Abstract
Background
Pancreas development in zebrafish shares many features with mammals, including the participation of epithelial progenitor cells expressing pancreas transcription factor 1a (ptf1a). However, to date it has remained unclear whether, as in mammals, ptf1a-expressing zebrafish pancreatic progenitors are able to contribute to multiple exocrine and endocrine lineages. To delineate the lineage potential of ptf1a-expressing cells, we generated ptf1a:creERT2 transgenic fish and performed genetic-inducible lineage tracing in developmental, regenerating, and ptf1a-deficient zebrafish pancreas.
Results
In addition to their contribution to the acinar cell lineage, ptf1a-expressing cells give rise to both pancreatic Notch-responsive-cells (PNCs) as well as small numbers of endocrine cells during pancreatic development. In fish with ptf1a haploinsufficiency, a higher proportion of ptf1a lineage-labeled cells are traced into the PNC and endocrine compartments. Further reduction of ptf1a gene dosage converts pancreatic progenitor cells to gall bladder and other non-pancreatic cell fates.
Conclusions
Our results confirm the presence of multipotent ptf1a-expressing progenitor cells in developing zebrafish pancreas, with reduced ptf1a dosage promoting greater contributions towards non-acinar lineages. As in mammals, loss of ptf1a results in conversion of nascent pancreatic progenitor cells to non-pancreatic cell fates, underscoring the central role of ptf1a in foregut tissue specification.
Keywords: ptf1a, pancreas, pancreatic Notch-responsive cells, endocrine, lineage tracing
Introduction
The zebrafish has emerged as a highly informative system for the study of pancreatic organogenesis and has provided new insights regarding cellular signaling, lineage hierarchies and other developmental mechanisms including β-cell specification during pancreas morphogenesis (Yee et al., 2005; Kinkel and Prince, 2009; Tiso et al., 2009).
Pancreas development in zebrafish shares many features with mammalian pancreas development. The mammalian pancreas develops in two stages. During the first wave of development, referred to as the “primary transition”, there is early formation of an endocrine population (Rall et al., 1973; Herrera et al., 1991). Starting around embryonic day 13.5 (E13.5) in mouse, dramatic morphogenetic changes occur. This process is referred to as the “secondary transition” and is characterized by rapid cellular proliferation and differentiation. Mature endocrine cell populations also start to appear at this stage (Kemp et al., 1972; Pictet et al., 1972).
Pancreas transcription factor 1a (Ptf1a) is one of the earliest genes expressed in the pancreatic field (Hald et al., 2008). Lineage analysis in mice has shown that Ptf1a marks a multi-lineage progenitor pool that generates both endocrine and exocrine progeny during early pancreatic organogenesis. Between E13.5 to E18.5, the lineage contributions of the Ptf1a-expressing progenitor pool become progressively restricted, and by E18.5, Ptf1a exclusively labels cells that are destined to become acinar cells (Kawaguchi et al., 2002; Pan et al., 2013). In Ptf1a null mice, growth of the pancreas is severely retarded; there is a complete lack of acinar cells, and endocrine cells become redistributed to spleen (Krapp et al., 1998; Burlison et al., 2008). In addition, in the Ptf1a-deficient state, progenitor cells that would normally be directed to the pancreatic fate instead become incorporated into the duodenum or common bile duct (Kawaguchi et al., 2002; Burlison et al., 2008).
Similar to mammals, the zebrafish pancreas develops from two distinct anlagen arising from foregut endoderm. Although these have been termed the dorsal and ventral pancreatic buds (Field et al., 2003), their evolutionary relationship to the mammalian dorsal and ventral buds remain unknown. While in mammals these buds contain admixed endocrine and exocrine elements, there seems to early spatial segregation of zebrafish pancreatic lineages (Field et al., 2003; Ward et al., 2007). The fish dorsal bud develops first and gives rise to a primary endocrine cluster known as the principal islet, while the ventral bud appears later and has traditionally felt to be responsible for generating the exocrine ductal and acinar lineages as well as a later population of endocrine cells (Field et al., 2003; Lin et al., 2004; Zecchin et al., 2004; Hesselson et al., 2009; Wang et al., 2011). In zebrafish, the early dorsal bud-derived endocrine population may represent the equivalent of those endocrine cells that originate during the primary transition in the mammalian pancreas, perhaps corresponding to the “Brockman body” observed in jawless fishes (Epple and Brinn, 1975; Slack, 1995). In any case, these early cells have limited proliferation potential, and, the vast majority of mature hormone-producing cells are derived from the secondary transition (Hesselson et al., 2009; Parsons et al., 2009; Wang et al., 2011). With respect to progenitor cells responsible for generating differentiated pancreatic cell types, two populations have been defined in zebrafish: a Notch-responsive population and a ptf1a-expressing population (Lin et al., 2004; Wang et al., 2011). Lineage tracing studies have demonstrated that zebrafish Notch-responsive cells (PNCs) differentiate to ducts, centroacinar cells and endocrine cells during the secondary transition (Parsons et al., 2009; Wang et al., 2011).
In contrast, the precise contribution of ptf1a-expressing progenitor cells to different pancreatic lineages has yet to be determined. Previous work has clearly established that zebrafish ptf1a plays an important role in the establishment of acinar cell fate, equivalently to its mammalian counterpart (Lin et al., 2004; Zecchin et al., 2004). Furthermore, the early endocrine population arising in the “dorsal bud” is independent of ptf1a (Lin et al., 2004; Zecchin et al., 2004). However the role of ptf1a in the specification of endocrine cells arising during secondary transition remains unclear. Data has shown that a reduced level of ptf1a is more favorable for endocrine differentiation (Dong et al., 2008). Nevertheless, it is uncertain whether these endocrine cells are directly derived from a ptf1a-expressing progenitor population. To address these issues and to determine the fate of zebrafish ptf1a-expressing cells, we generated an inducible ptf1a:creERT2 transgenic fish line using BAC recombineering, and employed this line to complete lineage tracing studies. Early lineage labeling confirmed that ptf1a-expressing progenitors contribute primarily to the acinar cell lineage. Furthermore, we identified the contributions of ptf1a lineage to pancreatic Notch-responsive cells (PNCs) and endocrine cells during development. We also demonstrated that ptf1a lineage-labeled cells gave rise to newly formed β cells during regeneration. Interestingly, heterozygous ptf1asa126/wt mutant fish displayed enhanced contributions of ptf1a lineage-labeled cells to the PNC and endocrine cell fates. In addition, we observed that, in the absence of functional Ptf1a, ptf1a lineage-labeled cells were converted into gall bladder and other non-pancreatic cell types. In summary, we showed that early ptf1a-expressing cells in zebrafish pancreas displayed limited but demonstrable multipotency during development and regeneration. Under conditions where ptf1a dosage was reduced, the contribution of ptf1a-expressing progenitor cells to non-acinar lineages was increased. These findings confirm conservation of general mechanisms for pancreas development among vertebrates, while highlighting qualitative differences.
Results
Validation of the ptf1a:creERT2 driver line
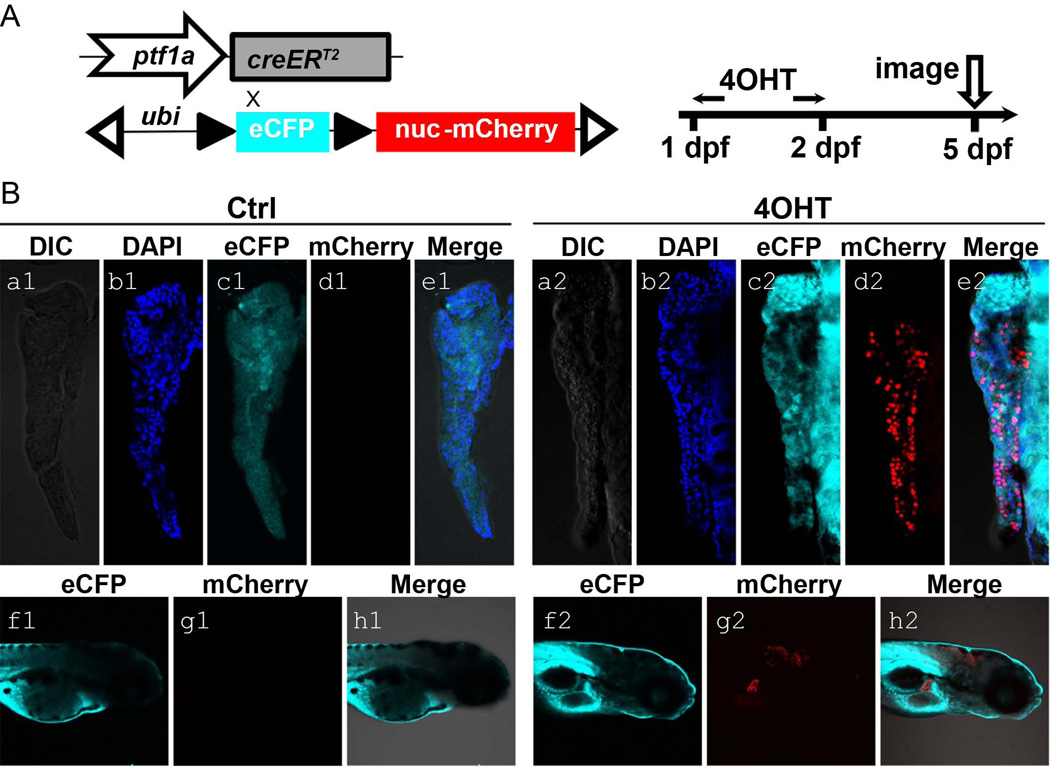
To lineage trace ptf1a-expressing cells in a temporarily controlled manner, we engineered a large genomic bacterial artificial chromosome (BAC) spanning the ptf1a locus. As similar BAC in which GFP replaced ptf1a coding sequence has been shown to faithfully recapitulate endogenous ptf1a expression in pancreas, hindbrain, retina and spinal cord (Park et al., 2008). We replaced the GFP sequence in the ptf1a:eGFP BAC with a DNA sequence encoding creERT2. To facilitate integration of the BAC construct into the genome, an additional cassette containing the inverted left and right arm of the Tol2 transposable element was used (Suster et al., 2011). Several independent F1 ptf1a:creERT2 transgenic founder lines were established and were crossed onto the ubi:loxP-CFP-loxP-nuc-mCherry reporter line.
In order to screen lines for inducible cre activity, ptf1a:creERT2; ubi:loxP-CFP-loxP-nuc-mCherry double transgenic larvae were treated for 24 hours with 5 µM 4-hydroxytamoxifen (4-OHT), beginning at 1 day post fertilization (dpf). Larvae were then fixed for imaging at 5 dpf (Fig. 1A). Untreated control larvae were used as comparisons. At 5 dpf, strong nuc-mCherry signal was observed in 4-OHT-treated larvae, indicating cre-dependent recombination (Fig.1B, d2 and g2). Rare to non-existent pancreatic or hindbrain nuc-mCherry signals were detected in larvae without 4-OHT treatment (Fig.1B, d1 and g1). The continued detection of CFP signal observed in Fig. 1B, c1 and f1) is due to the fact that our transgenic fish line carries multiple insertions of ubi:loxP-CFP-loxP-nuc-mCherry reporter, with not all copies undergoing cre-mediated recombination and CFP excision.
Figure 1. Generation of the ptf1a lineage tracing system and its initial characterization.
(A) Schematic diagram of ptf1a:creERT2 driver and responder ubi:loxP-CFP-loxP-nuc-mCherry. Double transgenic fish larvae were treated with 5 µM 4-OHT in E3 medium at 1 dpf for 24 hours and then fixed for imaging at 5 dpf. These larvae were raised along with untreated controls. (B) At 5 dpf, No pancreatic and hindbrain nuclear-mCherry signals were detected in control larvae (a1-e1, f1-h1). 4-OHT-treated larvae showed nuclear-mCherry signal (red) in exocrine pancreas (a2-e2) and hindbrain (f2-h2), indicating ptf1a-dependent Cre activity. CFP signal (cyan) could be detected ubiquitously (c1, f1, c2, f2). DAPI (blue) stains for nuclei (b1, b2).
A single transgenic line with the highest fluorescent intensity upon 4-OHT treatment and lowest incidence of recombination events without 4-OHT treatment (“leakage”) was selected and expanded for further use.
Detailed characterization of recombination efficiency in ptf1a:creERT2; ubi:loxP-CFP-loxP-nuc-mCherry transgenic embryos
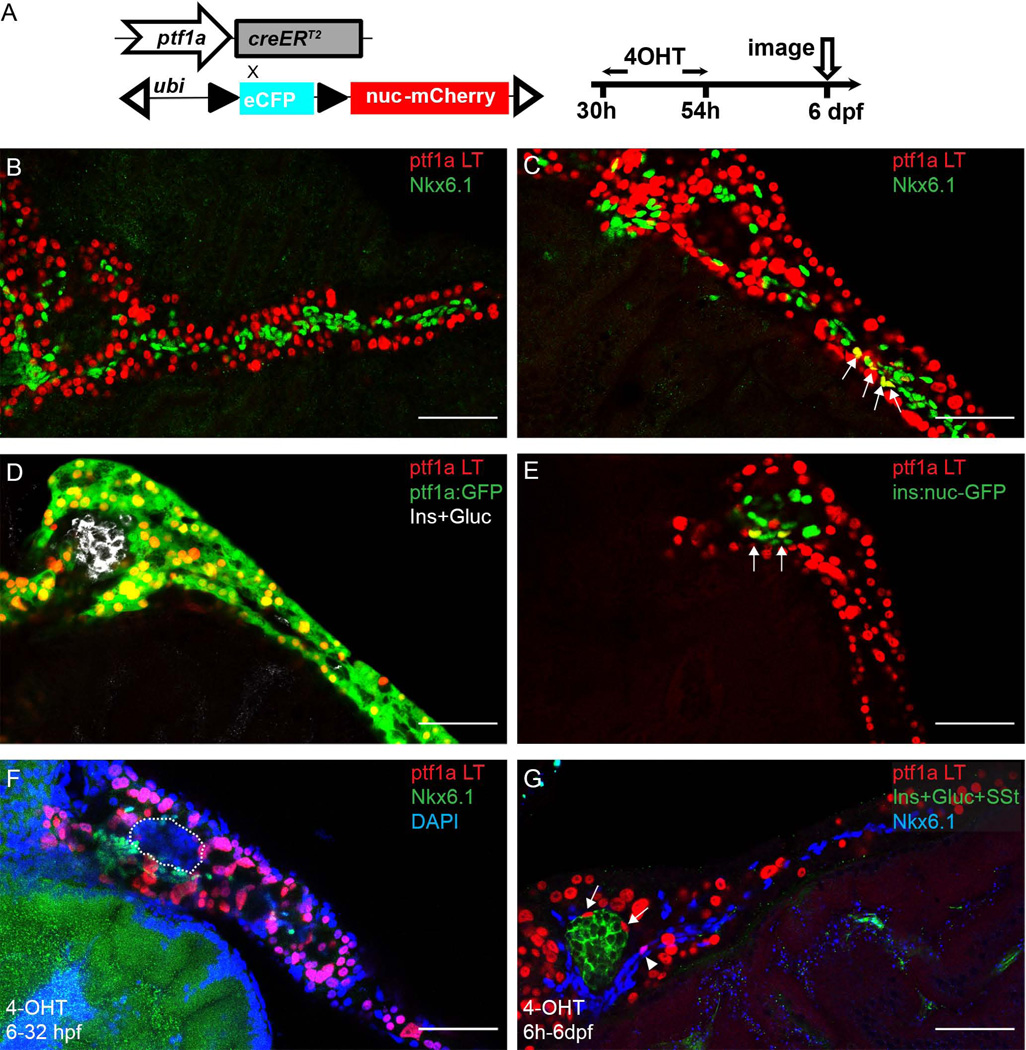
The expression of ptf1a is first detectable around 32 hours post fertilization (hpf). It has been reported that ligand-mediated recombination could be detected as early as 2 hours post 4-OHT treatment in transgenic zebrafish expressing creERT2 (Hans et al., 2009). We therefore treated ptf1a:creERT2; ubi:loxP-CFP-loxP-nuc-mCherry embryos with 4-OHT at 30–54 hpf in an effort to label the earliest pool ptf1a-expressing pancreatic progenitor cells (Fig. 2A). We subsequently imaged the fish at 6 dpf, when pancreatic architecture is fully formed.
Figure 2. The early ptf1a lineage generates a small fraction of PNC and endocrine cells.
(A) Experimental setup. (B–G) In all the panels, ptf1a lineage is indicated by nuclear-mCherry expression (red). (B, C) Immunofluorescence for Nkx6.1 (green) labels PNCs. (B) In some pancreata, the ptf1a lineage and PNCs show no overlap. (C) In some pancreata, ptf1a-lineage labeled cells are traced into PNCs. Arrows point to colabeling events. (D) In some pancreata, the ptf1a lineage has no demonstrable contribution to endocrine cell types. ptf1a:GFP transgene (green) shows current expression of ptf1a, which, at 6 dpf, is limited to acinar cells. Endocrine cells are labeled by a mixture of antibodies against Insulin (Ins) and Glucagon (Gluc), white. (E) In a few pancreata, some of the cells derived from the ptf1a lineage are co-labelled by the ins:nuc-GFP transgene (green), which is visualized in the nuclei of Insulin-secreting cells. Arrows point to co-labeling events. (F) 4-OHT treatment at 6h-32hpf. In the pancreas shown, there are no ptf1a lineage-labeled cells observed in the endocrine compartment (outlined) nor PNCs (Nkx6.1, green). (G) 4-OHT treatment at 6h-6dpf. Nkx6.1, blue. Arrows point to ptf1a lineage-labeled endocrine cells. Arrowhead points to ptf1a lineage-labeled PNCs. Scale bar, 50 µm.
In our selected transgenic line, the frequency of embryos displaying tamoxifen-independent creERT2 activity displayed significant clutch-to-clutch variability, ranging from 5% –30%. However, tamoxifen-independent recombination events were confined to very small numbers of cells. Among 32 fish with detectable 4-OHT-independent recombination, we observed an average of only 19 (±10) cells per fish with detectable nuc-mCherry signals, as assessed at 6 dpf. All cells displaying 4-OHT-independent reporter recombination also expressed markers of pancreatic acinar cell differentiation. In comparison, among 32 fish that had been treated with 4-OHT, there were on average 377 (± 82) lineage labeled cells per fish, representing 85% of total acinar cells. Stated differently, cells displaying 4-OHT-independent cre activity represented only a small proportion of total transgene expressing cells (<5%), and were limited to the acinar lineage. We therefore concluded that our study would not be significantly confounded by 4-OHT-independent recombination, especially as it related to the contribution of ptf1a-expressing cells to non-acinar lineages.
The early ptf1a lineage has a limited contribution to non-acinar cell fate in the zebrafish pancreas
We next sought to evaluate the contribution of ptf1a–expressing progenitor cells to different pancreatic lineages. By 6dpf, we observed broad contributions to the acinar cell lineage, confirming that ptf1a–expressing progenitor cells represent the major and perhaps exclusive source of these cells. We also observed lower magnitude contributions to the PNC and endocrine (principal islet) cell lineages. For the 36 fish we examined, 26 of them (72%), did not have any detectable ptf1a lineage contribution to PNCs (Fig. 2B; Fig. 5F, G). For the remaining 10 fish, there were on average 8 PNCs marked by the ptf1a lineage label (Fig. 2C; Fig. 5F, G). At this stage, there are an average 137 PNCs per fish, hence, cells of ptf1a lineage contribute to approximately 6% of the total PNC population.
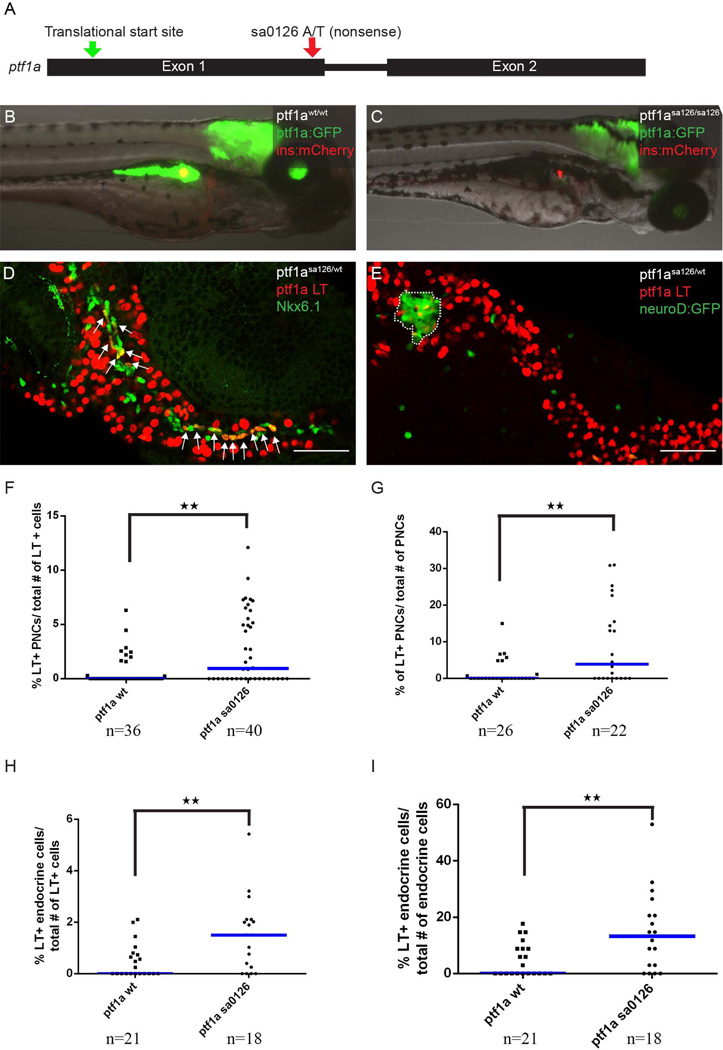
Figure 5. Ptf1a haploinsufficiency increases the the allocation of pancreatic progenitor cells to non-acinar lineages.
(A) The ptf1asa126 allele. This allele has an A to T transition in the first exon of ptf1a coding sequence, introducing a premature stop codon. (B) Wildtype larvae, with ptf1a:GFP (green) marking acinar cells and ins:mCherry (red) transgene marking β cells. (C) ptf1asa126/sa126 homozygous fish do not have exocrine pancreas. (D, E) The ptf1a lineage is labeled by nuclear-mCherry expression (red). (D) In ptf1asa125/wt fish, more lineage-labeled cells can be traced into Nkx6.1+ (green) PNCs. Arrows point to co-labeling events. (E) In pf1asa126/wt fish, more lineage-labeled cells can be traced into the neuroD:GFP+ endocrine compartment. The principal islet is outlined. (F, G) Quantification of ptf1a lineage-labeled cells contributing to PNCs, normalized by total number of ptf1a-lineage labeled cells (F), or by the total number of PNCs (G). (H, I) Quantification of ptf1a-lineage labeled cells contributing to the endocrine compartment, normalized by total number of ptf1a-lineage labeled cells (H), or by the total number of endocrine cells (I). (F-I) “n” indicates the number of fish quantified for each genotype. Blue bars show population median.
We subsequently surveyed the ptf1a lineage contribution to endocrine cells in the principal islet. At 6 dpf, the principal islet contains endocrine cells directly descended from the dorsal bud as well as cells differentiated during the secondary transition (Biemar et al., 2001; Hesselson et al., 2009). For the 21 larvae we assessed, 11 of them (52%) did not have any ptf1a lineage-labeled cells traced into the principal islet (Fig. 2D; Fig. 5H, I). For the 10 remaining larvae where we did observe contributions from ptf1a-expressing progenitors to the endocrine lineage, there were an average of 3.4 lineage labeled cells observable in the principal islet (Fig. 2E; Fig. 5H, I). This represents approximately 10% of the principal islet endocrine population at 6 dpf.
To confirm that the current 30–54 hpf 4-OHT treatment window does not preferentially label acinar cells, we compared lineage analysis results from different 4-OHT pulse intervals, varied from as early as 6 hpf, a time point when ptf1a is not yet expressed, to 6 dpf (Fig. 2F, G). In all cases, the lineage tracing results were comparable.
The ptf1a lineage contributes to endocrine cells in juvenile and adult zebrafish
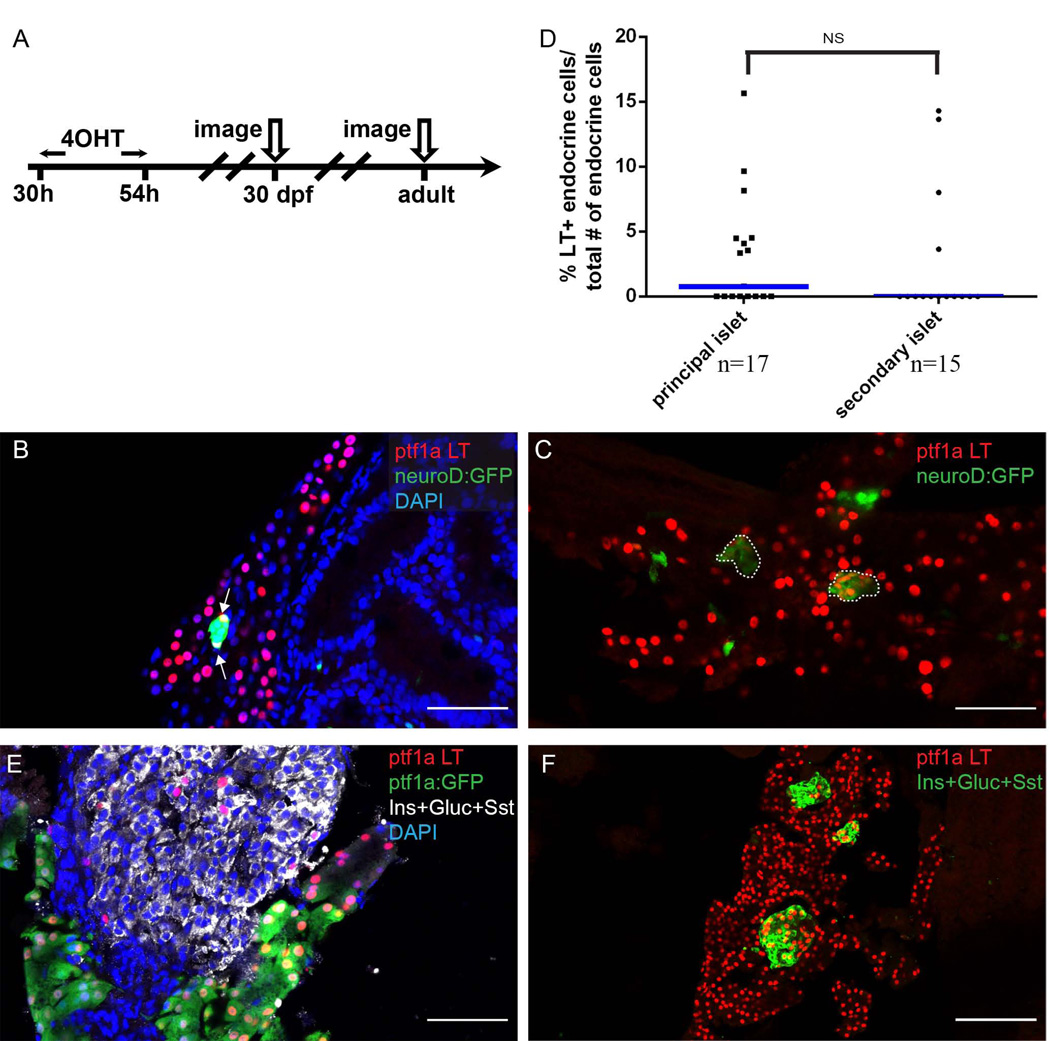
To determine the contribution of the ptf1a lineage to later arising endocrine cells, we pulsed 4-OHT again from 30–54 hpf, and further traced the ptf1a lineage into juvenile and adult fish (Fig. 3A). In 4-OHT treated fish, at 30 dpf, we saw widespread lineage labeling of acinar cells. In addition, we noted a contribution of ptf1a-expressing progenitors to endocrine cells within secondary islets (Fig. 3B, C). Interestingly, for some of the secondary islets in which we observed ptf1a lineage labeling, almost all endocrine cells within the islets were labeled (Fig. 3C). We theorized that the ptf1a lineage gave rise to the progenitors of those secondary islets, which subsequently had gone through clonal expansion process to form entire secondary islets. We further quantified the frequency of ptf1a lineage labeled cells based on their principal or secondary islet location (Fig. 3D). We observed no differences in the percentage of ptf1a-lineage labeled cells between these two populations (Fig. 3D). Similarly, in the adult pancreas, we observed isolated as well as clustered ptf1a lineage labeled cells located with islet tissue (Fig. 3E, F).
Figure 3. The early ptf1a lineage contributes to secondary islet and adult endocrine cells.
(A) Experimental setup. (B, C, E, F) The ptf1a lineage is labeled by nuclear-mCherry (red). (B, C) Sections from 30 dpf juvenile lineage-tracing fish. The secondary islet is labeled by the transgenic marker neuroD:GFP (green). (B) Arrows point to endocrine cells expressing the ptf1a lineage mark. Notice that not all nuclei of within the secondary islet are colabeled by the ptf1a lineage mark. DAPI, blue. (C) Secondary islets are labeled by neuroD:GFP (green) and outlined. The secondary islets consist of a clusters of ptf1a-lineage labeled cells. (D) Scatter plot shows frequency of ptf1a lineage labeled cells traced into principal or secondary islets. T-test shows no significance. Blue bars show population median. “n” indicates the number of slides quantified for each population. (E, F) Sections from adult lineage-tracing fish. The endocrine population is labeled by a mixture of antibodies against Insulin (Ins), Glucagon (Gluc) and Somatostatin (Sst). (E) Some of the endocrine cells express the ptf1a lineage label. The majority of ptf1a lineage-labeled cells co-express ptf1a:GFP transgene (acinar marker). (F) Selected islets consist of a cluster of ptf1a-lineage labeled cells. (F) DAPI, blue. Scale bar, 50 µm.
ptf1a lineage contributes to endocrine β-cell regeneration
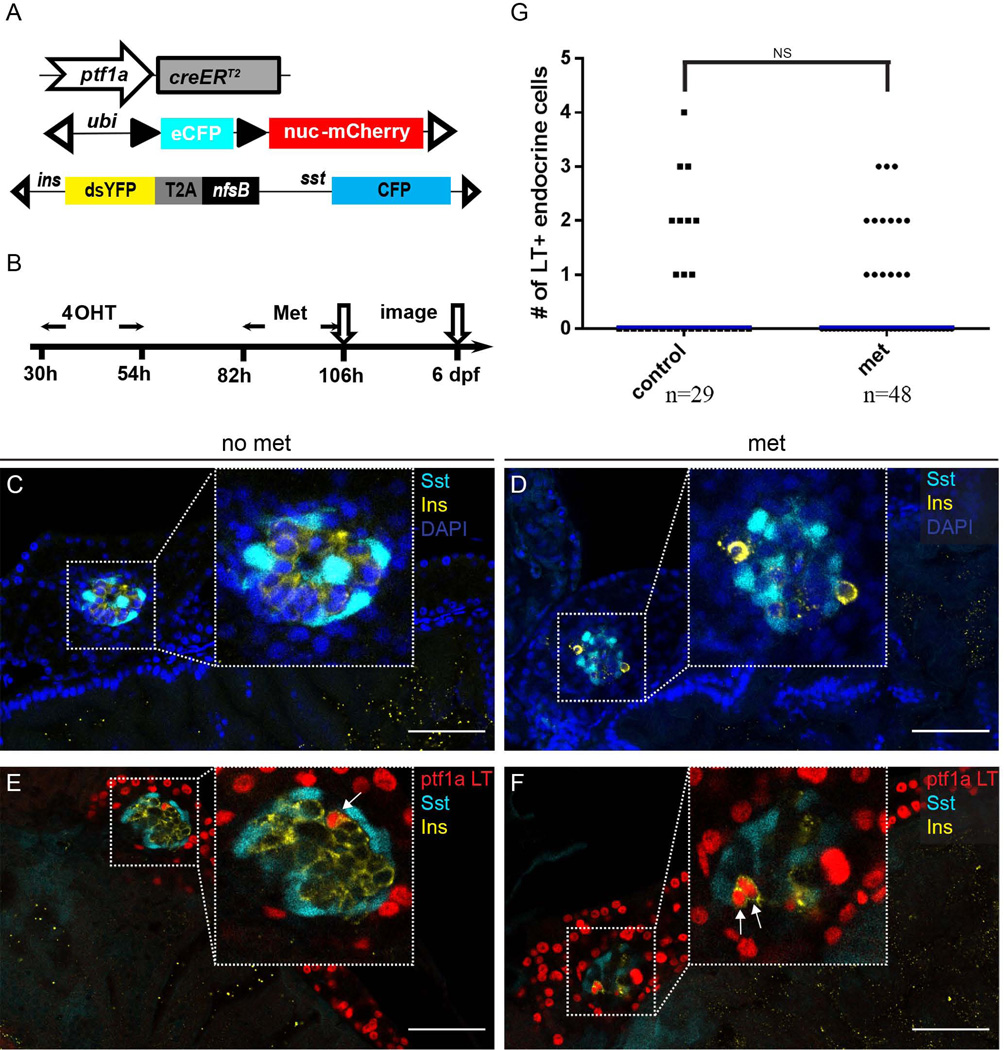
Having assessed the fate of ptf1a-expressing progenitor cells during normal pancreas development, we proceeded to determine whether these cells could contribute to β-cell regeneration. To this end, we employed a well-established nitroreductase (NTR) cell-ablation system to eliminate β cells (Pisharath et al., 2007). The NTR enzyme, encoded by the bacterial gene nfsb, can convert prodrugs such as metronidazole (Met) to cytotoxins (Lindmark and Muller, 1976; Anlezark et al., 1992). In our system, we expressed the nfsb gene under the control of insulin promoter. Subsequently, upon administration of Met, the insulin secreting β cells are ablated, with cell death occurring within 6 hours post Met treatment. Following β-cell ablation, regenerating insulin-expressing cells can be observed within 36 hours after Met removal (Pisharath et al., 2007). To determine the contribution of the ptf1a lineage to regenerating β-cells, we crossed ptf1a:creERT2; ubi:loxP-CFP-loxP-nuc-mCherry fish with sst:CFP; ins:dsYFP-2TA-nsfB fish (Fig. 4A). Besides the ptf1a lineage tracing constructs, the triple transgenic fish carries the nfsB transgene construct intended for β-cell ablation. In these fish, the insulin promoter drives expression of destabilized YFP (dsYFP) and nfsB, while the somatostatin (sst) promoter drives the expression of CFP (Fig. 4A).
Figure 4. The ptf1a lineage contributes to β-cell regeneration.
(A) Schematic depicting the triple transgenic line utilized in the experiment. (B) Experimental setup. (C) In control fish with no Metronidazole (Met) treatment, the principal islet has compact architecture. Insulin (Ins), yellow. Somatostatin (Sst), cyan. Nuclei are labeled by DAPI (blue). (D) Pancreatic region following 24h of Met treatment. The majority of Insulin-secreting cells are gone. Insulin (Ins), yellow. Somatostatin (Sst), cyan. DAPI, blue. (E, F) The ptf1a lineage is labeled by nuclear-mCherry expression (red) 48 h after removal of Met. (E) In untreated fish, there are rare ptf1a lineage cells traced into the principal islet. Insulin (Ins), yellow. Somatostatin (Sst), cyan. (F) In the Met treated fish, two regenerated β cells express the ptf1a lineage label. Insulin (Ins), yellow. Somatostatin (Sst), cyan. In (E) and (F), arrows point to Insulin+ cells that are labeled with the ptf1a lineage mark. (G) Scatter plot shows quantification of ptf1a lineage labeled cells within the endocrine compartment. No significant differences in the frequency of ptf1a lineage contributions are observed between normal and regenerating β cells. “n” indicates the number of fish quantified for each condition. Blue bars show population medium.
We treated the resulting triple transgenic fish with 4-OHT from 30–54 hpf and applied Met from 82–106 hpf (Fig. 4B). In the control pancreata, YFP-expressing β cells form a compact inner core, intermingled with somatostatin-secreting δ cells and surrounded by other endocrine cell types (Fig. 4C). In the Met-treated pancreata, much weaker YFP signals were detected within the principal islet (Fig. 4D). Some cells contained aggregates with high YFP fluorescence intensity. However, these cells were detached from the rest of endocrine cells and the fluorescence most likely represent cellular debris. (Fig. 4D). We assessed β-cell regeneration at 6 dpf, 48h after Met removal. At this time point, β cells started to regenerate but there was still a distinct reduction of β-cell numbers in the Met-treated fish (Fig. 4E, F). Interestingly, we observed that cells derived from the early ptf1a lineage contributed to the newly generated β cells (Fig. 4F). The level of contribution, however, was similar to the control pancreas (Fig. 4G), suggesting that ptf1a lineage was not the main source for new β cells. To be noted, in the current study, β-cell ablation was carried out from 82–106 hpf, a time when the endocrine population is rapidly proliferating. Our unpublished observation indicates that during this developmental time window, newly generated β cells mainly come from pre-existing endocrine cells and from the differentiation of PNC progenitors, likely explaining the modest contribution from ptf1a-expressing progenitor cells.
Reduction in ptf1a gene dosage enhances the contribution of ptf1a-expressing progenitor cells to non-acinar cell fates
In the mouse, pancreatic cells with different Ptf1a gene dosages seem to have altered cell fates (Fukuda et al., 2008; Pan and Wright, 2011). During early mouse pancreas development, it has been shown that intermediate expression of Ptf1a marks multipotent progenitor cells, whereas higher Ptf1a expression levels are observed in pancreatic progenitors undergoing restriction towards an acinar cell fate (Pan et al., 2013). Similarly, in zebrafish, studies have shown that reduced levels of ptf1a seem to promote endocrine differentiation (Dong et al., 2008). Furthermore, in the adult fish, reduction of ptf1a activity induces the expression of endocrine-specific genes in mature acinar cells (Hesselson et al., 2011). To formally characterize the ptf1a lineage allocation under conditions of reduced ptf1a dosage, we took advantage of the ptf1asa126/wt mutant fish generated by the Sanger Zebrafish Mutation Project (Kettleborough et al., 2013). The ptf1asa126 allele contains a nonsense mutation within exon1 of the ptf1a coding region (Fig. 5A). This mutation is predicted to result in either the expression of a truncated protein or the induction of nonsense-mediated decay (NMD) (Randlett et al., 2013).
We first characterized pancreatic morphology in ptf1asa126/wt heterozygous and ptf1asa126/sa126 homozygous fish. Homozygous ptf1a mutations phenocopied the previously reported ptf1a morpholino phenotype (Fig. 5B, C) (Lin et al., 2004). At 5 dpf, ptf1asa126/sa126 fish did not have an exocrine pancreas, as assessed by ptf1a:GFP transgene expression; whereas the principal islet appeared normal or slightly reduced in size (Fig. 5B, C). In contrast, the pancreatic morphology of ptf1asa126/wt heterozygous fish was undistinguishable from wildtype controls at 5dpf (Fig. 5D, E). These data are consistent with the prediction that the ptf1asa126 mutation represents either a hypomorphic or functionally null allele.
We next proceeded to perform lineage tracing experiments in ptf1asa126/wt heterozygous fish. Intriguingly, with reduced ptf1a dosage, more ptf1a lineage labeled cells were traced into PNCs and the principal islet (Fig. 5D, E). In the 40 fish examined, 19, (48%) did not have any ptf1a-lineage labeling in PNCs. The fraction of unlabeled fish is lower than in the wildtype condition, where the corresponding percentage is 72% (Fig. 5F, G). Among the 21 remaining ptf1asa126/wt fish where we observed ptf1a-lineage labeling in PNCs, an average of 17 cells per pancreas, or 13.5% of the total number of PNCs, were labeled as descendants of the ptf1a lineage. In comparison, in wildtype fish, we observed 5.8% of the PNCs were labeled by ptf1a lineage (Fig. 5F, G). These differences in the frequency of ptf1a-lineage contributions to PNCs were statistically significant.
Similarly, we observed that in ptf1asa126/wt heterozygotes, 22% had no ptf1a lineage-labeled cells within the principal islet (compared to 52% of wildtype fish) (Fig. 5H, I). In fish in which we did observe ptf1a-lineage labeling within the principal islet, there was a higher percentage of endocrine cells labeled (19% compared with 10% in the principal islet of ptf1a wildtype fish) (Fig. 5H, I). Each of these differences was statistically significant.
The total number of PNCs and total area of the endocrine population showed no difference between wildtype and ptf1asa126/wt heterozygous pancreas (data not shown). To exclude the trivial explanation that ptf1a lineage labeling was more efficient in ptf1asa126/wt heterozygotes compared to wildtype fish, we compared labeling efficiencies in the acinar population. We observed that, as is the case with wildtype fish, 85% of acinar cells were labeled in ptf1asa126/wt heterozygotes. All the above data suggest that haploinsufficiency for ptf1a promotes an enhanced contribution of ptf1a-expressing pancreatic progenitors to non-acinar cell fates, with higher level of regulatory mechanisms acting to determine the actual number of PNCs and endocrine cells.
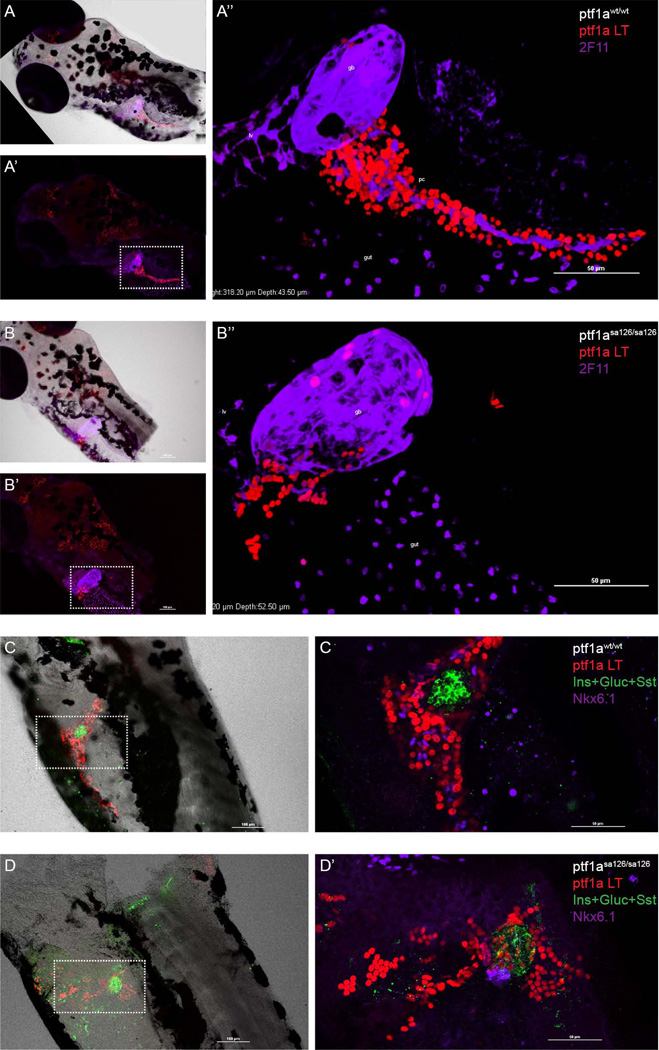
Further cell fate conversion in the ptf1asa126/sa126 homozygous fish
In homozygous ptf1asa126/sa126 fish, there are no detectable acinar cells and no secondary islets (Fig. 5C). Nonetheless, even in the absence of a morphologically discernible exocrine pancreas, we still observed a ptf1a lineage-labeled population in the region of the intestinal bulb. In order to characterize the identity of these cells, we performed immunolabeling using the 2F11 antibody that recognizes enteroendocrine cells, intrahepatic bile ducts, gall bladder, hepatopancreatic ducts and PNCs (Dong et al., 2007). 2F11 antibody staining indicated that some of the ptf1a lineage labeled cells were located within the gall bladder (Fig. 6A, B). Moreover, in the homozygous mutant fish, ptf1a lineage labeled cells could also be detected among PNCs as well as within the principal islet (Fig. 6C, D). Of note, in the ptf1asa126/sa126 fish, PNCs were properly specified (Fig. 6D). This observation was consistent with the notion that the ptf1a lineage and PNCs are independently specified (Wang et al., 2011). However, in the homozygous mutant fish, PNCs remained as a cluster of cells adjacent to the principal islets, and the total number of PNCs was significantly reduced (Fig. 6D). These data suggest that ptf1a lineage-derived acinar cells may provide a supporting cellular framework required for the proliferation and migration of PNCs and normal establishment of ductal network. The reduced number of PNCs is not likely to reflect a direct requirement for ptf1a in generating the PNC lineage, since under wildtype ptf1a conditions, only ~5% of PNCs were labeled by the ptf1a lineage mark. Alternatively, other cell types required for normal PNC expansion and migration might also be compromised in the ptf1asa126/sa126 fish.
Figure 6. ptf1a lineage analysis in homozygous ptf1asa126/sa126 fish.
In all panels, ptf1a lineage-labeled cells are shown in red. (A) In wildtype zebrafish, ptf1a lineage labeling can be observed in the hindbrain and pancreas (A’). A’’ is a high magnification, 3D-reconstructed view of the boxed region in A’. 2F11 (purple) labels gall bladder, liver, pancreatic PNCs, and some structures in the gut bulb. (B) In ptf1asa126/sa126 fish, there is no exocrine pancreatic structure. Some lineage labeled cells can be observed in the wall of gall bladder, as well as in another unidentified population adjacent to the gall bladder. B’’ is a zoomed-in view of the boxed region in B’. 2F11, purple. (C, D) A mixture of antibodies against Insulin (Ins), Glucagon (Gluc) and Somatostatin (Sst) (green) labels the endocrine cell population. Nkx6.1 (purple) labels PNCs. (C) In the wildtype zebrafish pancreas, the majority of lineage labeled cells are acinar cells. (D) In homozygous ptf1a mutant fish, the number of PNCs is reduced and they appear at an ectopic location. ptf1a lineage labeled cells can be detected in the principal islet and some of the PNCs. gb, gall bladder. lv, liver.
Discussion
In the present study, we generated a new ptf1a fate-mapping fish and carried out ptf1a lineage analysis in developing, regenerating and ptf1a-deficient pancreas. We observed a major contribution of the ptf1a progenitor lineage to later appearing acinar cells, and minor contributions to PNCs and endocrine cells during development. The ptf1a lineage also contributed to β cells during regeneration. Furthermore, ptf1a gene dosage was found to influence the magnitude of non-acinar lineage contributions. In heterozygous ptf1asa126/wt fish, we observed a larger contribution from the ptf1a progenitor lineage to the PNC and endocrine compartments. As in the mouse, a complete absence of ptf1a lead to trans-fating of ptf1a-expressing cells into gall bladder epithelium and other foregut tissues.
Limited contribution of the ptf1a lineage to non-acinar cell fates
In the current work, we observed limited contribution of the ptf1a lineage to non-acinar cell fates. One explanation for this observation might be that the earliest multi-lineage progenitor population may be characterized by only low-level expression of ptf1a, resulting in inefficient labeling of this population. However, when we changed our 4-OHT treatment time to 6–32 hpf, a time period that only covers the earliest low level expression of ptf1a (Lin et al., 2004; Zecchin et al., 2004) we still observed robust labeling (Fig. 2F). Similarly, when 4-OHT treatment was extended from 6h-6dpf, we did not observed any further enhancement of labeling (Fig. 2G). In addition, the ubi:loxP-CFP-loxP-nuc-mCherry reporter line has shown robust labelling in the PNC and endocrine population upon crossing with a TP1 lineage tracing fish (unpublished observation). Together, these results suggest that the low frequency contribution of the ptf1a lineage to non-acinar fates is unlikely to be due to low labeling efficiency for the progenitor population. Thus it is probable that pancreatic-Notch-responsive cells (PNCs) represent the dominant progenitor population required for generating non-acinar pancreatic cell types, functioning largely independent of ptf1a (Wang et al., 2011).
Clonal expansion of secondary islets
In some of the secondary islets where we detected ptf1a lineage labeling, clusters of lineage-traced cells were observed. The low probability of ptf1a lineage labeling in the endocrine population made it almost certain that these cell clusters were generated by clonal expansion, implying that a fraction of secondary islets originated from single ptf1a positive progenitors. This is consistent with previous studies showing that newly generated secondary islets can also arise from single PNCs, and that these cells are highly proliferative (Matsuda et al., 2013)
ptf1a lineage and Notch-responsive lineage
We used Nkx6.1 as a marker for PNCs in the current study. We have previously observed that in the zebrafish pancreas, immunofluorescent labeling for Nkx6.1 completely overlaps with PNCs marked directly by the transgene TP1:eGFP (Huang et al., 2014). In mouse, it has been shown that Ptf1a and Nkx6.1 reciprocally repress each other’s expression (Schaffer et al., 2010). This appears to represent a significant mechanism for determining the allocation of cells between acinar and endocrine cell fates. Ongoing expression of Ptf1a directs cellular programming towards the acinar fate, while Nkx6.1 directs cells towards a ductal/endocrine fate (Schaffer et al., 2010). Our data suggest that the same mechanism may be applicable in the zebrafish pancreas. Specifically, we observed that in the setting of ptf1a haploinsufficiency, there was a significantly greater ptf1a lineage contribution to the Nkx6.1-expressing PNC compartment. PNCs have been shown to be the precursors of the endocrine population in zebrafish, and inhibition of Notch signaling enhances endocrine differentiation (Parsons et al., 2009; Wang et al., 2011). These results may imply that the increased contribution of ptf1a-expressing progenitor cells to the endocrine compartment in ptf1a heterozygous fish may proceed through a PNC intermediate.
Different levels of ptf1a regulate the multi-lineage potential of ptf1a-expressing progenitors
With little-to-no ptf1a activity in the ptf1asa126/sa126 fish, we observed a complete absence of exocrine pancreas, while early endocrine cell differentiation and principal islet structure was maintained. This result is similar to the ptf1a morpholino phenotype (Lin et al., 2004), but different from observations in human and mouse in which low level of ptf1a also reduces endocrine cell numbers and alters their distribution (Stoffers et al., 1997; Burlison et al., 2008; Fukuda et al., 2008). This may reflect the different developmental programming of zebrafish pancreas (Tiso et al., 2009), as well as fundamental differences between the development of principal and secondary islets. Another possible scenario is that in both ptf1a morphant and ptf1asa126/sa126 homozygous fish, there is still enough expression and function of ptf1a to support proper endocrine lineage differentiation. Evidence that supports this hypothesis includes a recent study on the function of ptf1a in synaptic neuropil, where it was demonstrated that the combination of ptf1asa126/sa126 allleles and a ptf1a translation-blocking morpholino were required to reveal a true ptf1a null phenotype (Randlett et al., 2013). In the ptf1asa126/sa126 fish, we also observed that the ptf1a lineage became trans-fated to gall bladder and other non-pancreatic fates. This cell-fate conversion is in keeping with observations in mice (Burlison et al., 2008; Fukuda et al., 2008).
Overall, it is apparent from the current work that, as in mouse, different zebrafish pancreatic cell fates require different threshold levels of ptf1a, at least for a defined subset of cells. Further studies investigating intermediate dosages/activities of ptf1a will be needed to address the function of Ptf1a in a more quantitative way. This might be done by the combination of ptf1a mutant alleles varying amounts of morpholino against ptf1a; or, by combining the ptf1asa126 Sanger allele with the ptf1aakreas hypomorphic allele (Dong et al., 2008). Certainly, more work is required to characterize downstream mechanisms that are responsive to different levels of ptf1a.
Our findings have provided an important understanding on the mechanisms of pancreatic development in zebrafish, and further clarify both similarities and differences between pancreas development in fish and mammals. Notably, they also underscore fundamental differences between “dorsal bud” and “ventral bud” development in zebrafish, with the ventral bud displaying a more mammalian-like program involving ptf1a-dependent development and the participation of multi-lineage ptf1a-expressing progenitor cells.
Experimental Procedures
Generation of transgenic zebrafish lines
We followed established BAC recombineering methods (Suster et al., 2011) to generate the BAC transgene tg(ptf1a:creERT2), abbreviated to ptf1a:creERT2, by replacing GFP with creERT2 elements in the ptf1a:GFP construct. The ptf1a:GFP construct is derived from genomic BAC CH211-142H2 (Park et al., 2008; Sharan et al., 2009), which encompasses the zebrafish ptf1a coding sequence. The ptf1a:GFP transgene has been shown to accurately recapitulate the endogenous ptf1a expression (Park et al., 2008; Pashos et al., 2013).
The lineage responder line tg(ubi:loxP-CFP-loxP-nuc-mCherry), abbreviated to ubi:loxP-CFP-loxP-nuc-mCherry was modified from the previously published line Tg(T2Kβactin:loxP-stop-loxP-hmgb1-mCherry)jh15 (Wang et al., 2011), where the βactin promoter was replaced by the ubiquitin promoter (ubi) (Mosimann et al., 2011). CFP was also included in the construct to facilitate the identification of the transgenic fish.
Ptf1asa126/wt fish were obtained from the Sanger Institute Zebrafish Mutation Project (Kettleborough et al., 2013).
Stable sst:CFP; ins:dsYFP-2TA-nsfB transgenic fish was established to express CFP under somatostatin (sst) promoter, and nsfB encoding Nitroreductase was expressed under insulin (ins) promoter (Pisharath et al., 2007; Walker et al., 2012).
All fish were maintained under standard conditions. All procedures were performed under the approval of the Johns Hopkins University School of Medicine Animal Care and Use Committee guidelines.
Drug treatment
For the induction of creER activity, 4-Hydroxytamoxifen (4-OHT, T176, Sigma) treatment was performed as previously described (Wang et al., 2011). Briefly, 4-OHT was dissolved in 100% ethanol to create a stock solution of 10 mM. Embryos were placed in a 5 µM solution of 4-OHT in E3 medium from 30 to 54 hours post fertilization (hpf), unless otherwise stated in the text.
For the β-cell ablation experiment, Metronidazole (Met, Sigma, M3761) treatment was performed as previously described (Pisharath et al., 2007). Briefly, Met was dissolved in E3 medium at a concentration of 10mM. Embryos were incubated in this solution for 24 hours from 82 to 106 hpf. Embryo-containing petri dishes were kept in the dark.
Immunofluorescence
For whole mount immunofluorescent staining, larvae were fixed in 4% paraformaldehyde at 4°C overnight. Following fixation, the whole gut region was dissected out and blocked for 1 hour with PBS with 0.2% Triton (PBST) and 10% fetal bovine serum. For cryosections, fish were similarly fixed and the gut-intestine system was dissected. Tissues were then immersed in 30% sucrose/PBS, embedded in optimal cutting temperature (OCT) compound, frozen in liquid nitrogen, and sectioned in 10 µm thickness using a cryostat. Primary antibodies used in this study included: mouse anti-Nkx6.1 (Developmental Studies Hybridoma Bank, F55A12), 1: 100. Guinea pig anti-Insulin (Dako, A0564), 1: 500. Rabbit anti-Glucagon (Dako, A0565), 1:400. Rabbit anti-Somatostatin (Dako, A0566), 1:500. Rabbit anti-DsRed (Clontech, 642496), 1:100. Mouse anti-2F11 (Abcam, ab71286), 1:200. Primary antibodies were incubated at 4°C overnight. After washing with PBST, samples were incubated with secondary antibodies (Jackson Immunoresearch, 1:300) in blocking buffer. Fluorescent images were acquired with Nikon A1 scanning confocal microscope. Cell counting was carried out manually. Briefly, whole mount tissues were scanned by confocal microscope and maximum projections were assembled from Z-stacks. Cell numbers were counted from reconstructed images. Students t-test was implemented for statistical analysis.
Key findings.
In the zebrafish pancreas, early ptf1a-expressing progenitor cells are multipotent.
The ptf1a lineage contributes to new β cells during regeneration.
The ability of the ptf1a lineage to contribute to non-acinar cell types is enhanced when ptf1a dosage is reduced.
Acknowledgments
We would like to thank Dr. Steven Farber for his help with interpretation of the experimental result. We also thank Dr. Wei Huang for intellectual input and Dr. John Goodier, Dr. Hao Ho, Dr. Guangliang Wang for their critical reading of the manuscript. This work was supported by NIH grants DK056211 and DK097087 (to SDL) and DK080730 (to MJP). SDL was also supported by the Paul K. Neumann Professorship at Johns Hopkins University.
References
- Anlezark GM, Melton RG, Sherwood RF, Coles B, Friedlos F, Knox RJ. The bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954)--I. Purification and properties of a nitroreductase enzyme from Escherichia coli--a potential enzyme for antibody-directed enzyme prodrug therapy (ADEPT) Biochem Pharmacol. 1992;44:2289–2295. doi: 10.1016/0006-2952(92)90671-5. [DOI] [PubMed] [Google Scholar]
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Developmental Biology. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- Dong PD, Provost E, Leach SD, Stainier DY. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008;22:1445–1450. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple A, Brinn JE., Jr Islet histophysiology: evolutionary correlations. Gen Comp Endocrinol. 1975;27:320–349. doi: 10.1016/0016-6480(75)90201-4. [DOI] [PubMed] [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, Kawaguchi M, Terao M, Doi R, Wright CV, Hoshino M, Chiba T, Uemoto S. Reduction of Ptf1a gene dosage causes pancreatic hypoplasia and diabetes in mice. Diabetes. 2008;57:2421–2431. doi: 10.2337/db07-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald J, Sprinkel AE, Ray M, Serup P, Wright C, Madsen OD. Generation and characterization of Ptf1a antiserum and localization of Ptf1a in relation to Nkx6.1 and Pdx1 during the earliest stages of mouse pancreas development. J Histochem Cytochem. 2008;56:587–595. doi: 10.1369/jhc.2008.950675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS One. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera PL, Huarte J, Sanvito F, Meda P, Orci L, Vassalli JD. Embryogenesis of the murine endocrine pancreas; early expression of pancreatic polypeptide gene. Development. 1991;113:1257–1265. doi: 10.1242/dev.113.4.1257. [DOI] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A. 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Stainier DYR. Suppression of Ptf1a Activity Induces Acinar-to-Endocrine Conversion. Current Biology. 2011;21:712–717. doi: 10.1016/j.cub.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Wang G, Delaspre F, Vitery MD, Beer RL, Parsons MJ. Retinoic acid plays an evolutionarily conserved and biphasic role in pancreas development. Dev Biol. 2014 doi: 10.1016/j.ydbio.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CVE. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nature Genetics. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kemp JD, Walther BT, Rutter WJ. Protein synthesis during the secondary developmental transition of the embryonic rat pancreas. J Biol Chem. 1972;247:3941–3952. [PubMed] [Google Scholar]
- Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, Fenyes F, Mehroke S, Scahill C, Gibbons R, Wali N, Carruthers S, Hall A, Yen J, Cuppen E, Stemple DL. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel MD, Prince VE. On the diabetic menu: zebrafish as a model for pancreas development and function. Bioessays. 2009;31:139–152. doi: 10.1002/bies.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, Leach SD. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Developmental Biology. 2004;274:491–503. doi: 10.1016/j.ydbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Lindmark DG, Muller M. Antitrichomonad action, mutagenicity, and reduction of metronidazole and other nitroimidazoles. Antimicrob Agents Chemother. 1976;10:476–482. doi: 10.1128/aac.10.3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda H, Parsons MJ, Leach SD. Aldh1-expressing endocrine progenitor cells regulate secondary islet formation in larval zebrafish pancreas. PLoS One. 2013;8:e74350. doi: 10.1371/journal.pone.0074350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Bankaitis ED, Boyer D, Xu XB, Van de Casteele M, Magnuson MA, Heimberg H, Wright CVE. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140:751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Park SW, Davison JM, Rhee J, Hruban RH, Maitra A, Leach SD. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134:2080–2090. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashos E, Park JT, Leach S, Fisher S. Distinct enhancers of ptf1a mediate specification and expansion of ventral pancreas in zebrafish. Dev Biol. 2013;381:471–481. doi: 10.1016/j.ydbio.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pictet RL, Clark WR, Williams RH, Rutter WJ. An ultrastructural analysis of the developing embryonic pancreas. Dev Biol. 1972;29:436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E-coli nitroreductase. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall LB, Pictet RL, Williams RH, Rutter WJ. Early differentiation of glucagon-producing cells in embryonic pancreas: a possible developmental role for glucagon. Proc Natl Acad Sci U S A. 1973;70:3478–3482. doi: 10.1073/pnas.70.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randlett O, MacDonald RB, Yoshimatsu T, Almeida AD, Suzuki SC, Wong RO, Harris WA. Cellular requirements for building a retinal neuropil. Cell Rep. 2013;3:282–290. doi: 10.1016/j.celrep.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Developmental Cell. 2010;18:1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protocols. 2011;6:1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- Tiso N, Moro E, Argenton F. Zebrafish pancreas development. Mol Cell Endocrinol. 2009;312:24–30. doi: 10.1016/j.mce.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Walker SL, Ariga J, Mathias JR, Coothankandaswamy V, Xie X, Distel M, Koster RW, Parsons MJ, Bhalla KN, Saxena MT, Mumm JS. Automated reporter quantification in vivo: high-throughput screening method for reporter-based assays in zebrafish. PLoS One. 2012;7:e29916. doi: 10.1371/journal.pone.0029916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rovira M, Yusuff S, Parsons MJ. Genetic inducible fate mapping in larval zebrafish reveals origins of adult insulin-producing beta-cells. Development. 2011;138:609–617. doi: 10.1242/dev.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AB, Warga RM, Prince VE. Origin of the zebrafish endocrine and exocrine pancreas. Dev Dyn. 2007;236:1558–1569. doi: 10.1002/dvdy.21168. [DOI] [PubMed] [Google Scholar]
- Yee NS, Lorent K, Pack M. Exocrine pancreas development in zebrafish. Dev Biol. 2005;284:84–101. doi: 10.1016/j.ydbio.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, Peers B, Bortolussi M, Argenton F. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Developmental Biology. 2004;268:174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]