo-V have a lysine at a similar position as the one in (HhH)2 domain 10, but clearly not all these (HhH)2 domains are catalytic, as shown by the lack of AP lyase activity in the deletion mutants.
Structure of the second repair active site
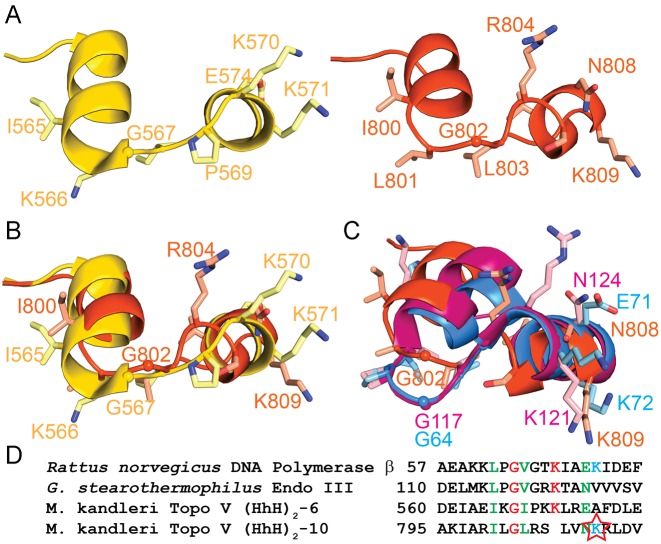
The approximate location of the second AP lyase active site of Topo-V was identified by a combination of deletion and site directed mutagenesis and mapped to the first helix of the first HhH motif belonging to (HhH)2 domain 10. Even though a lysine is expected to be the nucleophile for the AP lyase activity, the identity of the catalytic lysine could not be surmised from the site directed mutagenesis studies, as removing a single lysine does not abolish AP lyase activity completely. This is similar to what was observed previously when the first AP lyase site was identified and characterized (12). To narrow down the position of the active site lysine, structural and sequence comparisons of the region around the 10th (HhH)2 domain to the HhH motif of other AP lyases, such as Gs Endo III, Hs Ogg1, Pol β and the region between the 5th and 6th (HhH)2 domains of Topo-V, were performed (Figure 5A and C). For brevity, the region connecting two (HhH)2 domains comprising the last helix of the first (HhH)2 domain and the first helix of the second (HhH)2 domain including the intervening loop is designated the (HhH)2 junction. As was seen before for the (HhH)2 junction for domains 5 and 6 in Topo-V (12), there is good structural similarity between the junction for domains 9 and 10 (second AP lyase site of Topo-V) and the active site of other AP lyases. Structure-based sequence alignment of the (HhH)2 junction of domains 5 and 6 and domains 9 and 10 shows some conserved amino acids that are also present in other AP lyases, e.g. I565/I800, G567/G802 and E574/N808 (Figure 5B and D). Interestingly, the loop in the (HhH)2 junction is slightly different due to the presence of a proline (P569) in the first repair site that distorts the region and makes the loops in the two (HhH)2 junctions follow slightly different paths. In addition, the loop in the (HhH)2 junction of domains 9 and 10 is one residue shorter (GIP in domains 5–6 versus GL in domains 9–10). In the case of the first repair site, lysine 571 was identified as the catalytic lysine based on its structural similarity to the catalytic lysine in other AP lyases such as Gs Endo III and Hs Ogg1 (12). After structural alignment of the second Topo-V AP lyase site with the first site, it is clear that there is no lysine at an equivalent position in the second repair site (HhH)2 junction and that many of the mutated lysines are distant from the loop joining the two (HhH)2 domains (Supplementary Figure S7). The structural comparison also reveals that the spatially closest lysine to K571 is K809, which points in the same general direction. K809 of Topo-V also has the same orientation as the nucleophile of Gs Endo III (K121) and rat Pol β (K72) (Figure 5C). The Gs Endo III structure corresponds to that of a covalent complex of protein and abasic DNA and, as expected, K121 is directly involved in covalent bond formation with the DNA (35). Similarly, in the complex of Hs Ogg1 with abasic DNA (33) the active site lysine (K249) is in a similar position to K809 in Topo-97. Additionally, the structure-based sequence alignment shows that K809 of the second repair site of Topo-V is in an equivalent position to rat Pol β K72 (Figure 5C), which has previously been identified as the catalytic lysine (36). In addition to the catalytic lysine, several other residues are conserved in rat Pol β, Gs Endo III and the second repair site of Topo-V (Figure 5D). Thus, based on all these observations, K809 is the most likely nucleophile in the second AP lyase site of Topo-V. A main difference between the AP lyase sites of Endo III, Ogg1, Pol β and the ones in Topo-V is that, whereas the active site of the former group lies within an HhH motif, in Topo-V they are located in the junction between two (HhH)2 domains.
Figure 5.
Structure of the second AP lyase active site in Topo-V. (A) Schematic diagram of the structure of the first (left, yellow) and second (right, orange) AP lyase active sites in Topo-V. The two active sites are located in the (HhH)2 junctions of domains 5 and 6 and domains 9 and 10, respectively. For simplicity, only some of the side chains are drawn. (B) Superposition of the first and second AP lyase active sites. The two regions are structurally similar, although the loop connecting the two helices is different due to the absence of one amino acid in the second repair site. The catalytic lysine in the first repair site is K571, but there is no direct equivalent to this residue in the second site. Instead, K809 occupies a similar spatial position. Coloring as in A. (C) Superposition of the second repair active site of Topo-V (orange), GS Endo III (pink) and rat Pol β (blue). The overall structure of the three active sites is similar. Note that K809 in Topo-V, K121 in GS Endo III, and K72 in rat Pol β are all in a similar spatial position. K121 and K72 are the catalytic lysines in Endo III and polymerase β, respectively, whereas K809 is the likely active site lysine of the second repair site in Topo-V. (D) Sequence comparison of four different AP lyase repair active sites. The structural, sequence and biochemical analysis strongly suggest that K809 (star) is the nucleophile in the second repair active site of Topo-V.
Comparison of the structure of Gs Endo III and Hs Ogg1 in complex with DNA and the second repair site in Topo-V shows that Topo-V could accommodate DNA in a similar manner as other AP lyases with only minor conformational changes to prevent steric clashes (Supplementary Figure S9). The similarity between Topo-V and other lyases extends only to the region forming the active site, and hence it is not likely that the way these proteins interact with DNA is similar beyond the immediate vicinity of the active site (12). For this reason, the superimposed structures suggest a possible way for Topo-V to interact with DNA, but only at a very local level.
The reason for only a partial loss of AP lyase activity when the putative active site lysines in Topo-V are mutated is not clear. For every repair site, several lysines in the region had to be removed before activity was completely lost, even though some of them are relatively far away from the active site (Supplementary Figure S8). In the case of the first AP lyase site of Topo-V, a sodium borohydride-trapped complex was observed in all the single lysine mutants, suggesting that neighboring lysines could substitute for the active site lysine (12). This appears to be the case for the second and third repair sites, where single mutants did not abolish AP lyase activity completely.
Solution structure of Topo-97
In order to obtain information on the conformation of Topo-97 in solution, a SAXS experiment was done to produce a solution envelope for the molecule. Ab initio bead models of the molecule were obtained with the SAXS program package ATSAS (28). Twenty models were calculated and all showed remarkable agreement amongst themselves, with an NSD smaller than 0.92 Å in all cases. Averaging of all the models produced an envelope that shows an elongated molecule with a longest axis of ∼160 Å (Figure 6). The envelope clearly resembles the structure of Topo-97 although with the last three (HhH)2 domains in a different conformation. Manual fitting of the Topo-97 crystal structure to the envelope could be done by fitting separately the region corresponding to Topo-78 and the last three (HhH)2 domains in the fragment ((HhH)2 8–10) (Figure 6). The latter were placed in a conformation that is anti-parallel to the first six (HhH)2 domains, but a parallel conformation was also possible. The structures of Topo-78 and Topo-97 show that there is an unobserved, and probably mobile, region centered on domain 7 and this region was used as a hinge point between the two subsets of (HhH)2 domains. The model captures well the main features of the SAXS data (Supplementary Figure S2) even though there was no attempt to optimize the fit to the data. It would be possible to fit other models in the envelope by allowing extra mobility of the (HhH)2 domains, but this would require movements of the (HhH)2 domains relative to the topoisomerase domain. Since several crystal structures of Topo-V fragments show the same relative arrangement of the topoisomerase and (HhH)2 domains 1 to 6 (7,12,16), the Topo-78 region of the structure was moved as a rigid body into the SAXS envelop.
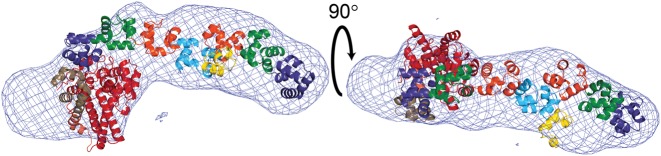
Figure 6.
SAXS envelope of Topo-97 shows an elongated molecule. Two orthogonal views of the SAXS envelope with a model of Topo-97 manually fit into the envelope. The SAXS envelope shows an elongated molecule that can accommodate the Topo-97 fragment. The model of Topo-97 based on the crystal structure was manually fit by breaking it into two parts, one comprising the topoisomerase domain and the first six (HhH)2 domains and the second comprising (HhH)2 domains 8–10.
The SAXS envelope captures well the anticipated shape of Topo-97. The full length Topo-V molecule has two additional (HhH)2 domains compared to Topo-97 and it is likely that these two (HhH)2 domains extend away from the 10th (HhH)2 domain. It is interesting to note that in solution the (HhH)2 domains appear to form a long rod and not bend sharply, as is observed in the Topo-97 crystal structure. This suggests that the bend observed in the crystal structure is due to a combination of the flexibility of the 7th (HhH)2 domain and also packing constraints in the crystal. It is thus likely that the conformation in solution is more similar to the one observed by SAXS. The SAXS model shows that the active sites are all far from each other. The distance between the topoisomerase active site and the first repair site is ∼69 Å in the Topo-78 and Topo-97 crystal structures and the SAXS model. In the SAXS model the distance to the second repair site from the topoisomerase active site is ∼70 Å. This shows that the three active sites are far from each other and it is difficult to envisage a situation where the three of them could interact with each other easily. If there is synergy between the active sites, it clearly has to be through changes that affect the overall shape of the molecule. Furthermore, it is not clear from the solution structure model how Topo-V could interact with DNA without changes in the conformation of the protein. Understanding the way Topo-V recognizes DNA lesions and changes the topology of DNA will require structural information of a complex with DNA.
CONCLUSIONS
Topo-V is the only topoisomerase known to have more than one catalytic activity and the only topoisomerase directly associated with dual topoisomerase and DNA repair activities. Topoisomerases appear to play an indirect role in DNA repair (3), a direct interaction between a topoisomerase and a DNA repair enzyme is not known. Topo-V was known to have at least two repair sites in addition to the topoisomerase active site. The experiments described here show that it has three AP lyase active sites, bringing the total number of active sites in the polypeptide to four. It is still not clear why this enzyme possesses two different functions that use four active sites in the same polypeptide. Removal of the topoisomerase site by mutation of the active site tyrosine does not impair repair activity and the repair sites can act independently of the topoisomerase domain (11). The converse is also true, the topoisomerase domain can relax DNA even in the absence of the repair sites (16). The presence of the (HhH)2 domains does improve topoisomerase activity, but this has been attributed to enhanced DNA binding in the presence of (HhH)2 domains (16) irrespective of whether the (HhH)2 domains carry repair function or not. These observations suggest that the repair and topoisomerase activities are to a large degree independent of each other, at least under in vitro conditions. As was pointed out before (12), the AP lyase reaction leads to the nicking of the DNA phosphodiester backbone and hence to the release of any torsional stress in the DNA. For this reason, it is unlikely that the AP lyase reaction precedes the relaxation reaction and suggests that the topoisomerase function may precede the AP lyase activity. In this scenario, DNA relaxation by the topoisomerase domain would facilitate efficient DNA repair. Furthermore, it is not clear why there are three apparently similar AP lyase sites performing the same reaction: processing of an abasic site. Some DNA repair enzymes have dual functions, glycosylase and AP lyase, where the glycosylase site recognizes and processes specific DNA lesions to produce an abasic site that is then acted upon by the AP lyase site. It is possible to imagine a scenario where the different repair active sites in Topo-V process different type of DNA lesions, but this does not appear to be the case based on the following observations. The structures of the two Topo-V AP lyase active sites show a strong similarity to the active sites of several AP lyases, including some dual specificity glycosylases, such as human Ogg1, but the structural similarity extends only to the HhH motifs carrying the catalytic lysine and does not extend to the regions involved in glycosylase activity. In Topo-V, the neighboring regions of the repair sites are also (HhH)2 domains, which are not known to be involved in glycosylase reactions. Thus, it appears that the three repair active sites are independent AP lyase sites. It is possible that the protein scans DNA searching for lesions and that having more than one AP lyase site is advantageous as several sites can be scanned simultaneously as the protein moves along. In this scenario, each of the repair sites would scan a complementary DNA strand; the protein could glide over the DNA scanning both strands without having to detach itself from the DNA or move from one strand to the other. For the latter scenario it may be required that the (HhH)2 domains are arranged in an anti-parallel fashion so that each AP lyase site interacts similarly with the complementary DNA strand. The model based on the SAXS data has the (HhH)2 domains arranged in an anti-parallel manner and using the 7th repeat not only as a hinge point, but as a turning point. The presence of three AP lyase sites does not necessarily imply that the lesions have to be equally spaced on the DNA; the different AP lyase sites could act on different lesions independently. In this regard, having independent sites that scan different strands or different DNA areas independently could make the repair much more efficient if the incidence of lesions is high, as it is expected in hyper-thermophiles. The presence of three repair sites clearly suggests that M. kandleri may need a very robust and efficient DNA repair activity, which might be due to the extreme environmental conditions that produce continuous DNA damage.
The location of the active sites is such that they are all distant from each other, suggesting that there is little possibility of direct interaction between them. Unless the (HhH)2 domains can re-arrange dramatically, it is not clear how the four active sites could interact with each other. It appears that DNA binding could trigger conformational changes in the (HhH)2 domains to provide access to the buried topoisomerase active site (16). Whether these changes would translate into some synergy between the active sites remains to be determined. In an open conformation of the protein, where the topoisomerase active site is not buried by the first two (HhH)2 domains, it is easy to envision a model where the DNA would run along the topoisomerase domain and extend to interact with the repair sites. Another possibility is that Topo-V could wrap around DNA, especially using the 7th (HhH)2 domain as a hinge region, which will bring the topoisomerase and AP lyse sites in closer proximity. In these models, the different active sites do not have to reside next to each other as the DNA would serve as the molecule that links the different active sites and provides synergy. Further experiments, including crystal structure determination of a Topo-V-DNA complex, will provide clues on how Topo-V engages with DNA, the role of the tandem (HhH)2 domains in DNA binding, an explanation for the enhancement of Topo-V activity as the number of (HhH)2 domains increases, and the possibility of synergy between the topoisomerase and AP lyase activities of Topo-V.
ACCESSION NUMBER
Coordinates for Topo-97 have been deposited in the PDB with accession number 5HM5.
Supplementary Material
Acknowledgments
We thank Steven Weigand of DND-CAT for his help and advice with the SAXS experiments, Bhaskar Chetnani for help with processing and analyzing SAXS data, and Alexei Slesarev for providing some of the Topoisomerase V plasmids used. We acknowledge staff and instrumentation support from the Keck Biophysics Facility, and the Center for Structural Biology at Northwestern University, and DND-CAT and LS-CAT at the APS at Argonne National Laboratory. LS-CAT was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. DND-CAT is supported by Northwestern University, E.I. DuPont de Nemours & Co., and The Dow Chemical Company. Use of the APS is supported by the Department of Energy (DOE). SAXS data were collected using an instrument funded by the National Science Foundation under Award Number 0960140. Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Keck Biophysics and Structural Biology Facilities is acknowledged.
Footnotes
Present address: Rakhi Rajan, Department of Chemistry and Biochemistry, University of Oklahoma, 101 Stephenson Parkway, Norman, OK 73019, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The authors declare no competing financial interests. R.R. was an American Heart Association postdoctoral fellow [10POST2600325]; National Institutes of Health [R01GM51350 to A.M.]. Funding for open access charge: National Institutes of Health (NIH) [R01GM51350 to A.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen S.H., Chan N.L., Hsieh T.S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 2.Schoeffler A.J., Berger J.M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 3.Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pommier Y., Leo E., Zhang H., Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker N.M., Rajan R., Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Taneja B., Patel A., Slesarev A., Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajan R., Osterman A.K., Gast A.T., Mondragon A. Biochemical characterization of the topoisomerase domain of methanopyrus kandleri topoisomerase V. J. Biol. Chem. 2014;289:28898–28909. doi: 10.1074/jbc.M114.590711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slesarev A.I., Stetter K.O., Lake J.A., Gellert M., Krah R., Kozyavkin S.A. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 10.Taneja B., Schnurr B., Slesarev A., Marko J.F., Mondragon A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belova G.I., Prasad R., Kozyavkin S.A., Lake J.A., Wilson S.H., Slesarev A.I. A type IB topoisomerase with DNA repair activities. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajan R., Prasad R., Taneja B., Wilson S.H., Mondragon A. Identification of one of the apurinic/apyrimidinic lyase active sites of topoisomerase V by structural and functional studies. Nucleic Acids Res. 2013;41:657–666. doi: 10.1093/nar/gks1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber R., Kurr M., Jannasch H.W., Stetter K.O. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110°C. Nature. 1989;342:833–834. [Google Scholar]
- 14.Slesarev A.I., Lake J.A., Stetter K.O., Gellert M., Kozyavkin S.A. Purification and characterization of DNA topoisomerase V. An enzyme from the hyperthermophilic prokaryote Methanopyrus kandleri that resembles eukaryotic topoisomerase I. J. Biol. Chem. 1994;269:3295–3303. [PubMed] [Google Scholar]
- 15.Belova G.I., Prasad R., Nazimov I.V., Wilson S.H., Slesarev A.I. The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J. Biol. Chem. 2002;277:4959–4965. doi: 10.1074/jbc.M110131200. [DOI] [PubMed] [Google Scholar]
- 16.Rajan R., Taneja B., Mondragon A. Structures of minimal catalytic fragments of topoisomerase V reveals conformational changes relevant for DNA binding. Structure. 2010;18:829–838. doi: 10.1016/j.str.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad R., Beard W.A., Strauss P.R., Wilson S.H. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Biol. Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 18.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 19.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vonrhein C., Blanc E., Roversi P., Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis I.W., Murray L.W., Richardson J.S., Richardson D.C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weigand S.J., Keane D.T. DND-CAT's new triple area detector system for simultaneous data collection at multiple length scales. Nucl. Instrum. Methods Phys. Res. A. 2011;649:61–63. [Google Scholar]
- 28.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;77:2896–2896. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano W.L. San Carlos: DeLano Scientific; 2002. [Google Scholar]
- 32.Thayer M.M., Ahern H., Xing D., Cunningham R.P., Tainer J.A. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fromme J.C., Bruner S.D., Yang W., Karplus M., Verdine G.L. Product-assisted catalysis in base-excision DNA repair. Nat. Struct. Biol. 2003;10:204–211. doi: 10.1038/nsb902. [DOI] [PubMed] [Google Scholar]
- 34.Shao X., Grishin N.V. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 2000;28:2643–2650. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fromme J.C., Verdine G.L. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad R., Beard W.A., Chyan J.Y., Maciejewski M.W., Mullen G.P., Wilson S.H. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase beta as revealed by site-directed mutagenesis. DNA binding and 5′-deoxyribose phosphate lyase activities. J. Biol. Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 37.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.