Abstract
In mammals, small RNAs are important players in post-transcriptional gene regulation. While their roles in mRNA destabilization and translational repression are well appreciated, their involvement in endonucleolytic cleavage of target RNAs is poorly understood. Very few microRNAs are known to guide RNA cleavage. Endogenous small interfering RNAs are expected to induce target cleavage, but their target genes remain largely unknown. We report a systematic study of small RNA-mediated endonucleolytic cleavage in mouse through integrative analysis of small RNA and degradome sequencing data without imposing any bias toward known small RNAs. Hundreds of small cleavage-inducing RNAs and their cognate target genes were identified, significantly expanding the repertoire of known small RNA-guided cleavage events. Strikingly, both small RNAs and their target sites demonstrated significant overlap with retrotransposons, providing evidence for the long-standing speculation that retrotransposable elements in mRNAs are leveraged as signals for gene targeting. Furthermore, our analysis showed that the RNA cleavage pathway is also present in human cells but affecting a different repertoire of retrotransposons. These results show that small RNA-guided cleavage is more widespread than previously appreciated. Their impact on retrotransposons in non-coding regions shed light on important aspects of mammalian gene regulation.
INTRODUCTION
In mammals, the best known small RNA targeting pathways include destabilization or translational repression of target mRNAs (1,2). A third mechanism, small RNA-guided endonucleolytic cleavage of target RNAs, is assumed to be very rare in animals, although it is prevalent in plants (3). Thus far, only a small number of microRNAs (miRNAs) were predicted to have this function in mammals (4–8), affecting a very small number of target genes. Endogenous small interfering RNAs (endo-siRNAs) are expected to induce target cleavage (9). However, their targetome is not yet well characterized.
The catalytic function of Ago2, which carries out the slicing reaction on mRNA targets, is highly conserved throughout mammals (10). This observation suggests that small RNA-directed cleavage may be an essential aspect of mammalian gene regulation and more widespread than currently appreciated. Three factors may have hindered progress in this research area. One is the possibility that small RNA-directed cleavage is highly cell type-specific. The specific cell types examined by previous studies may have failed to reveal the bulk of such events. Second, a diverse panel of small RNAs, not limited to miRNAs or siRNAs, may mediate mRNA cleavage, an aspect that has not been explored. Third, technical challenges, such as the enrichment of repetitive elements in the target sites or small RNAs, may have prevented discovery of the full spectrum of small RNA-mediated cleavage events.
Our study aimed to address the above challenges and better characterize small RNA-mediated cleavage in mammals. We analyzed a large amount of small RNA and Degradome Sequencing data (Deg-Seq, also known as PARE), with the latter capturing the 5′ ends of RNA degradation products (11,12), in mouse embryonic stem cells (mESCs), testis and cerebellum. This analysis allowed a systematic characterization of small cleavage-inducing RNAs (sciRNAs) and their targets simultaneously. Our bioinformatic method captures any type of sciRNAs, unlimited to known RNA classes, and accommodates existence of repetitive sequences in the RNA. As a result, we identified 398 sciRNAs and 810 cognate cleavage target genes, much more than previously known in the literature. Interestingly, about 40% of sciRNAs overlap known miRNAs, endo-siRNAs or piwi-interacting RNAs (piRNAs), revealing novel targets of these RNA regulators. This observation also indicates that sciRNAs, defined to conveniently refer to their function, may have diverse biogenesis pathways. sciRNAs demonstrated a high degree of cell type-specificity, developmental stage-specificity, and diversity in possible functional pathways. A striking feature of both sciRNAs and their target cleavage sites is their significant overlap with retrotransposable elements, providing evidence for the first time that retrotransposons in transcripts are leveraged as signals for gene targeting. Additionally, our analysis showed that the sciRNA pathway is also present in human cells but affecting a different repertoire of retrotransposons. Thus, sciRNA targeting is a conserved mechanism between human and mouse but involves different sciRNA molecules and targets, possibly reflecting the divergence of retrotransposons between the two genomes.
MATERIALS AND METHODS
Bioinformatic prediction of sciRNAs and their targets
Preprocessing
Deg-Seq and small RNA-Seq reads were trimmed with cutadapt (13) to remove adapters and PCR primers. For mESCs, Deg-Seq and small RNA-Seq data sets were acquired from GSE21975 (8) and GSE35368 (SRR402760, SRR402761, SRR402762, SRR402766) (14), respectively, while other data sets were generated in-house. 3′ end regions with quality less than 20 were also trimmed from Deg-Seq reads. A minimum length of 19 nt was required for small RNA-Seq since typical known small RNAs are longer than 19 nt. A minimum length of 25 nt was required for Deg-Seq reads to ensure specific mapping to the genome while retaining as many reads as possible. The first step of the pipeline was the exclusion of small RNA-Seq and Deg-Seq reads with low complexity since such reads tend to base-pair with each other by random chance. Low complexity reads were defined as those with tandem repeats of mono-, di-, tri- or quad-nucleotides of 5, 3, 2, 2, respectively. The length cutoffs were determined by examining repeat patterns of known functional small RNAs. Small RNA sequences were required to have length 19–24 nt and read count ≥20.
Gene annotation
To define a comprehensive set of annotated mRNAs, we merged the following gene annotation databases: Ensembl, UCSC knowngene, RefSeq, VegaGene, GENCODE, Pseudogene.org and NONCODEv4 (15).
Define significant peaks
Deg-Seq reads were aligned only to annotated regions (listed above) of genome mm10 or hg19 using Bowtie v.0.12.7 (16) requiring no mismatches and reporting up to 100 valid alignments. Reads that were mapped non-uniquely to the genome were counted as 1/n in calculating Deg-Seq coverage, where n is the total number of mapped loci. To identify Deg-Seq peaks (i.e. high coverage sites), we applied a binomial test to each continuous stretch of ≤4 nucleotides with ≥3 reads in each transcript. The expected probability of observing a Deg-Seq peak is 1/l where l is the total number of nucleotides in the transcript of interest with read coverage ≥1. A P-value cutoff was determined as the smaller of the Bonferroni-corrected P-value or 0.05. These significant peaks were considered candidate cleavage sites.
Small RNA-target alignment and parsing
The candidate cleavage sites with their upstream and downstream 25 nt were aligned to unique small RNA-Seq reads that passed the length and coverage filters. This alignment was conducted using miRanda (17) requiring a score of at least 60. miRanda was chosen as a convenient local alignment tool that aligns sequences by complementary (as opposed to matching) nucleotides and allows GU wobbles. However, the scoring option for miRNA seed match was not used because we require complementarity beyond the seed region for candidate sciRNAs. Additionally, the thermodynamic energy calculation was not used in order to minimize the number of assumptions we make and obtain a large initial list that can be later filtered using customized criteria. Nucleotides 9–11 relative to the 5′ end of the small RNA were required to match perfectly and overlap the Deg-Seq peak since this is required for cleavage-competent pairing (18). Gaps and G = U wobble base pairing were allowed, counting G = U base pairing as mismatch 0.5. Unique alignments with at most 4 mismatches were retained for further analyses, which we call ‘candidate sciRNAs’ and their targets.
100x shuffled sciRNAs
Given the large number of small RNAs and Deg-Seq peaks, control analyses were carried out to ensure that the base-pairing relationship was more significant than expected by chance. One hundred shuffled controls were generated for each candidate sciRNA, maintaining di-nucleotide frequencies in the sciRNA and masking simple repeats. Simple repeats were defined as tandem repeats of mono-, di-, tri- or quad-nucleotides (number of repeats > 3, 2, 2, 2, respectively). Unique controls were then aligned to the significant Deg-Seq peaks and their flanking regions followed by parsing as described above for the true small RNA-Seq data. Although it is desirable to use a larger number of shuffled controls, we found that the majority (mESCs, 73%; testis, 74%; cerebellum, 70%) of small RNAs had fewer than 100 unique shuffles due to low complexity and the constraints we imposed in shuffling (maintaining di-nt frequencies and simple repeats). Approximately half had less than 90 unique shuffled controls (mESCs, 50%; testis, 52%; cerebellum, 43%). These data suggest that the usage of 100 shuffled controls was a reasonable choice.
Calculate signal-to-noise ratio (SNR)
To identify sciRNAs with more targets than expected by chance, a signal-to-noise ratio was calculated using the true and control sciRNA-target alignments. First, an individual SNR (iSNR) was calculated for each candidate sciRNA at mismatch cutoffs ranging from 0 to 4 at 0.5 intervals. iSNR is defined as the ratio of total targets of the candidate sciRNA to the total targets of all shuffled small RNAs (plus a pseudocount) normalized by the total number of unique shuffled small RNAs (required to be >10). To avoid over-counting targets due to sequence similarity among small RNAs, those small RNAs sharing at least one common 17-mer were grouped together. In other words, for a given group, at least 2 small RNAs share a 17-mer. The results were not very sensitive to this parameter within the range of 15–18. For a range of iSNR cutoffs, a group SNR was calculated for each group of small RNAs as the ratio of total targets of candidate sciRNAs in the group to the total targets of all shuffled small RNAs in the group normalized by the total number of unique shuffled small RNAs. A minimum iSNR cutoff of 10 was chosen, although the resulting sciRNA-target predictions with less than 3 mismatches were insensitive to iSNR cutoffs. Finally, an average SNR was calculated for a given data set as the average of all group SNRs. The output of this pipeline is the small RNAs that have significantly more targets compared to their controls, which we call ‘predicted sciRNAs,’ and their targets. A signal-to-noise ratio was chosen as an alternative to, for example, an empirical P-value using the 100 shuffled controls as a null distribution. The SNR method affords higher resolution to detect highly confident sciRNA-target pairs, as most empirical P-values were very small.
Total RNA samples
Total RNA samples for whole brain embryo E10, cerebellum embryo E14, cerebellum embryo E18, cerebellum post-natal (PN) 3 weeks, cerebellum PN 6 months, testis embryo E14, testis embryo E18, testis PN 3 weeks and testis PN 6 months were purchased from Zyagen. All RNAs were obtained from the same BALB/C mouse strain. Total RNA of H1 cells was isolated using Trizol (Life Technologies). Additional column purification and DNaseI treatment were applied using Direct-zol RNA kit (Zymo Research).
Construction of small RNA sequencing libraries
Spike-in RNAs (Exiqon) were added into 1 μg total RNA before library construction. Small RNA sequencing libraries were generated using NEBNext Small RNA library Prep kit and NEBNext multiplex oligos for Illumina according to the manufacturer's instructions (NEB). The final libraries were purified from 6% PAGE gel, and their concentrations were measured using Qubit fluorometric assay (Life Technologies).
Construction of RNA sequencing libraries
rRNA was depleted using RiboMinus Trnascriptome ioslatoin kit (Life Technologies) from 10 μg total RNA. ERCC Spike-in RNA (Life Technologies) was added to 500 ng of rRNA depleted RNA. mRNA was isolated using the NEBNext Poly(A) mRNA magnetic isolation module. mRNA sequencing libraries were generated using the NEBNext Ultra Directional RNA library Prep kit and NEBNext multiplex oligos for Illumina according to the NEB. Final libraries were examined using the Qubit fluorometric assay (Life Technologies) and Bioanalyzer (Agilent) for quality confirmation. Note that RNA sequencing was carried out for testis samples, not cerebellum, given the low number of sciRNAs predicted in the latter.
Construction of Degradome sequencing libraries
To generate the degradome sequencing libraries, we used the global 5′ RACE library preparation method (8) with some modifications. Briefly, poly(A)+ mRNA was isolated from 400–500 μg of total RNA using Dynabead Oligo(dT) (Life Technologies) according to the manufacturer's instructions. This procedure was repeated to increase the effectiveness of poly(A) selection. The NEBNext Small RNA library Prep kit was used for 5′ adapter ligation, followed by reverse transcription using random hexamer primers containing the 3′ SR adapter sequence 5′-AGACGTGTGCTCTTCCGATCTNNNNNN. PCR was conducted in 25 cycles at 94°C 15 s, 60°C 30 s, 70°C 1 min. The final libraries were purified from 6% PAGE gel followed by AMPure XP Beads size selection (Beckman Coulter).
RESULTS
Prediction of sciRNA-mediated RNA cleavage events
We first examined whether the Deg-Seq peaks identified in our study could be artifacts due to PCR amplification bias. Since PCR amplification bias is known to be associated with or reflected in biases in GC content, nucleotide composition and read length (19), we examined these features for reads overlapping Deg-Seq peaks and reads outside of Deg-Seq peaks. We further separated each group of reads into those that have unique sequences (compared to other reads in the same peak) and those that are duplicated. Overall, there appears to be very little PCR amplification bias (Supplementary Figure S1).
Next, to evaluate whether it is potentially feasible to identify RNA cleavage events using Deg-Seq and small RNA sequences, we aligned a set of predicted endo-siRNAs (20) to the ±25 nt flanking sequence of significant Deg-Seq peaks in mESCs (8). At nucleotides 9–11 from the 5′ end of the endo-siRNA, there was an enrichment of Deg-Seq reads in wild type (WT) mESCs (Supplementary Figure S2, red) and a depletion of reads in Ago2-/- mESCs (Supplementary Figure S2, grey). This result is consistent with the known biochemical properties of Ago2 which cleaves the phosphodiester bond corresponding to bases 10–11 of the small RNA (18,21), suggesting that combined usage of Deg-Seq and small RNA-Seq with appropriate controls may enable identification of functional sciRNAs and their targets.
To achieve the above goal, we analyzed Deg-Seq and small RNA-Seq data as illustrated in Figure 1A (see Materials and Methods). This analysis was carried out for three cell types: mESCs, adult mouse cerebellum post-natal 6 months (6M PN) and adult mouse testis (6M PN) (see Supplementary Table S1 for all data sets in this study). Cerebellum and testis were chosen in order to compare and contrast sciRNA-mediated RNA cleavage in a tissue containing mature non-dividing cells with a tissue containing frequently dividing germ cells, respectively. mESCs were included because previously published small RNA-Seq and Deg-Seq data were available. Since complementary base pairing along the small RNA is likely required to induce cleavage (4), mismatches were counted in the entire small RNA-target alignment rather than the seed region alone. Across all data sets, the optimal mismatch cutoff corresponding to the highest average SNR was at most 1.5 (Figure 1B, Supplementary Figure S3). Thus, we allowed up to 1.5 mismatches for all downstream analyses.
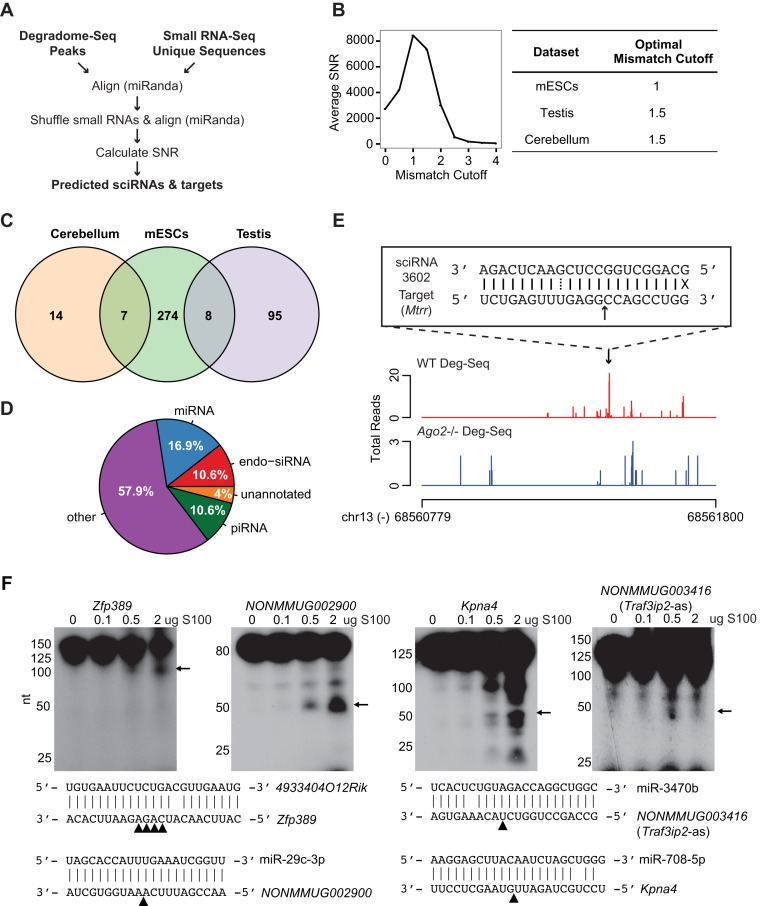
Figure 1.
Prediction of sciRNA-mediated mRNA cleavage events. (A) Bioinformatic pipeline schematic. (B) Left panel: average SNR for each mismatch cutoff in mESCs; right panel: optimal mismatch cutoff corresponding to the maximum average SNR for each data set. (C) Venn diagram of sciRNAs identified in mESCs, testis 6M PN, and cerebellum 6M PN. (D) Pie chart of sciRNA annotations (total = 398, combining sciRNAs from 3 cell types). (E) An example of predicted sciRNA-mediated cleavage. Read distributions are shown for the 3′ UTR of the Mtrr gene in Deg-Seq data of wild type (WT, red) and Ago2 knockout (blue) mESCs. Alignment of the sciRNA to the Deg-Seq peak is shown in a box, where a solid line indicates a base pair match, dotted line indicates a G = U wobble, X indicates a mismatch, and black arrow indicates location of the Deg-Seq peak. (F) Experimental validation of target RNA cleavage mediated by small RNAs. A total of 200 ng of Kpna4, NONMMUG002900, Zfp389 or NONMMUG003416 (Trafip2-as) RNA were incubated with different amount of HeLa S100 loaded with 50 nM synthetic sciRNA at 37°C for 30 min. Arrows indicate cleaved 5′ RNA fragments (whose sizes are consistent with predicted cleavage products). Small RNA/target RNA sequences are shown with arrowheads indicating the predicted cleavage sites (4 sites identified as Deg-Seq peaks in Zfp389).
Table 1 summarizes the total small RNAs predicted to induce cleavage and the set of Deg-Seq peaks that they target. Notably, many sciRNAs had more than one target, resulting in more than 1000 total sciRNA-target pairs. Supplementary Table S2 describes these sciRNAs and targets in detail. The relative scarcity of sciRNAs and targets in cerebellum is unlikely due to low sequencing depth since testis has the lowest number of Deg-Seq peaks and unique small RNA species in the initial sequencing data (Supplementary Table S1). The vast majority of sciRNAs were identified in only one cell type (Figure 1C), suggesting either a high degree of cell type-specificity or that there are more sciRNAs to be discovered. In addition, about 40% of sciRNAs were known miRNAs, endo-siRNAs or piRNAs (Figure 1D), with additional sciRNAs being novel small RNA species. Figure 1E shows an example of a novel small RNA inducing Ago2-dependent cleavage of Mtrr. These results suggest that small RNA-mediated target cleavage in mouse may be much more widespread than previously appreciated.
Table 1. Summary of the final sets of predicted sciRNAs, targeted cleavage sites and their combinations.
| Mismatches allowed | Predicted sciRNAs | Predicted sciRNA cleavage sites | Total sciRNA-target pairs | |
|---|---|---|---|---|
| Cerebellum | 1.5 | 21 | 8 | 30 |
| Testis | 1.5 | 103 | 599 | 1772 |
| mESC wild-type | 1.5 | 289 | 315 | 1108 |
| mESC Ago2-/- | 1.5 | 53 | 23 | 58 |
Experimental and genomic validations of sciRNA-target predictions
To provide experimental support, we carried out in vitro cleavage assays for four predicted cleavage events using HeLa S100 extracts and synthetic sciRNAs (Figure 1F, Supplementary Table S3). These events were picked to represent different types of target genes: a protein-coding gene (Kpna4) and non-coding genes including a lncRNA (NONMMUG002900), a pseudogene (Zfp389), and an antisense transcript of Traf3ip2. We observed an increasing amount of cleavage products with increasing S100, confirming the validity of the predicted targets.
To further validate our predictions, we applied the pipeline to Deg-Seq of Ago2-/- mESCs (8). This analysis yielded only 58 sciRNA-target pairs, about 5% of those predicted using WT Deg-Seq (Table 1). This result is consistent with the expectation that Ago2 is the main executor of target RNA cleavage and serves as validation of our method. The false discovery rate of our method is at most 5%, which could be an over-estimate since the above 58 sciRNA-target pairs likely include true cleavage events mediated by proteins other than Ago2.
To complement this analysis, we next examined whether sciRNAs were frequently bound by Ago2 in mESCs using Ago2 CLIP-Seq data (22). Compared to control small RNAs (see Supplementary Methods), sciRNAs were bound by Ago2 more often in wild type mESCs (Figure 2A). To rule out the possibility that canonical miRNAs were driving the observed sciRNA association with Ago2, we excluded miRNAs from the pool of sciRNAs. The remaining sciRNAs were still enriched in Ago2 CLIP (Figure 2B). Again, these data confirm that sciRNA function is dependent on Ago2.
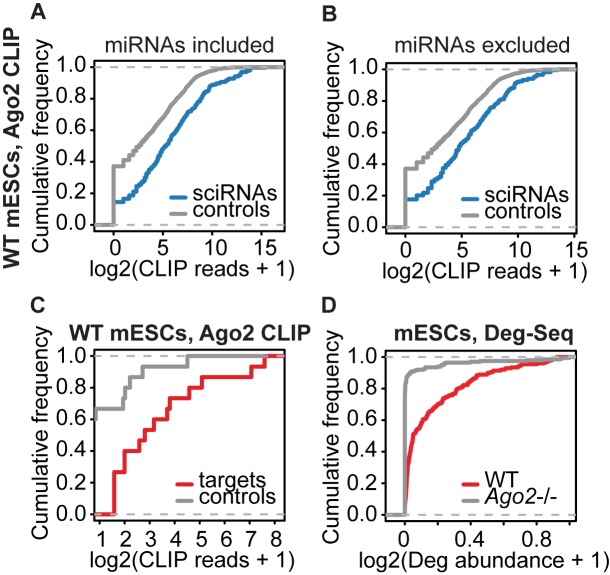
Figure 2.
Genomic data supporting the validity of sciRNA-target predictions. (A) Empirical cumulative frequency of abundance of Ago2 CLIP-Seq reads containing sciRNA sequences (blue) or control sequences randomly picked from Dicer-independent Dgcr8-independent small RNAs (grey) in WT mESCs (P = 2.2e-16, two-sided Kolmogorov–Smirnov (KS) test, same below). (B) Similar to (A), excluding miRNAs (P = 2.1e-10). (C) Similar to (A), comparing abundance of CLIP reads covering Deg-Seq peaks in target genes (red) or controls (grey, see Supplementary Methods) (P = 0.003). (D) Deg-Seq peak abundance in wild type (WT) and Ago2 knockout mESCs (P < 2.2e-16).
We next asked whether the cleavage sites in predicted target genes were associated with Ago2. Compared to controls with similar read coverage (see Supplementary Methods), we observed a highly significant enrichment of Ago2 CLIP-Seq reads for the target sites (Figure 2C). In addition to Ago2-association, we examined whether the Deg-Seq abundance of the target sites was dependent on Ago2 using Deg-Seq of Ago2-/- mESCs (8). We observed that sciRNA targets sites had significantly reduced Deg-Seq abundance in Ago2-/- mESCs compared to wild type cells (Figure 2D). Together, these results strongly support the validity of the predicted sciRNAs and their cleaved targets.
Small RNAs from diverse classes function as sciRNAs
Since sciRNAs are defined based on a common function (i.e. target cleavage), we hypothesized that a universal pathway may not explain their biogenesis. Rather, sciRNAs may include multiple types of annotated or novel small RNAs. To better understand sciRNA biogenesis, we examined their (i) annotation, (ii) dependence on the microprocessor in mESCs (20) and (iii) long hairpin RNA (hpRNA) structure (see Supplementary Methods, Figure 3A).
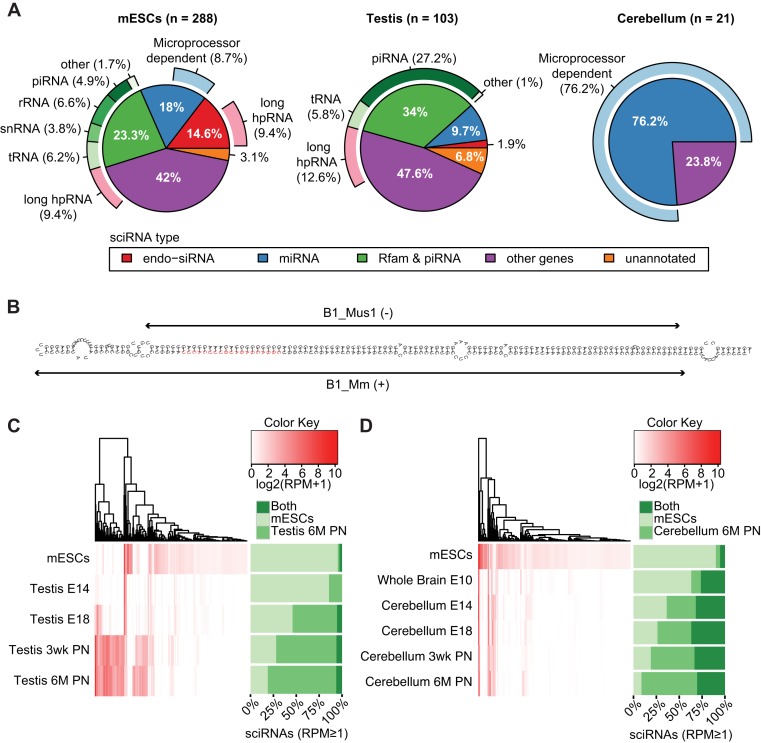
Figure 3.
Characterization of sciRNAs. (A) Categorization of sciRNAs in mESCs, testis and cerebellum according to: (i) annotation (inner circle), (ii) dependence on the microprocessor (outer circle, light blue) and (iii) long hpRNA structure (outer circle, pink). In mESCs, 1 unmapped sciRNA was excluded. (B) RNAfold structure of sciRNA 24792 (red) and flanking regions (within the Ccdc30 gene). This sciRNA was predicted in mESCs. Two inverted B1 repeats (Repeatmasker) are labeled. (C) Hierarchical clustering of sciRNA expression levels (reads per million, RPM) in mESCs and different stages of testis development. E14: embryonic day 14; E18: embryonomic day 18; 3wk PN: 3-week postnatal; 6M PN: 6-month postnatal. In the heatmap, RPM values of all sciRNAs that were identified originally in mESCs or 6M PN testis were visualized for each sample. Stacked bars on the right show the percentage of sciRNAs (among those with RPM ≥ 1) specific to mESCs (defined as those that were only identified in mESCs by the pipeline in Figure 1A, but not in the testis 6M PN data), testis 6M PN (similarly as defined above) or common to both. Note that some sciRNAs predicted originally in mESCs or testis may be excluded in the stacked bars due to low RPM. (D) Similar to (C), for cerebellum development. E10: embryonic day 10.
In mESCs and testis, miRNAs only explained 18% and 9.7% of sciRNAs, respectively, whereas 76.2% of sciRNAs were miRNAs in cerebellum (Figure 3A). In mESCs and cerebellum, many miRNAs had canonical microprocessor dependence (Dicer- and Dgcr8-dependent) based on data derived from mESCs (20). In contrast, no miRNAs in testis had the canonical microprocessor signature. These may be incorrectly annotated miRNAs, miRNAs generated by non-canonical pathways or canonical miRNAs in testis but with no microprocessor dependence in mESCs (since microprocessor dependence was evaluated using data from mESCs). The three cell types also differed dramatically in the number of predicted endo-siRNAs (Dicer-dependent and Dgcr8-independent), with mESCs having the most endo-siRNAs. Many (64%) of these endo-siRNAs in mESCs had long hairpin structure, consistent with their biogenesis model. Notably, an additional 9.4% and 12.6% of sciRNAs in mESCs and testis, respectively, also had predicted long hairpin structure (Figure 3A), thus are likely endo-siRNAs. Figure 3B illustrates an example of sciRNA-hosting long hairpin structure generated by inverted B1 sequences in mESCs.
Another category of sciRNAs consists of those that appear to be shorter forms of full-length non-coding RNAs from Rfam and piRNABank databases. For example, in testis, a large fraction (27.2%) of sciRNAs overlapped piRNA sequences, consistent with the high abundance of piRNAs in this tissue. piRNAs appeared to be trimmed from the 5′ end, 3′ end or both, to generate sciRNAs (Supplementary Figure S4), indicating existence of additional processing mechanisms. Similarly, tRNAs, snRNAs and rRNAs were also identified as possible sciRNA-generating RNAs, all of which were reported previously to produce small RNAs (23,24). The last category of sciRNAs aligned to annotated genes that are not miRNA/endo-siRNA/Rfam/piRNA genes (‘other genes’ in Figure 3A). Their biogenesis mechanisms remain unknown.
sciRNA expression varies during testis and cerebellum development
Since sciRNA populations in mESCs, adult testis and adult cerebellum were largely distinct, we examined the divergence process of sciRNA profiles from mESCs to the adult cells during development. We obtained small RNA sequencing data to examine sciRNA expression in several developmental stages of testis and cerebellum. We then compared expression profiles of sciRNAs between mESCs and testis (Figure 3C), or between mESCs and cerebellum (Figure 3D). Specifically, sciRNAs identified in mESCs or the adult tissue (testis 6M PN or cerebellum 6M PN) were labeled as mESC-specific (if predicted in mESC data only), adult tissue-specific (if predicted in adult tissue only) or common to both. Interestingly, we observed reciprocal changes in the relative enrichment of expressed mESC-specific and adult tissue-specific sciRNAs during the development of both testis and cerebellum. Thus, mESC-specific sciRNAs were gradually replaced by tissue- and adult-specific sciRNAs as the cells mature.
Notably, cerebellum and testis demonstrated different patterns of sciRNA expression during development. A considerable portion (25–30%) of sciRNAs in cerebellum was also present in mESCs, which was a general observation for all developmental stages (‘Both,’ Figure 3D). In contrast, sciRNAs common to both mESCs and testis were rare in all developmental stages (‘Both,’ Figure 3C). In testis stages embryonic day 18 (E18) and later, the majority of sciRNAs were testis-specific. On the other hand, there were few testis-specific sciRNAs at E14. This time point approximately precedes the development and proliferation of prospermatogonia (25). Thus, it is possible that sciRNAs in testis are primarily generated during spermatogenesis and largely distinct from those in mESCs or other tissues (e.g. brain).
In striking contrast to sciRNAs, miRNA profiles (excluding sciRNAs) were much more stable across all developmental stages included in this study (Supplementary Figure S5). A much larger fraction of miRNAs was common to mESCs and different stages of testis or cerebellum. Additionally, the difference between testis and cerebellum was not as pronounced as that observed for sciRNAs. The considerable distinction in the developmental- and tissue-specific profiles of sciRNAs and non-sciRNA miRNAs indicates that these two classes of small RNAs may have distinct cellular functions.
sciRNAs target non-coding regions of genes spanning diverse functional categories
Target genes in the three cell types demonstrated little overlap, with only 40 genes in common between any two samples (Figure 4A). This apparent tissue specificity is mainly due to the tissue-specific expression of sciRNAs. The number of sciRNAs expressed in a particular tissue but not predicted to induce cleavage was a small minority (Supplementary Table S4).
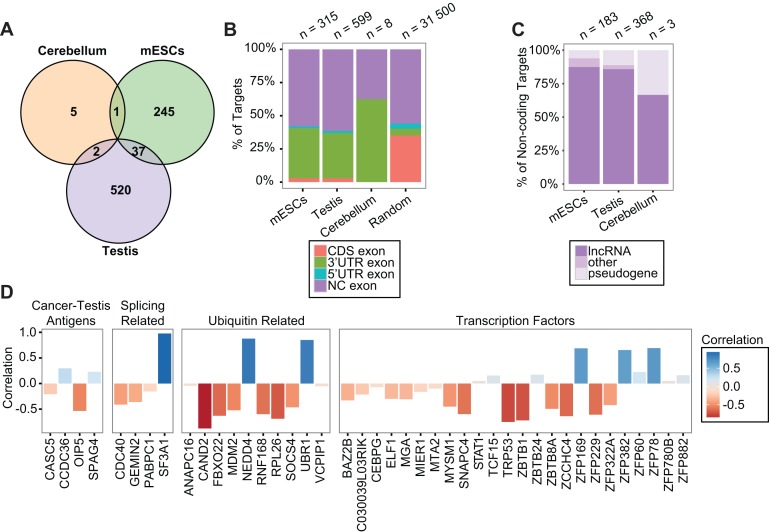
Figure 4.
Characterization of sciRNA targets. (A) Venn diagram of target genes predicted in mESCs, testis and cerebellum. (B) Distribution of target cleavage sites (Deg-Seq peaks) in different types of regions of the transcriptome. CDS: coding sequence; NC exon: exon of non-coding transcript. Random: random positions from random transcripts (see Supplementary Methods). (C) Types of non-coding transcripts among sciRNA targets, prioritized as pseudogene > lncRNA > other. (D) Pearson correlation of target mRNA expression and sciRNA expression for four example categories of target genes (see Supplementary Methods).
Strikingly, the majority of cleavage sites within coding genes was located in 3′ UTRs in all cell types, much more than expected by chance (Figure 4B). Since our search of cleavage sites was across the entire mRNAs, this 3′ UTR enrichment strongly testifies to the validity of our results. It should be noted that miRNAs are primarily known to target 3′ UTRs (26), which could arguably be partially due to the intense focus on 3′ UTRs in prediction algorithms and the usage of evolutionary conservation as a requirement of target sites. Thus, our study supports 3′ UTR targeting by small RNAs in an unbiased manner.
Besides the non-coding 3′ UTRs, many of the sciRNA targets are non-coding transcripts, derived from lncRNAs, pseudogenes or other non-coding RNAs in GENCODE and NONCODE annotations (Figure 4B and C). In all cell types, lncRNAs account for the majority of non-coding targets. A relatively large fraction of targets in testis and cerebellum was regulated by miRNAs (Supplementary Figure S6), whereas novel small RNAs derived from other genes account for the majority of targeting in mESCs.
Among the predicted sciRNA target genes, many are associated with important functional relevance. Figure 4D shows a subset of such genes grouped into transcription factors (27), ubiquitin related genes, splicing related genes and cancer-testis antigens (28). Importantly, most of these target genes demonstrated negative correlation in gene expression levels (measured by RNA-Seq of testis samples at different developmental stages) relative to their corresponding sciRNA expression (Supplementary Methods), further confirming the predicted functional relationship of sciRNAs and targets.
We also carried out pathway, ontology and Ingenuity network analyses for protein-coding and non-coding target genes to obtain a comprehensive view of functional relevance (Supplementary Table S5). Overall, sciRNA targets are involved in a diverse spectrum of functional categories, enriched with developmental-related processes and basic cellular function (cellular assembly and organization, cell morphology and cell cycle).
sciRNAs and target genes are enriched with repetitive elements
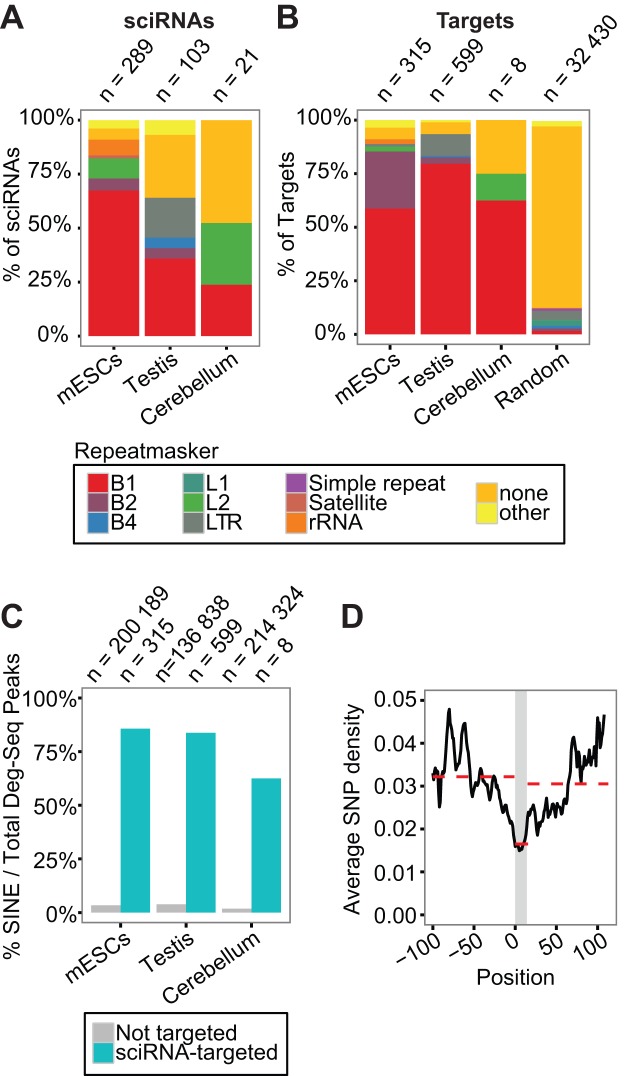
Although the biogenesis pathways of sciRNAs appear diverse, a unifying feature of the sciRNAs and their targets is their substantial overlap with repetitive elements. The majority of sciRNAs in mESCs and testis are repetitive, with most aligned to SINE elements, especially the B1 subclass (comparable to human Alus) (Figure 5A). Repetitive sciRNAs often target more RNAs than non-repetitive sciRNAs (Supplementary Figure S7A). Furthermore, we observed that B1-derived sciRNAs mapped to specific sub-regions of the consensus B1 sequence (Repbase (29)) in both sense and antisense orientations (Supplementary Figure S7B). Thus, many sciRNAs may be derived from pairs of inverted B1 repeats, as shown for Ccdc30 (Figure 3B). Since the above observation applies to both mESCs and testis, sciRNA biogenesis likely shares similar pathways and genomic features in the two cell types despite the involvement of different sciRNA species.
Figure 5.
Small RNA guided endonucleolytic cleavage targets retrotransposons. (A) Distribution of sciRNAs in different types of repeats (Repeatmasker). If more than one Repeatmasker annotation was identified, the following prioritization was used: B1 > B2 > B4 > others. Random: similar to Figure 4B. (B) Similar to (A), for target cleavage sites. (C) Percentage of Deg-Seq peaks overlapping SINE regions among all Deg-Seq peaks. Two groups of Deg-Seq peaks are shown: those targeted by sciRNAs (sciRNA-targeted) or otherwise (Not targeted). (D) Average SNP density per position in all B1 sequences bound by sciRNAs and their ±100 nt flanking region (chi-square P-value P < 2.2e-16). The y-axis is the average SNP density per nucleotide (see Supplementary Methods). A smoothing window of 10nt was applied to all data points. The grey region indicates the sciRNA-binding region. It ranges from 0–14 because the maximum length of targeted region was 24 and the smoothing window spanned 10 nt. Red dashed lines indicate the average SNP density of the three corresponding regions.
Similar to sciRNAs, the majority of target cleavage sites were in B1 elements (Figure 5B), and their ±5 nt sequences mapped to similar regions of the consensus B1 sequence as sciRNAs (Supplementary Figure S7B versus S7C). Because the majority of sciRNA cleavage sites are located in SINEs, we next tested whether this is a unique feature of sciRNA-directed degradation or common in the global Degradome. In contrast to the significant enrichment of SINEs in sciRNA-targeted cleavage sites, the remaining cleavage sites in the rest of the Degradome were rarely in SINE regions (Figure 5C). The fraction in repetitive regions only slightly increased when all types of repeats were considered, suggesting that SINE elements are driving this phenomenon (Supplementary Figure S7D).
To ensure that the relative enrichment of SINEs in target sites was not artificially inflated as a result of non-unique mapping of the Deg-Seq reads, we examined the sequence uniqueness of the flanking regions of predicted cleavage sites. The majority of target sequences were unique among all predicted targets of a specific cell type regardless of the length of flanking regions, although targets in testis had the smallest level of uniqueness (Supplementary Figure S7E). We then reexamined the overlap between target cleavage sites and repetitive elements after removing redundant target sequences. B1 elements were still enriched, confirming that SINE elements are enriched in the target pool (Supplementary Figure S7F). Thus, SINE-targeting, especially B1-targeting, is a unique feature of sciRNA-mediated cleavage in mouse.
Repetitive elements as signals for sciRNA targeting
It was previously speculated that SINEs are used as signals for miRNA targeting (30–32). However, other studies presented evidence against this postulation, showing that canonical miRNA targeting avoided Alu elements (33). Here, we suggest that B1 elements in mice serve as signals for small RNA targeting through endonucleolytic cleavage instead of the canonical miRNA pathway. If this speculation holds, then Ago2 should bind to sciRNA targets in B1 regions more often than to predicted canonical miRNA targets in B1 regions. To test this hypothesis, for miRNAs expressed in mESCs, we focused on their predicted canonical targets (as defined in microrna.org (34)) where the target sites are located in B1 elements. These targets were separated into two groups: those with target sites overlapping our predicted sciRNA target, and those that neither overlapped a sciRNA target nor contained a Deg-Seq peak (Supplementary Methods). It should be noted that only 2% (68 005 / 3 316 252) of the predicted canonical miRNA targets were in B1 regions. We observed that Ago2 binds to the first group more often than the second (Supplementary Figure S7G). The above results support our hypothesis that B1 elements are likely signals for sciRNA-mediated cleavage.
Next, we asked whether sciRNA-mediated targeting of B1 elements is under evolutionary selection. Since repetitive regions are poorly conserved across species, a conventional multi-species sequence conservation analysis was not feasible. Instead, we conducted an analysis of SNP enrichment in sciRNA target sites using known mouse SNPs (Supplementary Methods). Strikingly, we observed that SNPs were significantly depleted in sciRNA-targeted B1 sequences compared to the flanking B1 regions (Figure 5D), suggesting that sciRNA targets are under selection for sequence conservation. This finding also indicates that sciRNA-mediated regulation has potential functional significance.
Small RNA-guided endonucleolytic cleavage in human ESCs also targets retrotransposons
To investigate sciRNA-guided cleavage in human cells, we obtained small RNA-Seq and Deg-Seq data from human H1 ESCs (Supplementary Table S1) and conducted the same analysis as for the mouse data sets. A total of 34 sciRNAs and 23 target genes were identified (allowing up to two mismatches in the alignment), with about 50% sciRNAs being annotated miRNAs (Supplementary Figure S8A). The lower numbers of sciRNAs and targets compared to mESCs could be explained by lower depth of small RNA sequencing in human (Supplementary Table S1). Alternatively, differences in the repetitive sequences and their distribution in human and mouse genomes may also account for this difference. Nevertheless, these results allow an examination of the global properties of human sciRNAs and targets. Similar to their mouse counterparts, they were enriched with sequences overlapping retrotransposons (Supplementary Figure S8B and S8C). However, in addition to SINE (Alu) elements, LINE (L2) elements were considerably enriched among human sciRNAs and their target sites. Similar to mouse sciRNA targets, human target sites were often located in non-coding genes or 3′ UTRs of coding genes (Supplementary Figure S8D). Furthermore, functional analysis of human targets revealed similar categories as for mouse targets (Supplementary Figure S8E, Supplementary Table S5).
Despite the above high similarities in general properties of sciRNA targeting between human and mouse, the specific types of retrotransposons enriched in the human data are different from those in mouse. This is likely explained by the apparent difference in abundance, sequence composition and activity of retrotransposons across the two species (35,36). Thus, the sciRNA pathway is a conserved mechanism between human and mouse but leverages different sciRNA molecules, possibly to adapt to the divergence of retrotransposons between the two genomes.
DISCUSSION
We report a global analysis of endonucleolytic RNA cleavage events in mouse ESCs, testis and cerebellum. In mammals, mRNA cleavage was not previously considered a major pathway for small RNA-guided mRNA degradation, with a small number of genes predicted as targets of this mechanism (4–8). Our analysis revealed an expanded repertoire of hundreds of sciRNAs and their corresponding target genes in mouse and human, suggesting that this regulatory pathway is conserved and relatively prevalent in a cell-type specific manner. Given the potential functional significance of the target genes in development and essential cellular processes, sciRNA-mediated cleavage may have a much more profound impact on gene regulation and cellular function than previously appreciated.
We defined sciRNAs based on a unifying function, i.e. those that are predicted to cleave target RNAs via near perfect sequence complementarity. Thus, it is not surprising to find sciRNAs potentially reflecting diverse biogenesis mechanisms and overlapping known small RNAs of different categories (miRNAs, siRNAs, piRNAs). Despite this diversity, sciRNA expression appears to be under close regulation, as manifested by their striking expression specificity to developmental stages and cell types in contrast to all miRNAs (Figure 3C and D, Supplementary Figure S5). In addition to known categories of small RNAs, many sciRNAs were novel, with unknown biogenesis mechanisms and derived from genomic regions of known genes. These data suggest that the biogenesis and regulated expression of sciRNAs need further investigation.
Despite their heterogeneity in biogenesis, a salient feature of sciRNAs and their target regions is the enrichment of repetitive sequences, especially of the B1 class in mouse. Retrotransposons are very prevalent in mammals, accounting for more than 40% of the human and mouse genomes. However, little is known regarding the functional implication of their presence within genes. It was speculated that miRNAs or other small RNAs may target SINE elements embedded in mRNAs, and therefore the SINEs are used as signals for gene targeting (30–32). Yet, supporting data for this speculation was lacking. Studies that imposed the canonical miRNA targeting rules (requiring seed matching) predicted that Alu elements avoid targeting by miRNAs, thus providing data against the above speculation (33). Our results reconcile the seemingly conflicting hypotheses and data by supporting that B1 elements within murine RNA transcripts serve as signals for small RNA targeting, but through the endonucleolytic cleavage pathway instead of the canonical miRNA targeting based on seed matches alone. As retrotransposable elements spread across the genome and into non-coding regions of genes, sciRNA-mediated regulatory mechanisms may have evolved to leverage the abundant repetitive elements as signals for gene targeting, although this hypothesis remains to be tested.
Capturing such targeting events may have been difficult due to the repetitive nature of the small RNAs and their target sites. Non-uniquely mapped reads in sequencing data analysis are often excluded because they are difficult to interpret. In this study, non-unique alignments of Deg-Seq reads were retained, with their abundance normalized by total number of non-unique matches to the genome (Materials and Methods). Nevertheless, we observed that the majority of cleavage site-flanking sequences were unique, suggesting that enrichment of repetitive targets was not overestimated (Supplementary Figure S7E and F). The recognition of retrotransposons in RNA transcripts allows for targeting of multiple repeat-containing transcripts by a single sciRNA (Supplementary Figure S7A). However, it should be noted that the number of targets of a typical sciRNA is much smaller than that of canonical miRNAs. sciRNAs are still highly specific to their respective targets given their extended sequence complementarity and the high degree of divergence and uniqueness among retrotransposable elements (37).
It should be noted that our method imposed stringent criteria in predicting sciRNA-target relationships. In using the SNR approach, we assumed that sciRNAs should have more targets than expected by chance. Due to the requirement of extended sequence complementarity, many true sciRNAs may only target a small number of genes. As a result, a true sciRNA may not have a high SNR. Thus, it is possible that many more sciRNAs exist than presented in our study.
In summary, we report the discovery of a large number of sciRNAs and their cognate targets in mouse and human cells. This mode of gene regulation was previously poorly characterized in mammals. We demonstrate that this pathway mainly targets retrotransposons in mammalian genomes, and likely plays essential roles in gene regulation in a developmental stage- and cell type-specific manner.
ACCESSION NUMBERS
The data sets supporting the results of this article are available in the GEO repository, GSE68254.
Supplementary Material
Acknowledgments
We thank Dr Zefeng Wang and members of the Xiao laboratory for helpful discussions and comments on this work as well as the ENCODE Consortium for generating some of the data sets.
Footnotes
Present address: Jae-Hyung Lee, Department of Life and Nanopharmaceutical Sciences, Department of Maxillofacial Biomedical Engineering, School of Dentistry, Kyung Hee University, Seoul, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (NIH) [R01HG006264, U01HG007013 to X.X]; National Science Foundation [1262134 to X.X]; National Institute of Health predoctoral training grant [T90DE022734 to A.A.C]. Funding for open access charge: NIH [R01HG006264].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis B.P., Shih I., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 3.Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 4.Yekta S., Shih I.-H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 5.Davis E., Caiment F., Tordoir X., Cavaillé J., Ferguson-Smith A., Cockett N., Georges M., Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 6.Bracken C.P., Szubert J.M., Mercer T.R., Dinger M.E., Thomson D.W., Mattick J.S., Michael M.Z., Goodall G.J. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011;39:5658–5668. doi: 10.1093/nar/gkr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin C., Nam J.-W., Farh K.K.-H., Chiang H.R., Shkumatava A., Bartel D.P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karginov F. V, Cheloufi S., Chong M.M.W., Stark A., Smith A.D., Hannon G.J. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamura K., Lai E.C. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Carmell M. a, Rivas F.V., Marsden C.G., Thomson J.M., Song J.-J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science (80-.). 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 11.German M. a, Pillay M., Jeong D.-H., Hetawal A., Luo S., Janardhanan P., Kannan V., Rymarquis L. a, Nobuta K., German R., et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 12.Addo-Quaye C., Eshoo T.W., Bartel D.P., Axtell M.J. Endogenous siRNA and miRNA targets identified by sequencing of the arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- 14.Toedling J., Servant N., Ciaudo C., Farinelli L., Voinnet O., Heard E., Barillot E. Deep-sequencing protocols influence the results obtained in small-RNA sequencing. PLoS One. 2012;7:e32724. doi: 10.1371/journal.pone.0032724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., Zhu W., Wu W., Chen R., Zhao Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:1–6. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbashir S.M., Martinez J., Patkaniowska a, Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz S., Oren R., Ast G. Detection and removal of biases in the analysis of next-generation sequencing reads. PLoS One. 2011;6:e16685. doi: 10.1371/journal.pone.0016685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung A.K.L., Young A.G., Bhutkar A., Zheng G.X., Bosson A.D., Nielsen C.B., Sharp P. a. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Ender C., Meister G., Moore P.S., Chang Y., John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787–6799. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips B.T., Gassei K., Orwig K.E. Spermatogonial stem cell regulation and spermatogenesis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:1663–1678. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function genomics. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H.-M., Chen H., Liu W., Liu H., Gong J., Wang H., Guo A.-Y. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40:D144–D149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almeida L.G., Sakabe N.J., de Oliveira A.R., Silva M.C.C., Mundstein A.S., Cohen T., Chen Y.T., Chua R., Gurung S., Gnjatic S., et al. CTdatabase: A knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz J. SINEs as driving forces in genome evolution. Genome Dyn. 2012;7:92–107. doi: 10.1159/000337117. [DOI] [PubMed] [Google Scholar]
- 31.Lehnert S., Van Loo P., Thilakarathne P.J., Marynen P., Verbeke G., Schuit F.C. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS One. 2009;4:e4456. doi: 10.1371/journal.pone.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smalheiser N.R., Torvik V.I. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Hoffman Y., Dahary D., Bublik D.R., Oren M., Pilpel Y. The majority of endogenous microRNA targets within Alu elements avoid the microRNA machinery. Bioinformatics. 2013;29:894–902. doi: 10.1093/bioinformatics/btt044. [DOI] [PubMed] [Google Scholar]
- 34.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalopin D., Naville M., Plard F., Galiana D. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015;7:567–580. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sela N., Mersch B., Gal-Mark N., Lev-Maor G., Hotz-Wagenblatt A., Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umylny B., Presting G., Efird J.T., Klimovitsky B.I., Ward W.S. Most human Alu and murine B1 repeats are unique. J. Cell. Biochem. 2007;102:110–121. doi: 10.1002/jcb.21278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.